Abstract
Pancreatic cancer (PDAC) is one of the deadliest types of human cancers, owing to late stage at presentation and pervasive therapeutic resistance. The extensive tumour heterogeneity, as well as substantial crosstalk between the neoplastic epithelium and components within the microenvironment are the defining features of PDAC biology that dictate the dismal natural history. Recent advances in genomic and molecular profiling have informed on the genetic makeup and evolutionary patterns of tumour progression, leading to treatment breakthroughs in minor subsets of patients with specific tumour mutational profiles. The nature and function of tumour heterogeneity, including stromal heterogeneity, in PDAC development and therapeutic resistance, are increasingly being elucidated. Deep insight has been gained regarding the metabolic and immunological deregulation, which further sheds light on the complex biology and the observed treatment recalcitrance. Here we will summarize these recent achievements and offer our perspective on the path forward.
Keywords: Pancreatic cancer, Genetics, Ttumour heterogeneity, Cancer metabolism, Immune, Evasion, Immunotherapy
Pancreatic ductal adenocarcinoma (PDAC) has become the third leading cause of cancer-related death in the United States, and is on pace to become the second within the next decade [1,2]. PDAC remains the most lethal type of human cancer with the 5-year survival rate gaining incremental increase from 6% to 10% during the past five years, largely owing to the improvement in neoadjuvant and adjuvant therapies. Breakthroughs have been made in the past few years for the treatment of a small population of PDAC patients with inherited deficiencies in DNA damage repair. Unfortunately, for majority of the PDAC patients, our knowledge on the genetics and biology of this disease has not been translated to a leap in patient survival yet. Here we summarize the most recent advancement in several selected fields of PDAC research where we believe the next wave of breakthroughs will emerge to tackle this deadly disease.
1. The genomic landscape of pancreatic cancer
1.1. Clinically actionable genetic alterations of PDAC
Signature mutations of human PDAC include oncogenic mutations of KRAS present in over 90% of cases, and the frequent inactivation of TP53, SMAD4 and CDKN2A tumour suppressors [3]. Next generation sequencing efforts have identified a ‘long tail’ of additional recurrent mutations/alterations in PDAC with individual incidence below 10% [3]. It should be noted that many of genes with low frequency mutations belong to a handful of common pathways, including RAS signalling, TGFβ pathway, cell cycle control, WNT signalling, NOTCH signalling, epigenetic regulation, and DNA damage repair [3]. Additionally, recurrent non-coding mutations have also been identified in PDAC, which are enriched in transcriptionally active regions of the genome, implicating the role of these non-coding mutations in the regulation of expression programs in tumour cells [4].
Some of the genetic alterations offer therapeutically actionable targets that have already been translated into clinical application. Small molecule inhibitors targeting KRASG12C, a mutation present in ∼1.5% PDAC patients, is showing promising anti-tumour effect in clinical trials [5]. In addition, about 1% of human PDACs carry somatic inactivation of mismatch repair (MMR) genes, such as MLH1 and MSH2, and are characterized with a unique hypermutated genome with >10 somatic mutations/megabase (Mb), in contrast to the ∼1.1–1.8 mutation per Mb in most sporadic PDACs [6]. These MMR deficient tumours carry high neoantigen load and showed significantly improved responses to PD-1 blockade, which is now approved by the FDA for the treatment of this specific patient population [7]. Lastly, mutations in genes involved in homologous recombination repair (HRR), such as BRCA1, BRCA2 and PALB2, affect DNA double strand break repair and result in chromosomal aberrations characterized with big deletions with overlapping microhomology at breakpoint junctions and short tandem duplications [3]. These HRR genes are also mutated in the germline of patients with familial PDAC [8]. Loss of function BRCA1 or BRCA2 mutations are synthetic lethal to the inhibition of PARP, an enzyme critical for single-strand DNA damage repair. Indeed, recent phase 3 trial of olaparib, a PARP inhibitor, showed significant improvement in progression-free survival in germline BRCA-mutated metastatic PDAC patients who are sensitive to first-line platinum-based chemotherapy [9], implicating the potential of PARP inhibitor-based maintenance therapy in HRR-defective PDACs that exhibit similar ‘BRCAness’. However, an important caveat is that germline BRCA1 and BRCA2 mutations are per se not reliable biomarkers for sensitivity to PARP inhibitors unless the mutations are bi-allelic (i.e., accompanied by a somatic alteration in the other allele), thus resulting in an “unstable genome” phenotype, which confers sensitivity to DNA damage reagent such as cisplatin.
1.2. The genetic evolution of PDAC
Recent advances in next-generation sequencing coupled with multi-region sampling have provided critical insights into the genetic evolution of PDAC. Phylogenetic modelling of mutations identified from multiple PanIN, primary tumour and metastatic lesions from the same patients indicated that it takes years, if not decades, for the development of invasive PDAC from founder clones [10], implicating a relative long window for early detection. The clonal nature of the shared mutations among PanINs and advanced tumours supports the stepwise-progression model of pancreatic cancer , although multiple somatic alterations may occur simultaneously in a subset of tumours due to a single chromosomal catastrophe termed chromothripsis [11]. It is possible that a single chromothripsis event may lead to the neoplastic transformation of precursor cells if it leads to the simultaneous generation of multiple driver alterations. In this case, the trajectory of PDAC progression could be much shorter than we initially estimated, though such assumption needs to be validated in relevant in vivo models. In addition, while intra-tumoral genetic heterogeneity is defined by the existence of multiple subclones with distinctive driver or passenger mutations, recent analysis indicated that such subclonal mutations in untreated tumours are likely to be functionally irrelevant compared to the clonal driver mutations [12]. Moreover, analysis of metastatic PDAC and several other solid tumours revealed high uniformity of driver mutations in all metastatic lesions from the same patient [13,14]. Although it is likely that different subclones of the primary tumour give rise to the multiple metastatic lesions, they all share the same clonal driver mutations [14]. These findings hold significant clinical implications. The long latency and the conservation of clonal driver mutations during PDAC development suggest that liquid biopsy of the core genetic alterations could serve as a valuable tool for early detection. Importantly, the functional dominance of clonal driver mutations indicates that combinatory strategies targeting these genetic alterations should elicit similar responses in both primary tumour and metastases, although therapeutic resistance are expected to arise due to subclonal mutations [12].
2. Transcriptomic subtypes of pancreatic cancer with potential clinical relevance
2.1. Molecular subtypes of PDAC
Recent global transcriptomic analyses have defined human PDAC into several subtypes, which largely share comparable molecular features between different studies despite distinctive nomenclatures [15]. The original analysis described three subtypes – classical, quasimesenchymal and exocrine-like [15]. Such classification was largely confirmed by the International Cancer Genome Consortium (ICGC) study which updated the subtype names to progenitor (similar to the classical subtype), squamous (similar to the quasimesenchymal) and ADEX (Aberrantly Differentiated Endocrine eXocrine, similar to the exocrine-like subtype) respectively [15]. Moreover, a new immunogenic subtype was identified, which was partially overlapping with the previously described classical subtype given the enrichment of immune signature [15]. It has been suggested from these studies that the squamous/basal-like tumours are associated with an adverse prognosis and are more resistant to chemotherapy regimens compared to other subtypes [15]. A TCGA study later showed that the molecular signatures of the ADEX and immunogenic subtypes are mostly derived from acinar cell contamination and immune infiltration respectively, indicating the molecular signature of neoplastic epithelial cells largely dichotomize into either progenitor or squamous subtypes [16]. This is consistent with findings using a ‘virtual microdissection’ approach, digitally separating the neoplastic epithelial and stromal components, which identified two tumour cell-specific groups, including classical (overlapping with the progenitor subtype) and basal-like (overlapping with the squamous subtype) subtypes [15]. However, exocrine-like subtype has been identified in PDX-derived primary cultures [17], indicating that tumour purity may not be the only determinant for the appearance of this subtype. Moreover, it seems that such molecular subtypes are not fully recapitulated at the single cell level. Recent single cell RNAseq analysis of primary human PDAC identified distinct subtypes of neoplastic epithelial cells [18,19]. While some progenitor/classical signature genes are specifically expressed in certain subclusters, the squamous/basal-like signature is not consistently enriched in subclusters of cancer cells [18,19]. Several issues would need to be addressed for future analysis to further clarify the intra-tumoral heterogeneity, including limited sample size, single lesion instead of multi-region sampling, and analysis bias toward ductal marker-positive tumour cells.
