Abstract
Photobiomodulation (PBM) stimulates different types of stem cells to migrate, proliferate, and differentiate in vitro and in vivo. However, little is known about the effects of PBM on the differentiation of embryonic stem cells (ESCs) toward the otic lineage. Only a few reports have documented the in vitro differentiation of ESCs into inner-ear hair cells (HCs) due to the complexity of HCs compared with other target cell types. In this study, we determined the optimal condition to differentiate the ESCs into the otic organoid using different culture techniques and PBM parameters. The efficiency of organoid formation within the embryoid body (EB) was dependent on the cell density of the hanging drop. PBM, using 630 nm wavelength light-emitting diodes (LEDs), further improved the differentiation of inner-ear hair cell-like cells coupled with reactive oxygen species (ROS) overexpression. Transcriptome analysis showed the factors that are responsible for the effect of PBM in the formation of otic organoids, notably, the downregulation of neural development-associated genes and the hairy and enhancer of split 5 (Hes5) gene, which inhibits the differentiation of prosensory cells to hair cells. These data enrich the current differentiation protocols for generating inner-ear hair cells.
Keywords: embryonic stem cell, tissue regeneration, Notch, genomics, inner ear, photobiomodulation
Graphical Abstract
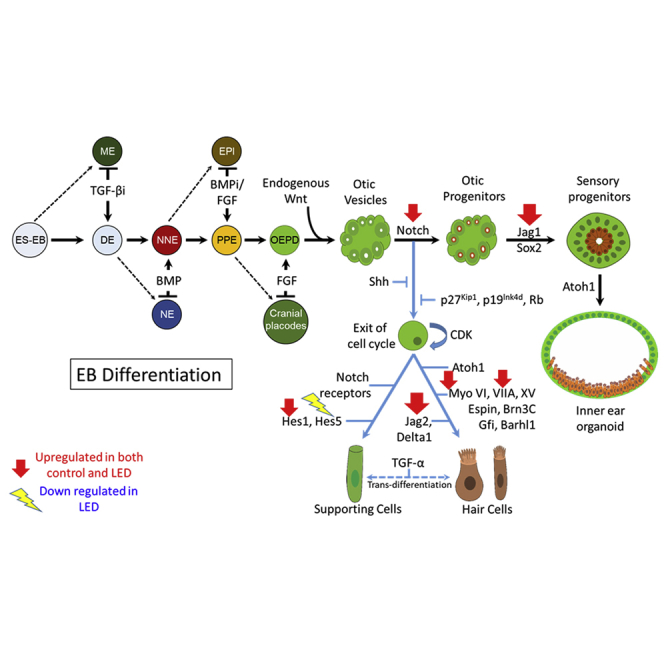
More effective and specified differentiation is required for stem cell differentiation into inner-ear hair cell-like cells. Photobiomodulation is known to enhance the differentiation in other target cell lineage. In this study, we determined more effective and specified otic-lineage differentiation by photobiomodulation and uncovered underlying molecular processes.
Introduction
Photobiomodulation (PBM) therapy, also known as low-level laser therapy, has existed for over half a century, but its cellular mechanisms of action are only recently being unraveled. PBM therapy uses a low-power light source (laser or light-emitting diode [LED]) in the red or near-infrared region (NIR) of the spectrum, with wavelengths of 600–700 and 780–1,100 nm, respectively, and output power of 1–500 mW.1 Recent advances in the adoption of PBM in stem cell therapy have shown that stem cells and progenitor cells respond favorably to light. PBM stimulates different types of stem cells to migrate, proliferate, and differentiate in vitro2, 3, 4, 5, 6 and in vivo.7, 8, 9 However, little is known about the effects of PBM on the differentiation of embryonic stem cells (ESCs) toward the otic lineage.
Only a few reports have documented the in vitro differentiation of ESCs into inner-ear hair cells (HCs), due to the complexity of HCs compared with other target cell types. The differentiation of stem cells into HCs is a complex physiological process that is regulated by the cascading expression of systemic hormones and exogenous bioactive molecules. The most promising outcomes for successfully differentiating ESCs into HC-like cells10, 11, 12, 13 or inner-ear organoids14, 15, 16 have used chemically defined conditions that mimic the early stages of embryonic development. These studies have revealed that initiated ESCs undergo ectodermal differentiation, followed by induction toward the non-neural ectoderm, followed by the preplacodal ectoderm. Self-guided organogenesis forms otic vesicles as organoid bodies that contain the sensory epithelia. However, only a few studies have replicated these results, and the efficacy of differentiation, especially in vivo, remains questionable. Therefore, it is important to find additional inexpensive and harmless interventions to accelerate the differentiation process and increase the yield of differentiated HCs.
In this study, we investigated the effects of PBM on the differentiation of ESCs into inner-ear HC-like cells. The effects of cell density and culture technique on in vitro differentiation were also studied. Finally, transcriptome analysis was used to identify factors responsible for the effects of PBM in the formation of otic organoids.
Results
EB Formation and Culture Techniques
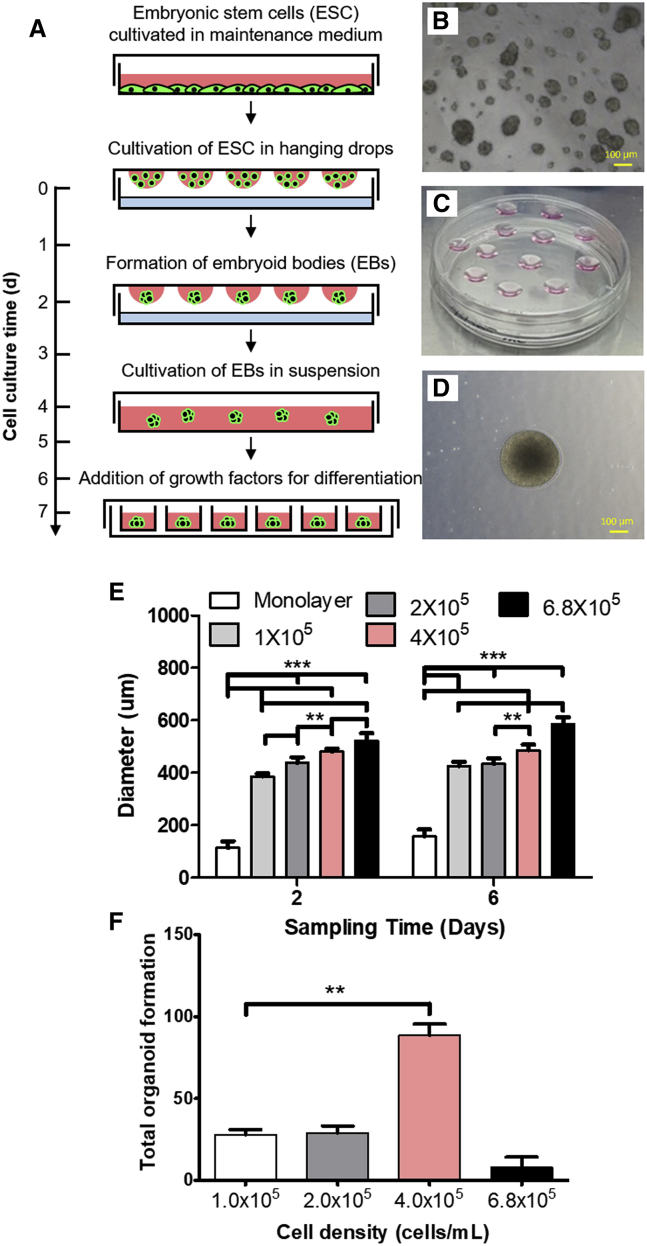
To test whether the culture technique can affect embryoid body (EB) formation, two different techniques were compared: a monolayer culture technique using Matrigel (cell adherence molecule) and the hanging-drop technique. The hanging-drop technique generates cell clusters using gravity by loading drops of cell culture media and cells onto the cover of cell culture dishes (Figure 1). With the use of the monolayer culture technique (cell concentrations = 9 × 104 cells/mL), the size of each EB was smaller compared with those generated using the hanging-drop technique. The EB diameter was quantified at differentiation days 2 and 6. Statistically significant increases in the diameter of EBs generated using the hanging-drop technique (cell concentrations > 1 × 105 cells/mL) were observed. In addition, most EBs generated using the monolayer culture technique were not maintained during the entire differentiation process. Next, the hanging-drop technique was used to assess whether cell density affects the size of EBs and the rate of successful organoid generation. ESCs were grown at four different densities (1, 2, 4, and 6.8 × 105 cells/mL). At both time points (days 2 and 6), the diameter of the EBs was greater, with a higher cell density (two-way ANOVA; p < 0.0001; statistical significance after Bonferroni post hoc analysis is shown as ∗∗p < 0.01 and ∗∗∗p < 0.001 in Figure 1E). The rate of organoid formation did not increase with increasing cell density but was not different between incubation periods. Organoids were observed starting at day 14, and the highest rate of organoid formation was observed with an ESC density of 4 × 105 cells/mL. A significantly increased number of organoids was observed with a cell density of 4 × 105 cells/mL compared with 1 × 105 cells/mL (two-tailed Mann-Whitney U test; n = 7, p = 0.0020, U = 0.0, power = 1.0, β-value = 0.0) (Figure 1F). Despite the increased EB size with a higher density of ESCs, the optimal density for generating organoids was 4 × 105 cells/mL.
