Abstract
The Streptococcus pyogenes CRISPR/Cas9 (SpCas9) system is now widely utilized to generate genome engineered mice; however, some studies raised issues related to off-target mutations with this system. Herein, we utilized the Campylobacter jejuni Cas9 (CjCas9) system to generate knockout mice. We designed sgRNAs targeting mouse Tyr or Foxn1 and microinjected into zygotes along with CjCas9 mRNA. We obtained newborn mice from the microinjected embryos and confirmed that 50% (Tyr) and 38.5% (Foxn1) of the newborn mice have biallelic mutation on the intended target sequences, indicating efficient genome targeting by CjCas9. In addition, we analyzed off-target mutations in founder mutant mice by targeted deep sequencing and whole genome sequencing. Both analyses revealed no off-target mutations at potential off-target sites predicted in silico and no unexpected random mutations in analyzed founder animals. In conclusion, the CjCas9 system can be utilized to generate genome edited mice in a precise manner.
Keywords: Genetically engineered mouse, Campylobacter jejuni Cas9, CRISPR/Cas9, Gene editing
Highlights
-
•
Generate genetically engineered mice using the Campylobacter jejuni Cas9 (CjCas9) system.
-
•
CjCas9 showed reasonably high biallelic InDel mutation rate (up to 50%) in newborn mice.
-
•
CjCas9 system showed relatively higher specificity compared to SpCas9 system.
The CRISPR/Cas9 system has opened a new era for the production of genetically engineered mice (GEM). Simple microinjection [1] or even electroporation [2] of the CRISPR/Cas9 system can produce knockout and/or knockin mice easily and efficiently, instead of the time- and labor-consuming gene targeting of embryonic stem cells followed by chimeric mice generation. Potential limitation of the CRISPR/Cas9 system in terms of GEM production are unintended off-target mutations and that the target site should be determined by the protospacer adjacent motif (PAM) sequence. Until now, most previous studies have used the Cas9 system derived from Streptococcus pyogenes (SpCas9) for GEM production. For the SpCas9 system, target site should contain 5′-NGG-3′ which is the PAM sequence for SpCas9. For more precise genome editing, other Cas orthologs with different PAM sequences can be utilized for GEM production.