2.2. Determinants for the molecular subtypes
So far, no obvious genomic alterations have been associated with the transcriptomic subtypes, although the squamous/basal-like subtype exhibits more TP53 mutations [15], indicating non-genetic mechanisms underlying the formation of different tumour subgroups. It is also possible that genetic changes in the non-coding regions account for the observed molecular heterogeneity. In this case, these non-coding mutations may still function through the control of gene expression [4]. Several transcriptional programs have been recently indicated as the driver for the subgroups of tumour cells, including GATA6 for the progenitor/classical subtype [20,21], and KDM6A loss, and upregulation of ΔNp63/YAP1/GLI2 for the squamous/basal-like subtype [22], [23], [24], [25]. Interestingly, KDM6A and GLI2 can function as negative and positive regulators for p63 expression respectively [23,26], while YAP1 was reported to drive GLI2 expression [27]. Moreover, functional interaction between p63 and YAP1 is important for basal stem cell renewal [28]. Therefore, it is possible that these genes function convergently in PDAC to drive the squamous/basal-like subtype.
Besides the cell-autonomous mechanisms, the tumour microenvironment (TME) is also involved in orchestrating the transcription programs of tumour cells. Co-culture with cancer-associated fibroblasts (CAFs) are able to switch the expression signatures of tumour cells toward a proliferative or/and an epithelial-to-mesenchymal transition (EMT) phenotype in some cases [29]. Moreover, it seems that the tumour infiltrating myeloid cells are also capable of shaping the transcription program of tumour cells towards the squamous (basal-like) phenotype [30]. Depletion of these myeloid cells by targeting CSF1R or CXCR2 switches the expression signature of tumour cells away from the squamous subtype [30,31], implicating the dynamic nature of tumour expression programs and underscoring the importance of the TME in defining tumour molecular signatures.
2.3. Stromal heterogeneity
Desmoplastic stroma is a defining feature of PDAC, which can comprise as much as 90% of the total tumour volume. Similar to the classification of tumour cells, digital deconvolution of bulk PDAC transcriptomic data has also identified two distinct stromal subgroups, a ‘normal’ subtype resembling myofibroblast or pancreatic stellate cells (PSCs) and an ‘activated’ subtype characterized by inflammatory signatures [32]. Importantly, tumours of the activated stroma subtype are associated with significantly worse prognosis [32], underscoring the importance of stromal heterogeneity in defining PDAC biology. Interestingly, factors highly expressed in activated stroma, such as WNT5A ligand, has recently been shown to activate YAP1 oncogene and induce a squamous (basal-like) subtype of PDAC [25,32], further underscoring the importance of heterotypic tumour-stroma crosstalk in defining tumour heterogeneity. Recent studies, including single cell transcriptomic analysis, have further delineated the intra-tumoral CAF heterogeneity in PDAC, including identification of a myofibroblast (“myCAF”), an inflammatory fibroblast (“iCAF”), and an antigen-presenting CAF (“apCAF”) population, observed in both patients and in credentialed murine models [18]. These CAF populations are highly dynamic and their phenotypes can be determined by their proximity to tumour cells and the paracrine factors released by adjacent tumour cells [33]. TGFβ from tumour cells induce the formation of neighbouring myCAFs, which are characterized by high level of collagen deposition and are believed to restrict tumour growth, whereas IL-1 activates leukemia inhibitory factor (LIF) expression in more distal CAFs, which leads to the formation of tumour-promoting iCAFs through autocrine mechanism [33]. Notably, LIF and the interleukin IL-6, which are also highly expressed by iCAFs, have been shown to promote PDAC growth through paracrine mechanisms [34,35]. Therefore, the intra-tumoral heterogeneity of CAFs is likely the reason for the seemingly contradictory anti-tumour and pro-tumour functions of PDAC stroma. These findings also indicate that PDAC treatment could benefit from specifically targeting the tumour promoting function(s) of CAFs. Indeed, instead of the enhanced tumour growth following complete depletion of CAFs [36,37], inhibition of iCAF-specific LIF activity or reprogramming of CAFs into a “quiescent” state with a Vitamin D receptor agonist can mitigate tumour growth and sensitize tumours to gemcitabine in preclinical models [34,38], providing rationale for clinical trials to test such combinations (NCT03490669, NCT03520790, NCT02030860, NCT03415854).
3. Altered metabolic programs in PDAC
3.1. Regulation of glucose and glutamine metabolism
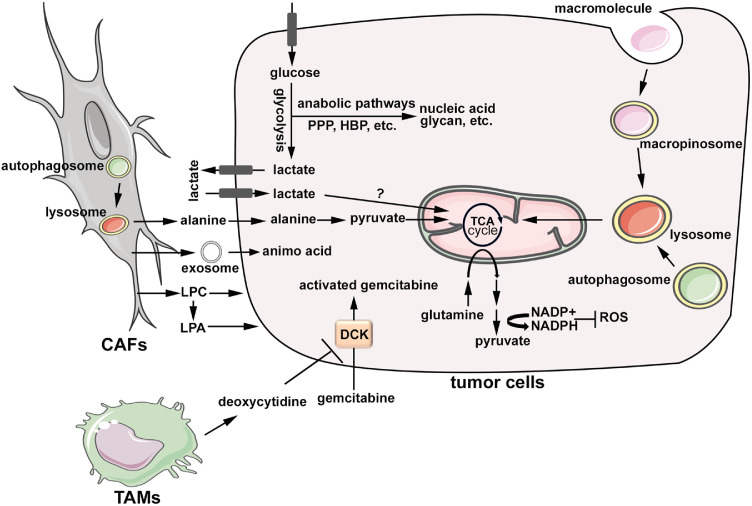
Human PDAC have limited access to nutrients due to the dense stroma and poor perfusion. Therefore, PDAC metabolism is reprogrammed in a way to support tumour biology under such a nutrient-deficient background. The metabolic reprogramming in PDAC cells is largely driven by a combination of genetic and microenvironment factors. As the dominant genetic alteration in PDAC, mutant KRAS is a powerful driver for glucose uptake and glycolysis, allowing growth under low glucose conditions (Fig. 1) [3]. Oncogenic RAS induces the transcription of glucose transporter, GLUT1, and multiple key glycolysis enzymes, including HK1/2, PFK1 and LDHA [3]. This is achieved at least partially through the induction of MYC, a key dependence in KRAS-driven tumours [3]. Moreover, the PI3K-RAC1 axis, a key effector of the RAS pathway in PDAC, has been shown to mobilize cytoskeletal aldolase through disruption of actin filaments, and thus enhance glucose flux through glycolysis [39]. The activation of glycolysis is further strengthened by hypoxia, a defining feature of human PDAC. It should be noted that, although most of glucose-derived pyruvate in PDAC cells is converted to lactate instead of feeding into the mitochondrial TCA cycle, recent studies revealed significant flux of glucose to the TCA cycle in vivo, which is mediated by the direct contribution of circulating lactate to the TCA cycle (Fig. 1) [40].
