Abstract
Viral respiratory diseases such as severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) always pose a severe threat to people. First identified in late December 2019, a novel coronavirus (2019-nCoV; SARS-CoV-2) has affected many provinces in China and multiple countries worldwide. The viral outbreak has aroused panic and a public-health emergency around the world, and the number of infections continues to rise. However, the causes and consequences of the pneumonia remain unknown. To effectively implement epidemic prevention, early identification and diagnosis are critical to disease control. Here we scrutinise a series of available studies by global scientists on the clinical manifestations, detection methods and treatment options for the disease caused by SARS-CoV-2, named coronavirus disease 2019 (COVID-19), and also propose potential strategies for preventing the infection.
Keywords: 2019 novel coronavirus, 2019-nCoV, SARS-CoV-2, Mechanism, Treatment, Detection
1. Introduction
At the end of 2019, several patients were diagnosed with pneumonia of unknown cause, epidemiologically associated with the same seafood market. Alongside the Spring Festival exodus, an outbreak seemed unavoidable. This condition attracted the attention of the Chinese Center for Disease Control and Prevention (CDC), who immediately launched an emergency response. The World Health Organization (WHO) also responded promptly and declared the outbreak a public health emergency of international concern (PHEIC). The causative agent of the unidentified pneumonia has been confirmed as a novel coronavirus by sequencing and aetiological investigations by several independent laboratories in China. Following the isolation of the new coronavirus, it was found to be distinct both from Middle East respiratory syndrome coronavirus (MERS-CoV) and severe acute respiratory syndrome coronavirus (SARS-CoV) [1,2]. Coronaviruses are single-stranded RNA viruses belonging to the family Coronaviridae that can cause various diseases with enteric, respiratory, hepatic and neurological symptoms [3]. The new coronavirus, originally denoted 2019 novel coronavirus (2019-nCoV) and officially renamed severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by the International Committee on Taxonomy of Viruses, and the disease it causes, namely coronavirus disease 2019 (COVID-19), has quickly become of tremendous concern worldwide. There have been significant outbreaks in many regions of China as well as global expansion including to Asia, Europe, North America, South America, Africa and Oceania. The disease is potentially zoonotic, with an estimated mortality rate of 2–5%. Person-to-person transmission may occur through contact and respiratory transmission or possibly by the faecal–oral route. Currently, the number of confirmed infections has been increasing daily but there is no definite treatment for COVID-19 pneumonia, although some potential drugs are under investigation. For the last two decades, outbreaks of coronaviruses and intermittent worldwide public health emergences remind us that coronaviruses are still a severe global health threat that cannot be ignored. According to the latest research, useful information for control of the disease is urgently required and is highly essential.
2. Virology
Coronaviruses are named for the crown-like (or corona in Latin) spikes of the virus protruding to the periphery, with a diameter of 60–160 nm observed by electron microscopy. Each viral particle is enveloped and contains a single-stranded positive-sense RNA genome of 27–32 kb with a 5ʹ-cap structure and 3ʹ-poly A tail that interacts with the nucleoprotein. All coronaviruses have similarities in the organisation and expression of the genome, and the genome size of coronaviruses is the largest among all RNA viruses. Sixteen non-structural proteins (nsp1 to nsp16), encoded by open reading frame (ORF) 1a/b at the 5ʹ-end, are followed by the nucleocapsid (N), spike (S), envelope (E) and membrane (M) structural proteins, which are encoded by other ORFs at the 3ʹ-end [4]. The envelope includes three proteins: the M protein binds the nucleocapsid and enhances viral assembly and budding; the E protein is involved in viral morphogenesis, release and pathogenesis; and the S protein contributes to homotrimeric spikes that recognise the cell receptor, thus helping the virus invade target cells [5]. Since the outbreaks of SARS in 2002 and MERS in 2012, the possibility of coronavirus transmission from animals to humans had been proven. Coronaviruses are ubiquitous pathogens in nature for humans and animals, usually causing gastrointestinal or respiratory infections and sometimes involving important organs such as the liver, kidney, heart and brain. They are sensitive to ultraviolet rays and heat. Lipid solvents such as ether, 75% ethanol, chlorine-containing disinfectant, peracetic acid and chloroform can effectively inactivate the virus.
Coronaviruses comprise the largest group belonging to the Nidovirales order, which contains the families Coronaviridae, Arteriviridae and Roniviridae. The family Coronaviridae is composed of large, single-stranded, positive-sense RNA viruses that are isolated from several species and were previously known to induce common colds and diarrhoeal illnesses in humans [5]. The subfamily Coronavirinae is one of two subfamilies in the Coronaviridae family, the other being the subfamily Torovirinae. The subfamily Coronavirinae is further subdivided into four groups, namely the alpha, beta, gamma and delta coronaviruses. The viruses were initially classified into groups according to serology but are now labelled by phylogenetic clustering [6]. Only alpha and beta coronavirus are of interest for human and clinical virologists [7]. Based on epidemiological data before 2019, only six coronaviruses were proven to cause human respiratory diseases: (i) HKU1, HCoV-NL63, HCoV-OC43 and HCoV-229E, which lead to only mild upper respiratory disease, rarely bring about severe disease in humans; and (ii) SARS-CoV and MERS-CoV, which attack the lower respiratory tract and induce severe respiratory syndrome. Sequence analysis shows that SARS-CoV-2 has a typical genome structure of coronaviruses and belongs to the cluster of Betacoronaviruses that include SARS-CoV and MERS-CoV. It forms a clade within the subgenus Sarbecovirus, Orthocoronavirinae subfamily. It is the seventh member of the family of coronaviruses that infect humans so far [8].
3. Potential hosts
Evolutionary models and phylogenetic analysis deserve attention for helping to estimate genetic variability and the evolutionary rate, in turn providing important implications for disease progression, drug trials and vaccine development. Zhou et al. reported that the full-length viral genome sequences from patients in the early outbreak were almost identical to each other and shared 79.5% sequence similar to SARS-CoV [2]. In addition, SARS-CoV-2 was 96% identical at the whole-genome level to a bat coronavirus. Wu et al. almost simultaneously and independently discovered the novel RNA virus from the family Coronaviridae [1]. Phylogenetic analysis of the complete viral genome revealed that the coronavirus was most closely related (89.1% nucleotide similarity) to a group of SARS-like coronaviruses previously sampled from bats in China. Benvenuto et al. built a phylogenetic tree including the whole-genome sequences of SARS-CoV-2 and highly similar available whole-genome sequences in GenBank [9]. The phylogenetic tree showed that SARS-CoV-2 clustered with a bat SARS-like coronavirus sequence isolated in 2015, whilst fast unconstrained Bayesian approximation (FUBAR) analysis revealed mutations in the spike glycoprotein and nucleocapsid protein. Therefore, SARS-CoV-2 was probably transmitted from bats or other hosts, from where it gained the ability to infect humans. Ramaiah et al. speculated that the novel virus closely related to a bat coronavirus, which reminded us that this novel strain might be evolved from the bat coronavirus by accumulating favourable genetic alterations for human infection [10]. Epitopes in SAR-CoV-2 structural proteins were differentially recognised by HLA-DR alleles, which suggested that a subunit vaccine including the eight immunodominant HLA-DR epitopes might induce an effective antiviral immune responses in various populations. Paraskevis et al. found that SAR-CoV-2 closely resembled the bat coronavirus RaTG13 sequence throughout the genome (similarity 96.3%) [11]. The latter does not provide the exact variant that caused the outbreak in humans, but the hypothesis that SAR-CoV-2 has originated from bats is possible. Xu et al. also reported that SAR-CoV-2 shared with the SARS/SARS-like coronaviruses a common ancestor that related to the bat coronavirus HKU9-1 [12]. They concluded that the SAR-CoV-2 and SARS-CoV S proteins shared an almost identical three-dimensional structure in the receptor-binding domain (RBD), thus maintaining similar van der Waals and electrostatic properties at the interaction interface with human angiotensin-converting enzyme 2 (ACE2) molecules despite the sequence diversity.