The Campylobacter jejuni Cas9 (CjCas9) system was reported recently as a type II-C CRISPR/Cas9 orthologue [3,4]. The CjCas9 gene is 2.95 kb in size encoding 984 amino acids, allowing it to be packaged into a single adeno-associated virus (AAV) vector with its sgRNA. Therefore, CjCas9 holds potential to be utilized for AAV-mediated in vivo gene editing [5]. The CjCas9 system uses 5′-NNNNACAC-3′ or 5′-NNNNRYAC-3′ as the PAM sequence [5,6], and can be utilized for GEM generation targeting specific sequences that cannot be edited by the SpCas9 system.
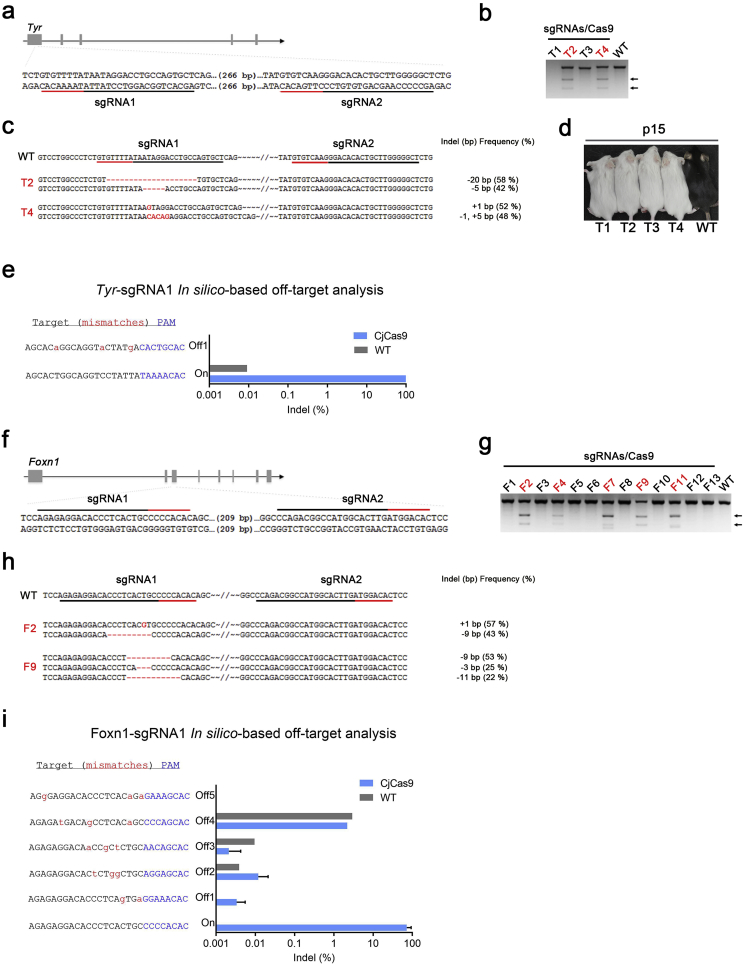
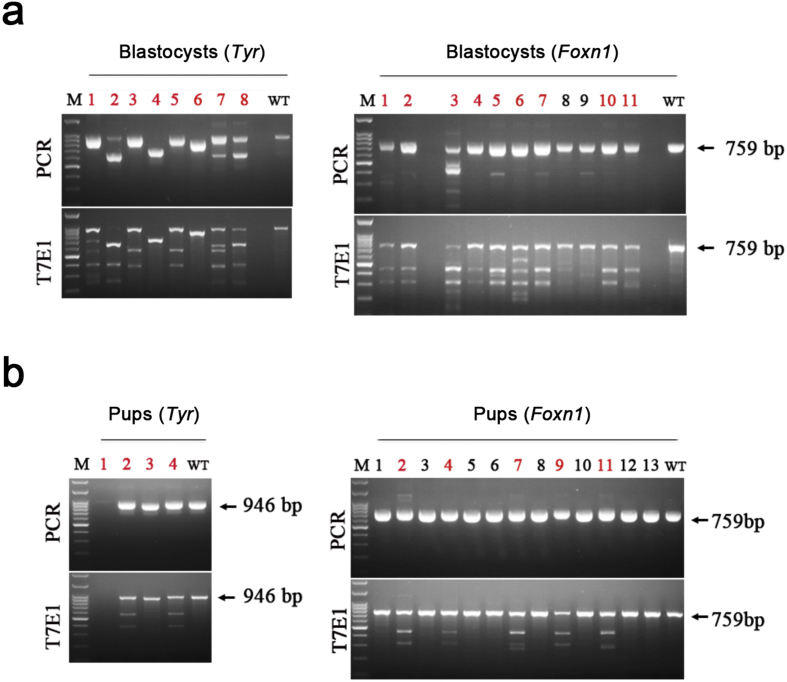
In the current study, we utilized the CjCas9 system for generating knockout mice. We targeted 2 genes, Tyrosine (Tyr) and Forkhead box protein N1 (Foxn1) for this study. We designed 2 sgRNAs for each gene targeting coding region (Table S1, Fig. 1a and f). Dual sgRNAs, each targeting either Tyr or Foxn1 along with the mRNA encoding CjCas9 were microinjected into mouse zygotes. After few days of in vitro culture, a T7E1 assay on genome DNA (gDNA) extracted from blastocysts of microinjected embryos was performed to analyze the genome editing efficiency of CjCas9 in murine embryos. The data revealed high on-target gene editing efficiency of CjCas9 in embryos at 100% (8 out of 8) and 81.8% (9 out of 11) for Tyr and Foxn1 genes, respectively (Fig. S1a). Upon confirming the high on-target gene editing efficiency of CjCas9, we transferred the microinjected embryos into surrogate dams and obtained 4 and 13 founder mice for Tyr and Foxn1 targets, respectively. The T7E1 assay of gDNA extracted from the founders revealed that 2 out of 4 (50%) or 5 out of 13 (38.5%) founders showed targeted mutation in the Tyr or Foxn1 gene, respectively (Fig. 1b and g). Targeted deep sequencing showed mutations in both alleles (~100% indel frequency; Fig. 1c and h, and Fig. S1b). We also observed functional Tyr protein deficiency from the CjCas9-mediated gene knockout, which was confirmed by detecting albino phenotypes in Tyr knockout founders (Fig. 1d). In previous report [7], gene editing efficiency of SpCas9 for generating Tyr knockout GEM was 26–77% when 1 or 2 sgRNA were used. Therefore, we conclude that gene editing efficiency of CjCas9 for GEM production is comparable to the SpCas9 system.
Fig. 1.