Fig. 1.
Metabolic reprogramming of neoplastic cells and crosstalk with the tumour microenvironment. Glucose uptake and glycolysis are activated in PDAC cells, which promotes the flux of glycolysis intermediates into biosynthetic pathways, including pentose phosphate pathway and hexose biosynthesis pathway, for the production of nucleic acid and glycan structures, amongst others. Tumour cells also uptake circulating lactate, which is further fed into TCA cycle through yet unknown mechanisms. Glutamine metabolism in PDAC cells is rewired to support the production of NADPH in the cytosol to maintain redox balance. PDAC cells are characterized by enhanced nutrient salvage, including the induction of macropinocytosis and autophagy, which provide substrate for energy production and anabolism in mitochondria. Autophagy in stromal fibroblasts provides alanine, which feeds into mitochondrial TCA cycle in tumour cells. Amino acids are also transferred from fibroblasts to tumour cells through exosomes. In addition, stromal fibroblasts provide phospholipids to support the proliferation of tumour cells. Tumour-associated macrophages excrete pyrimidines, in particular deoxycytidine, which competitively inhibit DCK in tumour cells to prevent gemcitabine activation and thus leading to chemoresistance. CAFs: cancer-associated fibroblasts; LPC: lysophosphatidylcholine; LPA: lysophosphatidic acid; TAMs: tumour-associated macrophages; DCK: deoxycytidine kinase; PPP: pentose phosphate pathway; HBP: hexose biosynthesis pathway; ROS: reactive oxygen species.
The induction of glycolysis in PDAC cells facilitates the flux of glycolysis intermediates through biosynthetic pathways to support tumour growth (Fig. 1). Oncogenic RAS and the hypoxic environment have been shown to induce the expression of key enzymes, GFPT1 and GFPT2, thus promoting glucose utilization through the hexosamine biosynthesis pathway (HBP) to sustain cell viability and proliferation [3,41]. Similarly, oncogenic RAS and hypoxia also enhance the flux of glycolysis intermediates into the non-oxidative arm of pentose phosphate pathway (PPP), likely through the induction of non-oxidative PPP-specific genes such as RPIA, RPE and TKT, to generate ribose-5-phosphate for de novo nucleotide biosynthesis and support cell proliferation [3].
Besides glucose, glutamine is another major carbon source for PDAC cells. While glutamine usually fuels the TCA cycle for oxidative phosphorylation and macromolecular biosynthesis, PDAC cells rely on non-canonical utilization of glutamine involving cytosolic malic enzyme (ME1) to maintain their ROS homeostasis (Fig. 1) [42]. Here, glutamine is converted sequentially by a pair of aspartate aminotransferases, GOT2 (mitochondrial) and GOT1 (cytosolic), into oxaloacetate, which is further converted to malate by malate dehydrogenase (MDH1) [42]. The subsequent utilization of malate for NADPH generation through the action of ME1 is critical for the redox homeostasis and tumour growth of PDAC [42]. In addition to its unique utilization, glutamine biosynthesis is also activated in PDAC. It was recently reported that glutamate ammonia ligase (GLUL), the enzyme for de novo glutamine biosynthesis, is overexpressed in both mouse and human PDAC and plays an essential role in transferring the terminal amide nitrogen to support nucleotide and hexosamine biosynthesis, anabolic pathways critical for tumour growth [43].
3.2. The role and regulation of nutrient salvage in PDAC
The depletion of glucose and glutamine in the PDAC microenvironment indicates that tumour cells need to employ alternative nutrient utilization strategies. Indeed, recent studies have revealed hyper-activation of nutrient scavenging mechanisms, including autophagy and macropinocytosis, in tumour cells, which has been recognized as the hallmark of PDAC metabolism (Fig. 1). Autophagy is a self-scavenging process for the recycling of cellular components to provide resources for macromolecular biosynthesis and bioenergetics. Consistent with the essential role of autophagy in RAS-induced cellular transformation [44], genetic disruption of autophagy with Atg5 deletion or blockade of mitophagy, a major form of selective-autophagy, through Bnip3l (NIX) deletion, suppressed tumour progression in autochthonous PDAC models driven by oncogenic KRAS [45,46]. Importantly, genetic inhibition of autophagy with the expression of dominant negative Atg4BC74A or pharmacologically with chloroquine treatment decreased the growth of fully formed PDAC in genetically engineered mouse (GEM) models, as well as in KRAS mutant patient derived xenograft (PDX) models, further supporting the critical role of autophagy for tumour maintenance [47]. Although autophagy is usually tightly controlled by nutrient and oxygen availability, it seems that basal autophagy in human PDAC cells remains constitutively active even under nutrient-rich conditions [44]. As the effector of the autophagic pathway, lysosome activity is also concordantly constitutively elevated in PDAC cells, likely due the nuclear sequestration of MiT/TFE proteins, major transcription factors mediating lysosome biogenesis [48]. While oncogenic RAS has been shown to activate autophagy, including mitophagy [45], recent studies showed that autophagy is further activated, and is required for viability, upon depletion of oncogenic KRAS with shRNA or inhibition of RAS downstream MEK/ERK signalling in PDAC cells [49,50]. Importantly, co-targeting autophagy with MEK/ERK inhibitors showed promising results in preclinical models [49,50], providing the rationale for clinical trials concurrently targeting autophagy and the MEK/ERK pathway that are underway.
In addition to recycling intracellular substrates, PDAC cells also actively scavenge extracellular nutrients through macropinocytosis. Abundant extracellular proteins, such as albumin and collagen, are taken up by tumour cells through macropinocytosis and delivered to lysosome for degradation to fuel tumour growth [51,52]. RAS oncogene is a potent inducer of macropinocytosis and a recent study identified the cell surface proteoglycan, Syndecan-1 (SDC1), which functions downstream of RAS, in promoting macropinocytosis through the activation of RAC1 [53]. Additionally, macropinocytosis can also be activated in nutrient-depleted tumour regions, likely through the activation of EGFR-PAK signalling following glutamine starvation [54].
