Abstract
There is a desire to engineer mammalian host cell lines to improve cell growth/biomass accumulation and recombinant biopharmaceutical protein production in industrially relevant cell lines such as the CHOK1 and HEK293 cell lines. The over-expression of individual subunits of the eukaryotic translation factor eIF3 in mammalian cells has previously been shown to result in oncogenic properties being imparted on cells, including increased cell proliferation and growth and enhanced global protein synthesis rates. Here we report on the engineering of CHOK1 and HEK cells to over-express the eIF3i and eIF3c subunits of the eIF3 complex and the resultant impact on cell growth and a reporter of exogenous recombinant protein production. Transient over-expression of eIF3i in HEK293 and CHOK1 cells resulted in a modest increase in total eIF3i amounts (maximum 40% increase above control) and an approximate 10% increase in global protein synthesis rates in CHOK1 cells. Stable over-expression of eIF3i in CHOK1 cells was not achievable, most likely due to the already high levels of eIF3i in CHO cells compared to HEK293 cells, but was achieved in HEK293 cells. HEK293 cells engineered to over-express eIF3i had faster growth that was associated with increased c-Myc expression, achieved higher cell biomass and gave enhanced yields of a reporter of recombinant protein production. Whilst CHOK1 cells could not be engineered to over-express eIF3i directly, they could be engineered to over-express eIF3c, which resulted in a subsequent increase in eIF3i amounts and c-Myc expression. The CHOK1 eIF3c engineered cells grew to higher cell numbers and had enhanced cap- and IRES-dependent recombinant protein synthesis. Collectively these data show that engineering of subunits of the eIF3 complex can enhance cell growth and recombinant protein synthesis in mammalian cells in a cell specific manner that has implications for the engineering or selection of fast growing or high producing cells for production of recombinant proteins.
Keywords: Cell engineering, mRNA translation, Eukaryotic initiation factor eIF3, CCT, Chaperonin containing T-polypeptide 1, c-Myc
Highlights
-
•
We have engineered the overexpression of eIF3i and eIF3c in CHOK1 and HEK293 cells.
-
•
HEK293 cells overexpressing eIF3i had faster growth and increased c-Myc expression.
-
•
Direct stable overexpression of eIF3i in CHOK1 cells was not achievable.
-
•
Overexpression of eIF3c in CHOK1 cells resulted in an increase in eIF3i.
-
•
eIF3c overexpressing CHOK1 cells had enhanced recombinant protein synthesis.
1. Introduction
There remains a desire to improve recombinant biopharmaceutical protein production from cultured mammalian cells (Budge, 2020), particularly in industrially relevant cell lines such as the CHOK1 and HEK293 cell lines (Kim et al., 2012; Lin et al., 2015; Godfrey et al., 2017; Jossé et al., 2018). Both these cell lines are particularly robust, amenable to genetic manipulations and have fast growth and proliferation rates that reflect their high protein synthetic capacity. Improvements in vector design for recombinant protein production in mammalian cells are such that transcription is, in most cases, no longer considered rate-limiting for synthesis of these proteins. Initiation of translation, on the other hand, remains a tightly controlled, potentially rate-limiting step in mammalian cell protein synthesis. A commonly adopted strategy in the large-scale production of therapeutic proteins in mammalian cells is to lower the culture temperature towards the end of rapid growth to mildly hypothermic levels (30–33 °C) (Wagstaff et al., 2013). This has been found empirically to prolong the productive life of the culture by delaying apoptosis and nutrient depletion and, for some proteins, this strategy also improves the yield of correctly folded protein (Al-Fageeh and Smales, 2006a, 2006b; Underhill and Smales, 2007).
Previously we have shown that although global protein synthesis is reduced during mammalian cell culture under mildly hypothermic conditions (Underhill et al., 2005), recombinant mRNA levels can be increased at reduced temperature (27–32 °C) due to increased stability and this, together with increased protein stability and enhancement of correct protein folding, can positively affect recombinant protein yields from mammalian cell lines (Marchant et al., 2008). In the course of a proteomic screen of protein synthesis changes occurring as cells are exposed to mildly sub-physiological temperatures, we identified eIF3i as one protein whose synthesis rate decreased significantly more than the global attenuation observed upon cooling and was then increased above the global increase upon rewarming (Roobol et al., 2009). This suggested that overexpression of eIF3i in CHO and HEK cells might enhance protein synthesis and compensate, at least in part, for the decrease in eIF3i under mildly hypothermic conditions. Other studies have also shown the relevance of translational control in mammalian cells to recombinant protein synthesis e.g. (Hargreaves et al., 2012, 2015; Roobol et al., 2015; Jossé et al., 2016; Vito and Smales, 2018), hence demonstrating the potential for engineering translational control to enhance cellular growth and recombinant protein production capability.
eIF3i is a component of the eukaryotic initiation factor 3 (eIF3) complex, the largest and most complex eukaryotic mRNA translation factor in terms of the number of its protein components. Mammalian eIF3 contains a single copy of 12 different subunits, 5 of which, a, b, c, g and i, are conserved and essential in vivo from yeasts to mammals (Hinnebusch, 2006; Pestova et al., 2007; Valášek et al., 2017). Mass spectrometry of intact and salt-dissociated subcomplexes of eIF3 has provided a subunit interaction map of the complex (Zhou et al., 2008) while several cryoEM studies (des Georges et al., 2015; Hashem et al., 2013; Querol-Audi et al., 2013; Smith et al., 2016) have delineated the overall shape of the complex, the locality of each of its subunits within this, and the positioning of the complex bound to the 40S ribosomal subunit. The eIF3 complex forms a 5-lobed structure that binds to the solvent-exposed side of the 40S ribosomal subunit. The various eIF3 subunits also contribute several additional binding sites for a number of other translation initiation factors (Aitken et al., 2016; Valášek, 2012) and stimulate mRNA binding to the 40S (Villa et al., 2013). The eIF3 complex can therefore be viewed as a scaffold that facilitates the bringing together, in the appropriate orientation, of key protein machinery components required to form the 43S pre-initiation complex. For cap-dependent translation binding between eIF3 and eIF4G promotes binding of the 43S complex with the eIF4F complex at the mRNA cap structure, resulting in the formation of the 48S preinitiation complex which can then scan to the start AUG codon (Hershey et al., 2000). There is also evidence that the association of eIF3 with the ribosome persists for the first few rounds of translation elongation and is thought to aid resumption of scanning after uORFs (Hronová et al., 2017) and has been shown to play a role in translation termination (Beznosková et al., 2015).