Figure 1.
Comparison of Diameter of EB between Culture Technique Monolayer Culture and Hanging Drop and the Number of Organoids with Different Cell Density
(A) Illustration showing the process of hanging drop. (B and D) EB formed by hanging drop (D) is much larger than EBs formed by monolayer culture (B). (C) The process of generating EBs with hanging-drop technique. A higher density of cells resulted in a larger diameter of EBs at both time points, and a statistically significant diameter increase over concentration was confirmed. EBs by hanging drops were statistically larger than EBs by the monolayer at both time points (E). The largest number of organoids was observed at the density of 4.0 × 105. Organoids were observed starting at day 14, and a statistically larger number of organoids at the density of 4.0 × 105 compared to the density of 1.0 × 105 were observed (F). Scale bars, 100 μm. NS, not significant. Error bars were expressed in standard deviation. ∗∗p < 0.01 and ∗∗∗p < 0.001.
EB Differentiation into Organoid
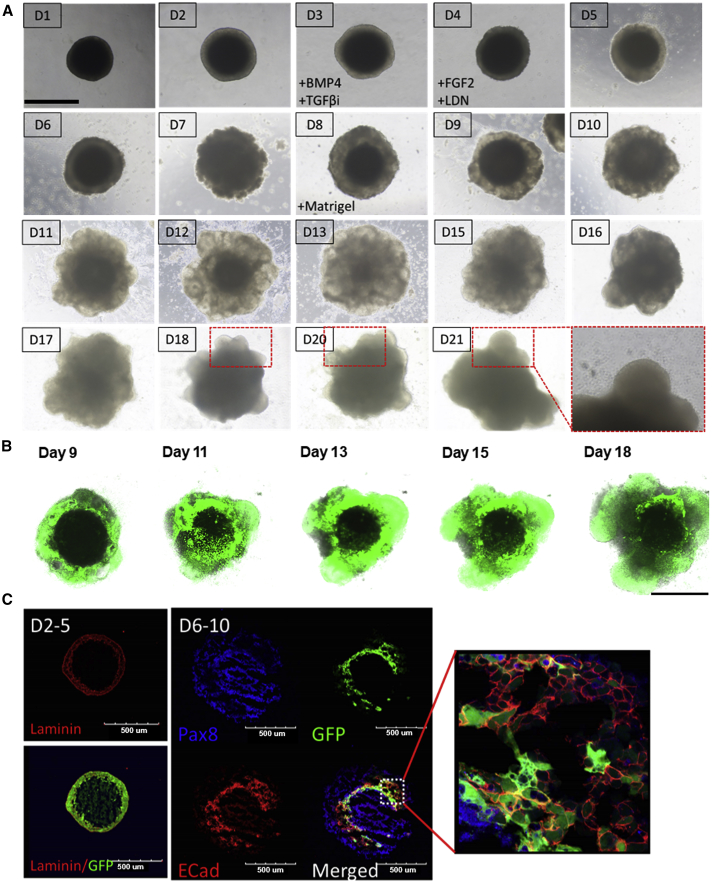
EBs were differentiated using specific factors. During this process, a specific portion of the EB begins to protrude and produce cystic lesions, which may include otic progenitors. With the use of confocal microscopy, EBs were monitored during the 18 days of culture. Peripheral growth of GFP-positive tissues and the existence of a central GFP-negative area were serially observed, suggesting outward growth of organoid during differentiation, along with internal loss of cells, which may lead to cavity formation. Confocal microscopy images captured during days 2 to 5 revealed laminin-positive cells, indicating the formation of basal epithelium. During days 6 to 10, PAX8-positive cells were used to identify the neural ectoderm, whereas ECAD-positive cells were indicative of the non-neural ectoderm containing the sensory epithelium of the otic vesicle (Figure 2).
Figure 2.
Differentiation of ES Cells into EBs
(A) Macro images of EBs using brightfield microscopy during the 21 days of culture. Image numbers indicate the day of differentiation following initiation of the hanging-drop process from day 1 to 21 (D1-D21). Specific factors added during the differentiation process, such as BMP4, FGF2, and Matrigel, are shown. Specific potions of interest within the EB are outlined with a red-dotted rectangle. Zoomed-in images of these regions are shown, revealing protruding cystic lesions that could include otic progenitors. (B) Macro images of the EBs using confocal microscopy during the 18 days of culture. Peripheral growth of GFP-positive tissues and the existence of a central GFP-negative area are serially observed. (C) Confocal microscopy images captured during days 2 to 5 of differentiation reveal laminin-positive cells indicating the formation of basal epithelium. PAX8-positive cells observed from days 6 to 10 mark the neural ectoderm, whereas ECAD-positive cells represent the non-neural ectoderm containing the sensory epithelium of the otic vesicle. Scale bars, 500 μm.
Assessing Differentiation into Myosin VIIa-Positive Cells
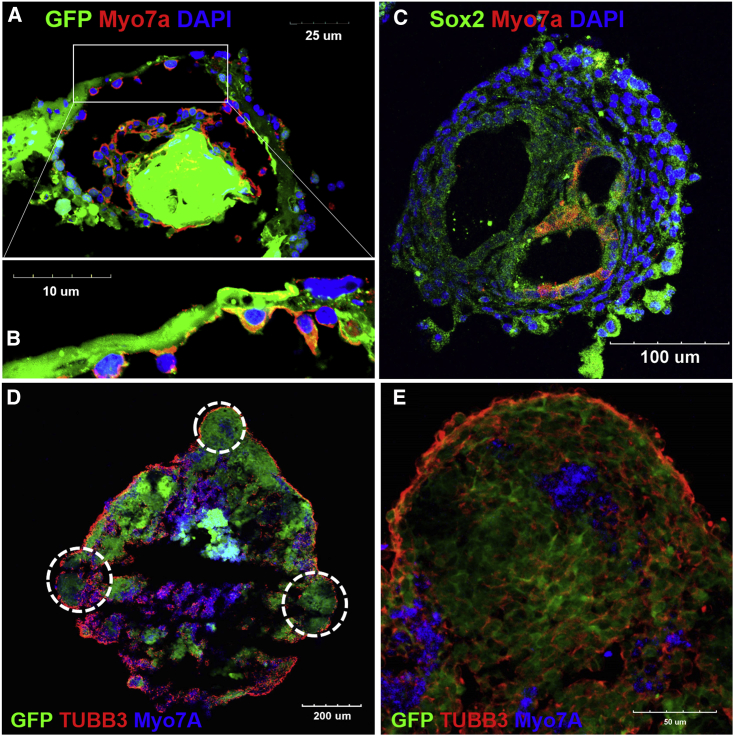
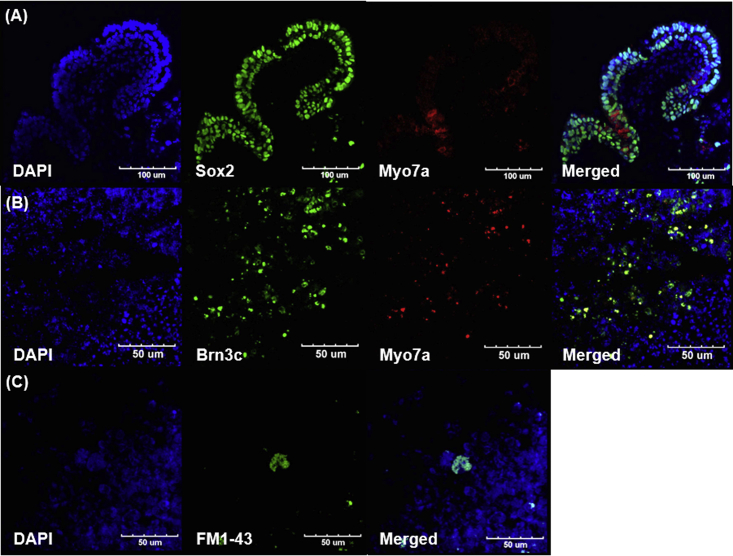
EBs were differentiated for 18 days and analyzed for possible otic cell differentiation. After fixation, the organoids were dissected (Figure S1) and sectioned using a cryocut (Figure 3). Protruding, cystic-like areas of EBs, which were presumed to be organoids, were observed. At lower magnification, these areas exhibited acellular regions that were surrounded by MYO7A-positive cells, which were likely HC-like cells that differentiated from mouse ESCs (mESCs) (Figures 3A and 3B). SOX2-positive cells, which were thought to be differentiated supporting cell-like cells, were observed adjacent to the MYO7A-positive cells at higher magnification (Figure 3C). Nerve fibers surrounding the organoid were observed (Figures 3D and 3E), which are primarily located near the MYO7A-positive cells. Because MYO7A is also expressed in the retina, we performed additional staining for the photoreceptor marker rhodopsin. However, no rhodopsin expression was observed in the differentiated organoids, which discounts the possibility of optic differentiation (Figure S2).