Although many scientists believe that bats are the intermediate host, most species of bats live in tropical or subtropical rain forests and caves far from human populations, thus the probability of bats transmitting the virus directly to humans is unlikely. It is generally known that bats may spread viruses to other intermediate hosts such as wild animals or livestock, from which the viruses are then transmitted to humans. Ji et al. carried out a comprehensive sequence analysis and comparison in conjunction with relative synonymous codon usage bias among different animal species based on the SARS-CoV-2 sequence [13]. The result suggested that SARS-CoV-2 appeared to be a recombinant virus between the bat coronavirus and a coronavirus of unknown origin. They considered that snake was a most probable wildlife animal reservoir, but the conclusion was controversial. Guo et al. introduced VHP (Virus Host Prediction) to predict the potential hosts of viruses by deep learning algorithm and predicted that bat coronaviruses were assigned with more similar infectivity patterns with SARS-CoV-2 [14]. The results illustrated that bat and mink might be two candidate reservoirs of SARS-CoV-2. Lam et al. identified pangolin-associated coronaviruses belonging to two sublineages of SARS-CoV-2-related coronaviruses, including one very closely related to SARS-CoV-2 in the RBD by metagenomic sequencing [15]. Hence, pangolins should be considered as possible intermediate hosts and should be removed from markets to prevent such zoonotic transmission.
4. Clinical manifestations
As of 21 February 2020, more than 80 000 cases of COVID-19 have been confirmed and most of them had a history of close contact with the epidemic area or with confirmed patients. Major initial symptoms include fever, most of which are high fevers that occur within several days and are not alleviated by routine anti-infective drugs, as well as cough, headache and muscle pain or fatigue [16]. Other clinical symptoms observed at a lower frequency include elevated troponin levels, diarrhoea, myalgia and myocarditis [16]. It should be emphasised that some asymptomatic persons are infected with SARS-CoV-2 [17], therefore the presence of asymptomatic carriers requires due attention. In one study, nearly 20% of patients appeared to have co-morbidities with regard to dysfunction of other organs, primarily renal impairment, and patients with underlying cardiovascular diseases often demonstrated co-morbid heart failure [18]. Patients gradually develop initial symptoms in the cardiovascular system, digestive system and nervous system, which increases the difficulty of diagnosis [16]. In this study, the median interval from the start of initial symptoms to significant symptom aggravation such as dyspnoea or the appearance of acute respiratory distress syndrome (ARDS) was 7 days (range 1–20 days), which is consistent with previous reports [18]. According to the newly released pneumonitis diagnosis and treatment plan for novel coronavirus infection, severe patients often have dyspnoea and/or hypoxaemia 1 week after the onset of the illness. Serious cases can quickly progress to ARDS, septic shock, irreformable metabolic acidosis, coagulopathy and multiple organ failure. Nearly 80% of patients have normal or decreased white blood cell counts, and 72.3% have lymphocytopenia [16]. Lung involvement is present in all cases [19,20], with most chest computed tomography (CT) images showing lesions in multiple lung lobes, some of which are dense. Ground-glass opacity co-existing with consolidation shadows or cord-like shadows are observed. Since respiratory support is administered to most patients, oxygen saturation can be maintained above 90% as indicated by pulse oximetry monitoring [16]. It is reported that severe and critically ill patients have moderate to low fever, or even without obvious fever. Mild patients show only low fever, mild fatigue and no pneumonia. Judging from the current cases, most patients have a good prognosis, but it is poor for the elderly and those with chronic underlying diseases. Symptoms in children are relatively mild.
Regarding the current situation, the source of infection is mainly infected patients, but the possibility of asymptomatic infection should not be ignored. Respiratory droplets and close contacts are the key routes of transmission. The possibility of aerosol transmission in a relatively closed environment for a long-time exposure to high concentrations of aerosol also exists, but still requires scientific evidence. Recently, SARS-CoV-2 RNA has been detected in the faeces of some confirmed patients with pneumonia, indicating that SARS-CoV-2 is likely to be transmitted through the faecal–oral route. Zhang et al. found that the ACE2 cell receptor is highly expressed on type II alveolar epithelial cells, oesophageal epithelium, stratified epithelial cells, and even in absorptive intestinal epithelial cells from the ileum and colon [21]. Their bioinformatics analysis based on single-cell transcriptomes suggests that the digestive tract may serve as an infection pathway for SARS-CoV-2. The spread and infection of the virus are complex problems requiring co-operation from multiple perspectives including medicine, biology and fluid mechanics to give a complete answer.
5. Detection
5.1. General examination
Mild patients may have no positive signs, whilst severe patients may have shortness of breath with moist rales of both lungs, weakened breath sounds, dull percussion, and enhanced or weakened vocal fremitus on palpation [22].
5.2. Chest imaging
Suspected or confirmed cases should undergo chest radiography as early as possible and a chest CT scan when necessary [23]. In the early phase of the disease, chest images show interstitial changes and multiple small plaques, especially in the lung periphery. The changes then deteriorate further bilaterally and are mainly distributed in the middle and outer zones of the lung with multiple infiltrating shadows and/or ground-glass opacities. Patients may have a single lobe or multiple lobes involved. When the condition improves, a fibrous stripe may appear [24]. Conversely, lung consolidation may occur in severe cases in whom pleural effusions are rarely seen.
5.3. Laboratory examination
5.3.1. Haematological examination
In the early stage, the white blood cell count is normal or decreased, with a decreased lymphocyte count. If the absolute lymphocyte count is <0.8 × 109/L or the CD4+ and CD8+ T-cell counts are significantly decreased, this requires extreme attention. It is generally recommended to re-check the routine blood changes after 3 days [22]. In some patients, muscle enzymes, liver enzymes and myohaemoglobin levels are increased. Otherwise, troponin is increased in some critical patients. Most patients display an elevated erythrocyte sedimentation rate (ESR) and C-reactive protein (CRP) level, with normal procalcitonin level. Severe cases show progressively decreased blood lymphocyte counts and high D-dimer levels. Inflammatory factors are often increased in severe and critical patients.