eIF3i is a 36 kDa protein containing 7WD repeat sequences which fold as a 7-bladed β-propeller (Herrmannová et al., 2012; Smith et al., 1999). WD repeat proteins commonly form such rigid, circular structures that serve as a stable platform for protein-protein interactions. The importance of this structure to eIF3i function has been confirmed in yeast by the severe effects on cell proliferation and global protein synthesis of mutations in the WD repeats of eIF3i, when compared with mutations elsewhere in the protein (Verlhac et al., 1997). However, based upon pull-down assays to determine subunit interactions within the eIF3 complex (Valasek et al., 2002) and from the analysis of eIF3 subcomplexes by mass spectrometry (Zhou et al., 2008), it appears that eIF3i is not centrally located within the complex and its association with the complex is labile. Its only direct binding partner within the complex is the large scaffolding subunit eIF3b (Verlhac et al., 1997; Fraser et al., 2004) though there is evidence for the spectrin domain of eIF3a being involved in the formation of an a-b-i-g complex (Dong et al., 2013) Although conserved and essential in vivo, eIF3i is not necessary for in vitro reconstitution of a pre-initiation complex that can scan to the start AUG (Matsutani et al., 2007). The role of eIF3i within the eIF3 complex may therefore be more related to its mediating essential in vivo regulatory inputs into the translation initiation process rather than its being an essential structural component within the pre-initiation complex.
eIF3i is one of 5 eIF3 components (a, b, c, h and i) that, when stably overexpressed in 3T3 cells, induce an oncogenic phenotype with increased growth rate, increased protein synthetic rate, attenuated apoptosis and increased anchorage-independent growth (Zhang et al., 2007). However, in contrast to overexpression of the large core subunits a, b and c, stable overexpression of eIF3i does not upregulate the expression of the remaining eIF3 components. Stable overexpression of eIF3c or eIFF3h selectively increases the translation of several mRNAs encoding proteins involved in cell growth and proliferation (Zhang et al., 2007). Expression of the encoded proteins is normally tightly regulated and/or restricted to specific parts of the cell cycle, so their overexpression leads to unregulated growth. With the goal of enhancing protein synthesis in CHO and HEK cells, we here examine the effect of transient and stable overexpression of eIF3i in these cell lines.
2. Materials and methods
2.1. Cells and cell culture
CHOK1, HEK293 and NIH3T3 cells were maintained as described elsewhere (Roobol et al., 2009). The Invitrogen FlpIn system was used to generate stable CHOK1 and HEK293 cell lines according to manufacturer's instructions. In all cases cells were maintained in a 5% CO2 environment. Cell proliferation was in some cases measured by growing cells in E-plates monitored using the xCELLigence System (ACEA Biosystems). Metabolic labelling with 35S-labelled amino acids was carried out in cysteine and methionine deficient DMEM (Sigma, D0422) containing Tran[35S] label (MP Radiochemicals) at 380 kBq/ml for 1 h. Exposure to mildly hypothermic conditions (32 or 27 °C), 24 h post-transfection, was for a further 24 h.
2.2. Plasmids and transfections
For transient over-expression of eIF3 subunits in mammalian cells, cDNAs encoding human eIF3 subunits were amplified by RT-PCR from HeLa cell RNA with primers (Table 1) flanked by suitable restriction sites for insertion into pcDNA3.1V5His (Invitrogen), with or without the natural stop codon, the latter permitting read through to a V5 tag on the distal vector arm, or, for generating stable cell lines, into pcDNA5-FRT (Invitrogen). The plasmids used in attempts to generate the CHO cell lines expressing HA-tagged human eIF3c or HA-tagged eIF3i were a kind gift from Dr. L.Zhang (University of California, Davis). For transient overexpression of CCT oligomer, cDNAs encoding the 8 CHO CCT subunits were generated by RT-PCR (primers in Table 2) then inserted separately into the pcDNA3.1 vector. Cells were transiently transfected 24 h after seeding 3 x 105 viable cells per well in 6-well plates, generally with 2 μg DNA per well using Optimem (Invitrogen) as a diluent and either Lipofectamine 2000 or LTX Lipofectamine with Plus reagent (Invitrogen) as transfection vehicle according to the manufacturer's instructions. For co-transfection of eIF3i and CCT subunits, 2 μg/well of eIF3i plasmid and 1 μg/well of each CCT plasmid were used. Based on transfections with a pcDNA3.1 construct encoding GFP, typical transfection efficiencies of ~50% were obtained.
Table 1.
Primers used for amplification of cDNAs encoding human eIF3 subunits (restriction sites added for insertion into particular vectors are not shown).
| Gene Target | Sequence |
|---|---|
| 3c forward | TCGCCATGTCGCGGTTTTTC |
| 3c reverse | TCAGTAGGCCGTCTGAGACTGC |
| 3g forward | TTTGCGATGCCTACTGGAGACTTTG |
| 3g reverse | TTAGTTGGTGGACGGCTTGG |
| 3h forward | AAGATGGCGTCCCGCAAGGAAG |
| 3h reverse | TTAGTTGTTGTATTCTTGAAGAGCC |
| 3i forward | GGGATGAAGCCGATCCTACTGC |
| 3i reverse | TTCTTAAGCCTCAAACTCAAATTCG |
Table 2.
Primers used for amplification of cDNAs encoding CHO cell CCT subunits.
| Gene Target | Sequence |
|---|---|
| CCT1 forward | AAGGATCCGTTCCCCGCTGTGGTGGC |
| CCT1 reverse | GCGAGCACAGAATTAATACGACT |
| CCT2 forward | TATGGATCCACTTCCGCCTTCCTTTCCCC |
| CCT2 reverse | AGTTTAAGTAACTGGGTTTTTATTTTAATACAGTCTGAG |
| CCT3 forward | TATGGATCCCTCTTCTCTCCAGAAGCTTCTGCC |
| CCT3 reverse | CAAATGACTTAGATTTAATCATTGGAAGC |
| CCT4 forward | TATGGATCCTCAGCTTCCGCCTCCTCTGC |
| CCC4 reverse | GCAGTTTGGAATCAAATATTTAATATTTTCATTAAATC |
| CCT5 forward | TATGGATCCTAGTCCCGAGCGGTCCGTG |
| CCT5 reverse | TTAACAGTTTCCAAACAGCTGCTTTTATTCCAGC |
| CCT6 forward | TATGGATCCACGTTGTTCTCGGCCTCTCC |
| CCT6 reverse | CTCATCTTAATATGTAATTTTTGTGTCAATGTAG |
| CCT7 forward | TATGGATCCGAGCATTGTGGGCTTGGGTG |
| CCT7 reverse | ATGAGAAAAACCATGACCAAATAATCTTAAG |
| CCT8 forward | TCTTCCTCCGAGCGCGTGAG |
| CCT8 reverse | CGCGGATCCGAATTAATACGACTCACTATAGG |
2.3. Antibodies
Proteins containing the V5 epitope were detected with a mouse monoclonal anti-V5 (Sigma, V8012) and HA-tagged proteins with a rabbit anti-HA (Sigma H6908). The rabbit polyclonals against eIF3i and eIF3h were sourced from Proteintech Group (11287-1-AP) and Sigma (AY50491), respectively. Other anti-eIF3 subunit antibodies used were; anti-eIF3a, Cell Signaling (#2538); anti-eIF3b, Santa Cruz (sc16377); anti-eIF3c, Sigma (E6408). A rabbit polyclonal against an N-terminal sequence of c-Myc was from Abcam (ab11917). Subunit-specific rabbit polyclonals against individual CCT subunits and an antibody to RPL7a have been characterized elsewhere (Roobol and Carden, 1999). α-tubulin was detected with the TAT mouse monoclonal (Woods et al., 1989) which was a kind gift from Prof. Keith Gull (University of Oxford, UK). β-actin was detected with the mouse monoclonal AC-15 (Sigma). For western blot detection of pulldown proteins a protein A-HRP conjugate (Millipore) was used, otherwise anti-rabbit, -mouse or -goat-HRP was used as secondary.