Figure 3.
Observation of Myosin VIIa (MYO7A)- and β-Tubulin Isotype 3A (TUBB3)-Positive Cells in the Organoid
At 18 days of differentiation, tissues were fixed and prepared for the epifluorescence analysis. (A) The whole organoid was mounted in cryogenic medium and cut into sections. The tissue within the rectangle is magnified in (B). Epifluorescence analysis of the tissue revealed MYO7A-positive cells (red; nuclei were stained by DAPI) inside the organoid (A). At the higher magnification, expression of both GFP and MYO7A was identified in these cells (B). SOX2-positive (green; nuclei were stained by DAPI) and MYO7A-positive cells (red) are shown in (C). (D and E) Different organoids at the same stage grown following staining for various neural markers (red, nerve fibers stained by TUBB3; blue, MYO7A; and green, GFP from stem cells). (D) Dotted circles demarcate organoid formation. MYO7A-positive cells are observed in both low (D) and high magnification (E). Under high magnification, nerve fibers are shown to be densely populated near MYO7A-positive cells compared to non-MYO7A-positive cells (E). Scale bars differ among images, with individual scales denoted in each image.
The EBs used to generate cystic protruding organoids were analyzed by qRT-PCR to confirm the expression of several markers. There was a statistically significant increase in M expression in differentiated specimens and HEI-OC1 cells compared with negative control cells (two-tailed Mann-Whitney U test; HEI-OC1 versus maintenance cells, n = 12, p = 0.0166, U = 30.0, power = 0.961, β-value = 0.039; differentiated cells versus maintenance cells, n = 12–18, p < 0.0001, U = 0.0, power = 1.0, β-value = 0.0; Figure S3A). There was a statistically significant increase in Oct4 expression in maintenance cells compared with HEI-OC1 and differentiated cells (two-tailed Mann-Whitney U test; HEI-OC1 versus maintenance cells, n = 12, p < 0.0001, U = 0.0, power = 0.999, β-value = 0.001; differentiated cells versus maintenance cells, n = 12–18, p = 0.0118, U = 48.0, power = 0.993, β-value = 0.007; Figure S3B). There was a statistically significant increase in Sox2 expression in maintenance cells compared with HEI-OC1 (two-tailed Mann-Whitney U test; HEI-OC1 versus maintenance cells, n = 6–10, p = 0.0002, U = 0.0, power = 0.999, β-value = 0.001; differentiated cells versus maintenance cells, n = 6–18, p = 0.1717, U = 33.0, power = 0.186, β-value = 0.814; Figure S3C). Sox2 expression in differentiated cells was ambiguous with no statistically significant differences among groups, possibly due to the expression of Sox2 by differentiated supporting cells.
These results suggest that it is possible to generate the cystic organoid, which has several markers of inner-ear HCs when used in combination with previously introduced protocols. However, the efficiency of cystic organoid formation was quite low, and other specific costaining of multiple specific HC markers was not identified.
Effects of PBM Using LED Irradiation on Mouse ESC Differentiation into Organoids
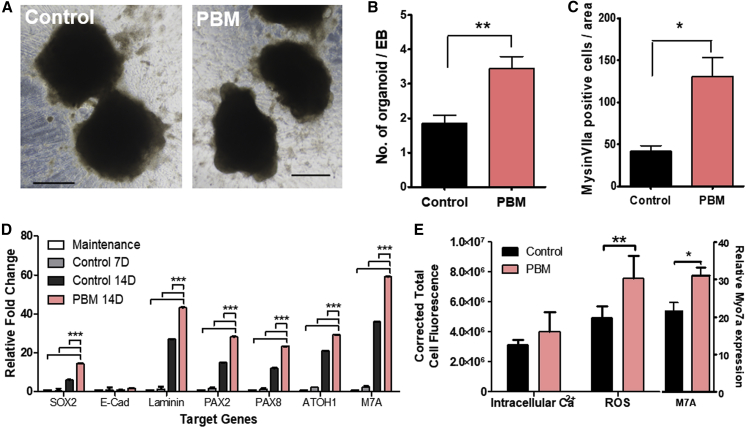
To improve the efficiency and specificity of the differentiation process, LED irradiation was performed during the maturation phase, and the rate of organoid formation at 14 days (with cell density of 4 × 105 cells/mL) was evaluated. A statistically significant increase in organoid formation was observed in EBs irradiated at 630 nm (total 110 organoids per 32 EBs) compared to control (total 52 organoids per 28 EBs) (two-tailed Mann-Whitney U test; p = 0.0011, U = 230.0; Figures 4A and 4B). The diameter of EB did not statistically increase after PBM (Figure S4). The number of MYO7A-positive cells between two groups was compared at 18 days. Organoids were randomly selected and stained for MYO7A, and the number of MYO7A-positive cells within the image (300 × 300 μm) was counted. Statistically larger numbers of MYO7A-positive cells were found in the PBM group (independent t test, two-tailed; t = 3.714, p = 0.0340) (Figure 4C).
Figure 4.
Effects of PBM on the Differentiation of Mouse ESC Organoids
Effects of PBM on the mESC differentiation process. (A) EBs at differentiation day 14. PBM-exposed EBs had a greater number of protruding organoid-like structures, relative to controls. (B) Statistically significant increases in organoid-like protrusions were observed in PBM-exposed EBs. (C) A higher number of MYO7A-positive cells were observed in PBM-exposed samples. (D) A higher expression of genes associated with HC differentiation was observed in PBM-exposed samples, compared to undifferentiated and differentiated but unexposed samples. (E) With the use of epifluorescence microscopy, intensities of ROS and intracellular calcium expressions were analyzed and show that PBM does not affect intracellular calcium levels but significantly increases ROS expression. Black scale bar, 500 μm. Error bars were expressed in standard deviation. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Next, the gene expression of HC differentiation markers was assessed using qRT-PCR with differentiated cells without LED irradiation as a control (exactly 1 μg of RNA was collected from each group using various numbers of EBs). Genes related to HC differentiation were expressed at higher levels in PBM-exposed samples, relative to undifferentiated cells, as well as differentiated cells not exposed to PBM (Figure 4D). PBM treatment resulted in increased expression of all serial markers associated with otic differentiation, not limited to specific steps. Detailed results are given in Table 1.
Table 1.
Statistical Analysis of qRT-PCR Results for Levels of mRNA Expression for Genes Related to HC Differentiation
| Target Genes | Two-Way ANOVA (p Value) | Maintenance versus Control 7D | Maintenance versus Control 14D | Maintenance versus PBM 14D | Control 7D versus Control 14D | Control 7D versus PBM 14D | Control 14D versus PBM 14D |
|---|---|---|---|---|---|---|---|
| Sox2 | ∗∗∗p < 0.001 | ns | ∗∗∗ | ∗∗∗ | ∗∗∗ | ∗∗∗ | ∗∗∗ |
| Ecad | p > 0.05 | ns | ns | ns | ns | ns | ns |
| Laminin | ∗∗∗p < 0.001 | ns | ∗∗∗ | ∗∗∗ | ∗∗∗ | ∗∗∗ | ∗∗∗ |
| Pax2 | ∗∗∗p < 0.001 | ns | ∗∗∗ | ∗∗∗ | ∗∗∗ | ∗∗∗ | ∗∗∗ |
| Pax8 | ∗∗∗p < 0.001 | ns | ∗∗∗ | ∗∗∗ | ∗∗∗ | ∗∗∗ | ∗∗∗ |
| Atoh1 | ∗∗∗p < 0.001 | ∗∗ | ∗∗∗ | ∗∗∗ | ∗∗∗ | ∗∗∗ | ∗∗∗ |
| Myo7a | ∗∗∗p < 0.001 | ∗∗∗ | ∗∗∗ | ∗∗∗ | ∗∗∗ | ∗∗∗ | ∗∗∗ |
7D, 7 days; 14D, 14 days.