3.3. Metabolism heterogeneity and crosstalk
While metabolic reprogramming and nutrient salvage activation are overarching commonalities, there is also substantial underlying heterogeneity driven by both cell autonomous mechanisms and microenvironmental cues. For example, mutant KRAS dosage has been shown to affect metabolism phenotype, with tumours harbouring loss of the wild type allele and concurrent amplification of the mutant allele exhibiting enhanced glycolysis [55]. In addition, it seems that different molecular subtypes of PDAC exhibit distinctive metabolic features, with the squamous subtype enriched for a glycolytic phenotype, characterized by elevated MCT4 expression, while the classic subtype tumours are more lipogenic [56,57]. The ambient tumour microenvironment also contributes towards metabolic heterogeneity in PDAC. For example, it has been shown that tumour cells in the hypoxic regions undergo EMT and upregulate glycolysis [41]. MCT4 is highly expressed in hypoxic tumour cells to mediate the efflux of lactate, which is then utilized by neighbouring tumour cells through MCT1, another lactate transporter exclusively expressed in the normoxic region [41]. Analogous metabolic symbiosis also exists between tumour cells and their surrounding stroma. It was demonstrated that cancer associated fibroblasts (CAFs) release non-essential amino acids (mostly alanine) through enhanced autophagy, which in turn fuel TCA cycle of tumour cells to support anabolic needs (Fig. 1) [58]. In addition, CAFs also supply amino acids to tumour cells through exosomes which in turn promotes the utilization of glucose through glycolysis and the generation of lipogenic acetyl-CoA from glutamine through reductive carboxylation in the mitochondria (Fig. 1) [59]. A recent study also discovered that CAFs release abundant lysophosphatidylcholine (LPC) to support tumour growth (Fig. 1) [60]. However, instead of directly feeding into tumour cell metabolism, the CAF-derived LPC is converted into a signalling lipid, lysophosphatidic acid (LPA), in the extracellular space by autotaxin released from adjacent tumour cells, which in turn exerts mitogenic signals through the LPA-receptor on tumour cell surface (Fig. 1) [60].
In contrast to our understanding on the metabolic programs in primary tumours, little is known on how metastatic sites fuel their metabolic needs. Recent characterization of PDX cell lines derived from metastatic tumours and matched primaries indicates that the former specifically overexpress 6-phosphogluconate dehydrogenase (PGD), an enzyme of the oxidative arm of PPP [61]. Importantly, PGD depletion preferentially suppressed the tumorigenicity of metastatic tumours [61], providing a compelling rationale to target the unique metabolic dependencies in metastatic PDAC.
3.4. Metabolism reprogramming and therapy resistance
The profound metabolic reprogramming also contributes to the extensive therapy resistance, another hallmark of PDAC biology. The selective activation of non-oxidative PPP and subsequent nucleotide biosynthesis pathway in PDAC, in particular under hypoxic conditions, has been shown as a confounding factor leading to the resistance to gemcitabine treatment [62]. In addition, deregulated fatty acid metabolism, such as enhanced de novo fatty acid biosynthesis due to overexpression of FASN or increased cholesterol uptake through LDLR, also contributes to chemotherapy resistance [63,64]. Consistent with the chemo-resistance of cancer stem cell (CSC), a subpopulation of tumour cells with self-renewal, differentiation, and tumour propagating capacity, the resistance to gemcitabine induced by fatty acid is also associated with enhanced stemness of tumour cells [64]. Notably, recent studies demonstrated that PDAC CSCs are associated with decreased glycolysis and enhanced mitochondrial respiration, as well as unique dependence on oxidative phosphorylation (OXPHOS) for survival [65,66]. It possible that targeting OXPHOS may prevent the emergence of chemo-resistance through the depletion of PDAC CSCs. In addition to the cell autonomous mechanisms, the PDAC TME also contribute to the development of chemo-resistance. For example, tumour associated macrophages (TAMs) specifically release pyrimidines, including deoxycytidine, which inhibits gemcitabine uptake by tumour cells and depletion of TAMs sensitizes PDACs to gemcitabine treatment (Fig. 1) [67], implicating the potential of combining macrophage targeted therapy with chemotherapy for PDAC treatment.
4. Immune evasion in PDAC and strategies for effective immunotherapy
4.1. Mechanisms for the immune evasion in PDAC
The ability to evade immune surveillance has been recognized as a hallmark of cancer. Among solid tumours, PDAC is an immunologically “cold” tumour, characterized by sparse T cell infiltrates. In contrast to immunologically “hot” tumours such as melanoma, with high neoantigen load and robust T cell infiltrates, human PDAC express moderate range of neoantigens [68,69]. Although T cell cytolytic activity does not per se correlate with increased tumour mutational burden or neoepitope load in PDAC [68], neoantigen number combined with abundance of CD8+ T-cell infiltration, as well as the topographic proximity of cytotoxic T cells to tumour cells, are associated with patient survival [69,70]. Importantly, study of long-term PDAC survivors have revealed the neoantigen quality as a key determinant for anti-tumour immunity [69].
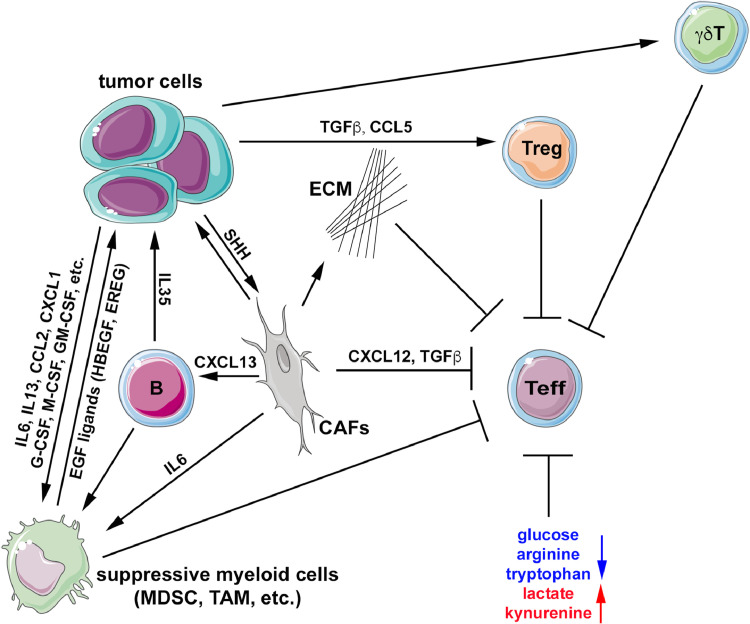
The major factor contributing to the non-immunogenic characteristic of PDAC is the TME, including the stromal compartment. Excessive extracellular matrix (ECM) deposition by CAFs creates a physical barrier preventing T cells from migrating toward tumour cells. In addition, multiple components of TME are capable of dampening anti-tumour immunity, including diverse populations of CAFs, endothelial cells, subsets of myeloid cell, suppressive B cells, regulatory T cells (Tregs), and gamma delta (γδ) T cells, amongst others [71]. It should be noted that tumour cells themselves, in particular oncogenic KRAS, play instrumental roles in shaping the immunosuppressive TME. It has been shown in GEM models that oncogenic KRAS activates CAFs in the TME through the induction of Hedgehog ligands, which lead to the deposition of dense extracellular matrix (ECM) and the exclusion of T cells from tumour TME (Fig. 2) [72]. Expression of CXCL12 by CAFs is involved in immunosuppression (Fig. 2) as blocking CXCL12 interaction with its receptor CXCR4 with mAb lead to CD8+ T cells accumulation [72], and emerging data in PDAC patients suggests this is being recapitulated in the clinic as well.
Fig. 2.
Immune evasion orchestrated by tumour cells and the tumour microenvironment. PDAC cells release cytokines and recruit immunosuppressive cells, including myeloid cells, regulatory T cells and γδT cells, which inhibit the function of effector T cells. Tumour cells also promote the activation of stromal fibroblasts by producing ligands, such as SHH. Activated fibroblasts not only enhance the growth of tumour cells, but also inhibit effector T cells through several different mechanisms, including the deposition of extracellular matrix to impede T cell trafficking, excretion of cytokines to suppress T cells activity, and induction of immunosuppressive myeloid cells directly with cytokines or indirectly through the recruitment of suppressive B cells. Some of the immune infiltrates, including suppressive B cells and myeloid cells, also produce growth factor ligands or cytokines to directly stimulate tumour growth. Metabolite changes in the tumour microenvironment also contribute to the inhibition of T cell activation, including depletion of glucose, arginine and tryptophan and the accumulation of lactate and kynurenine. Treg: regulatory T cells, ECM: extracellular matrix, CAFs: cancer-associated fibroblasts, Teff: effector T cells, MDSC: myeloid-derived suppressor cell; TAM: tumour-associated macrophage.