2.4. Cell extraction, SDS-PAGE, western blotting, immunoprecipitation, immunofluorescence microscopy and luciferase assays
Protein was extracted from mammalian cells after the cells had been washed with PBS at the appropriate temperature, by lysis in the culture flask or plate with ice-cold 20 mM HEPES-NaOH, pH 7.2, containing 100 mM NaCl, 10 mM Na β-glycerophosphate, 1% (w/v) Triton X-100, 50 mM NaF, 1 mM activated Na orthovanadate, 0.2 mM PMSF, 10 μg/ml leupeptin and 2 μg/ml pepstatin. SDS-PAGE and western blotting were undertaken as previously described (Roobol et al., 2009). Immunoprecipitation under native conditions and immunofluorescence microscopy were undertaken as described by Roobol et al. (1999). For immunoprecipitation under denaturing conditions, the Triton-100 in the above lysis buffer was replaced by 1% NP40 + 0.5% deoxycholate +0.1% SDS. For luciferase measurement, cells were transfected with a pcDNA3.1 vector encoding firefly luciferase and 24 h later cells were lysed and luciferase activity determined using the SteadyLite Plus reagent according to manufacturer's instructions. To assess cap- and IRES-dependent translation, cells were transfected with the bicistronic reporter construct pRMF (Stoneley et al., 2000) with which cap-dependent translation was determined by Renilla luciferase activity while IRES-dependent translation was determined by Firefly luciferase activity using the Promega Dual-Glo kit. Protein half-life determinations were as in (Roobol et al., 2015). Protein was quantified by the method of Bradford (1976).
3. Results
3.1. The levels of transiently over-expressed eIF3i achievable in CHOK1 and HEK293 cells are limited and transient over-expression does little to enhance protein synthesis
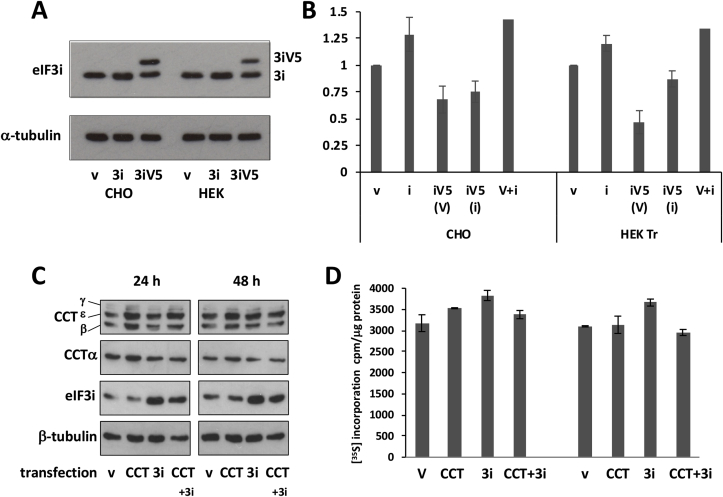
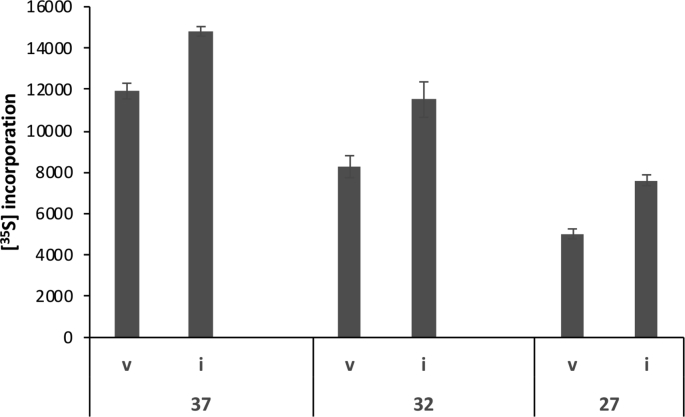
CHOK1 and HEK cells transiently transfected with eIF3i-pcDNA3.1 showed only a limited overall increase (30–40%) in total eIF3i expression 24 h after transfection (Fig. 1A and B). In the case of V5-tagged eIF3i, for which the endogenous and transfected eIF3i polypeptides could be resolved, the over-expression of eIF3i-V5 suppressed expression of the endogenous protein to the extent that the total level of eIF3i was not greater than ~40% above the amount in mock transfected cells (Fig. 1A and B). Although the eIF3i transient over-expression amounts achieved were similar to that reported for NIH3T3 cells stably over-expressing eIF3i (Zhang et al., 2007), in contrast to that stable cell line, CHOK1 cells transiently over-expressing eIF3i showed a relatively low enhancement (<20%) of protein synthesis as determined by [35S]-methionine incorporation (e.g. Fig. 1D). We note, however, that the effect of overexpressed eIF3i on [35S] incorporation in CHO cells was enhanced by exposing the cells to mild hypothermia (Supplementary Fig. 1). These data suggested that in CHOK1 cells transiently overexpressed eIF3i levels, though similar to those reported for stably transfected NIH 3T3 cells, had only a minor impact on enhancing protein synthesis. Collectively these data suggest that the total amount of eIF3i in both CHOK1 and HEK293 cells is tightly regulated.
Fig. 1.
Transient expression of eIF3i in CHOK1 cells is limited in extent and with little effect on global protein synthesis levels. A. Immunoblot detection of eIF3i in CHOK1 and HEK cell lysates prepared from cells transiently transfected with eIF3i in pcDNA3.1V5 with (3iV5) or without (3i) read-through to the V5 tag (v, vector control) and maintained at 37 °C for 24 h. B. Immunoblots generated from 3 independent experiments of the type shown in Fig. 1A were quantified with NIH ImageJ. (V) is the upper band of the 3iV5 samples in (A) and (i) the lower band of the 3iV5 samples in (A) whilst V + I is the sum of these two bands in the immunoblots of V5-tagged 3i transfections. Error bars represent ± 1SD. C. Co-expression of all 8 components of the molecular chaperone CCT with eIF3i did not improve the level of eIF3i expression. CHOK1 cells were co-transfected with 2 μg vector encoding human eIF3i and 1 μg each of vectors encoding each of the CHO cell CCT subunits with or without 35S-labelling for 1 h at the end of the 24 or 48 h period. Western blots of cell lysates were probed for the indicated CCT subunits, eIF3i and tubulin as a loading marker while (D) 35S-labelled samples were processed for scintillation counting, error bars represent ± 1 SD from 3 measurements.