In quantitative epifluorescence analyses (measurement of epifluorescence intensity), increased MYO7A expression was observed in 630 nm LED-irradiated organoids (Figure 4E) relative to differentiated control organoids without LED irradiation (two-tailed Mann-Whitney U test; n = 10–18, p = 0.0414, U = 47.0, power = 0.786, β-value = 0.214; Figure 4D). Significant increases in reactive oxygen species (ROS) levels were observed compared to controls (two-tailed Mann-Whitney U test; p = 0.0012, U = 0.0; Figure 4E), whereas no significant increase in intracellular calcium levels was observed. Increased levels of ROS could be related to increased metabolic activity due to increased differentiation.
Next, the morphology of organoids generated after PBM irradiation was assessed. In these organoids, a well-organized structure was observed, as evidenced by SOX2-positive cell layers (presumably supporting cells), along with intermittent MYO7A-positive cells (presumably hair cell-like cells; Figure 5A). Double expression of MYO7A and BRN3C (an inner-ear HC-specific marker) was observed (Figure 5B). Expression of N-[3-triethylammoniumpropyl]-4-[p-di-butylaminostyryl] pyridinium dibromide (FM1-43) dye, which enters the hair cell via a mechano-transduction channel, was also observed in PBM-treated organoids, suggesting the presence of active mechano-transduction in inner-ear HCs (Figure 5C). Together, these results demonstrate specific, quantitative increases in otic differentiation in response to PBM.
Figure 5.
Detection of Inner-Ear HC Markers in Organoids Treated with PBM
ES-J1 cells were used to generate otic organoids, after which, EBs were fixed and prepared for triple immunostaining analysis. (A) Immunofluorescence showing PBM-exposed cells expresses SOX2 and myosin VIIa (MYO7A) after 21 days of 3D cell culture. (B) MYO7A-positive cells also express another HC marker, BRN3C. (C) Representative HCs after FM1-43 treatment. Scale bars differ among images, with individual scales denoted in each image.
Transcriptome Sequencing Analysis to Identify DEGs in Inner-Ear HC-like Cells Irradiated at 630 nm
To obtain a global view of the transcriptional responses of EBs to PBM, comparative RNA sequencing (RNA-seq) analyses were performed in control EBs and EBs irradiated at 630 nm. With the use of an Illumina HiSeq 2500, an average of 56.9 million raw reads was produced, with a read length of 100 bp. Original RNA-seq data of this study are available at Mendeley Data (http://dx.doi.org/10.17632/vchs6rtdxp.2). At least 5.51 Gb of clean data, accounting for more than 96.8% of the raw data, was prepared for further analysis. All filtered sequence reads were mapped to the mouse reference genome (GRCm38). An average of >84.7% of read pairs was uniquely mapped on the mouse genome (Table S1). After gene annotation using the Ensembl database (release 77), 20,958 and 20,731 expressed genes were detected in control and 630 nm-irradiated EBs, respectively. Of these, 19,474 genes were expressed in both groups, whereas 1,484 and 1,257 genes were unique to the control and 630 nm wavelength groups, respectively. Thus, 22,215 expressed genes were identified in at least one EB (Figure S5A).17 The genes expressed in both groups exhibited a uniform expression level, suggesting that there were no issues in sample preparation and data production (Figure S5B).
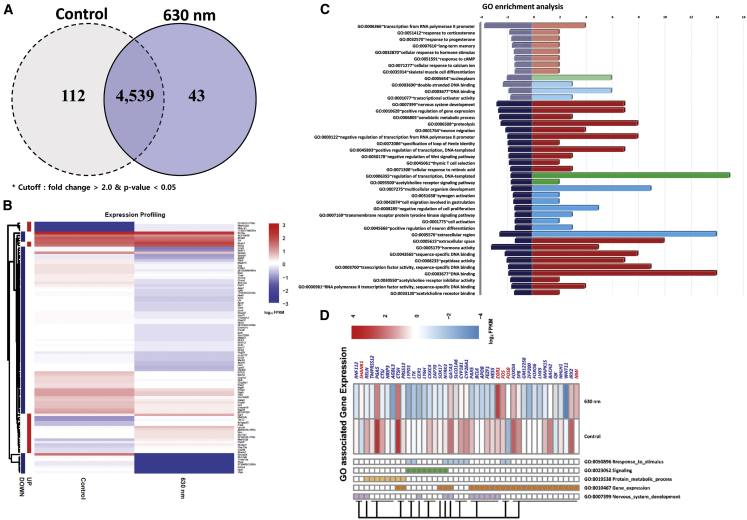
To screen all genes that were differentially expressed in different cell states, quantitative comparisons of gene expression were performed. According to set criteria (log2 fold change [FC] ≥ 1 and p < 0.05), a pairwise comparison was established between the control and 630 nm groups. A total of 155 differentially expressed genes (DEGs; 43 up- and 112 downregulated) were detected in the comparison of the control and 630 nm groups (Figure 6). Based on the similarity of gene expression, hierarchical clustering analysis was performed using the identified DEGs, excluding predicted transcripts. The expression pattern of the 155 DEGs revealed marked differences in EBs irradiated at 630 nm compared with control (Figures 6 and S6). Detailed information of all identified DEGs is listed in Table S2.
Figure 6.
Results of Transcriptome Analysis for EBs Affected by PBM Irradiation
(A) Venn diagram comparing the number of up- and downregulated genes (log2 FPKM ≥ 1, P-value < 0.05) between 630 nm PBM-treated and control EBs. Each group used in RNA-seq was conducted by pooling the five EBs cultivated under the same conditions. Overlapping areas represent the number of genes that is not significant. (B) Gene-expression profile of 102 DEGs between samples. The intensity of color indicates gene-expression levels normalized according to log10 FPKM values. (C) Gene ontology enrichment analysis and the expression profiles of 102 DEGs in EBs affected by PBM irradiation. Functional classification was performed using DAVID. The x axes on the left and right side denote the significance level (scored as −log10 [p value]) and the number of genes that maps to the corresponding GO terms, respectively. The y axis denotes the related functional GO terms (biological process in red, cellular component in green, and molecular function in sky blue). The shaded bar and the full bar graphs represent the results of GO analysis using up- and downregulated DEGs, respectively. (D) Gene-expression profiles of DEGs associated with five relevant GO terms (response to stimulus, signaling, protein metabolic process, gene expression, and nervous system development). The color intensity indicates gene-expression levels normalized according to log2 FPKM values. Up- and downregulated genes are indicated in red and blue, respectively.
GO Enrichment Analysis of DEGs
To classify the functions of the 43 up- and 112 downregulated genes in the PBM groups systematically, Gene Ontology (GO) enrichment analysis was conducted using the Database for Annotation, Visualization and Integrated Discovery (DAVID) functional annotation tool, v6.8 (https://david.ncifcrf.gov/).18 Of the 155 DEGs, 55 were significantly associated with 42 GO terms (28 biological process [BP], three cellular component [CC], and 11 molecular function [MF]) (Figure 6; Table S3). Only nine DEGs upregulated in the PBM groups were involved in 12 significant GO terms: eight BP, one CC, and three MF. Forty-six downregulated DEGs were involved in 20 BP, two CC, and eight MF categories. The regulation of transcription and DNA-templated GO: 0006355 was associated with a higher number of DEGs, and transcription from the RNA polymerase II promoter (GO: 0006366) was significantly enriched with four genes: early growth response 1 (Egr1), Finkel-Biskis-Jinkins (FBJ) osteosarcoma oncogene (Fos), NMYC-interactor gene (Nmi), and FosB (Table S3). To further investigate the key biological processes and DEGs that might be associated with inner-ear HC development, the significant GO terms were reclassified into five major GO categories: “response to stimulus (GO: 0050896),” “signaling (GO: 0023052),” “protein metabolic process (GO: 0019538),” “gene expression (GO: 0010467),” and “nervous system development (GO: 0007399).” In total, 26 DEGs were involved in gene-expression processes, such as transcription from RNA polymerase II promoter, positive regulation of gene expression, negative regulation of transcription from RNA polymerase II promoter, positive regulation of transcription, DNA-templated regulation of transcription, and zymogen activation. As shown in Figure 6, 12 nervous system development processes, nine responses to stimulus, eight signaling, and eight protein metabolic process-associated genes were enriched. We focused on 55 of 155 DEGs that were involved in the biological processes significantly changed by PBM. Most DEGs associated with the selected GO terms were downregulated. Only eight genes were upregulated in EBs irradiated at 630 nm. Regulation of gene-expression and nervous system development-related genes exhibited significant expression changes in the PBM group (Figure 6C).