Generation and analysis of knockout mice using Campylobacter jejuni Cas9 system. Design of guide RNA sequence for targetting Tyr and Foxn1 genes (a, f). Targetted mutagenesis were analysed by T7E1 assay (b, g) and targetted deep sequencing (c, h). Phenotype changes in Tyr knockout mice (d). Off-target anaysis of the knockout mice (e, i).
Previously, Fuji et al. reported that GEM generation using CjCas9 system [8]. However, off-target analysis results from the report was limited (only 3 potential off-target sites were analyzed). In the present study, we analyze not only potential off-target sites but also mutations in whole genome level and chromosomal changes. To determine the potential off-target sites, we predicted off-target sites up to 3 bp mismatches in silico (Table S3). Targeted deep sequencing of these potential off-target sites revealed high specificity of the CjCas9 system targeting Tyr and Foxn1 genes in murine zygotes (Fig. 1e and i).
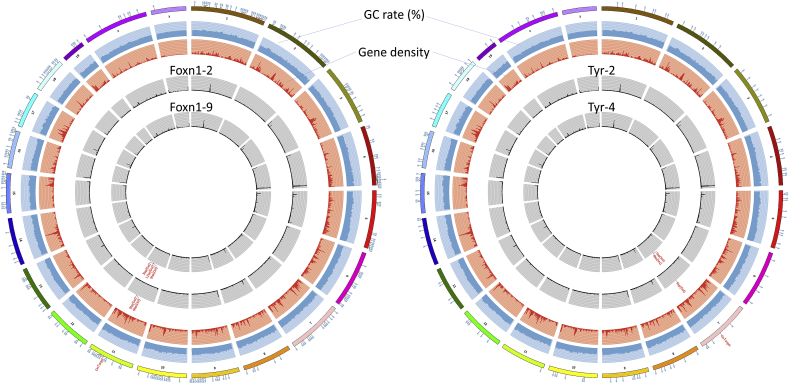
Having achieved knockout mouse colonies using CjCas9, we investigated the target specificities at the whole genome level. We selected 2 samples from Tyr and Foxn1 mutated founders, respectively (T2, T4, F2, and F9), and 2 sham control samples from CjCas9-treated offspring that showed wild-type (WT) genotypes (S1 and S2). We further utilized 2 WT control samples from WT mice obtained from same vendor/colony from where we ordered male mice to perform microinjections (WT1, WT2). These WT controls supposedly “inbred” with our founder mice, however, were produced from a different time/litter. Thus, we selected these mice as controls to show accumulated spontaneous mutations at the whole genome level compared to our founder mice. Our whole-genome sequencing (WGS) data revealed that SNP/InDel differences among all the CjCas9 treated groups (including sham controls) were less than 50; however, sequence differences between CjCas9-treated groups and WT groups were much higher (1277–1603). We also found no evidence of chromosomal changes such as translocation after cjCas9 treatment (data not shown). To identify any potential off-target indel or SNVs caused by CjCas9, WGS was performed for these samples at sufficient depth ( × 20–30) to detect more than 95% of heterozygous variants. We searched for indels at potential off-target sites (Table S6) from our WGS results and did not find any (Fig. 2). Altogether, our data suggest that indel and SNV changes in CjCas9-treated groups when compared to the WT group are more likely to result from spontaneous mutations rather than being CjCas9-induced.
Fig. 2.
Whole-genome sequencing analysis of founder knockout mice.
Generation of GEM with CjCas9 has several advantages. First, CjCas9 utilizes a unique PAM sequence compared to other orthologs allowing more target gRNA sequences to be utilized for a more comprehensive target gRNA choices from different orthologs. Furthermore, its long PAM sequence 5′-NNNNACAC-3′ or 5′-NNNNRYAC-3′ [5,6], may results in relatively higher specificity when compared with SpCas9. A recent elegant study showed that certain serotypes of AAV can penetrate the zona-pellucida [9]. As CjCas9 can be packaged into a single AAV vector with single or multi-sgRNAs and promoters, it is possible to generate genome-edited animals without the need for tools to manipulate zygotes such as microinjectors or electroporators. This may have a particular advantage in certain animals where zygote handling is difficult, such as in non-human primates.
In conclusion, we successfully produced 2 knock-out mouse lines using the CjCas9 system with high efficiency. All the mice generated with CjCas9 system has no off-target mutation and chromosomal changes.
