Abstract
The calcium-activated chloride channel, also known as TMEM16A, shows both calcium and membrane potential dependent activation. The channel is expressed broadly and contributes to a variety of physiological processes, and it is expected to be a target for the treatment of diseases such as hypertension, pain, cystic fibrosis and lung cancer. A thorough understanding of the structural characteristics of TMEM16A is important to reveal its physiological and pathological roles. Recent studies have released several Cryo-EM structures of the channel, revealed the structural basis and mechanism of the gating of the channel. This review focused on the understandings of the structural basis and molecular mechanism of the gating and permeation of TMEM16A channel, which will provide important basis for the development of drugs targeting TMEM16A.
Keywords: Ion channel, TMEM16A, CaCCs, Structure, Molecular mechanism, Drug target
1. Introduction
Calcium-activated chloride channels (CaCCs) were first found in the Xenopus oocytes [1], [2]. The protein is broadly expressed and its most important physiological function is to mediate and control anion permeation of the membrane in response to the increasing of intracellular Ca2+ [3], [4]. The extensive confusion and controversy surrounding the molecular identification of CaCCs hindered progress in the field for many years. In 2008, three laboratories demonstrated that CaCCs is encoded by TMEM16A [5], [6], [7]. One year later, TMEM16B was also confirmed to be CaCCs [8]. In vertebrates, the TMEM16 family consists of ten members with a high degree of sequence conservation. Among them, TMEM16A and TMEM16B are CaCCs, while most of the others are considered to serve as Ca2+-activated lipid scramblases with non-selective ion channel activity [9], [10], [11], [12], which catalyze lipid shuffling between leaflets within the bilayer in an ATP-independent manner. In addition, lipid scramblase nhTMEM16 and afTMEM16 were also found in eukaryotic cells from Nectria haematococca and Aspergillus fumigatus, respectively [13], [14].
As the most classical CaCCs, TMEM16A has typical characteristics of CaCC, including Ca2+ and voltage-dependent activation [15], [16], [17], [18]. Recent studies have found that TMEM16A is regulated by phosphatidylinositol 4, 5-bisphosphate [PI(4,5)P2] [19], [20], [21] in addition to calcium ion, voltage and anion regulation. Currently, TMEM16A has been identified in epithelial cells [3], neuronal cells [22], smooth muscle cells [23], vascular endothelial cells [24], and myocardial cells [25]. The wide distribution of TMEM16A in various tissues indicates its diversity in physiological functions [26]. Studies have shown that TMEM16A is involved in control neuronal signaling, airway and exocrine gland secretion, smooth muscle contraction, and rhythmic movements of the gastrointestinal system [4], [27], [28], [29].
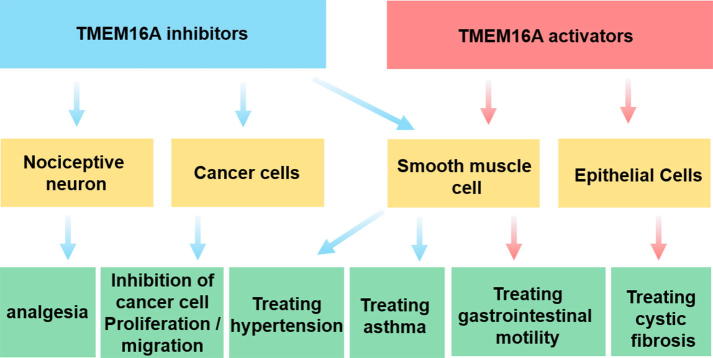
TMEM16A pharmacological modulators may have broad therapeutic applications (Fig. 1) [30], [31]. In epithelial cells, TMEM16A can act as a compensatory chloride channel of cystic fibrosis transmembrane conductance regulator, and activation of it can provide a way for chloride and bicarbonate secretion needed to ameliorate mucociliary clearance and restore anti-microbial activity [32]. It may be an attractive strategy for cystic fibrosis treatment. On the other hand, in small intestinal smooth muscle cells, TMEM16A is required to maintain the slow wave of smooth muscle interstitial cells of Cajal, which indicates that the modulators of TMEM16A channels have regulatory functions on gastrointestinal motility [33], [34]. In the nervous system, TMEM16A is capable of augmenting the excitability of DRG neurons under inflammatory or neuropathic conditions and thereby aggravates inflammation or tissue injury-induced pathological pain [35]. TMEM16A inhibitors have the potential to be developed as novel analgesics. In addition, TMEM16A is also associated with cancer. Studies have found that TMEM16A is abnormally upregulated in some cancer cells including head and neck squamous cell carcinoma [36], gastric cancer [37], gastrointestinal stromal tumors [38] and colon cancer [39], and inhibition of TMEM16A can inhibit proliferation, migration, and invasion of cancer cell [40], [41]. Therefore, TMEM16A channel modulators have potential applications in the treatment of the above diseases.
Fig. 1.
TMEM16A as a drug target. Scheme of possible pharmacological effects (blue: inhibitors; red: activators) of TMEM16A modulators. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Although several TMEM16A regulators have been discovered by high-throughput screening, to find the more specific and potent regulators is still in need [42], and it is urgent to understand the structure and functional relationship of TMEM16A. Fortunately, the structure of the TMEM16A channel with the alternatively-spliced segments a and c (named TMEM16(ac)) was revealed in 2017 [43], [44], [45], and with the development of Cryo-EM technology, more and more structures of TMEM16 family members were revealed, which can guide us to understand the structure–function relationship of TMEM16A. Here, we attempt to summarize the latest research on the structure of TMEM16A and the gating and permeation mechanism of this channel, and hope to provide a comprehensive understanding for drug design based on TMEM16A channel, which will help discover novel drugs, such as analgesics, anticancer drugs, and antihypertensive drugs.