Based on the prediction results obtained using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) system, network analysis was performed to predict protein–protein interactions among 43 DEGs implicated in five major GO categories (Figure S7).19 Among these, 25 proteins were clustered together. Notably, FOS, FOSb, and EGR-1 were located at the hub of this network and interacted with other proteins; these genes were the only upregulated genes in this network. Hairy and enhancer of split 5 (Hes5), which is a key gene that is inhibited during the transdifferentiation of supporting cells to HCs, was among the downregulated genes (Figure 6D). In addition to comparing the control and 630 nm laser groups, genes related to the HC sensory system were assessed; most genes, including Myo7a, were upregulated (Figure S8; Table S4).
Discussion
Currently, attempts to differentiate stem cells and induced pluripotent stem cells toward the otic lineage have used several methods, such as two-dimensional (2D) monolayer cultures with cocultured-inactivated feeder cells or exogenous tissue20, 21, 22 or 3D cultures of floating cell aggregates to form EBs.23, 24, 25 The present study is the first report the use of the hanging-drop method to form 3D spheroids of otic organoids. The hanging-drop method, a scaffold-free system, enables EB formation without any synthetic materials and force besides gravity. It was previously used to form organotypic cultures of different cell types and cell lines.26 The formation of 3D spheroids typically varies among different cell types.27 According to our observations, the initial cell density also affects EB formation with hanging drops; the optimal density for generating organoids in the current study was 4 × 105 cells/mL.
Epifluorescence analysis showed that the expression of myosin VIIa in cells surrounding the cavity-like structures in the organoid mirrored the development of otic vesicles in a developing embryo.28 Other studies that used the same serum-free floating culture of EB-like aggregates with a quick reaggregation protocol, as in the current study, showed that only a finite number of myosin VIIa-positive cells also expressed multiple HC markers.25 Furthermore, upregulation of >50% of genes related to the HC sensory system (Figure S7) supports the possibility of otic lineage differentiation.
In light of the apparent sensitivity of stem cells to lasers, several studies have evaluated the differentiation of stem cells from different sources using PBM.29, 30, 31, 32, 33 The time point of light energy irradiation was determined considering the fact that PBM has enhanced the differentiation into specific target cells other than otic differentiation.7, 8, 9 If this is true, then there could be a possibility that early PBM exposure, before the ESCs have been treated with the factors that facilitate into the otic differentiation, might lead to differentiation to other specific lineage. Thus, in the present study, the otic differentiation protocol, according to a previously confirmed method, was applied prior to LED irradiation to minimize the undesired effect of PBM. Cytochrome c oxidase (CCO) responds readily to red and NIR light29 via a mechanism thought to involve displacing inhibitory nitric oxide (NO); the subsequent increased CCO activity increases the mitochondrial membrane potential (MMP), thus allowing mitochondria to produce more ATP. The effect of red and NIR light in promoting stem cell differentiation is likely due to shifting the metabolic profile from glycolysis to oxidative phosphorylation due to the increased mitochondrial number and activity caused by light exposure.
Transcriptome sequencing analysis to identify DEGs in inner-ear HC-like cells with PBM irradiation showed that most genes related to nervous system development processes were downregulated in the PBM groups, including the Hes5 gene, which is an inhibitor of inner-ear HC transdifferentiation.34, 35, 36, 37 Hes5 represses Atoh1 expression by directly binding to the promoter region of Atoh1, thereby favoring the differentiation of supporting cells over HCs.38 The downregulation of development processes in the nervous system can be directly traced to the schedule of PBM treatment, because LED irradiation coincides with the time point at which the neural ectoderm and non-neural ectoderm divide. Downregulation of neural development might suggest that these EBs are favoring non-neural ectodermal differentiation, which could eventually lead to inner-ear differentiation. Meanwhile, genes with important roles as transcriptional regulators, such as Fos, FosB, and Egr1, were upregulated with PBM. Interestingly, a previous study reported that PBM pretreatment regulated the expression of FOS and EGR-1.39 FOS protein, which can dimerize with members of the JUN protein family, is an essential subunit of the transcription factor complex activator protein 1 (AP-1). The regulation/activation of transcription factors, such as AP-1 and nuclear factor (NF)-κB, is likely caused by PBM-induced ROS.40 Thus, protein–protein network analysis based on physical correlations among the identified DEGs revealed potentially similar functional networks between the transcriptome and protein interactome and allowed us to propose a hub network that explains gene regulation in PBM-irradiated EBs.
Other biological processes affected by PBM include increased Wnt signaling, which facilitates HC differentiation.23 Inhibited proteolysis, which could be the consequence of the ubiquitylation initiated by increased ATP production, also pushes ESCs toward differentiation into specific lineages.41 Negative regulation of the retinoic acid response and the acetylcholine receptor signaling pathway inhibits the neuronal differentiation of ESCs.42,43
In conclusion, the present study demonstrated that EBs were generated from ESCs using the hanging-drop technique and that differentiation into inner-ear HC-like cells was enhanced by PBM with LEDs. Furthermore, gene-expression studies identified the factors responsible for the enhancing effects of PBM on the formation of otic organoids. Although these findings are not useful for identifying clinical therapies for the regeneration of HCs, they provide critical information regarding disease modeling and enriching current differentiation protocols for generating inner-ear HCs.
Materials and Methods
Cells and Cell Culture
Undifferentiated mouse ESCs expressing enhanced green fluorescence protein (EGFP), donated by Prof. Hosup Shim (Dankook University, Cheonan, Korea),44 and ES cells from J1 mouse (ES-J1, ATCC, Manassas, VA, USA) were used. The cells were cultured in gelatin-coated plates without feeder cells and maintained in ESC medium consisting of high-glucose Dulbecco’s modified Eagle’s medium (DMEM; Sigma-Aldrich, St. Louis, MO, USA), supplemented with 15% (v/v) heat-activated fetal bovine serum (FBS; ATCC, Manassas, VA, USA), 0.1 mM β-mercaptoethanol (Gibco, Invitrogen, Carlsbad, CA, USA), 0.1 mM GlutaMAX (Gibco), 0.1 mM ES-qualified nonessential amino acid (NEAA; Welgene, Daegu, Korea), 1% penicillin-streptomycin (PS; ATCC), 1,000 U/mL leukemia inhibitory factor (Millipore, Merck, Burlington, MA, USA), 0.033% CHIR99021 (Tocris Bioscience, Bristol, UK), and 0.125% PD035901 (Tocris Bioscience) at 37°C in a humidified incubator with 5% CO2.
Ectodermal differentiation was initiated using Glasgow minimum essential medium (Gibco), supplemented with 1.5% knockout serum replacement (Gibco), 0.1 mM β-mercaptoethanol, 1 mM sodium pyruvate (Stem Cell Technologies, Vancouver, BC, Canada), 0.1 mM NEAA, and 1% PS. The maturation medium consisted of DMEM/F12 (Sigma-Aldrich), supplemented with 1% N2 supplement (Gibco), 1% GlutaMAX, and 0.1% Normocin (InvivoGen, San Diego, CA, USA).
Embryoid Body Formation
EBs were generated using monolayer cell cultures or the hanging-drop technique (Figures 1A and 1C). For monolayer cultures, undifferentiated ESCs were detached with 0.25% trypsin/ethylenediaminetetraacetic (TE) acid (ATCC), seeded in 96-well U-shaped plates (Nalge Nunc International, Roskilde, Denmark) at a density of 9,000 cells/mL, and cultured for 24 h in ectodermal differentiation media at 37°C in a humidified incubator containing 5% CO2. After removing one-half of the medium from the culture plate, an equal volume of differentiation media containing 2% Matrigel (v/v; BD Biosciences, Franklin Lakes, NJ, USA) was added, and cells were cultured for 48 h under the same conditions. For the hanging-drop technique,27 after dissociating ESCs with TE, drops containing varying cell densities (1 × 105, 2 × 105, 4 × 105, and 6.8 × 105 cells/mL) per 30 μL maintenance medium were inoculated onto the lid of the culture dish and cultured for 48 h under the same conditions described above for monolayer cultures. The resulting EBs were collected, washed with phosphate-buffered saline (PBS), plated in 24-well plates, and cultured in ectodermal differentiation media at 37°C in a humidified incubator containing 5% CO2.