5.3.2. Molecular diagnosis
Real-time reverse transcription PCR (RT-PCR)
Samples from suspected SARS-CoV-2 patients collected from the upper respiratory tract (nasopharyngeal and oropharyngeal), lower respiratory tract (expectorated sputum, endotracheal aspirate or bronchoalveolar lavage), blood and faeces can be diagnosed by RT-PCR [25]. Two sequence regions (ORF1b and N) designed based on the first public access sequence in GenBank have been selected for primer and probe designs, which are highly conserved amongst Sarbecoviruses. The N gene assay is ~10 times more sensitive than the ORF1b gene assay in detecting positive clinical specimens [26]. Existing PCR methods have very good specificity but low sensitivity, meaning that negative test results cannot exclude the presence of SARS-CoV-2. Moreover, laboratory sample contamination caused by lack of control can lead to false-positive results. In addition, RT-PCR tests may be falsely negative due to insufficient viral material or operational error. Some patients with negative RT-PCR results may present with positive chest CT findings for COVID-19, meaning that PCR results can assist clinical diagnosis and evaluation but the possibility of the disease cannot be confirmed or ruled out. For individuals with a high clinical suspicion but negative RT-PCR screening, a combination of CT scanning and repeated swab tests may be helpful [27].
SHERLOCK technique
The CRISPR-based SHERLOCK (Specific High-sensitivity Enzymatic Reporter UnLOCKing) technique allows portable, multiplexed and ultrasensitive detection of RNA or DNA from clinically relevant samples. SHERLOCK assays are set up with recombinase-mediated polymerase pre-amplification of DNA or RNA and subsequent Cas13- or Cas12-mediated detection via colorimetric read-outs and fluorescence that provide results in <1 h with a setup time of <15 min [28]. Based on the RNA sequence of the novel coronavirus, researchers carefully designed two guide RNAs, one recognising the S gene of the new coronavirus and the other recognising the Orf1ab gene. To maximise the accuracy of the detection, scientists have selected sequences that are most specific for the new coronavirus. In this way, interference from other respiratory virus genomes can also be minimised. Theoretically, as long as the RNA corresponds to the new coronavirus in the sample, the guide RNA can accurately recognise it and activate the Cas13a protein to bind to it. Cas13a is a very interesting enzyme; once activated, it will indiscriminately and intensely cut any other RNA molecules it encounters. In this way, by confirming whether these molecules have been cut off, it can detect the presence of the new coronavirus in the original sample. They consistently detected SARS-CoV-2 target sequences in a range between 20 and 200 aM (10–100 copies per microlitre of input) by using synthetic SARS-CoV-2 virus RNA fragments. The test can be read using a dipstick in <1 h without requiring elaborate instrumentation, but still needs to be confirmed with patient samples.
6. Pathogenic mechanisms
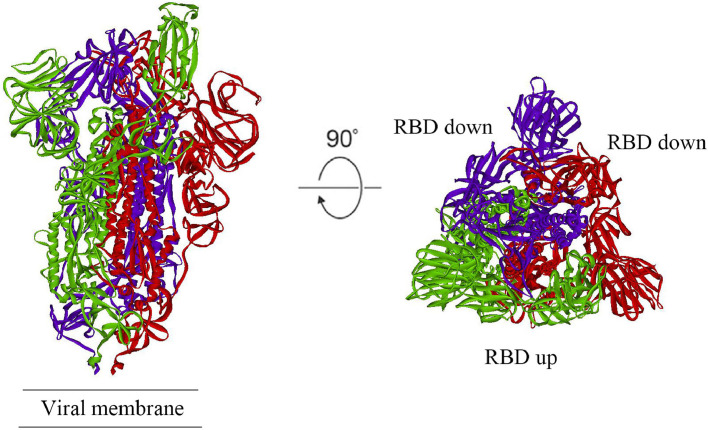
Ren et al. sequenced SARS-CoV-2 and phylogenetic analysis revealed that it belonged to the Betacoronaviruses with 79.0% nucleotide identity to SARS-CoV and 51.8% identity to MERS-CoV, meaning that it was closer to SARS-CoV than MERS-CoV [29]. Importantly, homology modelling revealed that the RBD structure of SARS-CoV-2 was similar to that of SARS-CoV [30]. From previous reports, the interaction with ACE2 was responsible for SARS-CoV entering human cells [31,32]. Dong et al. compared the structural similarity of the S protein in various viruses and speculated that SARS-CoV-2 could most likely use the same receptor as SARS-CoV [33]. Zhou et al. conducted virus infectivity studies using HeLa cells with or without ACE2 proteins obtained from humans or animals and found that SARS-CoV-2 could invade all ACE2 protein-expressing cells except for mouse cells [34]. In short, SARS-CoV-2 uses ACE2 as an entry receptor into ACE2-expressing cells, but not cells without ACE2, and SARS-CoV-2 is likely to bind to ACE2 receptor in humans just like SARS-CoV. Dimitrov reported that SARS-CoV infected human host cells by a basic interaction of its S glycoprotein and the receptor ACE2 on human cells [35]. Although the sequence of SARS-CoV-2 is a little different from SARS-CoV, its functionally important ORFs and major structural proteins, especially the spike (S) protein, are well described, of which the original identity is 76% [36]. The S protein has two regions (S2 and S1), with stronger affinity with the RBD in the S1 region interacting with ACE2 [26]. Similar to SARS-CoV, the SARS-CoV-2 virus may also engage the RBD to bind ACE2 in order to enter human host cells, but since many residues in S1 and S2 have been replaced in SARS-CoV-2, differences in the interactions with ACE2 on host cells exist [33,37]. Wrapp et al. determined the cryogenic electron microscopy (cryo-EM) structure of the SARS-CoV-2 S protein and the structure is presented in Fig. 1 [38]. These structures have been used to calculate the reconstruction of the S protein of SARS-CoV-2. Fig. 1 shows side and top views of the prefusion structure of SARS-CoV-2 S protein with a single receptor-binding domain (RBD) in the ‘up’ conformation. The two RBD ‘down’ protomers are shown in either purple or red, and the RBD ‘up’ protomer is shown in green. The result suggests that ACE2 binds to the SARS-CoV-2 S protein ectodomain with 15 nM affinity, which is approximately 10–20-fold higher than that of SARS-CoV, and this result is very surprising.
Fig. 1.
Structure of SARS-CoV-2 (2019-nCoV) spike (S) protein in the prefusion conformation. Side and top views of the prefusion structure of SARS-CoV-2 S protein with a single receptor-binding domain (RBD) in the ‘up’ conformation. The two RBD ‘down’ protomers are shown in either purple or red, and the RBD ‘up’ protomer is shown in green. Figure was modified from Wrapp et al. [38]. SARS-CoV-2 (2019-nCoV), severe acute respiratory syndrome coronavirus 2 (2019 novel coronavirus).
