Abstract
Herein we investigated whether inherent differences in mitochondrial activity in mouse pluripotent cells could be used to identify populations with an intrinsic ability to differentiate into primordial germ cells (PGCs). Notably, we determined that stem cells sorted based on differences in mitochondrial membrane activity exhibited altered germline differentiation capacity, with low-mitochondrial membrane potential associated with an increase in PGC-like cells. This specification was not further enhanced by hypoxia. We additionally noted differences between these populations in metabolism, transcriptome, and cell-cycle. These data contribute to a growing body of work demonstrating that pluripotent cells exhibit a large range of mitochondrial activity, which impacts cellular function and differentiation potential. Furthermore, pluripotent cells possess a subpopulation of cells with an improved ability to differentiate into the germ lineage that can be identified based on differences in mitochondrial membrane potential.
Keywords: Biological sciences, Primordial germ cell, PGC, Stem cell, Mitochondria, Stem cell research, Cell biology
Biological sciences; Primordial germ cell; PGC, Stem cell, Mitochondria; Stem cell research; Cell biology.
1. Introduction
There is a well-established link between cellular metabolism and pluripotency, with mitochondrial activity implicated in both stem cell reprogramming and differentiation (Lonergan et al., 2006; Woods, 2017). Naïve mouse ESCs (mESCs), derived from the embryonic inner cell mass (ICM) of the pre-implantation blastocyst, are energetically bivalent and rely on both glycolysis and oxidative phosphorylation for ATP production, while epiblast stem cells (EpiSCs), representing a more differentiated cell type, are distinctly glycolytic in comparison to their naïve ESC counterparts (Zhou et al., 2012). Additionally, activation of oxidative phosphorylation has been shown to be beneficial for efficient transition from a primed to a naïve state in mESCs, indicating there is a link between mitochondrial activity and mESC pluripotency (Sone et al., 2017). Beyond metabolic dependency during pluripotency and early stages of differentiation, mESCs with attenuated mitochondrial activity demonstrate a compromised differentiation response (Mandal et al., 2011) and mESCs sorted for mitochondrial activity based on membrane potential have altered capacity for mesoderm differentiation (Schieke et al., 2008).
Cellular metabolism is tightly regulated in primordial germ cells (PGCs) and both oxidative phosphorylation and glycolysis are required during differentiation from pluripotent stem cells (Hayashi et al., 2017). With recent advances, it is now possible to derive well-characterized primordial germ cell-like cells (PGCLCs) from mESCs and induced pluripotent stem cells (iPSCs), enabling careful observation of early stages of germline differentiation (Hayashi et al., 2011, 2012; Hikabe et al., 2016). Based on the conclusion that cellular metabolism is closely correlated with pluripotency and differentiation, we sought to examine the connection between mitochondrial activity in the differentiation of the mammalian germ lineage.
2. Results
2.1. Pluripotent stem cells are heterogeneous with regard to membrane potential
mESCs and miPSCs exhibit heterogeneity in mitochondrial activity within undifferentiated colonies when cultured in a feeder-free, 2iLIF culture system. Visualization of mitochondrial activity demonstrates that a portion of cells have elevated Δψm as compared to others, regardless of colony size (Figure 1A). Staining with tetramethylrhodamine methyl ester (TMRM) to specifically label active mitochondria that have intact membranes and with Mitotracker Green (MTG) labeling of the total mitochondrial pool as a counterstain, reveals a higher TMRM signal (red) within the center of the colonies, whereas green (indicative of low-membrane potential) predominates the colony edges (Figure 1A). Notably, a number cells in both mESC and miPSC cultures exhibit a yellow color, indicating intracellular mitochondrial heterogeneity (e.g. a combination mitochondria with high- and low-Δψm). To validate the TMRM signal, live mESCs and miPSCs labeled with MTG and TMRM were dissociated into single cells and analyzed by fluorescence activated cell sorting (FACS) in the presence or absence of the mitochondrial uncoupler carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP). Cells that were dual-labeled with MTG and TMRM displayed reduced TMRM signal in response FCCP, but no decrease in MTG signal, indicating that TMRM signal intensity is Δψm-dependent in both mESCs and miPSCs (Figure 1B). To evaluate potential differences in cellular function, mESC and iPSCs were analyzed by FACS and isolated for further culture in vitro, based on differences in Δψm (high- and low-Δψm; Figure 1B).
Figure 1.
Inherent differences in mitochondrial activity in pluripotent stem cells. (A) Imaging of undifferentiated mESC and miPSC colonies. Mitochondria with high membrane potential were stained by TMRM (red) and total mitochondria stained in MitoTracker Green FM (MTG, green), with nuclei stained in Hoechst (blue). Scale bars = 50 μm. (B) mESCs and miPSCs stained with TMRM and MTG were treated with FCCP (400 nM). TMRM staining alone was used to isolate viable cells from the highest 5% of TMRM signal (high-Δψm) and lowest 5% of TMRM signal (low-Δψm). Density plots and histograms represent n = 3 replicates, and a representative plot is shown for the TMRM gating strategy. Data represent mean ± SEM for n = 6 experiments with representative FACS plots shown for each sample set. Significant differences between populations was calculated by student's T-test, with P-values shown.
2.2. Mitochondrial membrane potential correlates to ROS and ATP production in pluripotent stem cells but does not impact pluripotency
To further explore differences between low- and high-Δψm populations of pluripotent cells, we next evaluated ATP production and the generation of reactive oxygen species (ROS) between undifferentiated mESCs and miPSCs, based on low- and high-Δψm. Not unexpectedly, high-Δψm mESCs and miPSCs produced significantly more ATP than low-Δψm cells following isolation via FACS based on differences in membrane potential (Figure 2A). Similarly, quantitative analysis of ROS using flow cytometric analysis of 2′,7′-Dichlorodihydrofluorescein diacetate (H2DCFDA), which converts to 2′,7′-dichlorofluorescein (DCF; green fluorescence) following oxidation, revealed a demonstrable increase in green fluorescence, indicative of elevated ROS, in high-Δψm cells as compared to low-Δψm cells (Figure 2B). To determine whether the observed alterations in mitochondrial activity between the two populations could be attributed to differences in mitochondrial number within the cell, we analyzed the average mitochondrial DNA (mtDNA) copy number by qPCR, using the mtDNA genes NADH dehydrogenase subunit 1 (Nd1) and NADH dehydrogenase subunit 4 (Nd4), normalized to the nuclear gene telomerase reverse transcriptase (Tert). There was no significant difference in the levels of Nd1 or Nd4 between groups, indicating mtDNA copy number does not differ based on Δψm in pluripotent stem cells (Figure 2C).