Differentiation into Inner-Ear-like Structures
ESCs were differentiated into inner-ear-like organoids, as described by Koehler et al.16 Briefly, on the second day of differentiation, non-neural ectoderm was induced by adding 10 ng/mL recombinant bone morphogenetic protein 4 (BMP4; Stemgent, Beltsville, MD, USA) and 1 μM SB431542 (Stemgent) to the cultured cells. On the 3rd day, preplacodal ectoderm was induced by adding 25 ng/mL fibroblast growth factor 2 (FGF2; Peprotech, Rocky Hill, NJ, USA) and 1 μM LDN-193189 (Stemgent). The cells were cultured for 2 days, and the medium was replaced with maturation medium containing 1% Matrigel on day 6. One-half of the medium was replaced with maturation medium without Matrigel every other day until day 18. The differentiation process is illustrated in Figure 2.
LED Irradiation
For PBM, LEDs (Wontech, Daejeon, Korea) with wavelengths of 630 nm were used to irradiate cells during maturation. The target cells were directly irradiated at an intensity of 40 mW for 750 s (total energy density = 30 J/cm2/day). Light irradiation was performed once a day from days 6 to 8 for 15 min. The power of the LED was measured at the bottom of the empty plate before the cells were irradiated with a SOLO 2 laser power meter (Newport Corporation, Irvine, CA, USA) and an XLP12-1S-H2-DO detector head (Newport Corporation). PBM information and parameters are shown in Table 2.
Table 2.
PBM Device Information and Parameters
| Parameters | Information |
|---|---|
| Manufacturer | Wontech, KOR |
| Light type | light-emitting diode |
| Number of array | 126 |
| Mode | continuous wave (CW) |
| Wavelength | 630 nm |
| FWHM | TYP 16.8 nm/LED |
| Electrical power | TYP 39.8 mW/LED |
| Irradiation time | 750 s/treatment, once a day for 3 days |
| Energy density | 40 mW/cm2 |
FWHM, full width at half maximum; TYP, typical.
EB Morphological Analysis
The size and morphological characteristics of the EBs formed using the different culture techniques were observed using brightfield microscopy (CKX53; Olympus, Tokyo, Japan); the EBs were photographed, and their diameters were measured on days 2 and 6 of differentiation. The average EB diameter obtained using monolayer and hanging-drop cultures was also compared on days 2 and 6 of differentiation. Meanwhile, the serial differentiation of organoid formation on the EBs and GFP expression was observed using epifluorescence from maturation phase at day 7 until day 21.
Epifluorescence Analysis
To prepare the EBs for imaging, cryosections were made using GFP-ESC and ES-J1 cells. GFP-ESC was used to monitor the differentiation into the otic lineage, and ES-J1 was used for the triple immunostaining analysis. Briefly, EBs were fixed with 4% paraformaldehyde in PBS overnight at 4°C and washed three times with cold PBS for 10 min. The cells were cryoprotected by incubating in 10%, 20%, and 30% (w/v) sucrose-PBS solutions for 30 min each and transferred to cyromolds. The sucrose solution was replaced with optical cutting temperature compound (Tissue-Tek, Torrence, CA, USA). The samples were snap frozen with dry ice to make a block and cut into 5 μm slices using a cryostat microtome (Leica, Wetzlar, Germany). The sectioned samples were mounted onto slides and stained for imaging.
After mounting, the cells were permeabilized with 0.25% Triton X-100 (Sigma-Aldrich) in PBS for 10 min at room temperature (RT) and blocked for 1 h at RT with 10% normal goat serum (NGS; Vector Laboratories, Burlington, ON, Canada) and 0.1% Triton X-100 to prevent nonspecific binding.45 To identify MYO7A-positive cells inside organoids, cells were incubated with primary antibody in PBS containing 3% NGS and 0.1% Triton X-100 overnight at 4°C. The cells were washed three times with PBS for 5 min each and then incubated with corresponding secondary antibody for 1 h. Nuclei were visualized using 4′,6-diamidino-2-phenylindole (DAPI). The primary antibodies are shown in Table S5.
For dissected organoids, EBs with protruding organoids were selected and stained. Before mounting, they were dissected using microneedles under a dissecting microscope. The epithelia were spread to flatten them out and mounted on slides for viewing. Representative images were acquired using a confocal microscope (Olympus).
qRT-PCR
To analyze the expression of inner-ear-like structural genes, exactly 1 μg of RNA was collected from each group using various numbers of EBs using GeneAll Hybrid-R (GeneAll Biotechnology, Songpa-gu, Korea), according to the manufacturer’s instructions. cDNA was synthesized using forward and reverse primers (Oligomer, Bioneer, Daejeon, Korea). Detailed data are given in Table 3.
Table 3.
Oligomers Associated with Otic Differentiation Marker
| Oligomer | Forward Sequence | Reverse Sequence |
|---|---|---|
| Atoh1 | 5′-TCCTATGAAGG AGGTGCGGG-3′ |
5′-TTAGGGCCCTG TCCTCGAAG-3′ |
| Ecad | 5′-TCTTAGGCACCC AGTAGGCC-3′ |
5′-TTCCAGGGAGA CTGCTAGGC-3′ |
| Gapdh | 5′-AGGTCGGTGTGA ACGGATTTG-3′ |
5′-GTAGACCATGT AGTTGAGGTCA-3′ |
| Laminin-β1 | 5′-CACCCCTAGCCA ACTTGCTG-3′ |
5′-CTTTGTTCTC CTCACCCGGC-3′ |
| Myo7a | 5′-CACCAAGGGAGA TTGTGGCC-3′ |
5′-CCTTGGACAC CATGACACGG-3′ |
| Pax2 | 5′-GACAGCACCAGAC AAGAGGC-3′ |
5′-TAGCCAAAAA GCCTCGGCAG-3′ |
| Pax8 | 5′-CTTTGCAGTCCC CAGCTCAG-3′ |
5′-GCCAAGTGCT CTCCTGTGTC-3′ |
| Sox2 | 5′-CACCCCTAGCC AACTTGCTG-3′ |
5′-CTTTGTTCTC CTCACCCGGC-3′ |
Gene expression was assessed using qRT-PCR on an AB Applied Biosystem 7500 Real-Time PCR System (Life Technologies, Carlsbad, CA, USA). Three individual experiments were carried out for each sample. Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) was used as a control to normalize target gene expressions.
ROS Production
The production of ROS and intracellular calcium ions was evaluated by confocal microscopy. Immediately after the third LED irradiation, EBs were prepared for epifluorescence analysis, as described above. After permeabilization and blocking, the sectioned samples were stained with 20 μM 2′,7′-dichlorodihydrofluorescein diacetate (H2DCF-DA; Invitrogen) for 15 min at RT and washed twice with PBS. H2DCF-DA is a nonfluorescent compound that is converted into highly fluorescent dichlorofluorescein by cellular peroxides. Calcium Green-1 acetoxymethyl (CG1-AM; Molecular Probes, Eugene, OR, USA) was used to detect calcium ions trapped in the cytoplasm by esterase hydrolysis within the cell. The samples were stained with 4 μM Calcium Green-1 AM for 1 h at 33°C with 5% CO2. Representative images from each staining procedure were acquired using a confocal microscope (Olympus) with emission at 488 nm under an equivalent set of image development conditions. With the use of ImageJ, v1.43u (NIH, Bethesda, MD, USA), the raw image was converted into a mask by image binarization. The mask is used to outline the regions to consider for analysis as regions of interest (ROIs). The area integrated density of the ROI was measured, along with several adjacent background readings. The total corrected total cellular fluorescence (CTCF) = integrated density – (area of selected cell × mean fluorescence of background readings) was calculated.
Sample Preparation and Library Construction for RNA Sequencing Analysis
The effects of PBM on the differentiation of EBs into inner-ear-like organoids were further analyzed via RNA-seq). Only irradiated EBs that showed significant results at 630 nm were used and compared against the control. Five EBs cultured at each condition were pooled, because not enough RNA could be obtained from a single EB. RNA was extracted from the pooled samples using the TRIzol reagent (Invitrogen), according to the manufacturer’s instructions. To increase the purity and avoid DNA contamination, the RNA was purified using an RNeasy Mini Kit in the presence of DNase I (QIAGEN, Hilden, Germany). The quality of the RNA was verified using the 28S/18S ratio and the RNA integrity number (RIN) using an Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). All RNAs extracted from pooled samples of the control and 630 nm-irradiated EBs had RINs > 8.1. Before construction of the cDNA library for RNA-seq, mRNA was enriched with oligo-dT magnetic beads using 2 μg RNA. Double-stranded cDNAs were synthesized immediately with SuperScript III reverse transcriptase (Thermo Fisher Scientific, Waltham, MA, USA). Then, a TruSeq RNA sample prep kit (Illumina, San Diego, CA, USA) was used to subject the synthesized cDNA samples sequentially to end repair, polyadenylation, and 5′ and 3′ adaptor ligation. The processed cDNA fragments were subjected to library enrichment by PCR. Then, appropriately sized fragments were separated on a BluePippin 2% agarose gel cassette (Sage Science, Beverly, MA, USA); the final library size was 400–500 bp. All constructed cDNA libraries were sequenced using the paired-end sequencing mode on an Illumina HiSeq 2500 sequencer.