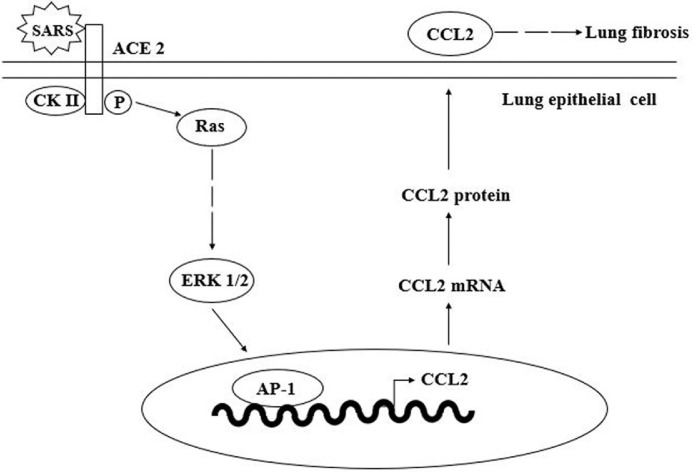
However, when SARS-CoV attempts to invade host cells, it is not as simple as ACE2 interacting with the S protein, as other pathways and cytokines are involved. Chemokine (C-C motif) ligand 2 (CCL2) is an important cytokine in the spike–ACE2 signalling pathway. CCL2 is a small CC chemokine that attracts memory T lymphocytes, monocytes and basophils. The receptor of CCL2 is CCR2, both of which are involved in the inflammatory reaction [39], therefore they are associated with some lung inflammatory disorders. Referring to previous reports, patients infected with SARS-CoV had pulmonary fibrosis due to CCL2. Cheung et al. observed the expression of CCL2 in SARS-CoV patients and found that expression of CCL2 was upregulated in the sera of patients [40]. To determine the relationship between the spike–ACE2 signalling pathway and CCL2, Chen et al. suggested an interaction between the S protein of SARS-CoV with ACE2 on human cells [41] (Fig. 2 ). Infected lung epithelial cells induce casein kinase II (CK II), which is able to phosphorylate ACE2 and involves ERK1/2 activation. CK II phosphorylates ACE2 at Ser-787, by which SARS-CoV binds to its ACE2 receptor and causes a conformational change of ACE2. It also activates the ACE2 downstream signal transduction pathways, including ERK1/2 and AP-1 [42]. These changes lead to the activation of ERK1/2 and AP-1 and upregulate CCL2. ACE2 also participates in an intracellular signalling pathway. It activates ERK1/2 by interacting with upstream factors such as Ras and protein kinase C. Finally, the elevated levels of CCL2 protein in the sera of SARS-CoV-infected patients account for the development of lung fibrosis.
Fig. 2.
Model of the angiotensin-converting enzyme 2 (ACE2) signalling pathway involved in SARS-CoV-induced CCL2 expression. Infection of lung epithelial cells with SARS-CoV induces casein kinase II (CK II)-mediated phosphorylation of the ACE2 receptor, leading to activation of ERK1/2 and AP-1 and upregulation of CCL2. The elevated level of CCL2 protein detected in the sera of SARS-CoV-infected patients may account for the development of lung fibrosis. Figure was modified from Chen et al. [41]. SARS-CoV, severe acute respiratory syndrome coronavirus; CCL2, chemokine (C-C motif) ligand 2.
7. Therapeutic targets
SARS-CoV-2 is characterised by high contagiousness, high morbidity and high mortality, but no specific drugs for COVID-19 have been developed so far. Many researchers are trying to find therapeutic targets of the virus to develop high-efficiency, low-toxicity targeted drugs. A number of epidemiological studies have shown that the transmission characteristics of SARS-CoV-2 appear to be similar to SARS-CoV [2,[43], [44], [45], [46]. These similarities give doctors and scientists a clue to look for targeted drugs. Although coronaviruses are subject to extensive mutations, some key proteins, particularly replication-related enzymes, are highly conserved [34], so drugs targeting conserved proteases are usually able to block replication and proliferation of the virus and exhibit a broad spectrum [47]. Specific inhibitors of key proteases involved in viral replication and proliferation are effective ways of killing viruses. Candidate compounds include RNA proteases, membrane proteins, spike glycoproteins, polymerases and viral envelope that act directly on the virus, as well as targets on the host such as receptors and proteases for virus entry and endocytosis [48], [49], [50]. Recently, scientists screened out four small-molecule drugs (prulifloxacin, tegobuvir, nelfinavir and bictegravir) with strong binding affinity to the main protease of SARS-CoV [47]. The first case of SARS-CoV-2-infected pneumonia in the USA experienced significant improvement in clinical symptoms after receiving an intravenous (i.v.) infusion of remdesivir on the seventh day of hospitalisation [51]. Remdesivir is a novel nucleotide analogue prodrug under development that inhibits viruses by inhibiting RNA-dependent RNA polymerase (RdRp) and that was originally used to treat Ebola although it was not very effective. Amazingly, it is effective against SARS and MERS [52], [53], [54], [55].
In addition to this, SARS-CoV-2 is unable to infect cells without ACE2 and cannot bind to other common receptors of coronavirus (APN, DPP4, etc.) [2,34,56,57]. After replacing four out of five important interface amino acid residues, the SARS-CoV-2 S protein maintains the core structure and interacts perfectly with the human ACE2 molecule [12]. These studies show that ACE2-targeted drugs are expected to be used to treat SARS-CoV-2. The S2 subunit of the SARS-CoV spike protein plays a key role in mediating fusion of the virus and host cell and host cell entry. Heptapeptide repeat 1 (HR1) and heptapeptide repeat 2 (HR2) can interact to form six-helix bundles (6Hb), which makes the virus and cell membrane tightly bound [58]. A variety of effective fusion inhibitors against SARS-CoV and MERS-CoV have been developed using S-HR1 and S-HR2, such as HR2P peptide [59]. Researchers have found that the HR1 and HR2 regions of SARS-CoV-2 could also interact to form 6Hbs, and they have designed a pan-coronavirus fusion inhibitor denoted EK1 that can significantly inhibit SARS-CoV-2 pseudovirus infection in a dose-dependent manner [60].
8. Treatment strategies
8.1. Antiviral therapy
At present, different institutions and organisations in China have issued several guidelines for the diagnosis and treatment of novel coronavirus pneumonia, none of which recommended specific drugs for COVID-19. According to the latest treatment protocol (Pilot Version 6) issued by the National Health Commission of China, antiviral therapy can be tried with interferon, lopinavir/ritonavir, chloroquine phosphate and umifenovir. These and other potentially effective drugs are described below.