2. The overall structure of the TMEM16A channel
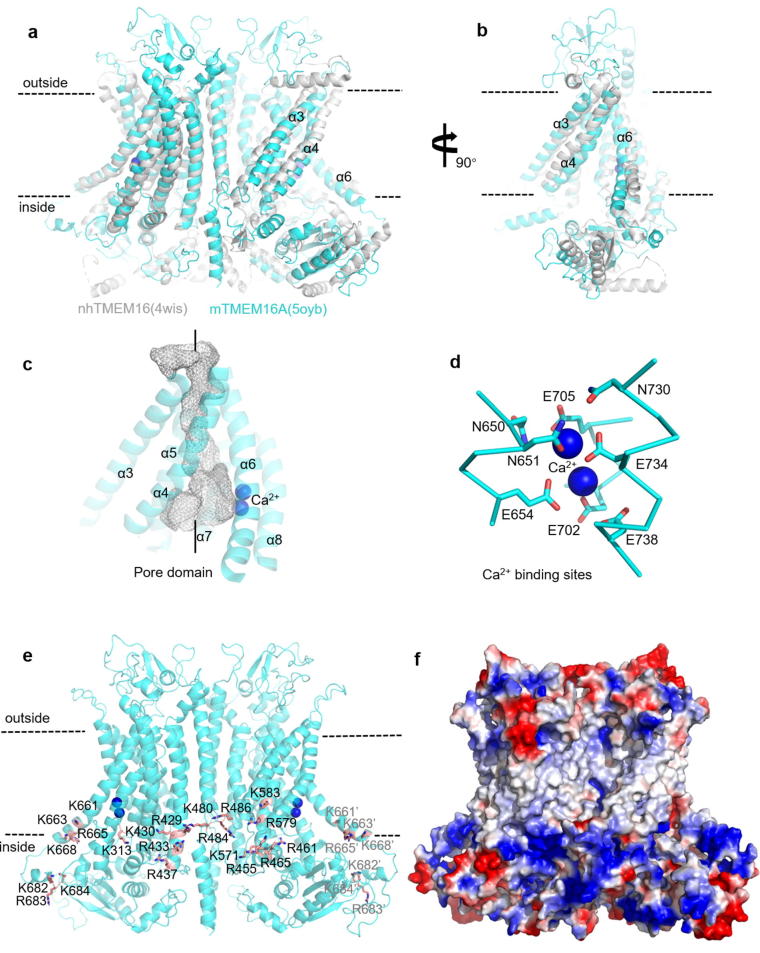
In 2014, Brunner et al. present the X-ray structure of a TMEM16 homologue from Nectria haematococca (nhTMEM16), which is the first high-resolution structure of TMEM16 family (Fig. 2a and b) [13]. The structure of nhTMEM16 is a dimer arranged in a bilobal ‘butterfly’ fold, with each subunit containing a two Ca2+-binding site and ten transmembrane (TM) helices. Each monomer has a hydrophilic, membrane-spanning groove that provides a route for lipid headgroups to move across membranes. Both termini are structured and located on the cytoplasmic side of the membrane. The α-helices and β-strands of the amino terminal domain are organized in a ferredoxin-like fold. The three α helices of the carboxy terminus are wrapped around the N-terminal domain of the adjacent subunit, thereby constituting a large part of the subunit interface [13]. Before the TMEM16A structure was revealed, the structure of nhTMEM16 provided important reference for us to understand the structure of TMEM16A, and provided the possibility for homology modeling of TMEM16A.
Fig. 2.
The molecular architecture of TMEM16A. (a-b) Cartoon representation of overlap of the Ca2+-bound structure of TMEM16A channel (cyan) with lipid scrambler (nhTMEM16; gray). (c) The pore domain of TMEM16A, the pores are shown by a gray grid. (d) Ca2+ binding sites. (e) PI(4,5)P2 binding sites. For the sake of clarity, key amino acids are marked as sticks and only the amino acids that can be displayed on this side are shown. (f) Electrostatic surface of TMEM16A. Red (−50 kT/e); blue (+50 kT/e). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Sequence of TMEM16A has at least four different alternatively-spliced segments a, b, c, and d, resulting in proteins having between 712 and 1006 amino acids [5]. Alternatively-spliced segment a (N-terminal first 116 amino acids) is very important forTMEM16A activity. Stepwise shortening of N-terminus caused an in parallel stepwise decrease in TMEM16A expression and function [46]. Segments b (22 amino acids localized in the N-terminus) and c (EAVK in the first intracellular loop) may be part of protein regions involved in voltage and Ca2+ sensing. Lacking segment b causes an important change in the Ca2+-dependent sensitivity of TMEM16A [47]. Deletion of segment c results in approximately 50-fold decreases in the apparent Ca2+ sensitivity, and enhances voltage-dependent activation of the channel [18]. Segment d (containing 26 amino acids) does not appear to alter TMEM16A channel activities [47]. However, there may be tight coordination between alternative splicing of segment b and d. On the one hand, the five tissues (liver, placenta, prostate, thyroid, and trachea) that contain the b segment tend to exclude the d segment from mature transcripts. On the other hand, skipping of segment b is associated with segment d inclusion in several but not all other tissues [47].
With the maturity of Cryo-EM technology, in May 2017, Paulino et al. first disclosed the TMEM16A structure (PDB ID: 5NL2) of Mus musculus with a resolution of 6.6 Å [43]. The biggest difference from nhTMEM16 is the subunit cavity site, which is sealed from the membrane to prevent lipid ingress and allows only ion permeation. This structure reveals for the first time the structural differences between lipid scramblase and ion channels in the TMEM16 family. In December 2017, two groups published two research papers in Nature independently. They revealed four high-resolution TMEM16A structures in different states, including the double Ca2+ binding state (5OYB, 6BGI); the single Ca2+ binding state (6BGJ), and the Ca2+ free state (5OYG) [44], [45]. Among them, 5OYB is the most complete crystal structure of TMEM16A with highest resolution (Fig. 2a and b). The overall construction of the mTMEM16A are very similar to that of the nhTMEM16. On the extracellular side, long stretches of amino acids that connect α-helices α1–α2, α5–α6 and α9–α10 interact to form a folded domain, consisting of coil regions with interspersed elements of secondary structure that are stabilized by four disulfide bridges. Ten transmembrane helices penetrate the cell membrane, including a closed ion channel and a conserved Ca2+ binding sites. On the intracellular side, the N-terminal domain displays a ferredoxin-like fold that resembles the equivalent part in the nhTMEM16 scramblase (Fig. 2a and b) [13], [44], [45].
3. Ca2+ binding sites
The Ca2+ binding plays a key role for Ca2+-dependent gating in TMEM16A [48], [49]. After years of arguing that TMEM16A is regulated by Ca2+ directly or indirectly, it is considered that the Calmodulin is not necessary in functional TMEM16A [50]. And then, it is a tortuous process to find the Ca2+ binding site because there is no classical motif as EF-hand in the sequence of TMEM16A. Xiao and Pang et al. identify a region (EEEEEAVK) in the first intracellular loop that is crucial for both Ca2+ and voltage sensing [18], [51]. Studies have shown that deletion of EAVK significantly reduces apparent Ca2+ affinity. Mutation of the EEEE/AAAA eliminates the intrinsic voltage dependence but does not alter the apparent Ca2+ affinity [18], [51]. Since this fragment is randomly curled and there is no calcium ion binding to it in the existing stereostructure, how the loop regulates the calcium dependent gating of the channel still needs to be clarified.