Transcriptome Alignment and Identification of Differentially Expressed Genes
Prior to sequencing, low-quality raw reads that may cause error in subsequent analyses were filtered out using FastQC. Specifically, reads, including >10% skipped bases, sequencing reads including >40% of bases with a quality score <20%, reads with an average quality score <20, and sequences with a quality score <20 in both ends of the filtered reads, were discarded to increase mapping quality.46 After filtering, the raw reads were aligned with the mouse (Mus musculus) genome using Ensembl release 77 with TopHat software, v.2.1.0.17,47 Only uniquely mapped read pairs were used in DEG analyses. To identify DEGs, gene expression was measured as fragments per kilobase of exon per million fragments (FPKM), and the expression levels were calculated using Cufflinks software, v.2.2.1.48 DEGs between two samples were determined using Cuffdiff in the Cufflinks pipeline. To compare the control with 630 nm, DEGs with a log2FC ≥1 and p <0.05 were investigated. It is necessary to consider that a quantitative analysis with limited statistical option was performed, since the pooled sample with five EBs has been used for DEG comparison. We conducted the DEG analysis using the “blind” option of Cuffdiff in cross-replicate dispersion estimation methods. GO and enrichment analyses were performed using DAVID, v6.8 (https://david.ncifcrf.gov/).20 The GO enrichment analysis workflow followed the following criteria. First, the gene list identified from DEG analysis was used as the input. Second, three main categories of gene functions (CC, MF, and BP) were extracted for GO annotation. Third, functional enrichment analysis was performed with default parameters (minimum overlap of 2, enrichment factor of 1.5, and p < 0.05) for filtering. Clustering analysis of the DEGs was performed based on the log2 FPKM values, and a heatmap was generated using Pretty Heatmaps (pheatmap) software (v1.0.8; available at http://cran.r-project.org/web/packages/pheatmap/index.html) with hierarchical clustering method (complete) functions. To investigate protein–protein interactions, STRING, an open-source bioinformatics tool, was used with the following parameters: organism, Mus musculus; confidence (score), 0.40; and interactors, none/query proteins only.19 Genes related to the GO terms of interest were searched and obtained from the AmiGO 2 database (http://amigo.geneontology.org/amigo).49
Statistical Analysis
All data from experimental and control samples are expressed as means ± standard deviations. At least five replicates per group were used for all experiments, unless stated otherwise. All data were analyzed using GraphPad Prism (GraphPad Software, La Jolla, CA, USA) or SPSS (IBM SPSS Statistics, Armonk, NY, USA) software. Kolmogorov-Smirnov tests were used to determine whether the data were parametric or nonparametric. To reject the null hypothesis, estimation of the power analysis was assessed by determining the probability of type II errors (beta value). p < 0.05 was considered statistically significant and was also defined as ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Author Contributions
S.-Y.C. and N.T.C. performed experiments, data analysis, interpretation, and manuscript writing. S.M. performed experiments. J.Y.J. executed data discussion, data analysis, and interpretation. P.-S.C. provided experimental design and data discussion. H.S. contributed study material. K.H. performed data analysis and interpretation. J.-C.A. and M.Y.L. conceived, designed, and wrote the manuscript and discussed, analyzed, and interpreted data.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
The English in this document has been checked by at least two professional editors, both native speakers of English. For a certificate, please see: http://www.textcheck.com/certificate/sreUk6. This research was supported by a grant from a Creative Materials Discovery Program through the National Research Foundation (2019M3D1A1078943), funded by the National Research Foundation of Korea (NRF) and supported by the Leading Foreign Research Institute Recruitment Program through the NRF, funded by the Ministry of Science and ICT (MSIT) (NRF-2018K1A4A3A02060572). This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number HI15C1524).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2020.03.010.
Contributor Information
Jin-Chul Ahn, Email: jcahn@dankook.ac.kr.
Min Young Lee, Email: eyeglass210@gmail.com.
Supplemental Information
References
- 1.Huang Y., Chi F., Han Z., Yang J., Gao W., Li Y. New ectopic vestibular hair cell-like cells induced by Math1 gene transfer in postnatal rats. Brain Res. 2009;1276:31–38. doi: 10.1016/j.brainres.2009.04.036. [DOI] [PubMed] [Google Scholar]
- 2.Arany P.R., Cho A., Hunt T.D., Sidhu G., Shin K., Hahm E., Huang G.X., Weaver J., Chen A.C., Padwa B.L. Photoactivation of endogenous latent transforming growth factor-β1 directs dental stem cell differentiation for regeneration. Sci. Transl. Med. 2014;6:238ra69. doi: 10.1126/scitranslmed.3008234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barboza C.A., Ginani F., Soares D.M., Henriques A.C., Freitas Rde.A. Low-level laser irradiation induces in vitro proliferation of mesenchymal stem cells. Einstein (Sao Paulo) 2014;12:75–81. doi: 10.1590/S1679-45082014AO2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kushibiki T., Tu Y., Abu-Yousif A.O., Hasan T. Photodynamic activation as a molecular switch to promote osteoblast cell differentiation via AP-1 activation. Sci. Rep. 2015;5:13114. doi: 10.1038/srep13114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wollman Y., Rochkind S., Simantov R. Low power laser irradiation enhances migration and neurite sprouting of cultured rat embryonal brain cells. Neurol. Res. 1996;18:467–470. doi: 10.1080/01616412.1996.11740454. [DOI] [PubMed] [Google Scholar]
- 6.Wu J.-Y., Wang Y.-H., Wang G.-J., Ho M.-L., Wang C.-Z., Yeh M.-L., Chen C.H. Low-power GaAlAs laser irradiation promotes the proliferation and osteogenic differentiation of stem cells via IGF1 and BMP2. PLoS ONE. 2012;7:e44027. doi: 10.1371/journal.pone.0044027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Min K.H., Byun J.H., Heo C.Y., Kim E.H., Choi H.Y., Pak C.S. Effect of Low-Level Laser Therapy on Human Adipose-Derived Stem Cells: In Vitro and In Vivo Studies. Aesthetic Plast. Surg. 2015;39:778–782. doi: 10.1007/s00266-015-0524-6. [DOI] [PubMed] [Google Scholar]
- 8.Park I.-S., Chung P.-S., Ahn J.C. Enhanced angiogenic effect of adipose-derived stromal cell spheroid with low-level light therapy in hind limb ischemia mice. Biomaterials. 2014;35:9280–9289. doi: 10.1016/j.biomaterials.2014.07.061. [DOI] [PubMed] [Google Scholar]
- 9.Soares D.M., Ginani F., Henriques Á.G., Barboza C.A.G. Effects of laser therapy on the proliferation of human periodontal ligament stem cells. Lasers Med. Sci. 2015;30:1171–1174. doi: 10.1007/s10103-013-1436-9. [DOI] [PubMed] [Google Scholar]
- 10.Li H., Liu H., Heller S. Pluripotent stem cells from the adult mouse inner ear. Nat. Med. 2003;9:1293–1299. doi: 10.1038/nm925. [DOI] [PubMed] [Google Scholar]
- 11.Rivolta M., Li H., Heller S. Generation of inner ear cell types from embryonic stem cells. Methods Mol. Biol. 2006;330:71–92. doi: 10.1385/1-59745-036-7:71. [DOI] [PubMed] [Google Scholar]