8.1.1. Nucleoside analogues
Remdesivir is a nucleoside analogue with antiviral activity developed by Gilead Sciences. Its anti-RNA virus activity has been confirmed, such as against Ebola virus, Marburg virus, Nipa virus and Hendra virus [61], [62], [63]. Furthermore, remdesivir has exhibited preventive and therapeutic effects against MERS-CoV and SARS-CoV in vitro [64,65]. Remdesivir has been observed to have the ability to reduce MERS-CoV replication, improve pulmonary function and reduce pulmonary lesions in Calu-3 cells and mouse models [54]. It has been reported that in the Vero E6 cell model, remdesivir inhibits the infection of SARS-CoV-2 virus, with an EC50 (half-maximal effective concentration) of 0.77 μM and a selectivity index of >129.87 [66]. Since SARS-CoV-2 and MERS-CoV possess a similar coronavirus structure [2,8], remdesivir has great potential against SARS-CoV-2. As reported previously, the first case of new coronavirus pneumonia treated in the USA received only supportive treatment for nausea, vomiting and other symptoms at the initial stage of admission, whilst the patient's symptoms improved significantly after receiving an i.v. injection of remdesivir for 1 day [51]. However, remdesivir has not been approved in any country and its safety and effectiveness must be confirmed. Thus, two randomised controlled, double-blind phase III trials have been initiated in February 2020 (ClinicalTrials.gov ID NCT04252664 and NCT04257656).
Ribavirin is a broad-spectrum antiviral nucleoside analogue that has been used in the treatment of hepatitis C virus and respiratory syncytial virus. The antiviral mechanism involves interacting with virus RdRp to inhibit RNA synthesis [67]. It has been shown in in vitro experiments that ribavirin inhibits the replication of MERS-CoV and HCoV-OC43, but the dosage that produces a significant effect is not within the range of typical human therapies [68]. Simultaneous use of interferon can reduce the dose of ribavirin [69]. In a primate model, clinical symptoms of MERS could be improved by the combination of ribavirin and type I interferon [70]. However, side effects of ribavirin, such as anaemia, limit its widespread use [71]. In addition, two meta-analyses of SARS and MERS case studies demonstrated limited efficacy in treating patients with coronavirus respiratory syndrome [72,73]. Therefore, further studies are needed to determine whether ribavirin can effectively treat novel coronavirus pneumonia (COVID-19), and ribavirin and interferon are still in clinical trials (Chinese Clinical Trials Registry ID R2000029387).
8.1.2. Protease inhibitors
Lopinavir/ritonavir (Kaletra, Aluvia) is a protease inhibitor used in combined therapy for human immunodeficiency virous (HIV) [74]. Lopinavir inhibits cleavage of the gag-pol protein, whilst ritonavir inhibits cleavage of the gag-pol protein precursor and lopinavir metabolism to increase the concentration of lopinavir [75]. It has been shown that lopinavir/ritonavir could inhibit the replication of MERS-CoV and SARS-CoV in vitro. In primate models, animals treated with lopinavir/ritonavir for MERS have a better prognosis than untreated animals [76]. A non-critical SARS-CoV-2-infected patient in South Korea received lopinavir/ritonavir (Kaletra; AbbVie) on the eighth day of admission, after which the clinical symptoms improved and the coronavirus load began to decrease until undetectable [77]. Animal models suggest that TMPRSS2 (type II transmembrane serine protease) plays an important role in coronavirus transmission [78]. TMPRSS2 activates the S protein of highly pathogenic human coronavirus, which binds to the receptor ACE2 and enters host cells [30,42,[78], [79], [80]. Thus, TMPRSS2 inhibitors are also considered as drugs for the treatment of novel coronavirus pneumonia [81].
8.1.3. Broad-spectrum antivirals
Interferon (IFN) is a type of glycoprotein that triggers the antiviral immune response in patients infected with MERS [82]. In animal models, IFN inhibits the replication of SARS-CoV and MERS-CoV. In addition, combination therapies of IFN with other antiviral drugs have been used to treat SARS or MERS patients and show synergistic effects [69,82].
Chloroquine also has a strong antiviral effect on SARS-CoV-infected cells. It interferes with virus–receptor binding by ACE2 terminal glycosylation [83,84]. In vitro, chloroquine can enhance the effects of other antiviral drugs [38].
8.2. Immune therapy
Due to the similar RBD structures of SARS-CoV-2 and SARS-CoV, screening anti-SARS-CoV antibodies will facilitate the rapid development of monoclonal antibodies and vaccines against SARS-CoV-2 [37]. Tian et al. reported that CR3022, a SARS-CoV-specific human monoclonal antibody, could effectively bind to the SARS-CoV-2 RBD [37]. CR3022 has the potential to be used alone or in combination with other neutralising antibodies for the prevention and treatment of SARS-CoV-2 infection [37]. However, the dose needs to be determined before applying monoclonal antibodies [85]. Using convalescent plasma to treat critically ill patients has been included in the latest Chinese treatment protocol (Pilot Version 6). One meta-analysis showed that glucocorticoids reduce the risk of ARDS [86]. However, different studies suggest that glucocorticoids slow viral clearance [87]. Clinical data are still needed to demonstrate the value of glucocorticoids for SARS-CoV-2 [75].
8.3. Other methods
Studies have shown that, similar to SARS-CoV, the S protein RBD of SARS-CoV-2 is human ACE2, which is one of the reasons for SARS-CoV-2 infecting humans [15,37]. Using cell experiments, Zhou et al. reported that ACE2 was the receptor of SARS-CoV-2, as SARS-CoV-2 could bind ACE2 receptors of civet, bat, pig and human origin, but cannot infect cells without ACE2 [2]. Therefore, application of ACE inhibitors (ACEIs) and angiotensin II receptor type 1 (AT1R) inhibitors under the condition of close monitoring of blood pressure is likely to reduce the damage in patients with SARS-CoV-2 infection [88]. XueBiJing is a traditional Chinese medicine injection commonly used to treat inflammation in severe cases. A randomised controlled trial shows that on the basis of Western medicine treatment, XueBiJing injection could significantly reduce the fatality rate of community-acquired pneumonia [89]. Further reliable evidence is needed for potential Chinese medicines for COVID-19.
9. Prevention
9.1. Traveller screening
Traveller screening is a way to limit the further spread of SARS-CoV-2 following its recent emergence, with the aim of curtailing the geographic spread of the infection [90]. Traveller symptom screening depends on the natural history of the infection. Individuals who are infected are likely to show detectable symptoms with increasing time since exposure. However, traveller screening is also limited. Gostic et al. performed tests to estimate the effectiveness of traveller screening and found, within the narrow range of the tests, that traveller screening outcomes were sensitive at a short mean incubation period [91]. However, for longer incubation periods, a larger proportion of departing travellers will not exhibit symptoms. After long incubation periods they still feel healthy enough to travel and do not realise they have been exposed to SARS-CoV-2, which is simultaneously difficult to detect [92].