Figure 2.
Low- and high-Δψm undifferentiated cells have altered ROS and ATP production, but similar mtDNA copy number and differentiation capacity. (A) ATP generation from high-Δψm and low-Δψm cells. (B) ROS detection by H2DCFDA dye incorporation and FACS analysis in high-Δψm and low-Δψm cells. Plots represent n = 3 and values shown are average mean fluorescence intensity ± SEM for each population with P-values. Insets show H2DCFDA signal for unstained cell populations. (C) mtDNA copy number was detected by quantitative PCR of mitochondrial genes Nd1 and Nd4 and normalized to nuclear gene Tert. Fold change was calculated in comparison to low-Δψm cells. (D) 0.5 × 106 cells from each population were injected subcutaneously in Matrigel into female 6-week old NOD/SCID mice to form teratomas (n = 4). Teratomas were removed after 22 or 25 days and processed for H&E staining to reveal cells from all three germ layers. Ectoderm: neural rosettes, endoderm: gland like/epithelium, mesoderm: cartilage. Scale bars = 50 μm. Data for ATP, ROS, and mtDNA copy number analyses represent mean ± SEM for triplicate replicates. Significant differences between populations were calculated by student's T-test, with P-values shown.
Further, we sought to evaluate the inherent or retained cellular plasticity between low- and high-Δψm pluripotent cells via teratoma assay. Undifferentiated mESCs and miPSCs were sorted by TMRM activity, and subsequently injected in a Matrigel plug subcutaneously into the flank of a NOD/SCID recipient mouse and incubated for up to 25 days. Each cell population generated tumors (100% formation rate), with no significant change in size or morphology (Figure S1). All tumors formed were teratomas, based on identification of each of the three germ layers (Figure 2D), indicating no inherent differences in the degree of plasticity between the cell types based on Δψm.
2.3. Transcriptomic profiling of low- and high- Δψm cells reveals distinct transcriptional profiles
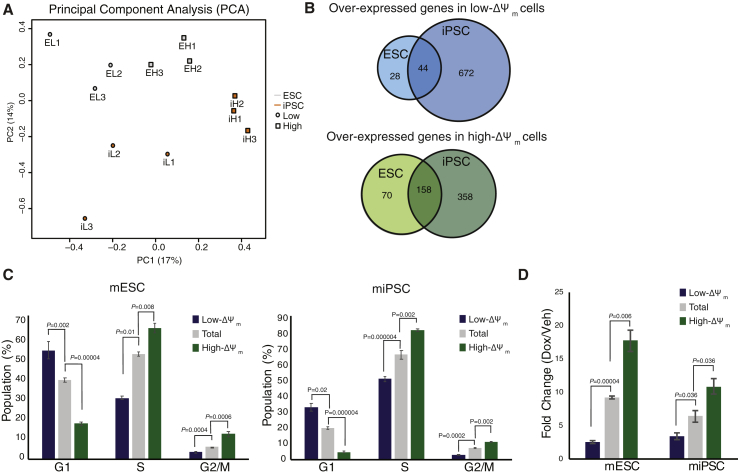
To determine if changes in transcriptome accompanied alterations in mitochondrial membrane potential, undifferentiated mESCs and miPSCs isolated by FACS based on low- and high- Δψm were analyzed by microarray. Principal component analysis (PCA) revealed samples separated by Δψm along the PC1 axis, accounting for 17% of the experimental variance, as well as by cell line along the PC2 axis, which accounted for 14% of the variance (Figure 3A). For ESCs, 302 total genes were differentially expressed between low- and high-Δψm cells, and 1,234 genes were differentially expressed for iPSCs (n = 3, FDR corrected, q threshold = 0.25). Although differences in pluripotency and germ cell markers were not evident (Table S1, Table S2), our data demonstrate transcriptional differences between the groups, as 44 genes were over-expressed in both low-Δψm ESCs and iPSCs, and 158 genes were over-expressed in both high-Δψm ESCs and iPSCs (Figure 3B, Table S5). Notably, genes commonly associated with pluripotency did not demonstrate quantitative changes in expression based on Δψm (Table S1), consistent with the findings of the teratoma assay (Figure 2D).
Figure 3.
Transcriptome changes in low- and high-Δψm cells reveal that mitochondrial activity correlates to cell cycle regulation. (A) Principal component analysis (PCA) was computed across all genes from microarray data of undifferentiated mESCs and miPSCs sorted by membrane potential. (B) Genes that were significantly over-expressed in low-Δψm and high-Δψm cells were compared to identify genes that were over-expressed in both cell lines. (C) Proliferation was assessed by BrdU incorporation and was used in conjunction with DAPI DNA staining to determine cell cycle by FACS. Mean ± SEM are shown for triplicate replicates. Representative FACS plots, controls, and gating strategy are shown in Figure S3. (D) Apoptosis was assessed by caspase 3/7 induction in vehicle (veh) or doxorubicin (dox) treated cells after sorting and 6 h of treatment. Fold change calculated as doxorubicin signal over vehicle for each condition. Mean ± SEM are shown for triplicate biological replicates.