- 12.Oshima K., Shin K., Diensthuber M., Peng A.W., Ricci A.J., Heller S. Mechanosensitive hair cell-like cells from embryonic and induced pluripotent stem cells. Cell. 2010;141:704–716. doi: 10.1016/j.cell.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Z., Luo X., Zhang L., Lu F., Dong F., Monsell E., Jiang H. Generation of human inner ear prosensory-like cells via epithelial-to-mesenchymal transition. Regen. Med. 2012;7:663–673. doi: 10.2217/rme.12.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Longworth-Mills E., Koehler K.R., Hashino E. Generating Inner Ear Organoids from Mouse Embryonic Stem Cells. Methods Mol. Biol. 2016;1341:391–406. doi: 10.1007/7651_2015_215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koehler K.R., Mikosz A.M., Molosh A.I., Patel D., Hashino E. Generation of inner ear sensory epithelia from pluripotent stem cells in 3D culture. Nature. 2013;500:217–221. doi: 10.1038/nature12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koehler K.R., Hashino E. 3D mouse embryonic stem cell culture for generating inner ear organoids. Nat. Protoc. 2014;9:1229–1244. doi: 10.1038/nprot.2014.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flicek P., Amode M.R., Barrell D., Beal K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fairley S., Fitzgerald S. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang D.W., Sherman B.T., Tan Q., Collins J.R., Alvord W.G., Roayaei J., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J., Hong F., Zhang C., Li L., Wang C., Shi H., Fu Y., Wang J. Differentiation and transplantation of human induced pluripotent stem cell-derived otic epithelial progenitors in mouse cochlea. Stem Cell Res. Ther. 2018;9:230. doi: 10.1186/s13287-018-0967-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamiya K., Karasawa K., Kobayashi K., Miwa A., Ikeda K. Differentiation of iPS Cells to Cochlear Cells are Regulated Depending on the Part of Cocultured Organs. J. Otol. Rhinol. 2015;S1:34–36. [Google Scholar]
- 22.Lahlou H., Lopez-Juarez A., Fontbonne A., Nivet E., Zine A. Modeling human early otic sensory cell development with induced pluripotent stem cells. PLoS ONE. 2018;13:e0198954. doi: 10.1371/journal.pone.0198954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeJonge R.E., Liu X.-P., Deig C.R., Heller S., Koehler K.R., Hashino E. Modulation of Wnt Signaling Enhances Inner Ear Organoid Development in 3D Culture. PLoS ONE. 2016;11:e0162508. doi: 10.1371/journal.pone.0162508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H., Roblin G., Liu H., Heller S. Generation of hair cells by stepwise differentiation of embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2003;100:13495–13500. doi: 10.1073/pnas.2334503100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ronaghi M., Nasr M., Ealy M., Durruthy-Durruthy R., Waldhaus J., Diaz G.H., Joubert L.M., Oshima K., Heller S. Inner ear hair cell-like cells from human embryonic stem cells. Stem Cells Dev. 2014;23:1275–1284. doi: 10.1089/scd.2014.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banerjee M., Bhonde R.R. Application of hanging drop technique for stem cell differentiation and cytotoxicity studies. Cytotechnology. 2006;51:1–5. doi: 10.1007/s10616-006-9001-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foty R. A simple hanging drop cell culture protocol for generation of 3D spheroids. J. Vis. Exp. 2011;51:2720. doi: 10.3791/2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitfield T.T. Development of the inner ear. Curr. Opin. Genet. Dev. 2015;32:112–118. doi: 10.1016/j.gde.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 29.de Freitas L.F., Hamblin M.R. Proposed Mechanisms of Photobiomodulation or Low-Level Light Therapy. IEEE J. Sel. Top. Quantum Electron. 2016;22:7000417. doi: 10.1109/JSTQE.2016.2561201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abramovitch-Gottlib L., Gross T., Naveh D., Geresh S., Rosenwaks S., Bar I., Vago R. Low level laser irradiation stimulates osteogenic phenotype of mesenchymal stem cells seeded on a three-dimensional biomatrix. Lasers Med. Sci. 2005;20:138–146. doi: 10.1007/s10103-005-0355-9. [DOI] [PubMed] [Google Scholar]
- 31.Li W.T., Leu Y.C., Wu J.L. Red-light light-emitting diode irradiation increases the proliferation and osteogenic differentiation of rat bone marrow mesenchymal stem cells. Photomed. Laser Surg. 2010;28(Suppl 1):S157–S165. doi: 10.1089/pho.2009.2540. [DOI] [PubMed] [Google Scholar]
- 32.Peng F., Wu H., Zheng Y., Xu X., Yu J. The effect of noncoherent red light irradiation on proliferation and osteogenic differentiation of bone marrow mesenchymal stem cells. Lasers Med. Sci. 2012;27:645–653. doi: 10.1007/s10103-011-1005-z. [DOI] [PubMed] [Google Scholar]
- 33.Soleimani M., Abbasnia E., Fathi M., Sahraei H., Fathi Y., Kaka G. The effects of low-level laser irradiation on differentiation and proliferation of human bone marrow mesenchymal stem cells into neurons and osteoblasts--an in vitro study. Lasers Med. Sci. 2012;27:423–430. doi: 10.1007/s10103-011-0930-1. [DOI] [PubMed] [Google Scholar]
- 34.Batts S.A., Shoemaker C.R., Raphael Y. Notch signaling and Hes labeling in the normal and drug-damaged organ of Corti. Hear. Res. 2009;249:15–22. doi: 10.1016/j.heares.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung J.Y., Avenarius M.R., Adamsky S., Alpert E., Feinstein E., Raphael Y. siRNA targeting Hes5 augments hair cell regeneration in aminoglycoside-damaged mouse utricle. Mol. Ther. 2013;21:834–841. doi: 10.1038/mt.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slowik A.D., Bermingham-McDonogh O. Hair cell generation by notch inhibition in the adult mammalian cristae. J. Assoc. Res. Otolaryngol. 2013;14:813–828. doi: 10.1007/s10162-013-0414-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abdolazimi Y., Stojanova Z., Segil N. Selection of cell fate in the organ of Corti involves the integration of Hes/Hey signaling at the Atoh1 promoter. Development. 2016;143:841–850. doi: 10.1242/dev.129320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahmoodian Sani M.R., Hashemzadeh-Chaleshtori M., Saidijam M., Jami M.S., Ghasemi-Dehkordi P. MicroRNA-183 Family in Inner Ear: Hair Cell Development and Deafness. J. Audiol. Otol. 2016;20:131–138. doi: 10.7874/jao.2016.20.3.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Natoli R., Zhu Y., Valter K., Bisti S., Eells J., Stone J. Gene and noncoding RNA regulation underlying photoreceptor protection: microarray study of dietary antioxidant saffron and photobiomodulation in rat retina. Mol. Vis. 2010;16:1801–1822. [PMC free article] [PubMed] [Google Scholar]
- 40.Chen A.C., Arany P.R., Huang Y.Y., Tomkinson E.M., Sharma S.K., Kharkwal G.B., Saleem T., Mooney D., Yull F.E., Blackwell T.S., Hamblin M.R. Low-level laser therapy activates NF-kB via generation of reactive oxygen species in mouse embryonic fibroblasts. PLoS ONE. 2011;6:e22453. doi: 10.1371/journal.pone.0022453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strikoudis A., Guillamot M., Aifantis I. Regulation of stem cell function by protein ubiquitylation. EMBO Rep. 2014;15:365–382. doi: 10.1002/embr.201338373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rochette-Egly C. Retinoic acid signaling and mouse embryonic stem cell differentiation: Cross talk between genomic and non-genomic effects of RA. Biochim. Biophys. Acta. 2015;1851:66–75. doi: 10.1016/j.bbalip.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 43.Shen J.X., Qin D., Wang H., Wu C., Shi F.D., Wu J. Roles of nicotinic acetylcholine receptors in stem cell survival/apoptosis, proliferation and differentiation. Curr. Mol. Med. 2013;13:1455–1464. doi: 10.2174/15665240113139990074. [DOI] [PubMed] [Google Scholar]
- 44.Ahn K.S., Jeon S.J., Jung J.Y., Kim Y.S., Kang J.H., Shin S., Choi T., Choi S.J., Chung P., Shim H. Isolation of embryonic stem cells from enhanced green fluorescent protein-transgenic mouse and their survival in the cochlea after allotransplantation. Cytotherapy. 2008;10:759–769. doi: 10.1080/14653240802419286. [DOI] [PubMed] [Google Scholar]
- 45.Lee M.Y., Kabara L.L., Swiderski D.L., Raphael Y., Duncan R.K., Kim Y.H. ROS Scavenger, Ebselen, Has No Preventive Effect in New Hearing Loss Model Using a Cholesterol-Chelating Agent. J. Audiol. Otol. 2019;23:69–75. doi: 10.7874/jao.2018.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin J.A., Wang Z. Next-generation transcriptome assembly. Nat. Rev. Genet. 2011;12:671–682. doi: 10.1038/nrg3068. [DOI] [PubMed] [Google Scholar]
- 47.Ghosh S., Chan C.K. Analysis of RNA-Seq Data Using TopHat and Cufflinks. Methods Mol. Biol. 2016;1374:339–361. doi: 10.1007/978-1-4939-3167-5_18. [DOI] [PubMed] [Google Scholar]
- 48.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carbon S., Ireland A., Mungall C.J., Shu S., Marshall B., Lewis S., AmiGO Hub. Web Presence Working Group AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.