9.2. Sesame oil
A folk method expressed that adding sesame oil into the nostrils can prevent the spread of SARS-CoV-2. To find theoretical reasons to support this method for preventing viral infection with SARS-CoV-2, Fan et al. discussed from the perspective of colloid and interface science that sesame oil had a low surface tension and was incompatible with water [93,94]. They investigated the epidemiological features of SARS-CoV-2 and found because of the low intermolecular attraction between adjacent two sesame oil molecules, pure sesame oil had a good wettability which could readily wet the surface of various solid and aqueous phases. In view of this feature, sesame oil might prevent the spread of SARS-CoV-2. Unfortunately, the mechanism of this method remains uncertain and it is not approved and verified by experimental and clinical studies. In future, scientists need to pay more scientific attention to these potentially useful clues from folk medicine methods.
9.3. Natural compounds
Traditional Chinese medicine herbs have been used for thousands of years. In 2003, glycyrrhizin, a traditional Chinese medicine, was suggested to be promising for treating SARS [95] and was considered to be effective and valuable owing to its availability and low toxicity. Searching the active compounds from Chinese herbal medicine to prevent SARS-CoV-2 could be a potential strategy. Chen and Du used molecular docking to find natural compounds and proposed five candidates, including scutellarin, baicalin, hesperetin, nicotianamine and glycyrrhizin, that were potential compounds targeting the ACE2 receptor and exerting an antiviral effect to prevent SARS-CoV-2 infection [96].
10. Conclusion
SARS-CoV-2 is driving China's urgent public health actions as well as international concern. Here we summarise recent progress in SARS-CoV-2 in the hope of providing potential interventions. Its spread is fast, with increasing numbers of infected patients nationwide and globally, and the future development of the disease is unclear but the public should pay attention to the virus since it may be very contagious.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China [contract nos. 31672290, 31100764 and 30901874], the Natural Science Foundation of Hunan Province, China [no. 2016JJ3180] and the Valuable Instrument and Equipment Fund of Central South University [nos. CSUZC2020043 and CSUZC2019046].
Competing interests: None declared.
Ethical approval: Not required.
References
- 1.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song S.G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benvenuto D., Giovanetti M., Salemi M., Prosperi M., De Flora C., Junior Alcantara L.C. The global spread of 2019-nCoV: a molecular evolutionary analysis. Pathog Glob Health. 2020:1–4. doi: 10.1080/20477724.2020.1725339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salata C., Calistri A., Parolin C., Palù G. Coronaviruses: a paradigm of new emerging zoonotic diseases. Pathog Dis. 2019;77 doi: 10.1093/femspd/ftaa006. pii: ftaa006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benvenuto D., Giovanetti M., Ciccozzi A., Spoto S., Angeletti S., Ciccozzi M. The 2019-new coronavirus epidemic: evidence for virus evolution. J Med Virol. 2020;92:455–459. doi: 10.1002/jmv.25688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramaiah A., Arumugaswami V. Insights into cross-species evolution of novel human coronavirus 2019-nCoV and defining immune determinants for vaccine development. bioRxiv. 2020 Feb 4 doi: 10.1101/2020.01.29.925867. [DOI] [Google Scholar]
- 11.Paraskevis D., Kostaki E.G., Magiorkinis G., Panayiotakopoulos G., Sourvinos G., Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect Genet Evol. 2020;79 doi: 10.1101/2020.01.26.920249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu X., Chen P., Wang J., Feng J., Zhou H., Li X. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji W., Wang W., Zhao X., Zai J., Li X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J Med Virol. 2020;92:433–440. doi: 10.1002/jmv.25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo Q., Li M., Wang C., Wang P., Fang Z., Tan J. Host and infectivity prediction of Wuhan 2019 novel coronavirus using deep learning algorithm. bioRxiv. 2020 Feb 2 doi: 10.1101/2020.01.21.914044. [DOI] [Google Scholar]
- 15.Lam T.T.-Y., Shum M.H.-H., Zhu H.-C., Tong Y.G., Ni X.B., Liao Y.S. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020 Feb 26 doi: 10.1038/s41586-020-2169-0. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 16.Liu K., Fang Y.Y., Deng Y., Liu W., Wang M.F., Ma J.P. Clinical characteristics of novel coronavirus cases in tertiary hospitals in Hubei Province. Chin Med J (Engl) 2020 Feb 7 doi: 10.1097/CM9.0000000000000744. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/s0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/s0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Y., Yang L., Han M., Huang M., Sun X., Zhen W. Clinical reports on early diagnosis of novel coronavirus (2019-nCoV) pneumonia in stealth infected patients. Preprints. 2020 doi: 10.20944/preprints202002.0156.v1. [DOI] [Google Scholar]
- 20.Liu Y., Yang Y., Zhang C., Huang F., Wang F., Yuan J. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci China Life Sci. 2020;63:364–374. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang H., Kang Z., Gong H., Xu D., Wang J., Li Z. The digestive system is a potential route of 2019-nCov infection: a bioinformatics analysis based on single-cell transcriptomes. bioRxiv. 2020 Jan 31 doi: 10.1101/2020.01.30.927806. [DOI] [Google Scholar]
- 22.Jin Y.H., Cai L., Cheng Z.S., Cheng H., Deng T., Fan Y.P. A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version) Mil Med Res. 2020;7:4. doi: 10.1186/s40779-020-0233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen K., Yang Y., Wang T., Zhao D., Jiang Y., Jin R. Diagnosis, treatment, and prevention of 2019 novel coronavirus infection in children: experts' consensus statement. World J Pediatr. 2020 Feb 7 doi: 10.1007/s12519-020-00343-7. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan Y., Guan H., Zhou S., Wang Y., Li Q., Zhu T. Initial CT findings and temporal changes in patients with the novel coronavirus pneumonia (2019-nCoV): a study of 63 patients in Wuhan, China. Eur Radiol. 2020 Feb 13 doi: 10.1007/s00330-020-06731-x. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu F., Du L., Ojcius D.M., Pan C., Jiang S. Measures for diagnosing and treating infections by a novel coronavirus responsible for a pneumonia outbreak originating in Wuhan, China. Microbes Infect. 2020;22:74–79. doi: 10.1016/j.micinf.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem. 2020 Jan 31 doi: 10.1093/clinchem/hvaa029. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020 doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. Erratum in: Nat Protoc 2020;15:1311. doi: 10.1038/s41596-020-0302-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren L.L., Wang Y.M., Wu Z.Q., Xiang Z.C., Guo L., Xu T. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin Med J (Engl) 2020 Feb 11 doi: 10.1097/cm9.0000000000000722. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/s0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaume M., Yip M.S., Kam Y.W., Cheung C.Y., Kien F., Roberts A. SARS CoV subunit vaccine: antibody-mediated neutralisation and enhancement. Hong Kong Med J. 2012;18(Suppl 2):31–36. [PubMed] [Google Scholar]