2.4. Analysis of cell-cycle and sensitivity to apoptosis reveals differential responsiveness to doxorubicin based on Δψm
While the microarray data did not demonstrate differences in pluripotency or germ line markers, genes associated with cell cycle were differentially expressed (Tables S2, S3, S4, S5). To determine whether or not Δψm was associated with cell cycle and proliferation rate in pluripotent stem cells, we analyzed cells for membrane potential, proliferation, and DNA content by flow cytometry. After a one-hour BrdU pulse, undifferentiated cells were stained with TMRM and sorted by Δψm as described above. Sorted cells were fixed and assessed for cell cycle stage by BrdU and DAPI labeling (Figure 3C). Analysis of BrdU and DNA content revealed distinct differences in cell cycle based on Δψm; cells that were sorted as high-Δψm had significantly more cells in G2/M and S phase, while the population of low-Δψm cells contained significantly fewer cells in G2/M and S phase. Conversely, the population of low-Δψm contained more cells in the G1 phase, with the total population control cells having an intermediate distribution. This finding was not entirely unexpected, due to our microarray data indicating that high-Δψm pluripotent cells express high-levels of Cyclin B1 (Ccnb1) and Cyclin dependent kinase 1 (Cdk1), as compared to those having low-Δψm. To evaluate apoptotic response to DNA damage, cells were sorted by TMRM activity, normalized by cell number, exposed to 10 μM doxorubicin for 6 h, and then assayed for caspase 3/7 activation as a measure for induction of apoptosis (Figure 3D). Consistent with our cell-cycle analysis, we found that in both mESCs and miPSCs, high-Δψm cells had significantly elevated caspase 3/7 activity in response to doxorubicin treatment as compared to the total cell population (mESC; P = 0.006, miPSC; P = 0.036), while low-Δψm cells had a lower fold change response to doxorubicin over vehicle compared to the total population control (mESC; P = 0.00004, miPSC; P = 0.036). This is expected since, while doxorubicin is classified as a cycle-nonspecific drug, it is most effective at inducing apoptosis during S and G2/M-phases (Ling et al., 1996; Potter et al., 2002).
2.5. Inherent differences in mitochondrial activity influences the ability of pluripotent stem cells to differentiate into primordial germ cell-like cells
It has previously been shown that altered metabolic state can modify germline profiles in differentiating pluripotent stem cell cultures, but a direct relationship between Δψm and PGC specification has not been evaluated (Hayashi et al., 2017). To investigate whether inherent differences in mitochondrial activity can influence PGCLC specification, pluripotent stem cells were sorted based on TMRM signal intensity by gating for the highest 5% TMRM (high-Δψm) or lowest 5% (low-Δψm) of cells and assessed for PGCLC formation following directed differentiation. Low- and high-Δψm pluripotent stem cells – both mESCs and miPSCs – exhibited no significant differences in viability detectable at day 6 of differentiation (Figure 4A) and no significant differences in average aggregate diameter (Figure 4B) as compared to total cell controls. However, FACS analysis of PGCLCs based on the well-characterized cell-surface markers CD61and Stage-Specific Embryonic Antigen-1 (SSEA-1) (Hayashi et al., 2011) revealed a significant increase in immunolabeling of CD61+/SSEA1+ cells differentiated from low-Δψm pluripotent stem cells as compared to those originating from high-Δψm cells (Figure 4C).
Figure 4.
Inherent differences in mitochondrial activity influences the ability of pluripotent stem cells to differentiate into PGCLCs. (A) After sorting by TMRM staining intensity and undergoing directed differentiation to PGCLCs, viability was assessed by DAPI incorporation and FACS analysis. (B) Aggregate diameter was compared for differentiated cells from high- or low-Δψm cells in comparison to the total population. (C) PGCLC differentiation was analyzed by assessing the proportion of SSEA1+/CD61 + cells by FACS. Data represent mean ± SEM for n = 6 experiments with representative FACS plots shown for each sample set. Significant differences between populations was calculated by student's T-test, with P-values shown.
2.6. Primordial germ cell-like cell-specification from pluripotent stem cells is dependent, at least in part, upon oxidative phosphorylation
Primordial germ cells are specified from the epiblast in the developing embryo (Ginsburg et al., 1990; Lawson and Hage, 1994), which relies primarily upon glycolysis for energy production (Leese and Barton, 1984). Culturing cells under hypoxic conditions by lowering the oxygen concentration to 5% O2 alters cellular metabolism by limiting mitochondrial OXPHOS and inducing cells to preferentially undergo glycolysis (Solaini et al., 2010). To determine if promoting a shift to glycolysis could drive specification of PGCLCs, we employed hypoxic culture conditions to undifferentiated and differentiating cultures of mESCs and iPSCs. In undifferentiated mESCs and iPSCs, hypoxic culture conditions significantly increased L-lactate production as compared to normoxic conditions (Figure 5A), and increased gene expression of glycolysis associated genes PDH Kinase 1 (Pdk1), phosphoglycerate kinase 1 (Pgk1), and glucose transporter type 1 (Slc2a1) (Figure 5B). Together, these data indicate that hypoxic culture conditions successfully promoted glycolysis. Undifferentiated cells were cultured under hypoxic conditions for 72 h prior to the initiation of differentiation and were maintained under hypoxia throughout the six-day PGCLC differentiation period. Hypoxia resulted in a slight improvement in cell viability, but no significant difference in aggregate size (Figure 5C). Interestingly, mESCs and miPSCs cultured under hypoxia displayed a significant reduction in the number of PGCLCs in comparison to normoxic control cultures, despite the induced glycolytic metabolic profile (Figure 5D), indicating that induction of PGCLCs is, at least in part, dependent upon OXPHOS.
Figure 5.
Hypoxic culture conditions to decrease mitochondrial activity does not increase germ cell differentiation. (A) L-Lactate production was assessed in undifferentiated cells that had been incubated under hypoxic conditions (5% O2) in comparison to control cells cultured in standard cell culture conditions (~21% O2). Cells were cultured with glucose analog 2-DG to detect cells with decreased L-Lactate production. (B) Glycolysis-related genes and common downstream targets of HIF1Awere evaluated for increase in response to hypoxic culture at 24, 48, and 72 h. (C) ESCs and iPSCs were cultured in normoxic or hypoxic conditions and underwent PGCLC differentiation maintained in each condition. The resulting cells were assessed for viability and imaged for aggregate morphology. (D) ESCs and iPSCs were assessed for PGCLC formation after culture under normoxic or hypoxic conditions. Data represent mean ± SEM for n = 3 replicates. Significant differences between populations was calculated by student's T-test, with P-values shown (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001).