CRediT authorship contribution statement
Jae Young Lee: Investigation, Methodology, Writing - original draft. Yoo Jin Jang: Investigation, Methodology, Writing - original draft. Ji Hyun Bae: Investigation. Yoon Hoo Lee: Investigation. Hee Sook Bae: Investigation. Seokjoong Kim: Conceptualization, Funding acquisition. Sin-Gi Park: Investigation, Data curation. Ok Jae Koo: Conceptualization, Writing - original draft, Writing - review & editing. Su Cheong Yeom: Conceptualization, Writing - review & editing, Supervision.
Declaration of competing interest
JYL, HSB, SJK and OJK are employees of ToolGen Inc, a company develops products based on CRISPR/Cas9 system.
SGP is employee of TheragenEtex Bio Inc, a company providing genome sequencing.
Acknowledgments
This work was supported by grants from ToolGen Inc., South Korea; the National Research Foundation, South Korea (No. 2015R1C1A1A01051949, 2017M3A9B4061404 and 2018M3A9H3020844); the Korea Food and Drug Administration (14182KFDA978), South Korea; and the Korean Health Technology R&D Project, Ministry of Health & Welfare, South Korea (HI16C0426).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2020.100752.
Contributor Information
Ok Jae Koo, Email: oj.koo@toolgen.com.
Su Cheong Yeom, Email: scyeom@snu.ac.kr.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
References
- 1.Jang D.E., Lee J.Y., Lee J.H., Koo O.J., Bae H.S., Jung M.H., Bae J.H., Hwang W.S., Chang Y.J., Lee Y.H., Lee H.W., Yeom S.C. Multiple sgRNAs with overlapping sequences enhance CRISPR/Cas9-mediated knock-in efficiency. Exp. Mol. Med. 2018;50:16. doi: 10.1038/s12276-018-0037-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hashimoto M., Takemoto T. Electroporation enables the efficient mRNA delivery into the mouse zygotes and facilitates CRISPR/Cas9-based genome editing. Sci. Rep. 2015;5 doi: 10.1038/srep11315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fonfara I., Le Rhun A., Chylinski K., Makarova K.S., Lecrivain A.L., Bzdrenga J., Koonin E.V., Charpentier E. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:2577–2590. doi: 10.1093/nar/gkt1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hooton S.P., Connerton I.F. Campylobacter jejuni acquire new host-derived CRISPR spacers when in association with bacteriophages harboring a CRISPR-like Cas4 protein. Front. Microbiol. 2014;5:744. doi: 10.3389/fmicb.2014.00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim E., Koo T., Park S.W., Kim D., Kim K., Cho H.Y., Song D.W., Lee K.J., Jung M.H., Kim S., Kim J.H., Kim J.H., Kim J.S. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017;8 doi: 10.1038/ncomms14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamada M., Watanabe Y., Gootenberg J.S., Hirano H., Ran F.A., Nakane T., Ishitani R., Zhang F., Nishimasu H., Nureki O. Crystal structure of the minimal Cas9 from Campylobacter jejuni reveals the molecular diversity in the CRISPR-cas9 systems. Mol. Cell. 2017;65:1109–1121. doi: 10.1016/j.molcel.2017.02.007. e1103. [DOI] [PubMed] [Google Scholar]
- 7.Zuo E., Cai Y.J., Li K., Wei Y., Wang B.A., Sun Y., Liu Z., Liu J., Hu X., Wei W., Huo X., Shi L., Tang C., Liang D., Wang Y., Nie Y.H., Zhang C.C., Yao X., Wang X., Zhou C., Ying W., Wang Q., Chen R.C., Shen Q., Xu G.L., Li J., Sun Q., Xiong Z.Q., Yang H. One-step generation of complete gene knockout mice and monkeys by CRISPR/Cas9-mediated gene editing with multiple sgRNAs. Cell Res. 2017;27:933–945. doi: 10.1038/cr.2017.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujii W., Ikeda A., Sugiura K., Naito K. Efficient generation of genome-modified mice using Campylobacter jejuni-derived CRISPR/Cas. Int. J. Mol. Sci. 2017;18 doi: 10.3390/ijms18112286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mizuno N., Mizutani E., Sato H., Kasai M., Ogawa A., Suchy F., Yamaguchi T., Nakauchi H. Intra-embryo gene cassette knockin by CRISPR/Cas9-Mediated genome editing with adeno-associated viral vector. iScience. 2018;9:286–297. doi: 10.1016/j.isci.2018.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.