In 2012, Yu et al. found that mutating E702 and E705 to glutamate greatly reduced the sensitivity of Ca2+ (mTMEM16A isofrorm 1 (NM-178642) is the basis of the numbering of amino acids) [52]. In 2014, Tien et al. identified several evolutionarily conserved acidic residues in TMEM16A (E654, E702, E705, E734, and D738) that are responsible for calcium activation [50]. Cryo-EM structure confirmed the previous results that acidic residues E654, E702, E705, E734 and D738 group together to form the Ca2+-binding sites in TMEM16A which can bind two calcium ions (Fig. 2d) [44], [45], [50], [52]. These key residues are distributed in the transmembrane α helix 6–8 which are highly conserved among members of the TMEM16 family [13], [50]. In addition to the five key acidic residues described above, mutagenesis studies indicate that the Ca2+ interacts with three polar residues (N650, N651, N730), and mutation of these residues to alanine lowers the potency of Ca2+, suggesting that N650, N651 and N730 are also helpful for Ca2+ binding (Fig. 2d) [45].
Recently, the stereostructure of TMEM16K has been disclosed. Interestingly, TMEM16K has a new Ca2+ binding site in TM10-α10. Examination of the mTMEM16A electron density map indicated that the density of this site was consistent with that of Ca2+, suggesting that TMEM16A may also have a new Ca2+ binding site (P887, D888) [53]. It is a common case that there are more than one ion binding sites in Ca2+ regulated protein like BK channel and Calmodulin [54], [55]. The function of this new Ca2+-binding site is unclear. The sites are conserved in mammalian, but not fungal TMEM16s. Moreover, the mutant of the sites did not alter lipid scrambling function of TMEM16K, we speculate that it may not affect the ion permeation of TMEM16A.
4. Pore domain
To understand how TMEM16A mediates and controls anion permeation, it is important to determine the location of the TMEM16A pore domain. The scramblase structures of TMEM16 family, including TMEM16K, TMEM16F, afTMEM16 and nhTMEM16 [53], [56], [57], [58], confirm that the “subunit cavity” of each subunit provides a suitable pathway for the polar lipid headgroups on their way across the hydrophobic core of the bilayer [58]. The structure of “subunit cavity” is the pore domain that mediates ion permeation in the mTMEM16A. However, this region, especially TM 3, 4 and 6, undergoes a conformational rearrangement relative to the TMEM16 lipid scramblase (Fig. 2b). Due to the rearrangement of the conformation, the 3–7 helix together form a closed channel [44], [45]. The shape of the pore is like an hourglass (Fig. 2c).
Mutagenesis and electrophysiological experiments identified some residues distributed in the pores. These residues are divided into two groups:one group is distributed along the entire pore and is important for anion selectivity and the other group is clustered together and is important for gating. It is reported that R515 on TM3, K603 on TM5–TM6 loop, K588 on TM5 play critical roles in anion selectivity. In addition, N546 and D554 on TM4, N591 and V599 on TM5, Q709 and F716 on TM7 and S639 on TM6 also affect the selectivity of the anion [44]. Considering that the positive charge on the basic residue may is responsible for the anion selectivity, Peters et al. identified four basic residues (R515, K603, R621 and R788) associated with anion selection [59]. On the other hand, mutagenesis studies found that seven residues within the pore may be involved in the calcium dependent gating of TMEM16A. Five mutations, N546A and I550A on TM4, Y593A and I596A on TM5, and F712A on TM7 increased apparent affinity of Ca2+. Otherwise, the V599A on TM5 and L643A on TM6 reduced apparent Ca2+ sensitivity [44]. During the activation of the channel by calcium ions, the neck region of the channel is speculated to undergo a conformational change, thereby allowing ion permeation. Therefore, these gating-related residues, sited in the neck of the channel, may be closely related to the allosteric process of the channel.
5. PI(4,5)P2 binding sites
Phosphatidylinositol (4,5)-bisphosphates [PI(4,5)P2] is an important signaling lipid [60] and a key regulator of many cation channels. However, relatively little is known about the regulation of anion channel by phosphoinositides [61]. In recent years, it has been found that TMEM16A is also regulated by PI(4,5)P2. De Jesús-Pérez et al. found that decreasing of intracellular PI(4,5)P2 resulted in a rundown of the TMEM16A current under whole-cell recording [62]. Ta et al. have reported that PI(4,5)P2 stimulates TMEM16A currents in excised patches [63]. In 2019, Le et al. found that PI(4,5)P2 can regulate activation and desensitization of the TMEM16A channel [19], and confirm the binding residues (R455, K465, R486, K571, R579 and K583) by simulation and mutagenesis experiments (Fig. 2e and f). They propose that the ion-permeable pores of TMEM16A consist of two modules. The “PI(4,5)P2 binding module” of the α helix 3–5 controls the channel desensitization, while the “Ca2+ binding module” of the α helix 6–8 controls Ca2+-dependent activation [19].
At the same time, Yu et al. identified three PI(4,5)P2binding sites in TMEM16A by experimental mutagenesis and molecular modeling [20] to reveal the mechanism by which PI(4,5)P2 regulates TMEM16A. These three sites are composed of 5 (R433, K430, R429, R437, K313), 7 (K659, R662, R665, R668, R682, R683, K684) and 3 (R461, K480, R484) basic residues respectively. It is experimental proved that these residues affect the response of TMEM16A to PI(4,5)P2 in different degrees. (Fig. 2e) [20]. These sites are uniformly distributed near the membrane-cytoplasmic interface where the electrostatic potential is high (Fig. 2f). This is because PI(4,5)P2 itself is negatively charged, its binding site usually contains 2 or more positively charged residues [61], [64]. It is noteworthy that the PI(4,5)P2 binding site is also distributed on TM6, which is the gating component of the TMEM61A channel [45], [65]. Yu et al. believe that PI(4,5)P2 may have an effect on TMEM16A by altering the ion-conducting pathway or affecting Ca2+ binding [20].