3. Discussion
Pluripotent stem cells are heterogeneous with regard to a number of parameters, including epigenetic status, transcriptional profiling, and mitochondrial activity (Chambers et al., 2007; Chang et al., 2008; Guo et al., 2010; Hayashi et al., 2008). Herein, we confirm that mESCs and iPSCs are heterogeneous with regard to mitochondrial membrane potential, which correlates with metabolic activity and alterations in gene expression. Moreover, we demonstrate that a subset of mESCs and iPSCs, identified and isolated by low-Δψm, preferentially differentiate into PGCLCs following initiation of a directed differentiation protocol. Additionally, these experiments were performed using 2iLIF culture conditions, as opposed to traditional conditions utilizing serum alone, which purportedly result in a dramatic reduction culture heterogeneity. Previous studies have demonstrated that pluripotent stem cells cultured in serum display heterogeneity with regard to morphology as well as expression of pluripotency-related genes, and that this heterogeneity is reduced through the utilization of the 2iLIF system, which incorporates the Mek inhibitor PD0325901 and the GSK3 inhibitor CHIR99021 (Marks et al., 2012; Yamaji et al., 2013; Ying et al., 2008). Indeed, our data support the use of the 2iLIF culture system with regard to pluripotency, however we did observe heterogeneity at the level of mitochondria, metabolism, cell-cycle, and non-pluripotency-related transcriptome.
Mitochondria are characterized as the ‘powerhouse of the cell’, and also participate in a number of fundamental cellular processes such as apoptosis, hormone biosynthesis, hormone signaling and responsiveness, ion flux, thermogenesis, heme synthesis, and processing of toxins, and have additional major roles during embryonic development (McBride et al., 2006; Woods et al., 2018). For example, preimplantation embryos display differential prevalence of high- and low- Δψm, as early as the first cell-fate decision, in the segregation of the trophectoderm (TE) and the inner cell mass (ICM) (Martin et al., 2019). In mice, the role of metabolism is further synchronized with the specification and development of PGCs. Initially, the specification of PGCs begins as early as 7.25 days post coitum (dpc) from the proximal epiblast, with PGCs identifiable initially as a small congregation of alkaline phosphatase-positive cells. At the conclusion of gastrulation, proliferation has increased this population to approximately 50–80 cells [reviewed in (Chiquoine, 1954; Ginsburg et al., 1990; Truman et al., 2017)]. Migration of PGCs from the hindgut to the genital ridge culminates with colonization of the indifferent gonad at approximately embryonic day e10.5 (Molyneaux and Wylie, 2004; Molyneaux et al., 2001). In females, this is followed by a burst of proliferation, in which PGC numbers reach approximately 20,000 (Speed, 1982; Tam and Snow, 1981), and then enter and arrest at the diplotene stage of prophase (Hilscher et al., 1974; Saitou and Yamaji, 2012; Speed, 1982). In males, PGCs arrest following migration to the genital ridge, and remain quiescent until approximately day 5 post-partum, when a subset is activated to become the spermatogonial stem cells (SSCs) of the testis (Saitou and Yamaji, 2012).
Using in vitro pluripotent stem cell models, metabolic profiles synchronous with early stage PGC competence and specification are emerging. As naïve mESCs move toward differentiation into epiblast-like cells (EpiLCs), cellular metabolism shifts from largely OXPHOS to predominantly glycolysis (Hayashi et al., 2017). Recently, it has been demonstrated that in this ‘primed’ glycolytic state, PGC transient developmental competence can be remarkably extended through the addition of alpha-ketoglutarate (αKG; Lu and Teitell, 2019; Tischler et al., 2019). Moreover, stabilization of hypoxia-inducible factor 1 alpha (HIF-1α) acts as a primary driver for the transition of mESCs into EpiSCs, consistent with the acquisition of glycolytic metabolism at this stage in the differentiation process (Zhou et al., 2012). As EpiSCs convert to PGCLCs, this trend is reversed, and OXPHOS increases while glycolysis declines (Lu and Teitell, 2019; Tischler et al., 2019). Importantly, our findings demonstrate that a ‘pre-primed’ state may exist within heterogeneous pluripotent stem cell populations, in which undifferentiated cells exhibiting predominantly low-Δψm and glycolytic metabolic phenotype have an increased potential to develop into PGCLCs. Furthermore, we demonstrate through culture in hypoxic conditions that induction of glycolytic metabolism is not sufficient to drive PGCLC specification. Rather, this population of cells requires OXPHOS to ultimately reach the PGCLC fate, consistent with the reported metabolic properties of the naïve-EpiSC-PGCLC trajectory of differentiation (Lu and Teitell, 2019; Tischler et al., 2019).
In addition to the difference in number of PGCLCs specified between the high- and low-Δψm mESCs, we identified several physiological variances between these two sub-populations of pluripotent stem cells identified based on differential mitochondrial activity. We first determined that cells characterized by higher TMRM generated more ATP per cell than their low-Δψm counterparts, and also generated more ROS per cell, confirming that cells with a predominance of high membrane potential mitochondria indeed represent cells with elevated mitochondrial metabolic activity at the time of sorting. We also determined that there was no significant change in mitochondrial DNA copy number per cell, which is frequently used as a proxy for mitochondrial number (D'Erchia et al., 2015). Because cellular metabolism has been linked to pluripotency (Zhang et al., 2012), we also established that both populations were capable of tri-lineage differentiation in teratoma formation assays, indicating that both populations are still broadly pluripotent despite an altered ability to differentiate to the germ lineage in this system. Prompted by the findings of a microarray analysis, we established that mitochondrial activity correlates to the apoptotic responsiveness and the cell cycle profile of each population of cells, further clarifying that the distinction in mitochondrial profiles is not a transient effect at the time of sorting, but rather identifies populations of PSCs with distinct cellular metabolism.
In the search to identify genes linked to altered mitochondrial activity between the two cell populations, we identified cell cycle control as the primary correlative of mitochondrial activity based on microarray data. Cell cycle phase has been previously linked to naïve ESC pluripotency in both mouse and human studies (Ballabeni et al., 2011; Coronado et al., 2013; Li et al., 2012; Liu et al., 2017; Pauklin and Vallier, 2013; Ter Huurne et al., 2017). Specifically, mouse ESCs are characterized by a short G1 phase which lengthens during differentiation (Coronado et al., 2013). Our findings that low-Δψm cells consist of significantly more cells in G1 phase, as well as an improved differentiation to the germ lineage, provide further evidence that control of germ cell differentiation is dependent on mitochondrial activity and cell cycle phase in the starting population.