We have previously shown that eIF3i requires processing through the molecular chaperone CCT for its correct folding and found that a 3T3 cell line stably overexpressing eIF3i which showed enhanced cell growth and proliferation also had elevated levels of CCT (Roobol et al., 2014). In contrast, the CCT content of CHOK1 cells transiently transfected with eIF3i was unchanged compared to the mock transfected cells (Fig. 1C) and co-transfection of eIF3i with the eight CCT subunits failed to enhance protein synthesis as measured by [35S]-methionine incorporation (Fig. 1D), though this latter may have been due to the heavy synthetic load imposed on the cell by overexpression of the eight CCT subunits together with eIF3i. An explanation for these findings is that in CHOK1 cells transiently transfected with eIF3i, the CCT level was insufficient to support the correct folding of eIF3i, resulting in eIF3i protein that could not incorporate into the eIF3 complex.
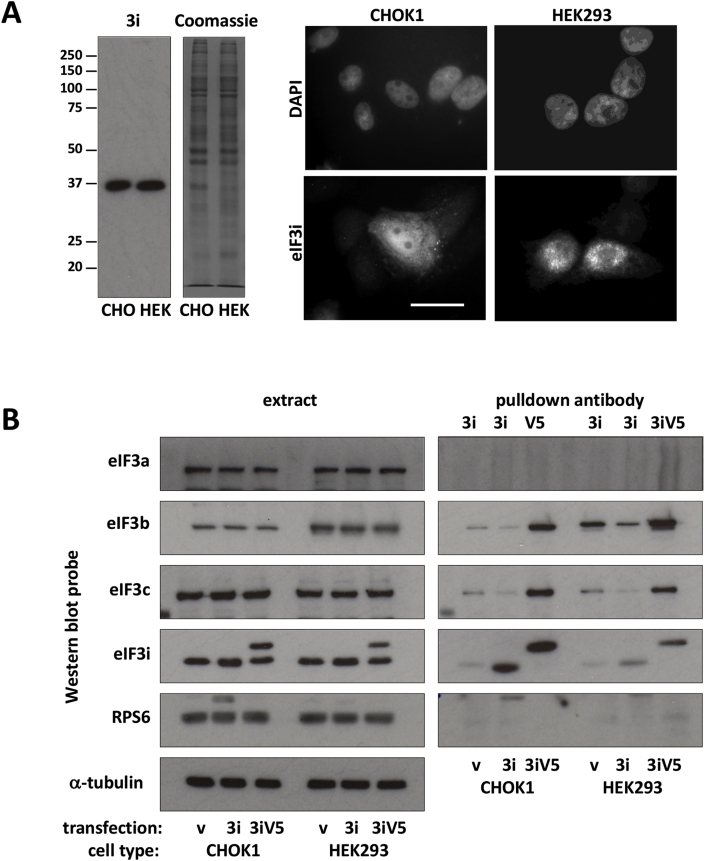
The intensity of immunostaining of transiently transfected cells with an eIF3i-specific antibody was greater than that of staining in surrounding non-transfected cells (Fig. 2A). To determine whether exogenous eIF3i was capable of incorporation into the eIF3 complex, proteins in eIF3i and eIF3iV5 transfected cell extracts were immunoprecipitated with an antibody to eIF3i or to the V5 epitope and analysed by western blot (Fig. 2B). The main proteins other than eIF3i and V5-tagged eIF3i (~40 kDa) in the pulldowns were identified by immunoblot as eIF3b and to a lesser extent eIF3c. However, it was noticeable that although the amount of eIF3i in the HEK eIF3i pulldown was less than in the CHO eIF3i pulldown, the co-precipitating eIF3b was higher in the HEK pulldown than in the CHO pulldown supporting the idea that human eIF3i incorporated into the HEK eIF3 complex better than into the CHO eIF3 complex. No eIF3a or RPS6 was detected in eIF3i pulldowns from either cell type transiently overexpressing eIF3i (Fig. 2B). These data suggest that the majority of the transiently overexpressed eIF3i did not incorporate into the eIF3 complex, though it might have incorporated to some extent into subcomplexes containing 3c or 3b.
Fig. 2.
Most transiently overexpressed eIF3i in CHOK1 cells may not incorporate into the endogenous eIF3 complex. A. Immunofluorescence detection of eIF3i in CHOK1 and HEK293 cells 24 h after transient transfection with eIF3i. Bar = 10 μ. Side panel: western blots of equal amounts of cell lysate protein from CHOK1 and HEK cells probed for eIF3i. Arrowed, a protein weakly detected by the anti-eIF3i antibody on western blot. B. Pulldowns (PDs) with the indicated antibodies were prepared from extracts of CHOK1 and HEK cells transiently transfected as indicated. Western blots of the extracts and PDs were probed with the indicated antibodies.
3.2. eIF3i can be stably overexpressed in HEK293 cells but not in CHOK1 cells
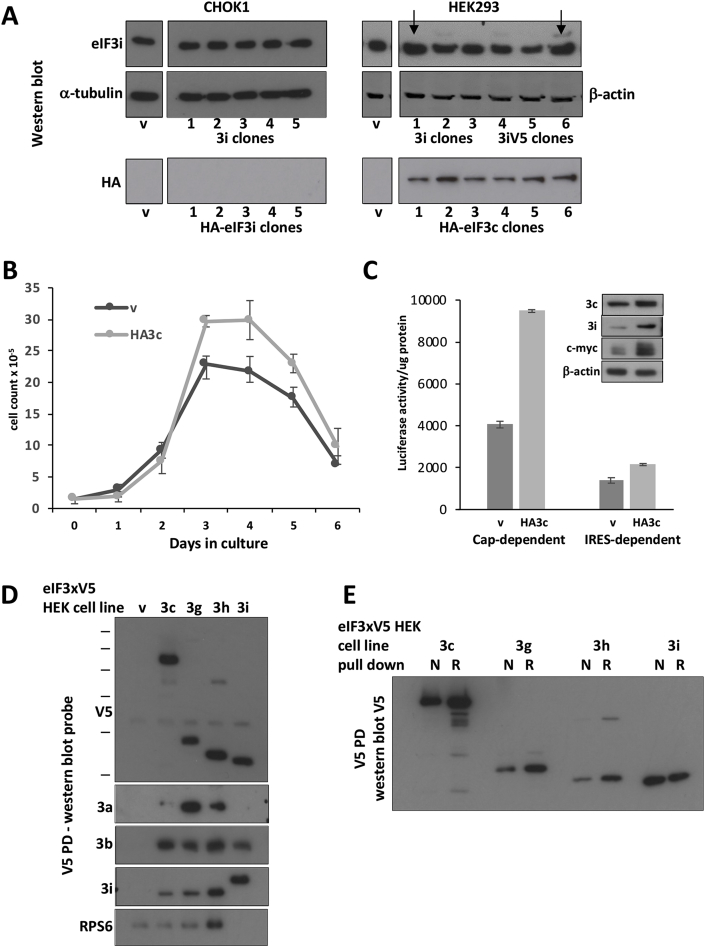
As reported previously (Zhang et al., 2007), a 3T3 cell line stably overexpressing eIF3i showed enhanced growth and protein synthesis levels at 37 °C when compared with the vector control cell line. Although we have successfully used the Flp-In system to generate a wide variety of stably over-expressing CHOK1 and HEK cell lines, including ones stably overexpressing eIF3c N-terminally tagged with the HA epitope and eIF3h (Roobol et al., 2014), we were unable to detect any CHOK1 clones over-expressing 3i or HA-tagged eIF3i (Fig. 3A).