PI(4,5)P2 plays a key regulatory role for TMEM16A, but the regulatory effects of other molecules of membrane lipid on TMEM16A are unclear. Therefore, the effects of the lipid environment may be worth studying. but it is not known whether there is a major site of action in so many sites. In addition, we don't know how these binding sites work together to make PI(4,5)P2 work, and how PI(4,5)P2 causes the conformation change of the TMEM16A channel. We expect more reports on the molecular mechanism concerning PI(4,5)P2 regulation of the TMEM16A channel, and hope to obtain the three-dimensional structure of the TMEM16A-PI(4,5)P2 complex.
6. Calcium-dependent conformational transition
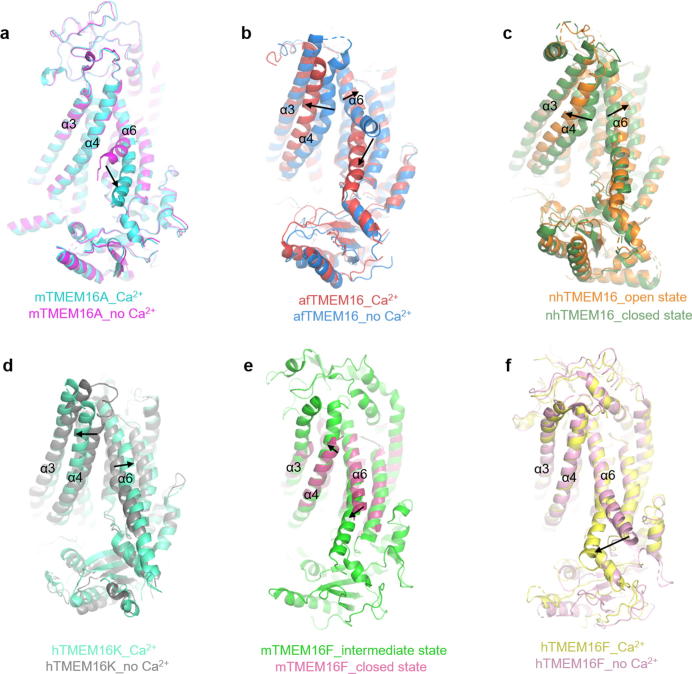
The structure of TMEM16A in different states provides potential information for understanding the allosteric mechanism of Ca2+ activation. The disclosure of a large number of structures of its homologue also provides an important and extensive reference for the investigation of the TMEM16A channel. Based on the TMEM16A structure in Ca2+ free (5OYG) and bound (5OYB) state, Paulino et al. provided an understanding of the activation mechanism of mTMEM16A (Fig. 3a) [45]. The most obvious difference between the two structures is in the inner half of TM6 which is used as a gating element during the activation process. The Ca2+ ions at the binding site attract E654, which causes the lower half of TM6 to reorient and results in the transition of this fragment from α-helix to π-helix. The G644 acts as a door shaft during this process. To further determine the role of TM6 in the gating process, Peters et al. identified key gating residues and Ca2+-binding sites in the sixth transmembrane segment by molecular dynamics simulation and experimental mutagenesis in 2018. The experimental results suggest that K645 at the lower end of TM6 is critical residue for regulating the dependence of channel gating on voltage, Ca2+, and anion [65]. In the same year, Lam et al. found that the bound Ca2+ affect the static of the narrow neck by long-distance coulomb interaction with the two residues K588 and K645 of the channel neck, thereby affecting the ion permeation. Among them, K645 has a stronger effect [66]. In summary, TM6 is a key component of TMEM16A channel gating, especially E654 and K645 on TM6 play a crucial role in Ca2+ binding and ion permeation.
Fig. 3.
Cartoon representation of overlap between different states of TMEM16 member. (a) Ca2+-free structure (magenta) of TMEM16A vs the Ca2+-bound structure (cyan) of it. (b) Ca2+-free structure (blue) of afTMEM16 vs the Ca2+-bound structure (red) of it. (c) Open state (orange) of nhTMEM16 vs the close state (forest) of it. (d) Ca2+-free structure (gray) of hTMEM16k vs the Ca2+-bound structure (greencyan) of it. (e) Close state (ruby) of mTMEM16F vs the intermediate state (green) of it. (f) Ca2+-free structure (lightpink) of hTMEM16F vs the Ca2+-bound structure (yellow) of it. The direction indicated by the arrow is the ion channel or lipid scramblases from closed to open. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
During the gating process, TM3 and TM4 were considered as another key component. Le et al. demonstrated that TM3, TM4 and TM5 are TMEM16A “PI(4,5)P2 Binding Modules”, which bind with PI(4,5)P2 to facilitate channel opening of TMEM16A [19]. Although TM3 and TM4 were observed little difference between the Ca2+-free and Ca2+-bound states of TMEM16A, TM4 showed significant differences between the two states in the structures of nhTMEM16 and afTMEM16 (Fig. 3c and d) [45], [53], [56], [58]. Falzone et al. proposed that opening of the lipid pathway is primarily controlled by two structural elements, TM4 and TM6, which are curved when the protein is Ca2+-free, sealing the pathway from the lipid membrane. The Ca2+ binding facilitates the transition of TM6 to a straight conformation and disengagement of it from TM4, allowing TM6 to move toward TM8 and complete the formation of the Ca2+-binding site [56]. Kalienkova et al. also found a similar process on the nhTMEM16 channel (Fig. 3b and c). In open state of the nhTMEM16, the TM4 and TM6 are separated from each other over their entire length, forming two opposite edges of the semi-circular pole groove exposed to the lipid bilayer [58]. The same rule occurs in hTMEM16k that open conformation of the groove is necessary for scramblase activity (Fig. 3d) [53].
In TMEM16A from mouse, TM4 and TM6 are stacked together to cause TM3-TM7 to form a closed water hole, unlike the grooves accessible by the membrane lipids on both sides of the nhTMEM16, afTMEM16 and hTMEM16K. Considering the homology between channel protein and lipid scramblase of TMEM16 family, it is necessary to clarify whether mTMEM16A will produce a similar conformation rearrangement during the process of permeable ions. Alvadia et al. proposed three lipids transport mechanisms according to the structure of mTMEM16F in closed state and the intermediate state revealed in 2019. They also believe that TM4 and TM6 are key gating components (Fig. 3e) [67]. In the same year, the human TMEM16F structure was also disclosed by Feng et al. and they proposed a transport mechanism that is different from mTMEM16F. They found that opening of the grooves is not necessary during membrane deformation and lipid scrambling (Fig. 3f) [57]. And lipid scrambling and ion permeation do not share the same pathway. This finding provides new insights into the structure and function of TMEM16 family ion channels and lipid scramblase.