The role of metabolism on cell-fate determination is an emerging field, and the role of mitochondrial function in PGC specification is particularly cryptic, given the paucity of existing information and limitations in cell quantity, which preclude in-depth analysis. However, emerging techniques in single cell analytics (Tischler et al., 2019), and even individual organelles (MacDonald et al., 2019) will continue to shed light on both the biological function of mitochondria within stem cells and PGCs, as well as improved strategies on generating fertilization-capable oocytes from stem cell sources (Truman et al., 2017); the latter of which is particularly important given the role of oocyte mitochondria in establishment of the next generation (Woods et al., 2018). The finding that founding mitochondria play a role as early as PGC specificaion could have important implications for the successful dertivation of gametes from PSC and other stem cell sources, and as methods to differentiate the human germline for oocyte production are further characterized (Akahori et al., 2019; Bothun and Woods, 2019; Yamashiro et al., 2018), interrogation of the role of mitochondria in germline development, including the mitochondrial bottleneck, will provide additional insights into the mitochondrial dynamics during gamete development as well as during embryogenesis.
4. Limitations of the study
This study was performed on mouse ESCs and mouse iPSCs, which may not directly relate to biologically similar processes in humans. Future studies could include expansion of this work to human ESCs and iPCSs. Additionally, while we have identified mitochondrial activity as a potential ‘pre-primed’ state for PGC specification, this could be evaluated for the specification of other cell types.
Here, we utilized a model of in vitro germline development to examine the dependency on mitochondrial activity for germline differentiation from mESCs and iPSCs to PGCLCs. We found that mitochondrial activity is heterogeneous at the cellular level within otherwise morphologically similar pluripotent stem cells. Stem cells with lower average mitochondrial membrane potential more efficiently derive into PGCLCs, while cells with high mitochondrial membrane potential represented a more proliferative cell population with lower propensity for germline differentiation. We demonstrate that mitochondrial membrane potential in stem cells correlates with cellular proliferation and cell cycle stage. Furthermore, while culture in hypoxic conditions induced a glycolytic phenotype, this was not sufficient to drive PGC lineage specification. Therefore, we conclude that mitochondrial activity innate to pluripotent stem cells plays a direct role in germline differentiation from pluripotent stem cells.
5. Methods
5.1. Stem cell culture
Female mouse embryonic stem cells (mESCs) (one line of C57Bl/6 origin) and mouse induced pluripotent stem cells (iPSCs) (one line transformed from tail tip fibroblasts of female C57BL/6 mice using lentiviral mediated transduction of Oct4, Sox2, Klf4, and cMyc) were cultured in serum-free, feeder-free media conditions in 2iLIF media on gelatin coated tissue culture treated plastic in humidified 37 °C, 5% CO2 incubators. 2iLIF medium consisted of N2B27 basal media (1:1 mixture of Neurobasal Medium with B27 Supplement and DMEM/F12 Medium with N-2 MAX Media Supplement) supplemented with 400 nM PD0325901 (BioGems Int), 3 uM CHIR99021 (Sigma), and 1,000 U/ml LIF (Shenandoah Biotech). Cells were routinely passaged using Accutase cell detachment solution (Stem Cell Technologies) and all cells were used at passage 45 or earlier. For hypoxia experiments, cells were cultured in a hypoxia chamber flushed with 5% CO2/5% O2/90% N2 gas in pre-equilibrated media.
5.2. Imaging
ESCs and iPSCs were cultured on Matrigel (Corning) coated, 35 mm tissue culture dishes with a glass coverslip bottom in 2iLIF media. Cells were stained with 50 nM tetramethylrhodamine methyl ester (TMRM, Marker Gene) and MitoTracker Green (MTG, Molecular Probes) for 15 min at 37 °C, 5% CO2 and Hoechst 33342 trihydrochloride, trihydrate (Molecular Probes) was added during the last 5 min at 5 μg/ml. Cells were washed and imaged live in PBS on a Zeiss AxioPlan Inverted fluorescent microscope.
5.3. Fluorescence activated cell sorting (FACS)
Undifferentiated stem cells were stained in 2iLIF with 50 nM TMRM for 15 min at 37 °C, 5% CO2. 50 nM MitoTracker Green (MTG, Molecular Probes) was added under the same conditions as needed. 400 nM carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone) (FCCP, Tocris) was added to stained cells and incubated at room temperature for 30 min to assess TMRM specificity for membrane potential. For analysis of reactive oxygen species (ROS), 5 μM 2′,7′,-Dichlorofluorescin Diacetate (H2DCFDA, EMD Millipore) was added for 20 min at 37 °C, 5% CO2. Cells were washed and suspended in FACS buffer (PBS, 1% FBS, 25 mM HEPES, and 1 mM EDTA) for FACS sorting on a BD FACS Aria III special order flow cytometer equipped with the following lasers and filter sets: Blue laser (488 nm) equipped with FSC and SSC detectors and a FITC filter (505LP; 530/40 nm), a Yellow-Green laser (561 nm) equipped with a PE filter (586/15 nm) and a PE-Cy5 filter (635LP; 670/30 nm), and a UV laser (355 nm) equipped with a DAPI filter (450/50 nm). DAPI (Sigma, 0.2 μg/ml) was added to cells immediately prior to sorting for viability analysis. Cells were gated from debris by FSC vs SSC plots, followed by gating for singlet events and lastly, gating for DAPI negative events to determine the viable cell population. Cells representing the top 5% of TMRM labeled cells (high-ΔΨm) or lowest 5% (low-ΔΨm) were sorted from the total viable population. Cells were collected into media and pelleted at 300xg for 5 min for subsequent culture or analysis.
5.4. Directed differentiations and analysis
Primordial germ cell (PGC) differentiations were performed as described previously (Hayashi et al., 2011, 2012). Briefly, 1 × 105 cells were seeded in one well of a fibronectin coated 12-well plate in N2B27 media with 20 ng/ml Activin A, 12 ng/ml bFGF, and 1% knockout serum replacement. Cells were cultured for 48 h, with a media change at 24 h. After 48 h, cells were dissociated and seeded at 2 × 104 cells per well of an ultra-low attachment 96-well plate in GK15 medium (GMEM, 15% knockout serum replacement, 1X non-essential amino acids, 1X sodium pyruvate, 1X pen/strep, L-glutamine, 0.09 mM beta mercaptoethanol) supplemented with 500 ng/ml BMP4, 500 ng/ml BMP8a, 100 ng/ml SCF, 1,000 units LIF, and 50 ng/ml EGF. Cells aggregated and were cultured undisturbed for 4 days.