- 32.Struck A.W., Axmann M., Pfefferle S., Drosten C., Meyer B. A hexapeptide of the receptor-binding domain of SARS corona virus spike protein blocks viral entry into host cells via the human receptor ACE2. Antiviral Res. 2012;94:288–296. doi: 10.1016/j.antiviral.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong N., Yang X., Ye L., Chen K., Chan E.W.C., Yang M. Genomic and protein structure modelling analysis depicts the origin and infectivity of 2019-nCoV, a new coronavirus which caused a pneumonia outbreak in Wuhan, China. bioRxiv. 2020 Jan 22 doi: 10.1101/2020.01.20.913368. [DOI] [Google Scholar]
- 34.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. bioRxiv. 2020 Jan 23 doi: 10.1101/2020.01.22.914952. [DOI] [Google Scholar]
- 35.Dimitrov D.S. The secret life of ACE2 as a receptor for the SARS virus. Cell. 2003;115:652–653. doi: 10.1016/s0092-8674(03)00976-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malik Y.S., Sircar S., Bhat S., Sharun K., Dhama K., Dadar M. Emerging novel coronavirus (2019-nCoV)—current scenario, evolutionary perspective based on genome analysis and recent developments. Vet Q. 2020;40:68–76. doi: 10.1080/01652176.2020.1727993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian X., Li C., Huang A., Xia S., Lu S., Shi Z. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gharaee-Kermani M., Denholm E.M., Phan S.H. Costimulation of fibroblast collagen and transforming growth factor β1 gene expression by monocyte chemoattractant protein-1 via specific receptors. J Biol Chem. 1996;271:17779–17784. doi: 10.1074/jbc.271.30.17779. [DOI] [PubMed] [Google Scholar]
- 40.Cheung C.Y., Poon L.L., Ng I.H., Luk W., Sia S.F., Wu M.H. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J Virol. 2005;79:7819–7826. doi: 10.1128/JVI.79.12.7819-7826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen I.Y., Chang S.C., Wu H.Y., Yu T.C., Wei W.C., Lin S. Upregulation of the chemokine (C-C motif) ligand 2 via a severe acute respiratory syndrome coronavirus spike–ACE2 signaling pathway. J Virol. 2010;84:7703–7712. doi: 10.1128/JVI.02560-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heurich A., Hofmann-Winkler H., Gierer S., Liepold T., Jahn O., Pohlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J Virol. 2014;88:1293–1307. doi: 10.1128/jvi.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riou J., Althaus C.L. Pattern of early human-to-human transmission of Wuhan 2019 novel coronavirus (2019-nCoV), December 2019 to January 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.4.2000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guan W., Xian J. The progress of 2019 novel coronavirus (2019-nCoV) event in China. J Med Virol. 2020;92:468–472. doi: 10.1002/jmv.25705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu J.T., Leung K., Leung G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395:689–697. doi: 10.1016/s0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y., Zhang J., Wang N., Li H., Shi Y., Guo G. Therapeutic drugs targeting 2019-nCoV main protease by high-throughput screening. bioRxiv. 2020 Jan 30 doi: 10.1101/2020.01.28.922922. [DOI] [Google Scholar]
- 48.Li H., Wang Y., Xu J., Cao B. Potential antiviral therapeutics for 2019 novel coronavirus [in Chinese] Zhonghua Jie He He Hu Xi Za Zhi. 2020;43:E002. doi: 10.3760/cma.j.issn.1001-0939.2020.0002. [DOI] [PubMed] [Google Scholar]
- 49.Liu Q., Wang X. Strategies for the development of drugs targeting novel coronavirus 2019-nCoV. Acta Pharmaceutica Sinica. 2020;2 doi: 10.16438/j.0513-4870.2020-0106. [DOI] [Google Scholar]
- 50.Xia B., Kang X. Activation and maturation of SARS-CoV main protease. Protein Cell. 2011;2:282–290. doi: 10.1007/s13238-011-1034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aal3653. pii: eaal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lucey D.R. New treatments for Ebola virus disease. BMJ. 2019;366:l5371. doi: 10.1136/bmj.l5371. [DOI] [PubMed] [Google Scholar]
- 54.Sheahan T.P., Sims A.C., Leist S.R., Schäfer A., Won J., Brown A.J. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat Commun. 2020;11:222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mulangu S., Dodd L.E., Davey R.T., Jr, Tshiani Mbaya O., Proschan M., Mukadi D. A randomized, controlled trial of Ebola virus disease therapeutics. N Engl J Med. 2019;381:2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Letko M., Munster V. Functional assessment of cell entry and receptor usage for lineage B β-coronaviruses, including 2019-nCoV. bioRxiv. 2020 Jan 22 doi: 10.1101/2020.01.22.915660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang F., Zhang C. What to do next to control the 2019-nCoV epidemic? Lancet. 2020;395:391–393. doi: 10.1016/s0140-6736(20)30300-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu S., Xiao G., Chen Y., He Y., Niu J., Escalante C.R. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363:938–947. doi: 10.1016/s0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu L., Liu Q., Zhu Y., Chan K.H., Qin L., Li Y. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat Commun. 2014;5:3067. doi: 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xia S., Zhu Y., Liu M., Lan Q., Xu W., Wu Y. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immunol. 2020 Feb 11 doi: 10.1038/s41423-020-0374-2. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lo M.K., Jordan R., Arvey A., Sudhamsu J., Shrivastava-Ranjan P., Hotard A.L. GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci Rep. 2017;7:43395. doi: 10.1038/srep43395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brown A.J., Won J.J., Graham R.L., Dinnon K.H., 3rd, Sims A.C., Feng J.Y. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antiviral Res. 2019;169 doi: 10.1016/j.antiviral.2019.104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Wit E., Feldmann F., Cronin J., Jordan R., Okumura A., Thomas T. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc Natl Acad Sci U S A. 2020;117:6771–6776. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9 doi: 10.1128/mBio.00221-18. pii: e00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Elfiky A.A. Ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020 doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen L., Yang Y., Ye F., Liu G., Desforges M., Talbot P.J. Safe and sensitive antiviral screening platform based on recombinant human coronavirus OC43 expressing the luciferase reporter gene. Antimicrob Agents Chemother. 