Fig. 3.
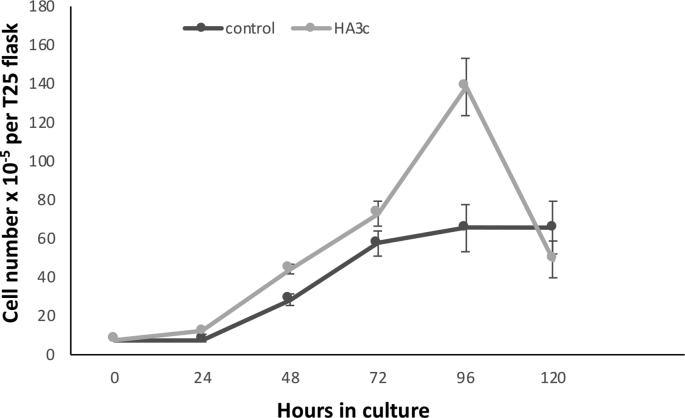
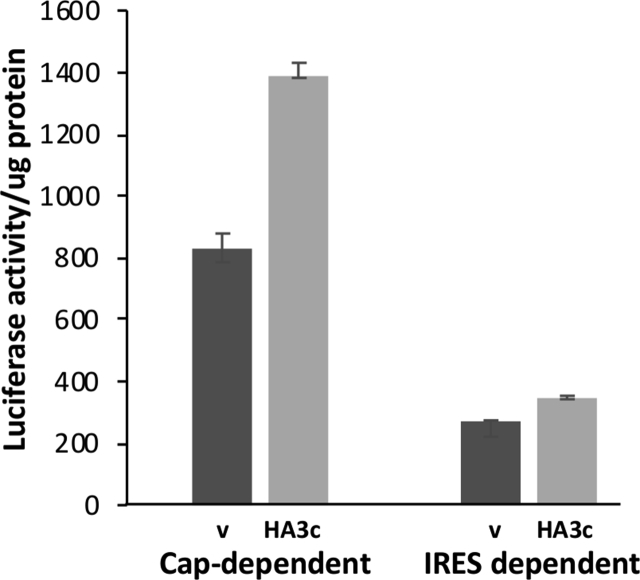
Although stable CHO cell lines overexpressing eIF3i or HAeIF3i were not detected, a CHO cell line expressing HAeIF3c and HEK cell lines individually expressing several eIF3 subunits including an eIF3i over-expressing HEK cell line were successfully generated. A. Cell line screens. The HEK cell lines chosen for further investigation are arrowed. B. A CHO cell line expressing HAeIF3c showed increased growth rate, error bars represent 1 SD from at least 3 separate measurements. Similar data was obtained for a second HAeIF3c clone investigated (Supplementary Fig. 2) and (C), increased cap- and IRES-dependent translation of the luciferase reporter pRMF (error bars represent 1 SD, 3 measurements) and increased expression of eIF3c, eIF3i and c-Myc as determined by western blot (inset). Supplementary Fig. 3 is a repeat of this experiment with a different cell line. D. Western blot of extracts prepared from HEK cells stably expressing the indicated eIF3 subunit (v = vector control) probed for the V5 epitope or the indicated proteins. E. Western blot of V5 PDs from extracts of HEK cells expressing the indicated V5-tagged eIF3 subunits and extracted under either native (N) or denaturing (D) conditions.
The CHOK1 HA-eIF3c cell line we generated showed similar properties (Fig. 3B and C and Supplementary 2 and 3) to those reported for the 3T3 HA-eIF3c cell line in that it showed enhanced growth rate, increased cap- and IRES-dependent translation and upregulated expression of eIF3i and c-Myc (Fig. 3C and Supplementary Fig. 3). The observed upregulation of the transcription factor c-Myc in the eIF3c over-expressing cell line likely contributes to the enhanced proliferation rate of the CHOK1 HA-eIF3c cell line. This shows that eIF3i expression levels can be elevated in CHOK1 cells when the cell also overexpresses the eIF3c subunit. Similarly, it has been shown in 3T3 cells (Zhang et al., 2007) that overexpression of only eIF3a or eIF3b or eIF3c was sufficient to upregulate the amount of complete eIF3 complex whereas overexpression of eIF3i did not.
Although the amount of overexpression achievable for eIF3i was also limited in HEK293 cells, stable clones were isolated in which eIF3i and eIF3iV5 expression was increased over the control cell line content (arrowed in Fig. 3A). In addition, stable HEK cell lines were generated which overexpressed V5-tagged eIF3c, eIF3g, and eIF3h. V5 pulldowns from 35[S]-labelled extracts of these cell lines identified several co-precipitating proteins in addition to the targeted protein (Fig. 3D). Immunoblot analysis of the pulldowns (Fig. 3E) revealed eIF3 core components eIF3a and eIF3b along with RPS6, this latter being most prominently in the eIF3hV5 pulldown. The eIF3iV5 pulldown however, did not show the presence of eIF3a and contained less eIF3b than the other cell line pulldowns. This suggests that the exogenous eIF3i was either not fully incorporated into the eIF3 complex or that the exogenous eIF3iV5 was not accessible when incorporated into the complete eIF3 complex. However, a small amount of eIF3iV5 was incorporated into a subcomplex containing eIF3b. The notion that exogenous eIF3i was not incorporated into the eIF3 complex was supported by a comparison of subunit availability to pulldown under native (N) and denaturing (D) conditions (Fig. 3E). For the eIF3c, eIF3g and eIF3h cell lines considerably more protein was pulled down under denaturing conditions than under native conditions. In contrast, the extent of eIF3i pulldown was similar under denaturing and native conditions. This would be consistent with eIF3c, eIF3g and eIF3h epitopes being less exposed than the eIF3i epitope under native conditions. This could arise either because the eIF3c, eIF3g and eIF3h epitopes were concealed by their incorporation into the eIF3 complex, or that correct folding of these subunits had obscured their epitopes. The availability of the exogenous eIF3i epitope for pulldown could reflect its failure to incorporate into the eIF3 complex and/or failure to fold correctly when overexpressed.