The calcium-dependent allosteric transition, also known as gating, and the chloride ions permeation of TMEM16A channel are key issues that needs to be solved urgently. Therefore, the detailed mechanism of how Ca2+ regulate channel gating deserves further study. We believe that Ca2+ can induce conformational changes of TM6, but how to transfer forces and which residues play a key role in the conformational changes of TM6 is inconclusive. In addition, how the two Ca2+ achieve synergistic regulation of the channel is still obscure.
7. Modulators of TMEM16A
Searching for more powerful modulators and studying the mechanism of their interaction with proteins has been a subject worthy of attention in this field of TMEM16A pharmacology. In the past decade, dozens of modulators of TMEM16A have been discovered, including a small number of activators and a large number of inhibitors [68]. Among them, activator ginsenoside Rb1 (GRb1), Resveratrol (RES) and Chitooligosaccharide (COS) can increase the amplitude and frequency of contractions form guinea pig ileum [69], [70], [71], inhibitor Dichlorophenol, Benzbromarone and Hexachlorophenol can be used as bronchodilators and prevent hypersecretion of mucus [72], and inhibitor T16Ainh-A01 and MONNA can reduce DRG neuron sensitivity and reducing neuropathic pain [73]. In addition, Matrine, a potent TMEM16A inhibitor was found to have anti-lung adenocarcinoma effect by our group [74].
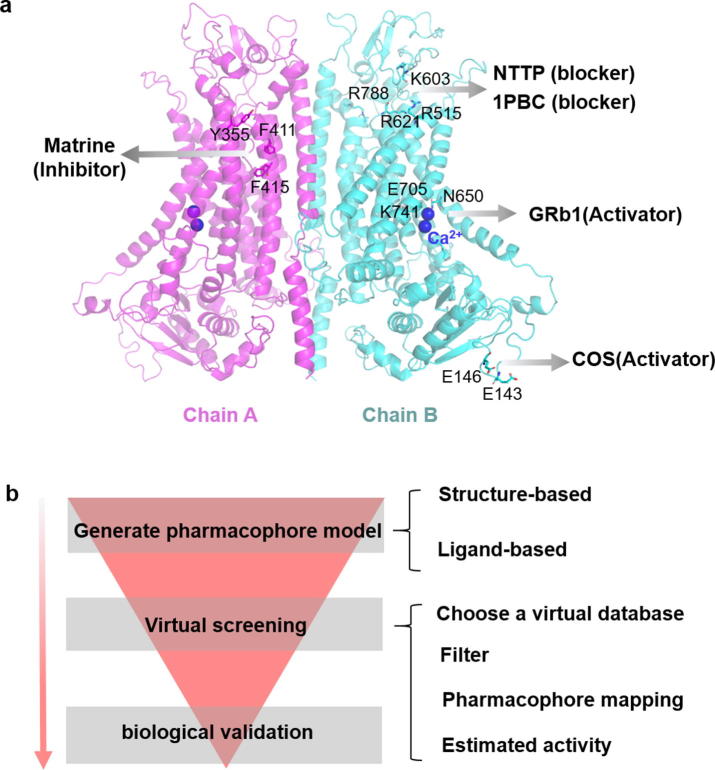
Currently, most researches on modulators mainly focus on the identification of modulators and the potential application of drugs. It is worth noting that the binding sites and regulatory mechanisms of most modulators are not clear, and the interaction mechanism between drugs and TMEM16A is rarely reported. In recent years, we have attempt to propose regulatory mechanisms for some modulators. The binding sites of activator GRb1 include: N650, A697, E705, L746, which E705 is part of the calcium ion binding site (Fig. 4a) [71]. In 2020, we identified two analogs of GRb1 (GRg2, GRf) from ginseng that can activate TMEM16A, and found that they have the same binding site as GRb1 [75]. Importantly, we found that electrostatic interactions and hydrophobic interactions together maintain the stability of the drug at the binding site, and the binding of the modulators may induce TM6 allosteric in the process of activating channels, which is similar to the calcium activation of TMEM16A mechanism. In addition, the binding site of COS is located in the intracellular (E143, E146), and electrostatic interaction is essential for the binding of COS to TMEM16A. However, the detailed mechanism of its activation channel is unknown [69]. The binding site of the inhibitor Matrine is between the first and second transmembrane helix and contains three aromatic amino acids (Y355, F411, F415) (Fig. 4a) [74]. We speculate that π-π stacking interaction may beneficial to binding of Matrine and TMEM16A.
Fig. 4.
Modulators of TMEM16A. (a) Modulator binding sites. Cartoon representation of TMEM16A in which key amino acids are marked as sticks. The two subunits of the TMEM16A are distinguished by magenta and cyan, respectively. The drug name is shown in bold font. (b) The flowchart of virtual screening. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
TMEM16A modulator drug discovery often uses high-throughput screening strategies [76]. This strategy took advantage of YFP fluorescence quenching by halide ions to identify compounds that perturb iodide influx through activated TMEM16A channels. In 2015, Peters et al. used this method to obtain two micromolar-grade TMEM16A channel blockers (NTTP, 1PBC) [59]. Although this is an effective strategy, the investigation is relatively expensive and the cycle is long. Consequently, virtual drug screening is considered an efficient and economical means of drug discovery [77]. The first step of this method is to use a three-dimensional quantitative structure–activity relationship (3D-QSAR) pharmacophore modeling method to construct build a pharmacophore model of the modulator. The second step requires a virtual screening of the drug to obtain candidate compounds. Finally, further biological validation of the candidate compounds would be necessary to absolutely determine the activity (Fig. 4b) [78]. In 2018, Lee et al. screened two potential candidate compounds of TMEM16A inhibitors by a ligand-based 3D-QSAR pharmacophore model and subsequent molecular docking methods [42]. In addition, a virtual screening strategy by constructing a structure-based 3D-QSAR pharmacophore model is also considered feasible, which requires reliable targeting of active sites to build pharmacophore models.
Based on the understanding of TMEM16A structure and the investigation of modulation mechanism, we believe that the upper entrance of the channel is a natural and excellent binding site of its blocker. Therefore, this site is suitable for the construction of a TMEM16A inhibitor pharmacophore model. However, the structure of this site is relatively flexible, and if it is used as a structural model of targeted binding sites, the biological functions of key residues in the pocket need to be further determined. Another important active site is the pocket for calcium binding sites. This site is suitable for the construction of a TMEM16A activator pharmacophore model. It is assumed that the drug can cause channels to opening in a manner similar to Ca2+-activated channels. In addition, some other modulator binding sites away from the pore domain can also be used to build pharmacophore models. However, these sites need to be carefully selected because the mechanism by which they affect the channel is unclear.