Aggregates were imaged by light microscopy and average aggregate diameter was measured as a proxy for aggregate size. Aggregates were dissociated in 0.05% trypsin for 8 min and single cell suspensions were washed. Cells were suspended in ice cold FACS buffer (PBS, 0.1% BSA) and labeled with anti-CD61 antibody conjugated to PE (eBiosciences, 1:100) and anti-SSEA1 antibody conjugated with DyLight 650 (Invitrogen, 1:100) on crushed ice for 15 min. Isotype only controls were included to assess non-specific labeling using Hamster IgG conjugated to PE (Invitrogen, 1:100) and Mouse IgM conjugated to DyLight 650 (Thermo, 1:10). Cells were washed in ice cold FACS buffer with DAPI (Sigma, 0.2 μg/ml) added for viability analysis, followed by analysis on a BD FACS Aria III Special Order Research Product. Intact cells were gated from debris by FSC vs SSC plots, followed by gating for singlet events and viable cells, and then gated by fluorescence minus one controls (see Figure S2 for gating strategy).
5.5. ATP and glycolysis cell based assays
Cells sorted by TMRM fluorescence were collected and assessed for ATP production using the ATP Bioluminescence Assay Kit CLS II (Roche). 30,000 cells were collected per biological replicate (n = 3) and each was assayed in technical triplicate so that each assay well had 7,500 cells. L-lactate production was assayed against an L-Lactate standard curve by a Glycolysis Cell-Based Assay Kit (Cayman Chemicals) and results were normalized to cell number. 50 μM 2-deoxy-D-glucose (2-DG, Acros Organics) was added to control cells 24 h prior to analysis. Plates were read on a Synergy H1 plate reader (BioTek).
5.6. qPCR
For mitochondrial copy number analysis, sorted cells were pelleted and lysed (10 mM EDTA, 0.5% SDS, 0.1 mg/ml Proteinase K) at 45 °C for 1 h. Lysate was diluted 1:100 in nuclease-free water and used directly in Taqman Fast Advanced Master Mix reactions (Applied Biosystems) using primers for Tert, VIC-MGB (Mm01352133_m1), Nd1, FAM-MGB (Mm04225274_s1), and Nd4, FAM-MGB (mm04225294_s1) on a StepOne Plus Thermocycler (Applied Biosystems). Nd1 and Nd4 detection levels were quantitated as fold change compared to an internal Tert control.
For RT-qPCR of glycolysis genes, RNA was extracted using the ReliaPrep RNA Miniprep System (Promega). Reverse transcription was performed using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) on DNase I digested RNA. PCR was performed using Fast SYBR Green Master Mix (Applied Biosystems) under standard fast cycling conditions on a StepOne Plus Thermocycler (Applied Biosystems) using the following primer sets: B2m Forward: TTCTGGTGCTTGTCTCACTGA Reverse: CAGTATGTTCGGCTTCCCATTC; Pdk1 Forward: GACTGTGAAGATGAGTGACCG Reverse: CAATCCGTAACCAAACCCAG, Pgk1 Forward: AACCTCCGCTTTCATGTAGAG Reverse: GACATCTCCTAGTTTGGACAGTG, Slc2a1 Forward: AGTTCGGCTATAACACTGGTG Reverse: GTGGTGAGTGTGGTGGATG.
5.7. Teratoma formation
Undifferentiated ESCs or iPSCs were sorted by TMRM as described above and cryopreserved before use. Cells were thawed, prepared in ice cold PBS, and diluted 1:1 with ice cold Matrigel (Corning) to a final volume of 5 × 106 cells in 200 μl Matrigel, which were then injected subcutaneously with a 22-gauge needle into the flank of NOD/SCID mice in n = 4 replicates per condition. Tumors were excised after 4 weeks and were fixed in 4% paraformaldehyde overnight. After processing, 6um sections were cut and stained by hematoxylin and eosin for imaging.
5.8. Microarray
RNA was isolated from sorted cells using the ReliaPrep RNA Miniprep System (Promega) and assayed on Mouse Gene 2.0 ST Arrays (Affymetrix). The arrays were normalized together using the Robust Multiarray Average (RMA) algorithm and a Chip Definition File (CDF) that maps probes on the array to unique Entrez Gene Identifiers. Principal Component Analysis (PCA) was performed across all genes and samples for z-normalized values.
Data were deposited to the GEO repository under the dataset identifier ID GSE128769. Note for review – dataset currently marked private. Link and token for review of dataset prior to publication are: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE128769:gzozisempdkvtqv.
5.9. Apoptosis and cell cycle analysis
For apoptosis analysis, cells were sorted by TMRM fluorescence, normalized by cell number, and plated in a white, 96-well plate. 10 μM doxorubicin was added to cells and they were incubated for 6 h prior to analyzing with the Caspase-Glo 3/7 Assay System (Promega) according to manufacturer's specifications. Luminescent signal was detected on a Synergy H1 plate reader (BioTek).
5-bromo-2 ft.-deoxyuridine (BrdU, Invitrogen eBioscience) was incorporated into undifferentiated cells at 20 μM for one hour at 37 °C, 5% CO2. Cells were collected, stained for TMRM and sorted for mitochondrial activity (see above), and fixed in 70% ethanol. DNA denaturation was performed in 2.0 M HCl for 30 min at room temperature and BrdU was labeled with APC conjugated anti-BrdU antibody (Invitrogen eBioscience) in PBS with 2% FBS for 30 min at room temperature. DNA staining was performed at 1 μg/ml in PBS with 0.1% Triton X-100 for 30 min on crushed ice. Cells were analyzed by bivariate flow cytometry on a BD FACS Aria III; gating was performed to exclude cell debris and isolate single cell events with exclusion of doublets (see Figure S3 for gating strategy). Gates for cell cycle were based on DNA content to identify cells in G1 and G2/M phase (2N and 4N peaks, respectively), with BrdU incorporated into replicating cells (representing the S phase and G2/M phase). Non-BrdU incorporated cells were stained with anti-BrdU antibody with and without DAPI as a control.
5.10. Statistical analyses
All data shown are represented as mean ± SEM for at minimum three biological replicates. Significance was determined by unpaired two-tailed Student's t test. Level of significance was set a P < 0.05.