PDAC TME is dominated with myeloid cells, including TAMs, granulocytes and inflammatory monocytes, which are actively recruited to the microenvironment during multistep pancreatic carcinogenesis. The recruitment of myeloid cells is orchestrated by oncogenic KRAS in the epithelial compartment, likely through the induction of various cytokines, including IL-6, IL-13, CCL2, G-CSF, M-CSF and GM-CSF [73]. It should be noted that tumour cells also recruit additional immunosuppressive cells other than myeloid cells, such as Tregs and γδT cells (Fig. 2) [73]. Interestingly, it was recently shown that tumour subclones that can recruit myeloid cells, likely through the CXCL1-CXCR2 axis, act dominantly to establish the overall immunosuppressive TME, even if they represent a minor fraction of the bulk neoplastic population [74], indicating that the immune microenvironment can also be defined by underlying tumour cell heterogeneity. In addition, CAFs also promote the immunosuppressive polarization of TAMs either directly through the secretion of IL-6 or indirectly through the recruitment of immunosuppressive B cell sub-population through CXCL13 (Fig. 2) [73,75]. Recent studies have also implicated the role of the gut and intra-tumoral microbiome (and more recently, the fungal mycobiome) in the formation of the immune environment in PDAC. Specifically, in preclinical models, distinct species of bacteria and fungi are enriched in both the gut and cancerous pancreas, and are associated with tumour progression, through the creation of a permissive immune milieu [76,77]. Conversely, in human PDAC patients, a unique microbiome signature has been described that is capable of inducing anti-tumour immunity and is associated with long-term survival [78].
It has been shown that the myeloid cell infiltration is critical for PDAC initiation [79]. While myeloid cells are implicated in the induction of immune checkpoint ligands on neoplastic cells and the mitigation of effector T cell function [73], myeloid cells may also directly promote the formation and maintenance of preneoplastic lesions, at least partially through the induction of EGF ligand, which amplifies MAP kinase signalling downstream of oncogenic RAS in epithelial cells (Fig. 2) [80].
In addition to the action of various cytokines, the local metabolic milieu also contributes to the immunosuppressive TME. The severe hypoxic environment of the PDAC TME can suppress anti-tumour immunity and enhance immune evasion. While mild hypoxia may promote effector T cell activation by activating glycolysis, sustained severe hypoxia, which is a characteristic of human PDAC, will enforce T cells to rely on glycolysis in a glucose-depleted microenvironment, leading to T cell dysfunction and the suppression of anti-tumour immunity [71]. Moreover, excessive lactate excretion from tumour cells due to enhanced glycolysis results in an immunosuppressive TME by inhibiting T cell and NK cell activation and promoting the immunosuppressive polarization of myeloid cells [81], [82], [83]. Additionally, amino acid availability in the TME, in particular arginine and tryptophan, are also critical determinants of anti-tumour immunity. Arginine plays an important role in T cell activation and memory T cell differentiation [84]. Indoleamine 2,3-dioxygenase (IDO), which catabolizes tryptophan into kynurenine, is overexpressed in PDAC [85]. The depletion of tryptophan in TME and production of kynurenine promotes the creation of a suppressive immune environment and attenuates anti-tumour T cell responses [71]. Increased serum kynurenine/tryptophan ratio is correlated with resistance to immune checkpoint blockade (ICB), implicating the potential of targeting IDO for combinatory immunotherapy [86]. Nonetheless, early clinical trials combining IDO inhibition with ICB in PDAC have not been efficacious, underscoring the multidimensionality of immune suppression existent within the PDAC TME.
4.2. Immunotherapeutic approaches and developments
The dominant immune suppressive environment renders PDAC to be largely resistant to ICB, with the exception of ∼1% patients with high mutation load due to mismatch repair deficiency [7]. Therefore, restoration of T cell-mediated immune surveillance remains a major goal for PDAC immunotherapy. One method to activate T-cells is to develop vaccines targeting tumour-associated antigens that are overexpressed in tumour cells compared to normal tissues, and/or tumour-specific neoantigens that are usually derived from mutational events. One of the most extensively evaluated vaccines in clinical trials is GVAX, which is a whole-cell vaccine composed of irradiated human allogeneic PDAC cell lines engineered to release GM-CSF at the vaccination site. While a phase 2 trial of GVAX in combination with CRS-207, a live-attenuated listeria vaccine expressing a PDAC-associated antigen mesothelin, showed no survival advantage over chemotherapy in patients with metastatic PDAC [87], an earlier phase 1 study for GVAX in combination with an anti-CTLA-4 antibody (ipilimumab) showed promising impact on the induction of anti-tumour T-cell response and overall patient survival [88], prompting additional GVAX clinical trials in combination with ICB and other modalities, including demethylating agents and radiation.
Adoptive transfer of T cells is another approach to target tumour antigens. TCRs reactive to KrasG12V and KrasG12D neoepitopes have been recently isolated from HLA-A*11:01 transgenic mice [89]. KRASG12D-specific T cells were also identified in colorectal cancer patients with the HLA-C*08:02 allele [90]. These findings implicate the potential for engineering anti-KRAS T cells for the fraction of patients who harbour HLA-A*11:01 or HLA-C*08:02. More recently, T cells recognizing additional neoantigens other than mutant KRAS have also been identified in PDAC patients [91], further expanding the repertoire of potential adoptive T cell therapies. Additional T cell therapies, including chimeric antigen receptor (CAR) T cells, are also under extensive clinical studies. While CART has shown clinical responses in haematological malignancies, little success has been achieved for similar approaches in solid carcinomas, including PDAC. Toxicity due to the expression of antigens in normal tissues can be one of the limiting factors. More importantly, T cell exhaustion due to chronic TCR signalling and multiple immunosuppressive mechanisms in the TME likely play dominant roles in the interfering T cell function [71].
Agents that alter the immune suppressive TME through reprogramming the myeloid compartment have recently emerged as a promising modality within the immunotherapy repertoire in PDAC. CD40 is mostly expressed on dendritic cells (DCs), B cells and myeloid cells. Activation of CD40 with an agonist antibody showed transient anti-tumour effect in PDAC GEM models through the reprogramming of tumour-associated macrophages, instead of effector T-cells [92]. Sustainable therapeutic responses have been achieved in preclinical models with additional chemotherapy, such as gemcitabine or nab-paclitaxel, prior to CD40 activation [93]. Importantly, a CD40 mAb (APX005M) in combination with gemcitabine, nab-paclitaxel and PD-1 mAb (nivolumab) showed promising anti-tumour effects in a phase 1b trial [94], raising the hope of achieving a sustained therapeutic response in additional clinical trials. Targeting receptors expressed on myeloid cell surface, including CXCR2, CCR2, CSF1R and dectin 1, has been shown to deplete myeloid cells, reduce tumour burden, improve anti-tumour immunity and sensitize tumour-bearing mice to ICB in preclinical models [31,[95], [96], [97]]. It was demonstrated that PI3Kγ is specifically expressed in tumour associated macrophages and is required for the polarization of macrophages toward immunosuppressive phenotype [98]. Genetic or pharmacological inhibition of PI3Kγ suppresses tumour growth and metastasis in KRAS-driven PDAC GEM models [98]. Additionally, low dose irradiation has been shown in PDAC preclinical models to reprogram tumour-associated macrophages to orchestrate T cell recruitment and promote anti-tumour immunity [99].