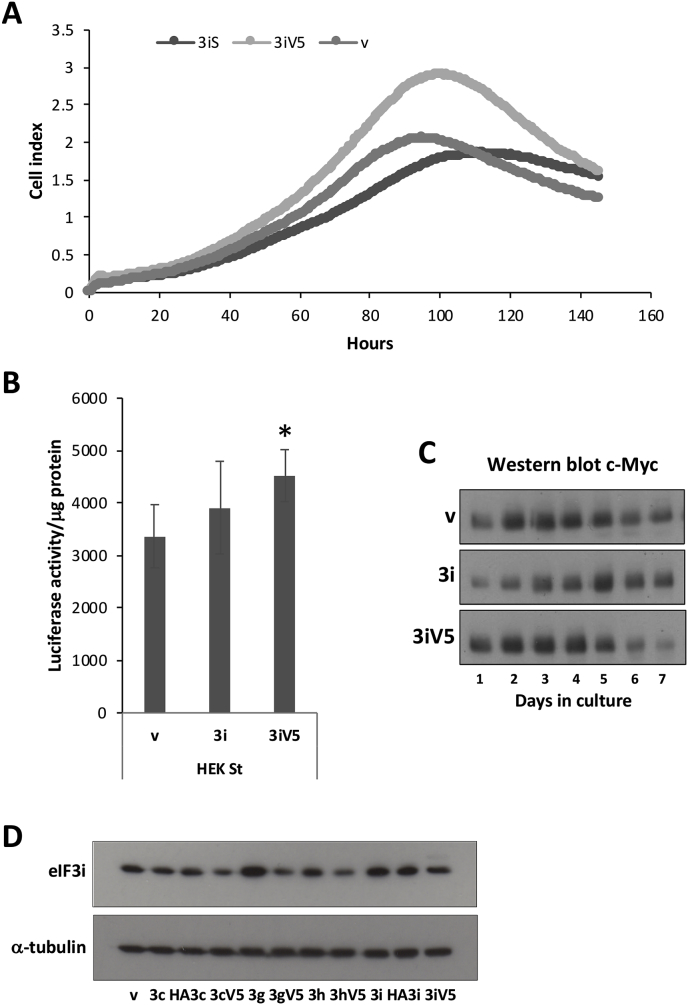
Nevertheless, a HEK cell line was established where the level of eIF3i expression was sufficiently elevated to impact on cell growth (Fig. 4A). A HEK cell line expressing eIF3iV5 had a faster growth rate and increased overall cell biomass compared to the control cell line (Fig. 4A). This 3iV5 HEK cell line also showed a significant increase in recombinant protein synthesis as measured by luciferase reporter protein expression (Fig. 4B). c-Myc expression in the eIF3iV5 cell line was higher, and elevated earlier in the growth phase, than in the control and untagged eIF3i cell line (Fig. 4C). Interestingly, the eIF3i expression levels were highest in the eIF3g-overexpressing cell line (Fig. 4D), consistent with previous data which suggest that expression of eIF3g and eIF3i are strongly interdependent with eIF3g stabilising eIF3i (Wagner et al., 2016). The likely explanation for this is that the elevated level of eIF3g in the eIF3g HEK cell line provides a binding partner for correctly folded eIF3i on its release from the CCT molecular chaperone. However, the generation of stable HEK cell lines over-expressing eIF3 subunits could not be attributed to an increase in CCT subunit levels in HEK cells. Three CCT subunits that had identical CHO and human immunogenic sequences were investigated in this regard and were found similar in the two parental cell lines (Fig. 5A).
Fig. 4.
Characterisation of the HEK eIF3i over-expressing cell lines. A. Cell growth of the indicated HEK cell lines as determined using the xCELLigence System. B. The eIF3iV5 HEK cell line showed significantly increased cap-dependent translation of a firefly luciferase reporter compared with the control cell line. Error bars are ± 1 SD and the difference between v and 3iV5 is statistically significant (p < 0.00044). C. Western blot detection of c-Myc in cell lysates of samples taken at sequential days in culture of the indicated cell lines. D. Western blot of cell extracts of the indicated panel of stable eIF3x HEK cell lines probed for eIF3i and α-tubulin as a loading marker.
Fig. 5.
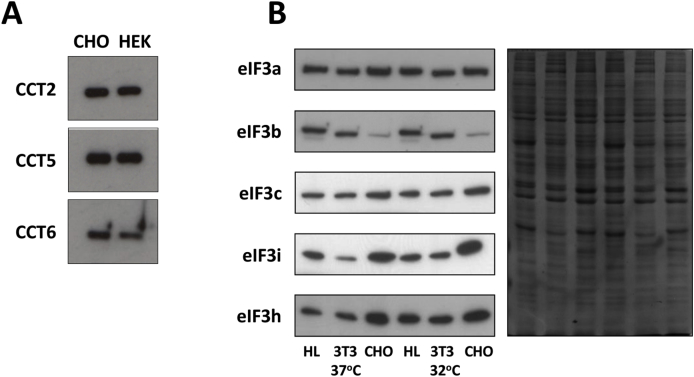
A. CHOK1 and HEK293 cells have similar CCT content. B. CHO cells contain higher levels of eIF3i compared to 3T3 and HEK293 cells. Western blots of lysates of the indicated cell lines probed for the indicated proteins. A Coomassie stained image of the different lanes is shown to the right of the western blot image to confirm equal loading of samples.
3.3. The natural level of eIF3i is high in CHOK1 cells compared to HEK cells
In the case of overexpression of eIF3i in CHOK1 and HEK293 cells, we found that even transient overexpression of this protein was quite limited and were unable to generate cell lines that stably expressed eIF3i alone above the natural level of this protein in CHOK1 cells. We therefore hypothesised that the level of eIF3i was already sufficiently high in CHOK1 cells relative to the other cell lines that stable overexpression of this protein could not be achieved. We investigated this by comparing ‘natural’ eIF3 subunit levels in CHOK1, NIH3T3 and HEK cells. The antibodies used for these immunoblot investigations were all raised against human eIF3 subunit proteins or peptide sequences, and so any increase in immunoblot detection in the non-human cells indicated an increase in the detected protein. A decrease in immunoblot detection relative to HEK cells could be accounted for by poor cross-reactivity of the antibody in the non-human cell lines; this probably accounts for the poor signal observed (Fig. 5B) for the eIF3b antibody in CHOK1 immunoblots. The immunoblot data clearly show that, when compared to HEK or 3T3 cells, CHOK1 cells express higher levels of eIF3i. In contrast, eIF3a and eIF3c were only marginally increased in CHOK1 cells relative to HEK and 3T3 cells. These cell-type related differences in eIF3i levels were maintained at 32 °C and, furthermore, eIF3i expression was increased at 32 °C when compared with 37 °C. The increase in eIF3i expression at 32 °C was in spite of its synthesis rate being lowered more than the average at 32 °C (Roobol et al., 2009) and may arise from improved folding of eIF3i at the lower temperature. This may also have arisen because, relative to eIF3a, eIF3b and eIF3c, eIF3i half-life at 37 °C is short (Roobol et al., 2014). At 32 °C, in general, protein half-lives are extended and this may allow accumulation of eIF3i at this temperature.
4. Discussion
From these data we conclude that the CHOK1 cell line has relatively high levels of eIF3i. As reported here, we show that the synthetic rate of exogenous eIF3i responds to existing eIF3i levels, and that this may account for our failure to stably overexpress this protein in CHOK1 cells. The stable overexpression of eIF3i in 3T3 cells (Zhang et al., 2007) promotes rapid growth phenotypes without upregulating the whole eIF3 complex and is dependent on phosphorylation by mTOR (Ahlemann et al., 2006) for this activity. The naturally high expression levels of the eIF3 subunit in CHOK1 and HEK293 cells compared to 3T3 cells might be anticipated to amplify this linkage of nutrient and growth factor status to protein synthesis and growth rates and so contribute to the high protein synthetic capacity of these cells, a property that has made these cell line one of choice in the biopharmaceutical industry.