Declarations
Author contribution statement
A. Bothun: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
D. Woods: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
D. Woods was supported by the National Science Foundation (1750996), and the National Institutes of Health (R01-HD091439).
Competing interest statement
The authors declare no conflict of interest.
Additional information
Data associated with this study has been deposited at GEO repository under the ascension number GSE128769.
Acknowledgements
We thank Yleashandra Maccow, Kaitlyn Izzo, and Hannah Alberico for excellent technical assistance in establishing protocols and collecting data.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Akahori T., Woods D.C., Tilly J.L. Female fertility preservation through stem cell-based ovarian tissue reconstitution in vitro and ovarian regeneration in vivo. Clin. Med. Insights Reprod. Health. 2019;13 doi: 10.1177/1179558119848007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballabeni A., Park I.H., Zhao R., Wang W., Lerou P.H., Daley G.Q., Kirschner M.W. Cell cycle adaptations of embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A. 2011;108:19252–19257. doi: 10.1073/pnas.1116794108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothun A.M., Woods D.C. Dynamics of WNT signaling components in the human ovary from development to adulthood. Histochem. Cell Biol. 2019;151:115–123. doi: 10.1007/s00418-018-1729-y. [DOI] [PubMed] [Google Scholar]
- Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Chang H.H., Hemberg M., Barahona M., Ingber D.E., Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiquoine A.D. The identification, origin, and migration of the primordial germ cells in the mouse embryo. Anat. Rec. 1954;118:135–146. doi: 10.1002/ar.1091180202. [DOI] [PubMed] [Google Scholar]
- Coronado D., Godet M., Bourillot P.Y., Tapponnier Y., Bernat A., Petit M., Afanassieff M., Markossian S., Malashicheva A., Iacone R. A short G1 phase is an intrinsic determinant of naive embryonic stem cell pluripotency. Stem Cell Res. 2013;10:118–131. doi: 10.1016/j.scr.2012.10.004. [DOI] [PubMed] [Google Scholar]
- D'Erchia A.M., Atlante A., Gadaleta G., Pavesi G., Chiara M., De Virgilio C., Manzari C., Mastropasqua F., Prazzoli G.M., Picardi E. Tissue-specific mtDNA abundance from exome data and its correlation with mitochondrial transcription, mass and respiratory activity. Mitochondrion. 2015;20:13–21. doi: 10.1016/j.mito.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Ginsburg M., Snow M.H., McLaren A. Primordial germ cells in the mouse embryo during gastrulation. Development. 1990;110:521–528. doi: 10.1242/dev.110.2.521. [DOI] [PubMed] [Google Scholar]
- Guo G., Huss M., Tong G.Q., Wang C., Li Sun L., Clarke N.D., Robson P. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev. Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Hayashi K., de Sousa Lopes S.M.C., Tang F., Lao K., Surani M.A. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Ogushi S., Kurimoto K., Shimamoto S., Ohta H., Saitou M. Offspring from oocytes derived from in vitro primordial germ cell-like cells in mice. Science. 2012;338:971–975. doi: 10.1126/science.1226889. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Ohta H., Kurimoto K., Aramaki S., Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146:519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- Hayashi Y., Otsuka K., Ebina M., Igarashi K., Takehara A., Matsumoto M., Kanai A., Igarashi K., Soga T., Matsui Y. Distinct requirements for energy metabolism in mouse primordial germ cells and their reprogramming to embryonic germ cells. Proc. Natl. Acad. Sci. U. S. A. 2017;114:8289–8294. doi: 10.1073/pnas.1620915114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikabe O., Hamazaki N., Nagamatsu G., Obata Y., Hirao Y., Hamada N., Shimamoto S., Imamura T., Nakashima K., Saitou M. Reconstitution in vitro of the entire cycle of the mouse female germ line. Nature. 2016;539:299–303. doi: 10.1038/nature20104. [DOI] [PubMed] [Google Scholar]
- Hilscher B., Hilscher W., Bulthoff-Ohnolz B., Kramer U., Birke A., Pelzer H., Gauss G. Kinetics of gametogenesis. I. Comparative histological and autoradiographic studies of oocytes and transitional prospermatogonia during oogenesis and prespermatogenesis. Cell Tissue Res. 1974;154:443–470. doi: 10.1007/BF00219667. [DOI] [PubMed] [Google Scholar]
- Lawson K.A., Hage W.J. Clonal analysis of the origin of primordial germ cells in the mouse. Ciba Found. Symp. 1994;182:68. doi: 10.1002/9780470514573.ch5. discussion 84—91. [DOI] [PubMed] [Google Scholar]
- Leese H.J., Barton A.M. Pyruvate and glucose uptake by mouse ova and preimplantation embryos. J. Reprod. Fertil. 1984;72:9–13. doi: 10.1530/jrf.0.0720009. [DOI] [PubMed] [Google Scholar]
- Li V.C., Ballabeni A., Kirschner M.W. Gap 1 phase length and mouse embryonic stem cell self-renewal. Proc. Natl. Acad. Sci. U. S. A. 2012;109:12550–12555. doi: 10.1073/pnas.1206740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Y.H., el-Naggar A.K., Priebe W., Perez-Soler R. Cell cycle-dependent cytotoxicity, G2/M phase arrest, and disruption of p34cdc2/cyclin B1 activity induced by doxorubicin in synchronized P388 cells. Mol. Pharmacol. 1996;49:832–841. [PubMed] [Google Scholar]
- Liu L., Michowski W., Inuzuka H., Shimizu K., Nihira N.T., Chick J.M., Li N., Geng Y., Meng A.Y., Ordureau A. G1 cyclins link proliferation, pluripotency and differentiation of embryonic stem cells. Nat. Cell Biol. 2017;19:177–188. doi: 10.1038/ncb3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonergan T., Brenner C., Bavister B. Differentiation-related changes in mitochondrial properties as indicators of stem cell competence. J. Cell. Physiol. 2006;208:149–153. doi: 10.1002/jcp.20641. [DOI] [PubMed] [Google Scholar]