Immunotherapy is likely to have its maximal impact in PDAC when combined with modulating the physical and functional stromal barriers to effective immune rejection. For example, decreasing the hydrostatic pressure in the dense PDAC stroma with PEGPH20, a pegylated hyaluronidase, has been shown to re-expand the tumour microvasculature and promote T cell infiltration when used in combination with GVAX in preclinical models [100,101]. Notably, a recently concluded pivotal phase 3 trial of PEGPH20 in combination with cytotoxic chemotherapy failed to improve overall survival in metastatic PDAC. While the failure could be partially due to the lack of appropriate patient stratification, it also reiterates that stromal targeting may have to be combined with immunotherapy to reap the full benefits of either approach. On the same lines, focal adhesion kinase (FAK1) activation in tumour cells induces fibrosis and excludes effector T cells infiltration in the TME. Inhibition of FAK alters cytokine production, reduces CAF activation and depletes immunosuppressive cells, such as myeloid-derived suppressor cell (MDSCs) and Tregs, in the TME [102]. Based on these findings, multiple clinical trials combining FAK inhibitor with ICB are currently ongoing in PDAC patients (NCT02758587, NCT02546531, NCT03727880). Additional trials are also underway to evaluate the impact of stromal remodelling with Vitamin D agonist with combination with immunotherapy (NCT03331562, NCT03519308). In the end, multipronged approaches will be needed to correct the multiple immune deficiencies in PDAC to invigorate and sustain the anti-tumour immunity.
5. Outstanding questions
The past decade has seen substantial advances in our understanding of PDAC genetics, including the molecular underpinnings of genomic and transcriptomic heterogeneity that impacts treatment responses and the natural history in patients. With the further development of various technologies, including single cell analysis and high-resolution imaging techniques, we expect to obtain additional compartment-specific delineation of alterations, and how these interplay within the ecosystem of the tumour microenvironment. While considerable progress has been made in preclinical model building including a diverse repertoire of genetically engineered mice, there continues to be an unmet need for developing ex vivo platforms that faithfully model the genetic, molecular and functional interaction between tumour subclones, as well as the interaction between tumour cells and various components of the TME. We also need to deeply characterize the impact of various therapeutic perturbations on the evolution of tumour subclones and the composition of heterogeneous TME components, through longitudinal monitoring and sampling of patients on clinical trials. These studies will greatly help us anticipate the resistance mechanisms that eventually lead to treatment failure, and proactively design combinatory targeting strategies.
6. Selection criteria
Pubmed search of articles: “genomics + pancreatic cancer”, “heterogeneity + pancreatic cancer”, “molecular subtype + pancreatic cancer”, “tumour metabolism + pancreatic cancer”. “immune evasion + pancreatic cancer”, “immunotherapy + pancreatic cancer”. Additional articles were selected based on articles in these searches and as suggested by reviewers.
Author contributions
W.Y., A.M. and H.Y. wrote the manuscript. W.Y. and H.Y. prepared the figures.
Declaration of competing interest
The authors claim no conflict of interest.
Acknowledgments/Funding
This work was supported by the NCI grant R01CA214793 to H.Y.; the Pancreatic Cancer Action Network (PanCAN)-AACR Pathway to Leadership Award and PanCAN-Translational grant to W.Y.; and R01 CA220236 and R01CA218004 to A.M.
Footnotes
This is the first in a Series of four papers about pancreatic cancer (For the other papers in this Series see www.thelancet.com/series/pancreatic-cancer).
References
- 1.Rahib L., Smith B.D., Aizenberg R., Rosenzweig A.B., Fleshman J.M., Matrisian L.M. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Ying H., Dey P., Yao W. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2016;30(4):355–385. doi: 10.1101/gad.275776.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feigin M.E., Garvin T., Bailey P. Recurrent noncoding regulatory mutations in pancreatic ductal adenocarcinoma. Nat Genet. 2017;49(6):825–833. doi: 10.1038/ng.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canon J., Rex K., Saiki A.Y. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 2019;575(7781):217–223. doi: 10.1038/s41586-019-1694-1. [DOI] [PubMed] [Google Scholar]
- 6.Humphris J.L., Patch A.M., Nones K. Hypermutation in pancreatic cancer. Gastroenterology. 2017;152(1) doi: 10.1053/j.gastro.2016.09.060. 68-74 e2. [DOI] [PubMed] [Google Scholar]
- 7.Le D.T., Durham J.N., Smith K.N. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts N.J., Norris A.L., Petersen G.M. Whole genome sequencing defines the genetic heterogeneity of familial pancreatic cancer. Cancer Discov. 2016;6(2):166–175. doi: 10.1158/2159-8290.CD-15-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golan T., Hammel P., Reni M. Maintenance olaparib for germline BRCA-Mutated metastatic pancreatic cancer. N Engl J Med. 2019;381(4):317–327. doi: 10.1056/NEJMoa1903387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makohon-Moore A.P., Matsukuma K., Zhang M. Precancerous neoplastic cells can move through the pancreatic ductal system. Nature. 2018;561(7722):201–205. doi: 10.1038/s41586-018-0481-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Notta F., Chan-Seng-Yue M., Lemire M. A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature. 2016;538(7625):378–382. doi: 10.1038/nature19823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reiter J.G., Baretti M., Gerold J.M. An analysis of genetic heterogeneity in untreated cancers. Nat Rev Cancer. 2019 doi: 10.1038/s41568-019-0185-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reiter J.G., Makohon-Moore A.P., Gerold J.M. Minimal functional driver gene heterogeneity among untreated metastases. Science. 2018;361(6406):1033–1037. doi: 10.1126/science.aat7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makohon-Moore A.P., Zhang M., Reiter J.G. Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat Genet. 2017;49(3):358–366. doi: 10.1038/ng.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collisson E.A., Bailey P., Chang D.K., Biankin A.V. Molecular subtypes of pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2019;16(4):207–220. doi: 10.1038/s41575-019-0109-y. [DOI] [PubMed] [Google Scholar]
- 16.TCGA Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 2017;32(2) doi: 10.1016/j.ccell.2017.07.007. 185-203 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noll E.M., Eisen C., Stenzinger A. CYP3A5 mediates basal and acquired therapy resistance in different subtypes of pancreatic ductal adenocarcinoma. Nat Med. 2016;22(3):278–287. doi: 10.1038/nm.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elyada E., Bolisetty M., Laise P. Cross-Species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov. 2019 doi: 10.1158/2159-8290.CD-19-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng J., Sun B.F., Chen C.Y. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019 doi: 10.1038/s41422-019-0195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aung K.L., Fischer S.E., Denroche R.E. Genomics-Driven precision medicine for advanced pancreatic cancer: early results from the compass trial. Clin Cancer Res. 2018;24(6):1344–1354. doi: 10.1158/1078-0432.CCR-17-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinelli P., Carrillo-de Santa Pau E., Cox T. GATA6 regulates EMT and tumour dissemination, and is a marker of response to adjuvant chemotherapy in pancreatic cancer. Gut. 2017;66(9):1665–1676. doi: 10.1136/gutjnl-2015-311256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adams C.R., Htwe H.H., Marsh T. Transcriptional control of subtype switching ensures adaptation and growth of pancreatic cancer. Elife. 