8. Summary and outlook
Recent progresses in the research of TMEM16A channel structure provide novel information for investigating its molecular mechanisms. And several new structures of its homologues provide an important reference for understanding the structure–function relationships of TMEM16A. Currently, more detailed gating and ion permeation mechanisms for the TMEM16A channel are need to be clarified, because this is the key to understanding the structure and functional relationship of the TMEM16A channel. In addition, TMEM16A is an important potential biomolecular target for neuropathic pain, asthma and lung cancer, and its structural understanding will facilitate the development of related drugs. However, there is relatively little research concerning the mechanism of TMEM16A modulator interaction with channels. We hope this review will provide readers with a comprehensive understanding for the structure of the TMEM16A channel.
CRediT authorship contribution statement
Sai Shi: Conceptualization, Writing - original draft. Chunli Pang: Writing - review & editing. Shuai Guo: Resources. Yafei Chen: Data curation. Biao Ma: Visualization. Chang Qu: Software. Qiushuang Ji: Writing - original draft. Hailong An: Supervision, Project administration.
Acknowledgments
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 11735006 to Y Zhan, 81830061 to HL An), the Natural Science Foundation of Tianjin of China (Grant No. 19JCYBJC28300 to HL An), the Natural Science Foundation of Hebei Province of China (Grant No. C2018202302 to YF Chen), the Youth Talent Support Program of Hebei Province of China (Grant No. 2013001 to YF Chen).
Competing interests
The authors declare no competing interests.
References
- 1.Barish M.E. A transient calcium-dependent chloride current in the immature Xenopus oocyte. J Physiol. 1983;342:309–325. doi: 10.1113/jphysiol.1983.sp014852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miledi R. A calcium-dependent transient outward current in Xenopus laevis oocytes. Proc R Soc Lond B Biol Sci. 1982;215(1201):491–497. doi: 10.1098/rspb.1982.0056. [DOI] [PubMed] [Google Scholar]
- 3.Huang F. Studies on expression and function of the TMEM16A calcium-activated chloride channel. Proc Natl Acad Sci U S A. 2009;106(50):21413–21418. doi: 10.1073/pnas.0911935106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oh U., Jung J. Cellular functions of TMEM16/anoctamin. Pflugers Arch. 2016;468(3):443–453. doi: 10.1007/s00424-016-1790-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caputo A. TMEM16A, a membrane protein associated with calcium-dependent chloride channel activity. Science. 2008;322(5901):590–594. doi: 10.1126/science.1163518. [DOI] [PubMed] [Google Scholar]
- 6.Schroeder B.C. Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell. 2008;134(6):1019–1029. doi: 10.1016/j.cell.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y.D. TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature. 2008;455(7217):1210–1215. doi: 10.1038/nature07313. [DOI] [PubMed] [Google Scholar]
- 8.Pifferi S., Dibattista M., Menini A. TMEM16B induces chloride currents activated by calcium in mammalian cells. Pflügers Arch Eur J Physiol. 2009;458(6):1023–1038. doi: 10.1007/s00424-009-0684-9. [DOI] [PubMed] [Google Scholar]
- 9.Gyobu S. A role of TMEM16E carrying a scrambling domain in sperm motility. Mol Cell Biol. 2016;36(4):645–659. doi: 10.1128/MCB.00919-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki J. Calcium-dependent phospholipid scramblase activity of TMEM16 protein family members. J Biol Chem. 2013;288(19):13305–13316. doi: 10.1074/jbc.M113.457937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki J. Calcium-dependent phospholipid scrambling by TMEM16F. Nature. 2010;468(7325):834–U135. doi: 10.1038/nature09583. [DOI] [PubMed] [Google Scholar]
- 12.Tian Y., Schreiber R., Kunzelmann K. Anoctamins are a family of Ca2+-activated Cl-channels. J Cell Sci. 2012;125(Pt 21):4991–4998. doi: 10.1242/jcs.109553. [DOI] [PubMed] [Google Scholar]
- 13.Brunner J.D. X-ray structure of a calcium-activated TMEM16 lipid scramblase. Nature. 2014;516(7530):207–212. doi: 10.1038/nature13984. [DOI] [PubMed] [Google Scholar]
- 14.Malvezzi M. Ca2+-dependent phospholipid scrambling by a reconstituted TMEM16 ion channel. Nat Commun. 2013;4(9):2367. doi: 10.1038/ncomms3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Contreras-Vite J.A. Revealing the activation pathway for TMEM16A chloride channels from macroscopic currents and kinetic models. Pflugers Arch. 2016;468(7):1241–1257. doi: 10.1007/s00424-016-1830-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cruz-Rangel S. Gating modes of calcium-activated chloride channels TMEM16A and TMEM16B. J Physiol. 2015;593(24):5283–5298. doi: 10.1113/JP271256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terashima H., Picollo A., Accardi A. Purified TMEM16A is sufficient to form Ca2+-activated Cl-channels. Proc Natl Acad Sci U S A. 2013;110(48):19354–19359. doi: 10.1073/pnas.1312014110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao Q. Voltage- and calcium-dependent gating of TMEM16A/Ano1 chloride channels are physically coupled by the first intracellular loop. Proc Natl Acad Sci U S A. 2011;108(21):8891–8896. doi: 10.1073/pnas.1102147108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le S.C. Molecular basis of PIP2-dependent regulation of the Ca(2+)-activated chloride channel TMEM16A. Nat Commun. 2019;10(1):3769. doi: 10.1038/s41467-019-11784-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu K. A network of phosphatidylinositol 4,5-bisphosphate binding sites regulates gating of the Ca(2+)-activated Cl(-) channel ANO1 (TMEM16A) Proc Natl Acad Sci U S A. 2019;116(40):19952–19962. doi: 10.1073/pnas.1904012116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Jesus-Perez J.J. Phosphatidylinositol 4,5-bisphosphate, cholesterol, and fatty acids modulate the calcium-activated chloride channel TMEM16A (ANO1) Biochim Biophys Acta Mol Cell Biol Lipids. 2018;1863(3):299–312. doi: 10.1016/j.bbalip.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Currie K.P., Wootton J.F., Scott R.H. Activation of Ca(2+)-dependent Cl-currents in cultured rat sensory neurones by flash photolysis of DM-nitrophen. J Physiol. 1995;482(Pt 2):291–307. doi: 10.1113/jphysiol.1995.sp020518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Large W.A., Wang Q. Characteristics and physiological role of the Ca(2+)-activated Cl-conductance in smooth muscle. Am J Physiol. 1996;271(1):435–454. doi: 10.1152/ajpcell.1996.271.2.C435. [DOI] [PubMed] [Google Scholar]