A variety of approaches to improving recombinant protein production have been investigated that include exposure to mildly hypothermic conditions in the later stage of culture (Lin et al., 2015), using elements of the unfolded protein response to expand the ER and Golgi (Bommiasamy et al., 2009; Tigges and Fussenegger, 2006), manipulation of cell signalling impinging on the process of translation (Jossé et al., 2016) initiation and upregulation of translation initiation (Zhang et al., 2007) and elongation (Dreesen and Fussenegger, 2011) by manipulating levels of selected components of these processes. In this report we have focussed on manipulating the levels of the translation initiation factors eIF3i and eIF3c, components of the large translation initiation factor eIF3, in the commercially relevant cell lines CHOK1 and HEK293 since it has been reported that overexpression of these translation initiation factors in 3T3 cells increased growth rate, increased protein synthetic capacity and delayed apoptosis (Zhang et al., 2007).
Although we demonstrate such objectives can be achieved, it is clear that endogenous cellular abundancies of specific eIF3 proteins should be taken into account. This includes establishing and considering (1) whether the natural level of expression of the targeted protein limits its overexpression, (2) whether the overexpression requires up-regulation of other proteins as in the case of eIF3i overexpression being most effective when there is also upregulation of its binding partner eIF3g and (3) whether overexpression of a complete complex can be achieved by overexpression of just one of its components as is the case for overexpression of eIF3c but not so for overexpression of eIF3i. These caveats, though here applied to the manipulation of eIF3 levels and functioning, may well apply to attempts to improve protein synthesis levels by manipulating the amounts of other proteins involved in translation initiation and elongation, particularly those that function in the context of a larger protein complex.
CRediT authorship contribution statement
Anne Roobol: Conceptualization, Investigation, Methodology, Writing - original draft, Writing - review & editing. Joanne Roobol: Methodology, Writing - original draft, Writing - review & editing. Matthew E. Smith: Methodology, Writing - review & editing. Martin J. Carden: Conceptualization, Writing - review & editing. John W.B. Hershey: Writing - review & editing. Anne E. Willis: Conceptualization, Funding acquisition, Methodology, Writing - review & editing. C. Mark Smales: Conceptualization, Funding acquisition, Methodology, Supervision, Writing - review & editing.
Declaration of competing interest
The authors declare no financial or commercial conflict of interest.
Acknowledgements
This work was partially supported by Grant BB/F018908/1 (to CMS) from the Biotechnology and Biological Sciences Research Council (BBSRC), UK and a Collaboration Award in Science Grant (2014)87/Z/16/Z from the Wellcome Trust (to AEW & CMS). We wish to thank Dr. Lili Zhang for her generous gift of NIH3T3 cell lines stably overexpressing eIF3 subunits and plasmids encoding human eIF3c and 3i with N-terminal HA tags.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ymben.2020.02.001.
Contributor Information
Anne E. Willis, Email: aew80@mrc-tox.cam.ac.uk.
C. Mark Smales, Email: c.m.smales@kent.ac.uk.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
SUPPLEMENTARY FIGURE 1.
The effect of mild hypothermia on general protein synthesis in CHOK1 cells transiently transfected with vector (v) or eIF3i (i).
SUPPLEMENTARY FIGURE 2.
A growth curve for a second CHOK1 clone expressing HAeIF3c. Error bars are ± 1SD from 4 measurements.
SUPPLEMENTARY FIGURE 3.
A repeat of the experiment shown in Fig. 3C with a second cell line.
References
- Ahlemann M. Carcinoma-associated eIF3i overexpression facilitates mTOR-dependent growth transformation. Mol. Carcinog. 2006;45:957–967. doi: 10.1002/mc.20269. [DOI] [PubMed] [Google Scholar]
- Aitken C.E. Eukaryotic translation initiation factor 3 plays distinct roles at the mRNA entry and exit channels of the ribosomal preinitiation complex. eLife. 2016;5 doi: 10.7554/eLife.20934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Fageeh M.B., Smales C.M. Control and regulation of cellular response to cold shock: the responses in yeast and mammalian systems. Biochem. J. 2006;397:247–259. doi: 10.1042/BJ20060166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Fageeh M.B., Smales C.M. The cold shock response in cultured mammalian cells: harnessing the response for improvement of recombinant protein production. Biotechnol. Bioeng. 2006;93:829–835. doi: 10.1002/bit.20789. [DOI] [PubMed] [Google Scholar]
- Beznosková P. Translation initiation factor eIF3 promotes programmed stop codon readthrough. Nucleic Acids Res. 2015;43:5099–5111. doi: 10.1093/nar/gkv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommiasamy H. ATF6α induces XBP1-independent expansion of the endoplasmic reticulum. J. Cell Sci. 2009;122:1626–1636. doi: 10.1242/jcs.045625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilising the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Budge J.D. Engineering of Chinese hamster ovary cell lipid metabolism results in an expanded ER and enhanced recombinant biotherapeutic protein production. Metab. Eng. 2020;57:203–216. doi: 10.1016/j.ymben.2019.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- des Georges A. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature. 2015;525:491–495. doi: 10.1038/nature14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z. Spectrin domain of eukaryotic initiation factor 3a Is the docking site for formation of the a:b:i:g subcomplex. J. Biol. Chem. 2013;288:27951–27959. doi: 10.1074/jbc.M113.483164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreesen I.A.J., Fussenegger M. Ectopic expression of human mTOR increases viability, robustness, cell size, proliferation, and antibody production of Chinese hamster ovary cells. Biotechnol. Bioeng. 2011;108:853–866. doi: 10.1002/bit.22990. [DOI] [PubMed] [Google Scholar]
- Fraser C.S. The j subunit of human translation initiation factor eIF3 is required for the stable binding of eIF3 and its subcomplexes to 40S ribosomal subunits in vitro. J. Biol. Chem. 2004;279:8946–8956. doi: 10.1074/jbc.M312745200. [DOI] [PubMed] [Google Scholar]
- Godfrey C.L. Polysome profiling of mAb producing CHO cell lines links translational control of cell proliferation and recombinant mRNA loading onto ribosomes with global and recombinant protein synthesis. Biotechnol. J. 2017;12 doi: 10.1002/biot.201700177. [DOI] [PubMed] [Google Scholar]
- Hargreaves E. Experimental and in silico modelling analyses of the gene expression pathway for recombinant antibody and by-product production in NS0 cell lines. PloS One. 2012;7 doi: 10.1371/journal.pone.0047422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves E. Biological insights into the expression of translation initiation factors from recombinant CHOK1SV cell lines and their relationship to enhanced productivity. Biochem. J. 2015;472:261–273. doi: 10.1042/BJ20150928. [DOI] [PubMed] [Google Scholar]