2019;8 doi: 10.7554/eLife.45313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andricovich J., Perkail S., Kai Y., Casasanta N., Peng W., Tzatsos A. Loss of KDM6A activates super-enhancers to induce gender-specific squamous-like pancreatic cancer and confers sensitivity to BET inhibitors. Cancer Cell. 2018;33(3) doi: 10.1016/j.ccell.2018.02.003. 512-26 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Somerville T.D.D., Xu Y., Miyabayashi K. TP63-Mediated enhancer reprogramming drives the squamous subtype of pancreatic ductal adenocarcinoma. Cell Rep. 2018;25(7) doi: 10.1016/j.celrep.2018.10.051. 1741-55 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tu B., Yao J., Ferri-Borgogno S. YAP1 oncogene is a context-specific driver for pancreatic ductal adenocarcinoma. JCI Insight. 2019 doi: 10.1172/jci.insight.130811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu M., Ingram L., Tolosa E.J. Gli transcription factors mediate the oncogenic transformation of prostate basal cells induced by a Kras-Androgen receptor axis. J Biol Chem. 2016;291(49):25749–25760. doi: 10.1074/jbc.M116.753129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernandez L.A., Northcott P.A., Dalton J. YAP1 is amplified and up-regulated in hedgehog-associated medulloblastomas and mediates Sonic hedgehog-driven neural precursor proliferation. Genes Dev. 2009;23(23):2729–2741. doi: 10.1101/gad.1824509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao R., Fallon T.R., Saladi S.V. Yap tunes airway epithelial size and architecture by regulating the identity, maintenance, and self-renewal of stem cells. Dev Cell. 2014;30(2):151–165. doi: 10.1016/j.devcel.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ligorio M., Sil S., Malagon-Lopez J. Stromal microenvironment shapes the intratumoral architecture of pancreatic cancer. Cell. 2019;178(1) doi: 10.1016/j.cell.2019.05.012. 160-75 e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Candido J.B., Morton J.P., Bailey P. CSF1R(+) macrophages sustain pancreatic tumor growth through T cell suppression and maintenance of key gene programs that define the squamous subtype. Cell Rep. 2018;23(5):1448–1460. doi: 10.1016/j.celrep.2018.03.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steele C.W., Karim S.A., Leach J.D.G. CXCR2 inhibition profoundly suppresses metastases and augments immunotherapy in pancreatic ductal adenocarcinoma. Cancer Cell. 2016;29(6):832–845. doi: 10.1016/j.ccell.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moffitt R.A., Marayati R., Flate E.L. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47(10):1168–1178. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Biffi G., Oni T.E., Spielman B. IL1-Induced JAK/STAT signaling is antagonized by TGFbeta to shape CAF heterogeneity in pancreatic ductal adenocarcinoma. Cancer Discov. 2019;9(2):282–301. doi: 10.1158/2159-8290.CD-18-0710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi Y., Gao W., Lytle N.K. Targeting LIF-mediated paracrine interaction for pancreatic cancer therapy and monitoring. Nature. 2019;569(7754):131–135. doi: 10.1038/s41586-019-1130-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Yan W., Collins M.A. Interleukin-6 is required for pancreatic cancer progression by promoting MAPK signaling activation and oxidative stress resistance. Cancer Res. 2013;73(20):6359–6374. doi: 10.1158/0008-5472.CAN-13-1558-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozdemir B.C., Pentcheva-Hoang T., Carstens J.L. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25(6):719–734. doi: 10.1016/j.ccr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rhim A.D., Oberstein P.E., Thomas D.H. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 2014;25(6):735–747. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherman M.H., Yu R.T., Engle D.D. Vitamin D receptor-mediated stromal reprogramming suppresses pancreatitis and enhances pancreatic cancer therapy. Cell. 2014;159(1):80–93. doi: 10.1016/j.cell.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu H., Juvekar A., Lyssiotis C.A. Phosphoinositide 3-Kinase regulates glycolysis through mobilization of aldolase from the actin cytoskeleton. Cell. 2016;164(3):433–446. doi: 10.1016/j.cell.2015.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hui S., Ghergurovich J.M., Morscher R.J. Glucose feeds the TCA cycle via circulating lactate. Nature. 2017;551(7678):115–118. doi: 10.1038/nature24057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guillaumond F., Leca J., Olivares O. Strengthened glycolysis under hypoxia supports tumor symbiosis and hexosamine biosynthesis in pancreatic adenocarcinoma. Proc Natl Acad Sci U S A. 2013;110(10):3919–3924. doi: 10.1073/pnas.1219555110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Son J., Lyssiotis C.A., Ying H. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496(7443):101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bott A.J., Shen J., Tonelli C. Glutamine anabolism plays a critical role in pancreatic cancer by coupling carbon and nitrogen metabolism. Cell Rep. 2019;29(5) doi: 10.1016/j.celrep.2019.09.056. 1287-98 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kimmelman A.C., Autophagy White E., Metabolism Tumor. Cell Metab. 2017;25(5):1037–1043. doi: 10.1016/j.cmet.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Humpton T.J., Alagesan B., DeNicola G.M. Oncogenic KRAS induces NIX-Mediated Mitophagy to promote pancreatic cancer. Cancer Discov. 2019 doi: 10.1158/2159-8290.CD-18-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang A., Rajeshkumar N.V., Wang X. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov. 2014;4(8):905–913. doi: 10.1158/2159-8290.CD-14-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang A., Herter-Sprie G., Zhang H. Autophagy sustains pancreatic cancer growth through both cell-autonomous and nonautonomous mechanisms. Cancer Discov. 2018;8(3):276–287. doi: 10.1158/2159-8290.CD-17-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perera R.M., Stoykova S., Nicolay B.N. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature. 2015;524(7565):361–365. doi: 10.1038/nature14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bryant K.L., Stalnecker C.A., Zeitouni D. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat Med. 2019;25(4):628–640. doi: 10.1038/s41591-019-0368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kinsey C.G., Camolotto S.A., Boespflug A.M. Protective autophagy elicited by RAF–>MEK–>ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat Med. 2019;25(4):620–627. doi: 10.1038/s41591-019-0367-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Commisso C., Davidson S.M., Soydaner-Azeloglu R.G. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497(7451):633–637. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olivares O., Mayers J.R., Gouirand V. Collagen-derived proline promotes pancreatic ductal adenocarcinoma cell survival under nutrient limited conditions. Nat Commun. 2017;8:16031. doi: 10.1038/ncomms16031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yao W., Rose J.L., Wang W. Syndecan 1 is a critical mediator of macropinocytosis in pancreatic cancer. Nature. 2019;568(7752):410–414. doi: 10.1038/s41586-019-1062-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee S.W., Zhang Y., Jung M., Cruz N., Alas B., Commisso C. EGFR-Pak signaling selectively regulates glutamine deprivation-induced macropinocytosis. Dev Cell. 2019;50(3) doi: 10.1016/j.devcel.2019.05.043. 381-92 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kerr E.M., Gaude E., Turrell F.K., Frezza C., Martins C.P. Mutant KRAS copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature. 2016 doi: 10.1038/nature16967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baek G., Tse Y.F., Hu Z. MCT4 defines a glycolytic subtype of pancreatic cancer with poor prognosis and unique metabolic dependencies. Cell Rep. 2014;9(6):2233–2249. doi: 10.1016/j.celrep.2014.11.025. [DOI] [PubMed] [Google Scholar]