- 24.Nilius B. Calcium-activated chloride channels in bovine pulmonary artery endothelial cells. J Physiol. 1997;498(Pt 2):381–396. doi: 10.1113/jphysiol.1997.sp021865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zygmunt A.C. Intracellular calcium activates a chloride current in canine ventricular myocytes. Am J Physiol. 1994;267(5 Pt 2):H1984–H1995. doi: 10.1152/ajpheart.1994.267.5.H1984. [DOI] [PubMed] [Google Scholar]
- 26.Loretta F. Regulation of TMEM16A chloride channel properties by alternative splicing. J Biol Chem. 2009;284(48):33360–33368. doi: 10.1074/jbc.M109.046607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartzell C., Putzier I., Arreola J. Calcium-activated chloride channels. Annu Rev Physiol. 2005;67:719–758. doi: 10.1146/annurev.physiol.67.032003.154341. [DOI] [PubMed] [Google Scholar]
- 28.Huang F., Wong X.M., Jan L.Y. International union of basic and clinical pharmacology. LXXXV: calcium-activated chloride channels. Pharmacol Rev. 2012;64(1):1–15. doi: 10.1124/pr.111.005009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pedemonte N., Galietta L.J. Structure and function of TMEM16 proteins (anoctamins) Physiol Rev. 2014;94(2):419–459. doi: 10.1152/physrev.00039.2011. [DOI] [PubMed] [Google Scholar]
- 30.Verkman A.S., Galietta L.J. Chloride channels as drug targets. Nat Rev Drug Discov. 2009;8(2):153–171. doi: 10.1038/nrd2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verkman A., Lukacs G., Galietta L. CFTR chloride channel drug discovery – inhibitors as antidiarrheals and activators for therapy of cystic fibrosis. Curr Pharm Des. 2006;12(18):2235–2247. doi: 10.2174/138161206777585148. [DOI] [PubMed] [Google Scholar]
- 32.Sondo E., Caci E., Galietta L.J. The TMEM16A chloride channel as an alternative therapeutic target in cystic fibrosis. Int J Biochem Cell Biol. 2014;52:73–76. doi: 10.1016/j.biocel.2014.03.022. [DOI] [PubMed] [Google Scholar]
- 33.Malysz J. Conditional genetic deletion of Ano1 in interstitial cells of Cajal impairs Ca(2+) transients and slow waves in adult mouse small intestine. Am J Physiol Gastrointest Liver Physiol. 2017;312(3):G228–G245. doi: 10.1152/ajpgi.00363.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh R.D. Ano1, a Ca2+-activated Cl- channel, coordinates contractility in mouse intestine by Ca2+ transient coordination between interstitial cells of Cajal. J Physiol. 2014;592(18):4051–4068. doi: 10.1113/jphysiol.2014.277152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee B. Anoctamin 1 contributes to inflammatory and nerve-injury induced hypersensitivity. Mol Pain. 2014;10(1):5. doi: 10.1186/1744-8069-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dixit R. TMEM16A/ANO1 is differentially expressed in HPV-negative versus HPV-positive head and neck squamous cell carcinoma through promoter methylation. Sci Rep. 2015;5:16657. doi: 10.1038/srep16657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu F. TMEM16A overexpression contributes to tumor invasion and poor prognosis of human gastric cancer through TGF-beta signaling. Oncotarget. 2015;6(13):11585–11599. doi: 10.18632/oncotarget.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Espinosa I. A novel monoclonal antibody against DOG1 is a sensitive and specific marker for gastrointestinal stromal tumors. Am J Surg Pathol. 2008;32(2):210–218. doi: 10.1097/PAS.0b013e3181238cec. [DOI] [PubMed] [Google Scholar]
- 39.Sui Y. Inhibition of TMEM16A expression suppresses growth and invasion in human colorectal cancer cells. PLoS ONE. 2014;9(12) doi: 10.1371/journal.pone.0115443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jia L. Inhibition of calcium-activated chloride channel ANO1/TMEM16A suppresses tumor growth and invasion in human lung cancer. PLoS ONE. 2015;10(8) doi: 10.1371/journal.pone.0136584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Z. Inhibition of Ca2+-activated chloride channel ANO1 suppresses ovarian cancer through inactivating PI3K/Akt signaling. Int J Cancer. 2019;144(9):2215–2226. doi: 10.1002/ijc.31887. [DOI] [PubMed] [Google Scholar]
- 42.Lee Y.H., Yi G.S. Prediction of novel anoctamin1 (ANO1) inhibitors using 3D-QSAR pharmacophore modeling and molecular docking. Int J Mol Sci. 2018:19(10). doi: 10.3390/ijms19103204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paulino C. Structural basis for anion conduction in the calcium-activated chloride channel TMEM16A. Elife. 2017:6. doi: 10.7554/eLife.26232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dang S. Cryo-EM structures of the TMEM16A calcium-activated chloride channel. Nature. 2017;552(7685):426–429. doi: 10.1038/nature25024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paulino C. Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM. Nature. 2017;552(7685):421–425. doi: 10.1038/nature24652. [DOI] [PubMed] [Google Scholar]
- 46.Sondo E. Non-canonical translation start sites in the TMEM16A chloride channel. Biochim Biophys Acta (BBA) – Biomembranes. 2014;1838(1):89–97. doi: 10.1016/j.bbamem.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferrera L. Regulation of TMEM16A chloride channel properties by alternative splicing. J Biol Chem. 2009;284(48):33360–33368. doi: 10.1074/jbc.M109.046607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boese S.H. Kinetics and regulation of a Ca2+-activated Cl- conductance in mouse renal inner medullary collecting duct cells. Am J Physiol Renal Physiol. 2004;286(4):F682–F692. doi: 10.1152/ajprenal.00123.2003. [DOI] [PubMed] [Google Scholar]
- 49.Qu Z., Wei R.W., Hartzell H.C. Characterization of Ca2+-activated Cl- currents in mouse kidney inner medullary collecting duct cells. Am J Physiol Renal Physiol. 2003;285(2):F326–F335. doi: 10.1152/ajprenal.00034.2003. [DOI] [PubMed] [Google Scholar]