2016;60:5492–5503. doi: 10.1128/aac.00814-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Falzarano D., de Wit E., Martellaro C., Callison J., Munster V.J., Feldmann H. Inhibition of novel β coronavirus replication by a combination of interferon-α2b and ribavirin. Sci Rep. 2013;3:1686. doi: 10.1038/srep01686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Falzarano D., de Wit E., Rasmussen A.L., Feldmann F., Okumura A., Scott D.P. Treatment with interferon-α2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nat Med. 2013;19:1313–1317. doi: 10.1038/nm.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.Y. Coronaviruses—drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morra M.E., Van Thanh L., Kamel M.G., Ghazy A.A., Altibi A.M.A., Dat L.M. Clinical outcomes of current medical approaches for Middle East respiratory syndrome: a systematic review and meta-analysis. Rev Med Virol. 2018;28:e1977. doi: 10.1002/rmv.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stockman L.J., Bellamy R., Garner P. SARS: systematic review of treatment effects. PLoS Med. 2006;3:e343. doi: 10.1371/journal.pmed.0030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Meynard J.L., Moinot L., Landman R., Morand-Joubert L., Besseghir A., Kolta S. Week 96 efficacy of lopinavir/ritonavir monotherapy in virologically suppressed patients with HIV: a randomized non-inferiority trial (ANRS 140 DREAM) J Antimicrob Chemother. 2018;73:1672–1676. doi: 10.1093/jac/dky055. [DOI] [PubMed] [Google Scholar]
- 75.van der Laan L.E., Garcia-Prats A.J., Schaaf H.S., Tikiso T., Wiesner L., de Kock M. Pharmacokinetics and drug–drug interactions of lopinavir–ritonavir administered with first- and second-line antituberculosis drugs in HIV-infected children treated for multidrug-resistant tuberculosis. Antimicrob Agents Chemother. 2018;62 doi: 10.1128/aac.00420-17. pii: e00420-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chan J.F., Yao Y., Yeung M.L., Deng W., Bao L., Jia L. Treatment with lopinavir/ritonavir or interferon-β1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J Infect Dis. 2015;212:1904–1913. doi: 10.1093/infdis/jiv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lim J., Jeon S., Shin H.Y., Kim M.J., Seong Y.M., Lee W.J. Case of the index patient who caused tertiary transmission of COVID-19 infection in Korea: the application of lopinavir/ritonavir for the treatment of COVID-19 infected pneumonia monitored by quantitative RT-PCR. J Korean Med Sci. 2020;35:e79. doi: 10.3346/jkms.2020.35.e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Iwata-Yoshikawa N., Okamura T., Shimizu Y., Hasegawa H., Takeda M., Nagata N. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J Virol. 2019;93 doi: 10.1128/jvi.01815-18. pii: e01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X. Clinical characteristics of coronavirus disease 2019 in China. N Engl Med. 2020 Feb 28 doi: 10.1056/NEJMoa2002032. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yan R., Zhang Y., Li Y., Xia L., Zhou Q. Structure of dimeric full-length human ACE2 in complex with B0AT1. bioRxiv. 2020 Feb 18 doi: 10.1101/2020.02.17.951848. [DOI] [Google Scholar]
- 81.Hoffmann M., Kleine-Weber H., Krüger N., Müller M., Drosten C., Pöhlmann S. The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. bioRxiv. 2020 Jan 31 doi: 10.1101/2020.01.31.929042. [DOI] [Google Scholar]
- 82.Arabi Y.M., Shalhoub S., Mandourah Y., Al-Hameed F., Al-Omari A., Al Qasim E. Ribavirin and interferon therapy for critically ill patients with Middle East respiratory syndrome: a multicenter observational study. Clin Infect Dis. 2019 Jun 25 doi: 10.1093/cid/ciz544. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vincent M.J., Bergeron E., Benjannet S., Erickson B.R., Rollin P.E., Ksiazek T.G. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol J. 2005;2:69. doi: 10.1186/1743-422x-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Savarino A., Di Trani L., Donatelli I., Cauda R., Cassone A. New insights into the antiviral effects of chloroquine. Lancet Infect Dis. 2006;6:67–69. doi: 10.1016/s1473-3099(06)70361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wan Y., Shang J., Sun S., Tai W., Chen J., Geng Q. Molecular mechanism for antibody-dependent enhancement of coronavirus entry. J Virol. 2020;94 doi: 10.1128/jvi.02015-19. pii: e02015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang J., Guo J., Li H., Huang W., Zhang T. Efficacy and safety of adjunctive corticosteroids therapy for patients with severe community-acquired pneumonia: a systematic review and meta-analysis. Medicine (Baltimore) 2019;98:e14636. doi: 10.1097/md.0000000000014636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Arabi Y.M., Mandourah Y., Al-Hameed F., Sindi A.A., Almekhlafi G.A., Hussein M.A. Corticosteroid therapy for critically ill patients with Middle East respiratory syndrome. Am J Respir Crit Care Med. 2018;197:757–767. doi: 10.1164/rccm.201706-1172OC. [DOI] [PubMed] [Google Scholar]
- 88.Sun M.L., Yang J.M., Sun Y.P., Su G.H. Inhibitors of RAS might be a good choice for the therapy of COVID-19 pneumonia [in Chinese] Zhonghua Jie He He Hu Xi Za Zhi [Chinese Journal of Tuberculosis and Respiratory Diseases] 2020;43:E014. doi: 10.3760/cma.j.issn.1001-0939.2020.0014. [DOI] [PubMed] [Google Scholar]
- 89.Editorial Board of Chinese Critical Care Medicine Xuebijing injection versus placebo for critically ill patients with severe community-acquired pneumonia: a randomized controlled trial: research results and clinical value [in Chinese] Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2019;31:1199–1203. doi: 10.3760/cma.j.issn.2095-4352.2019.10.004. [DOI] [PubMed] [Google Scholar]
- 90.Gostic K.M., Kucharski A.J., Lloyd-Smith J.O. Effectiveness of traveller screening for emerging pathogens is shaped by epidemiology and natural history of infection. Elife. 2015;4 doi: 10.7554/eLife.05564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gostic K., Gomez A.C.R., Mummah R.O., Kucharski A.J., Lloyd-Smith J.O.Estimated effectiveness of traveller screening to prevent international spread of 2019 novel coronavirus (2019-nCoV) medRxiv2020 Feb 3. doi: 10.1101/2020.01.28.20019224. [DOI]
- 92.Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020 Mar 10 doi: 10.7326/m20-0504. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fan W., Zeng J., Xu Y. A theoretical discussion of the possibility and possible mechanisms of using sesame oil for prevention of 2019-nCoV (COVID-19 coronavirus) from the perspective of colloid and interface science. ResearchGate. 2020 doi: 10.13140/RG.2.2.31786.98248. [DOI] [Google Scholar]
- 94.Fan W., Yan B., Wang Z., Wu L. Three-dimensional all-dielectric metamaterial solid immersion lens for subwavelength imaging at visible frequencies. Sci Adv. 2016;2 doi: 10.1126/sciadv.1600901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hoever G., Baltina L., Michaelis M., Kondratenko R., Baltina L., Tolstikov G.A. Antiviral activity of glycyrrhizic acid derivatives against SARS-coronavirus. J Med Chem. 2005;48:1256–1259. doi: 10.1021/jm0493008. [DOI] [PubMed] [Google Scholar]
- 96.Chen H., Du Q. Potential natural compounds for preventing SARS-CoV-2 (2019-nCoV) infection. Preprints. 2020 doi: 10.20944/preprints202001.0358.v3. [DOI] [Google Scholar]