- Hashem Y. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature. 2013;503:539–543. doi: 10.1038/nature12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmannová A. Structural analysis of an eIF3 subcomplex reveals conserved interactions required for a stable and proper translation pre-initiation complex assembly. Nucleic Acids Res. 2012;40:2294–2311. doi: 10.1093/nar/gkr765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey J.W.B., Merrick W.C. The pathway and mechanism of initiation of protein synthesis. In: Sonenberg N., Hershey J.W.B., Mathews M.B., editors. Translational Control of Gene Expression. Cold Spring Harbor Laboratory; 2000. pp. 33–88. [Google Scholar]
- Hinnebusch A.G. eIF3. A versatile scaffold for translation initiation complexes. Trends Biochem. Sci. 2006;31:553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Hronová V. Does eIF3 promote reinitiation after translation of short upstream ORFs also in mammalian cells? RNA Biol. 2017;14(12):1660–1667. doi: 10.1080/15476286.2017.1353863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossé L. MTORC1 signalling and eIF4E/4E-BP1 translation initiation factor stoichiometry influence recombinant protein productivity from GSCHOK1 cells. Biochem. J. 2016;473:4651–4664. doi: 10.1042/BCJ20160845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossé L., Zhang L., Smales C.M. Application of microRNA targeted 3'UTRs to repress DHFR selection marker expression for development of recombinant antibody expressing CHO cell pools. Biotechnol. J. 2018;13 doi: 10.1002/biot.201800129. [DOI] [PubMed] [Google Scholar]
- Kim J.Y., Kim Y.G., Lee G.M. CHO cells in biotechnology for production of recombinant proteins: current state and further potential. Appl. Microbiol. Biotechnol. 2012;93:917–930. doi: 10.1007/s00253-011-3758-5. [DOI] [PubMed] [Google Scholar]
- Lin C.Y. Enhancing protein expression in HEK-293 cells by lowering culture temperature. PloS One. 2015;10 doi: 10.1371/journal.pone.0123562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant R.J. Metabolic rates, growth rates and mRNA levels influence cell-specific antibody production levels from in vitro cultured mammalian cells at subphysiological temperatures. Mol. Biotechnol. 2008;39:69–77. doi: 10.1007/s12033-008-9032-0. [DOI] [PubMed] [Google Scholar]
- Matsutani M. Reconstitution reveals the functional core of mammalian eIF3. EMBO J. 2007;26:3373–3383. doi: 10.1038/sj.emboj.7601765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Lorsch J., Hellen C. North America; 2007. 4. The Mechanism of Translation Initiation in Eukaryotes. Cold Spring Harbor Monograph Archive; p. 48. [Google Scholar]
- Querol-Audi J. Architecture of human translation initiation factor 3. Structure. 2013;21:920–928. doi: 10.1016/j.str.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roobol A., Carden M.J. Subunits of the eukaryotic cytosolic chaperonin CCT do not always behave as components of a uniform hetero-oligomeric particle. Eur. J. Cell Biol. 1999;78:21–32. doi: 10.1016/S0171-9335(99)80004-1. [DOI] [PubMed] [Google Scholar]
- Roobol A. Disassembly of the cytosolic chaperonin in mammalian cell extracts at intracellular levels of K+ and ATP. J. Biol. Chem. 1999;274:19220–19227. doi: 10.1074/jbc.274.27.19220. [DOI] [PubMed] [Google Scholar]
- Roobol A. Biochemical insights into the mechanisms central to the response of mammalian cells to cold stress and subsequent rewarming. FEBS J. 2009;276:286–302. doi: 10.1111/j.1742-4658.2008.06781.x. [DOI] [PubMed] [Google Scholar]
- Roobol A. The chaperonin CCT interacts with and mediates the correct folding and activity of three subunits of translation initiation factor eIF3: b, i and h. Biochem. J. 2014;458:213–224. doi: 10.1042/BJ20130979. [DOI] [PubMed] [Google Scholar]
- Roobol A. p58(IPK) is an inhibitor of the eIF2α kinase GCN2 and its localisation and expression underpin protein synthesis and ER processing capacity. Biochem. J. 2015;465:213–225. doi: 10.1042/BJ20140852. [DOI] [PubMed] [Google Scholar]
- Smith T.F. The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- Smith M.D., Arake-Tacca L., Nitido A., Montabana E., Park A., Cate J.H. Assembly of eIF3 mediated by mutually dependent subunit insertion. Structure. 2016;24:1–11. doi: 10.1016/j.str.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneley M. Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res. 2000;28:687–694. doi: 10.1093/nar/28.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigges M., Fussenegger M. Xbp1-based engineering of secretory capacity enhances the productivity of Chinese hamster ovary cells. Met. Eng. 2006;8:264–272. doi: 10.1016/j.ymben.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Underhill M.F., Smales C.M. The cold shock response in mammalian cells: investigating the HeLa cell cold shock proteome. Cryotechnol. 2007;53:47–53. doi: 10.1007/s10616-007-9048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill M.F. eIF2a phosphorylation, stress perception and the shutdown of global protein synthesis in cultured CHO cells. Biotechnol. Bioeng. 2005;89:805–814. doi: 10.1002/bit.20403. [DOI] [PubMed] [Google Scholar]
- Valášek L.S. ‘Ribozoomin’ – translation initiation from the perspective of the ribosome-bound eukaryotic initiation factors (eIFs) Curr. Protein Pept. Sci. 2012;13:305–330. doi: 10.2174/138920312801619385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek L., Nielsen K.H., Hinnebusch A.G. Direct eIF2-eIF3 contact in the multifactor complex is important for translation initiation in vivo. EMBO J. 2002;21:5886–5898. doi: 10.1093/emboj/cdf563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valášek L.S. Embraced by eIF3: structural and functional insights into the roles of eIF3 across the translation cycle. Nucleic Acids Res. 2017;45:10948–10968. doi: 10.1093/nar/gkx805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlhac M.-H. Identification of partners of TIF34, a component of the yeast eIF3 complex, required for cell proliferation and translation initiation. EMBO J. 1997;16:6812–6822. doi: 10.1093/emboj/16.22.6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa N. Human eukaryotic initiation factor 4G (eIF4G) protein binds to eIF3c, -d, and -e to promote mRNA recruitment to the ribosome. J. Biol. Chem. 2013;288:32932–32940. doi: 10.1074/jbc.M113.517011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vito D., Smales C.M. Engineering of the cellular translational machinery and non-coding RNAs to enhance CHO cell growth, recombinant product yields and quality. Curr. Opin. Chem. Eng. 2018;22:199–208. [Google Scholar]
- Wagner S. Human eIF3b and eIF3a serve as the nucleation core for the assembly of eIF3 into two interconnected modules: the yeast-like core and the octamer. Nucleic Acids Res. 2016;44:10772–10788. doi: 10.1093/nar/gkw972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff J. 1H NMR spectroscopy profiling of metabolic reprogramming of Chinese hamster ovary cells upon a temperature shift during culture. PloS One. 2013;8 doi: 10.1371/journal.pone.0077195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 1989;93:491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]
- Zhang L., Pan X., Hershey J.W.B. Individual overexpression of five subunits of human translation initiation factor eIF3 promotes malignant transformation of immortal fibroblast cells. J. Biol. Chem. 2007;282:5790–5800. doi: 10.1074/jbc.M606284200. [DOI] [PubMed] [Google Scholar]
- Zhou M. Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc. Natl. Acad. Sci. U.S.A. 2008;105:18139–18144. doi: 10.1073/pnas.0801313105. [DOI] [PMC free article] [PubMed] [Google Scholar]