- Lu V., Teitell M.A. Alpha-ketoglutarate: a "magic" metabolite in early germ cell development. EMBO J. 2019;38 doi: 10.15252/embj.2018100615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald J.A., Bothun A.M., Annis S.N., Sheehan H., Ray S., Gao Y., Ivanov A.R., Khrapko K., Tilly J.L., Woods D.C. A nanoscale, multi-parametric flow cytometry-based platform to study mitochondrial heterogeneity and mitochondrial DNA dynamics. Commun Biol. 2019;2 doi: 10.1038/s42003-019-0513-4. Article number: 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal S., Lindgren A.G., Srivastava A.S., Clark A.T., Banerjee U. Mitochondrial function controls proliferation and early differentiation potential of embryonic stem cells. Stem Cell. 2011;29:486–495. doi: 10.1002/stem.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H., Kalkan T., Menafra R., Denissov S., Jones K., Hofemeister H., Nichols J., Kranz A., Stewart A.F., Smith A. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J.J., Woods D.C., Tilly J.L. Implications and current limitations of oogenesis from female germline or oogonial stem cells in adult mammalian ovaries. Cells. 2019;8 doi: 10.3390/cells8020093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride H.M., Neuspiel M., Wasiak S. Mitochondria: more than just a powerhouse. Curr. Biol. 2006;16:R551–R560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Molyneaux K., Wylie C. Primordial germ cell migration. Int. J. Dev. Biol. 2004;48:537–544. doi: 10.1387/ijdb.041833km. [DOI] [PubMed] [Google Scholar]
- Molyneaux K.A., Stallock J., Schaible K., Wylie C. Time-lapse analysis of living mouse germ cell migration. Dev. Biol. 2001;240:488–498. doi: 10.1006/dbio.2001.0436. [DOI] [PubMed] [Google Scholar]
- Pauklin S., Vallier L. The cell-cycle state of stem cells determines cell fate propensity. Cell. 2013;155:135–147. doi: 10.1016/j.cell.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter A.J., Gollahon K.A., Palanca B.J., Harbert M.J., Choi Y.M., Moskovitz A.H., Potter J.D., Rabinovitch P.S. Flow cytometric analysis of the cell cycle phase specificity of DNA damage induced by radiation, hydrogen peroxide and doxorubicin. Carcinogenesis. 2002;23:389–401. doi: 10.1093/carcin/23.3.389. [DOI] [PubMed] [Google Scholar]
- Saitou M., Yamaji M. Primordial germ cells in mice. Cold Spring Harb. Perspect. Biol. 2012;4 doi: 10.1101/cshperspect.a008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieke S.M., Ma M., Cao L., McCoy J.P., Jr., Liu C., Hensel N.F., Barrett A.J., Boehm M., Finkel T. Mitochondrial metabolism modulates differentiation and teratoma formation capacity in mouse embryonic stem cells. J. Biol. Chem. 2008;283:28506–28512. doi: 10.1074/jbc.M802763200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solaini G., Baracca A., Lenaz G., Sgarbi G. Hypoxia and mitochondrial oxidative metabolism. Biochim. Biophys. Acta. 2010;1797:1171–1177. doi: 10.1016/j.bbabio.2010.02.011. [DOI] [PubMed] [Google Scholar]
- Sone M., Morone N., Nakamura T., Tanaka A., Okita K., Woltjen K., Nakagawa M., Heuser J.E., Yamada Y., Yamanaka S. Hybrid cellular metabolism coordinated by Zic3 and Esrrb Synergistically enhances induction of naive pluripotency. Cell Metab. 2017;25:1103–1117. doi: 10.1016/j.cmet.2017.04.017. e1106. [DOI] [PubMed] [Google Scholar]
- Speed R.M. Meiosis in the foetal mouse ovary. I. An analysis at the light microscope level using surface-spreading. Chromosoma. 1982;85:427–437. doi: 10.1007/BF00330366. [DOI] [PubMed] [Google Scholar]
- Tam P.P., Snow M.H. Proliferation and migration of primordial germ cells during compensatory growth in mouse embryos. J. Embryol. Exp. Morphol. 1981;64:133–147. [PubMed] [Google Scholar]
- Ter Huurne M., Chappell J., Dalton S., Stunnenberg H.G. Distinct cell-cycle control in two different states of mouse pluripotency. Cell Stem Cell. 2017;21:449–455. doi: 10.1016/j.stem.2017.09.004. e444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischler J., Gruhn W.H., Reid J., Allgeyer E., Buettner F., Marr C., Theis F., Simons B.D., Wernisch L., Surani M.A. Metabolic regulation of pluripotency and germ cell fate through alpha-ketoglutarate. EMBO J. 2019;38 doi: 10.15252/embj.201899518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman A.M., Tilly J.L., Woods D.C. Ovarian regeneration: the potential for stem cell contribution in the postnatal ovary to sustained endocrine function. Mol. Cell. Endocrinol. 2017;445:74–84. doi: 10.1016/j.mce.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods D.C. Mitochondrial heterogeneity: evaluating mitochondrial subpopulation dynamics in stem cells. Stem Cells Int. 2017;2017:7068567. doi: 10.1155/2017/7068567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods D.C., Khrapko K., Tilly J.L. Influence of maternal aging on mitochondrial heterogeneity, inheritance, and function in oocytes and preimplantation embryos. Genes. 2018;9 doi: 10.3390/genes9050265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji M., Ueda J., Hayashi K., Ohta H., Yabuta Y., Kurimoto K., Nakato R., Yamada Y., Shirahige K., Saitou M. PRDM14 ensures naive pluripotency through dual regulation of signaling and epigenetic pathways in mouse embryonic stem cells. Cell Stem Cell. 2013;12:368–382. doi: 10.1016/j.stem.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Yamashiro C., Sasaki K., Yabuta Y., Kojima Y., Nakamura T., Okamoto I., Yokobayashi S., Murase Y., Ishikura Y., Shirane K. Generation of human oogonia from induced pluripotent stem cells in vitro. Science. 2018;362:356–360. doi: 10.1126/science.aat1674. [DOI] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Nuebel E., Daley G.Q., Koehler C.M., Teitell M.A. Metabolic regulation in pluripotent stem cells during reprogramming and self-renewal. Cell Stem Cell. 2012;11:589–595. doi: 10.1016/j.stem.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Choi M., Margineantu D., Margaretha L., Hesson J., Cavanaugh C., Blau C.A., Horwitz M.S., Hockenbery D., Ware C. HIF1alpha induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 2012;31:2103–2116. doi: 10.1038/emboj.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.