- 50.Tien J. A comprehensive search for calcium binding sites critical for TMEM16A calcium-activated chloride channel activity. Elife. 2014:3. doi: 10.7554/eLife.02772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pang C.L. Molecular simulation assisted identification of Ca(2+) binding residues in TMEM16A. J Comput Aided Mol Des. 2015;29(11):1035–1043. doi: 10.1007/s10822-015-9876-x. [DOI] [PubMed] [Google Scholar]
- 52.Yu K. Explaining calcium-dependent gating of anoctamin-1 chloride channels requires a revised topology. Circ Res. 2012;110(7):990–999. doi: 10.1161/CIRCRESAHA.112.264440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bushell S.R. The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K. Nat Commun. 2019;10(1):3956. doi: 10.1038/s41467-019-11753-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guan R.H. Allosteric-activation mechanism of BK channel gating ring triggered by calcium ions. PLoS ONE. 2017:12(9). doi: 10.1371/journal.pone.0182067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pang C. Combining fragment homology modeling with molecular dynamics aims at prediction of Ca(2)(+) binding sites in CaBPs. J Comput Aided Mol Des. 2013;27(8):697–705. doi: 10.1007/s10822-013-9668-0. [DOI] [PubMed] [Google Scholar]
- 56.Falzone M.E. Structural basis of Ca2+-dependent activation and lipid transport by a TMEM16 scramblase. Elife. 2019;8 doi: 10.7554/eLife.43229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feng, S., et al., Cryo-EM studies of TMEM16F calcium-activated ion channel suggest features important for lipid scrambling. Cell Rep, 2019. 28(2): p. 567–579 e4. [DOI] [PMC free article] [PubMed]
- 58.Kalienkova V. Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM. Elife. 2019;8 doi: 10.7554/eLife.44364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peters C.J. Four basic residues critical for the ion selectivity and pore blocker sensitivity of TMEM16A calcium-activated chloride channels. Proc Natl Acad Sci U S A. 2015;112(11):3547–3552. doi: 10.1073/pnas.1502291112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mclaughlin S. PIP(2) and proteins: interactions, organization, and information flow. Annu Rev Biophys Biomol Struct. 2002;31(1):151–175. doi: 10.1146/annurev.biophys.31.082901.134259. [DOI] [PubMed] [Google Scholar]
- 61.Logothetis D.E. Phosphoinositide control of membrane protein function: a frontier led by studies on ion channels. Annu Rev Physiol. 2015;77(1):81. doi: 10.1146/annurev-physiol-021113-170358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De Jesús-Pérez J.J. Phosphatidylinositol 4,5-bisphosphate, cholesterol, and fatty acids modulate the calcium-activated chloride channel TMEM16A (ANO1) Biochim Biophys Acta (BBA) – Mol Cell Biol Lipids. 2017 doi: 10.1016/j.bbalip.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ta C.M. Contrasting effects of phosphatidylinositol 4,5-bisphosphate on cloned TMEM16A and TMEM16B channels. Br J Pharmacol. 2017;174(18):2984–2999. doi: 10.1111/bph.13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suh B.C., Hille B. PIP2 is a necessary cofactor for ion channel function: how and why? Annu Rev Biophys. 2008;37(37):175–195. doi: 10.1146/annurev.biophys.37.032807.125859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peters, C.J., et al., The sixth transmembrane segment is a major gating component of the TMEM16A calcium-activated chloride channel. Neuron, 2018. 97(5): p. 1063–1077 e4. [DOI] [PMC free article] [PubMed]
- 66.Lam A.K., Dutzler R. Calcium-dependent electrostatic control of anion access to the pore of the calcium-activated chloride channel TMEM16A. Elife. 2018:7. doi: 10.7554/eLife.39122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alvadia C. Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F. Elife. 2019;8 doi: 10.7554/eLife.44365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ji Q. Recent advances in TMEM16A: structure, function, and disease. J Cell Physiol. 2019;234(6):7856–7873. doi: 10.1002/jcp.27865. [DOI] [PubMed] [Google Scholar]
- 69.Guo S. Entering the spotlight: chitosan oligosaccharides as novel activators of CaCCs/TMEM16A. Pharmacol Res. 2019;146 doi: 10.1016/j.phrs.2019.104323. [DOI] [PubMed] [Google Scholar]
- 70.Chai R. Identification of resveratrol, an herbal compound, as an activator of the calcium-activated chloride channel, TMEM16A. J Membr Biol. 2017;250(5):483–492. doi: 10.1007/s00232-017-9975-9. [DOI] [PubMed] [Google Scholar]
- 71.Guo S. Ginsenoside Rb1, a novel activator of the TMEM16A chloride channel, augments the contraction of guinea pig ileum. Pflugers Arch. 2017;469(5–6):681–692. doi: 10.1007/s00424-017-1934-x. [DOI] [PubMed] [Google Scholar]
- 72.Huang F. Calcium-activated chloride channel TMEM16A modulates mucin secretion and airway smooth muscle contraction. Proc Natl Acad Sci U S A. 2012;109(40):16354–16359. doi: 10.1073/pnas.1214596109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guadalupe G. Blockade of anoctamin-1 in injured and uninjured nerves reduces neuropathic pain. Brain Res. 2018;1696:38–48. doi: 10.1016/j.brainres.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 74.Guo S. Matrine is a novel inhibitor of the TMEM16A chloride channel with antilung adenocarcinoma effects. J Cell Physiol. 2019;234(6):8698–8708. doi: 10.1002/jcp.27529. [DOI] [PubMed] [Google Scholar]
- 75.Guo S. The molecular mechanism of ginsenoside analogs activating TMEM16A. Biophys J. 2020;118(1):262–272. doi: 10.1016/j.bpj.2019.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nagai T. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20(1):87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 77.Sliwoski G. Computational methods in drug discovery. Pharmacol Rev. 2014;66(1):334–395. doi: 10.1124/pr.112.007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Acharya C. Recent advances in ligand-based drug design: relevance and utility of the conformationally sampled pharmacophore approach. Curr Comput Aided Drug Des. 2011;7(1):10–22. doi: 10.2174/157340911793743547. [DOI] [PMC free article] [PubMed] [Google Scholar]