- 57.Daemen A., Peterson D., Sahu N. Metabolite profiling stratifies pancreatic ductal adenocarcinomas into subtypes with distinct sensitivities to metabolic inhibitors. Proc Natl Acad Sci U S A. 2015;112(32):E4410–E4417. doi: 10.1073/pnas.1501605112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sousa C.M., Biancur D.E., Wang X. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature. 2016;536(7617):479–483. doi: 10.1038/nature19084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao H., Yang L., Baddour J. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife. 2016;5:e10250. doi: 10.7554/eLife.10250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Auciello F.R., Bulusu V., Oon C. A stromal Lysolipid-Autotaxin signaling axis promotes pancreatic tumor progression. Cancer Discov. 2019;9(5):617–627. doi: 10.1158/2159-8290.CD-18-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McDonald O.G., Li X., Saunders T. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat Genet. 2017;49(3):367–376. doi: 10.1038/ng.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shukla S.K., Purohit V., Mehla K. MUC1 and HIF-1alpha signaling crosstalk induces anabolic glucose metabolism to impart gemcitabine resistance to pancreatic cancer. Cancer Cell. 2017;32(1) doi: 10.1016/j.ccell.2017.06.004. 71-87 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guillaumond F., Bidaut G., Ouaissi M. Cholesterol uptake disruption, in association with chemotherapy, is a promising combined metabolic therapy for pancreatic adenocarcinoma. Proc Natl Acad Sci U S A. 2015;112(8):2473–2478. doi: 10.1073/pnas.1421601112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tadros S., Shukla S.K., King R.J. De Novo lipid synthesis facilitates gemcitabine resistance through endoplasmic reticulum stress in pancreatic cancer. Cancer Res. 2017;77(20):5503–5517. doi: 10.1158/0008-5472.CAN-16-3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sancho P., Burgos-Ramos E., Tavera A. MYC/PGC-1alpha balance determines the metabolic phenotype and plasticity of pancreatic cancer stem cells. Cell Metab. 2015;22(4):590–605. doi: 10.1016/j.cmet.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 66.Viale A., Pettazzoni P., Lyssiotis C.A. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514(7524):628–632. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Halbrook C.J., Pontious C., Kovalenko I. Macrophage-Released pyrimidines inhibit gemcitabine therapy in pancreatic cancer. Cell Metab. 2019;29(6) doi: 10.1016/j.cmet.2019.02.001. 1390-9 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Balli D., Rech A.J., Stanger B.Z., Vonderheide R.H. Immune cytolytic activity stratifies molecular subsets of human pancreatic cancer. Clin Cancer Res. 2017;23(12):3129–3138. doi: 10.1158/1078-0432.CCR-16-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Balachandran V.P., Luksza M., Zhao J.N. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 2017;551(7681):512–516. doi: 10.1038/nature24462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carstens J.L., Correa de Sampaio P., Yang D. Spatial computation of intratumoral T cells correlates with survival of patients with pancreatic cancer. Nat Commun. 2017;8:15095. doi: 10.1038/ncomms15095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Anderson K.G., Stromnes I.M., Greenberg P.D. Obstacles posed by the Tumor microenvironment to T cell activity: a case for synergistic therapies. Cancer Cell. 2017;31(3):311–325. doi: 10.1016/j.ccell.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Feig C., Jones J.O., Kraman M. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc Natl Acad Sci U S A. 2013;110(50):20212–20217. doi: 10.1073/pnas.1320318110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dougan S.K. The pancreatic cancer microenvironment. Cancer J. 2017;23(6):321–325. doi: 10.1097/PPO.0000000000000288. [DOI] [PubMed] [Google Scholar]
- 74.Li J., Byrne K.T., Yan F. Tumor cell-intrinsic factors underlie heterogeneity of immune cell infiltration and response to immunotherapy. Immunity. 2018;49(1) doi: 10.1016/j.immuni.2018.06.006. 178-93 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mace T.A., Ameen Z., Collins A. Pancreatic cancer-associated stellate cells promote differentiation of myeloid-derived suppressor cells in a STAT3-dependent manner. Cancer Res. 2013;73(10):3007–3018. doi: 10.1158/0008-5472.CAN-12-4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aykut B., Pushalkar S., Chen R. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature. 2019 doi: 10.1038/s41586-019-1608-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pushalkar S., Hundeyin M., Daley D. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 2018;8(4):403–416. doi: 10.1158/2159-8290.CD-17-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Riquelme E., Zhang Y., Zhang L. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. 2019;178(4) doi: 10.1016/j.cell.2019.07.008. 795-806 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liou G.Y., Doppler H., Necela B. Macrophage-secreted cytokines drive pancreatic acinar-to-ductal metaplasia through NF-kappaB and MMPs. J Cell Biol. 2013;202(3):563–577. doi: 10.1083/jcb.201301001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Y., Yan W., Mathew E. Epithelial-Myeloid cell crosstalk regulates acinar cell plasticity and pancreatic remodeling in mice. Elife. 2017;6 doi: 10.7554/eLife.27388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Colegio O.R., Chu N.Q., Szabo A.L. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 2014;513(7519):559–563. doi: 10.1038/nature13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fischer K., Hoffmann P., Voelkl S. Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood. 2007;109(9):3812–3819. doi: 10.1182/blood-2006-07-035972. [DOI] [PubMed] [Google Scholar]
- 83.Husain Z., Huang Y., Seth P., Sukhatme V.P. Tumor-derived lactate modifies antitumor immune response: effect on myeloid-derived suppressor cells and nk cells. J Immunol. 2013;191(3):1486–1495. doi: 10.4049/jimmunol.1202702. [DOI] [PubMed] [Google Scholar]
- 84.Geiger R., Rieckmann J.C., Wolf T. L-Arginine modulates T cell metabolism and enhances survival and anti-tumor activity. Cell. 2016;167(3) doi: 10.1016/j.cell.2016.09.031. 829-42 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Witkiewicz A.K., Costantino C.L., Metz R. Genotyping and expression analysis of IDO2 in human pancreatic cancer: a novel, active target. J Am Coll Surg. 2009;208(5):781–787. doi: 10.1016/j.jamcollsurg.2008.12.018. discussion 7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li H., Bullock K., Gurjao C. Metabolomic adaptations and correlates of survival to immune checkpoint blockade. Nat Commun. 2019;10(1):4346. doi: 10.1038/s41467-019-12361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Le D.T., Ko A.H., Wainberg Z.A. Results from a phase 2b, randomized, multicenter study of GVAX pancreas and CRS-207 compared to chemotherapy in adults with previously-treated metastatic pancreatic adenocarcinoma (ECLIPSE study) Journal of Clinical Oncology. 2017;35(4_suppl):345. doi: 10.1158/1078-0432.CCR-18-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Le D.T., Lutz E., Uram J.N. Evaluation of ipilimumab in combination with allogeneic pancreatic tumor cells transfected with a GM-CSF gene in previously treated pancreatic cancer. J Immunother. 2013;36(7):382–389. doi: 10.1097/CJI.0b013e31829fb7a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang Q.J., Yu Z., Griffith K., Hanada K., Restifo N.P., Yang J.C. Identification of T-cell receptors targeting KRAS-Mutated human tumors. Cancer Immunol Res. 2016;4(3):204–214. doi: 10.1158/2326-6066.CIR-15-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tran E., Ahmadzadeh M., Lu Y.C. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science. 2015;350(6266):1387–1390. doi: 10.1126/science.aad1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gros A., Tran E., Parkhurst M.R. Recognition of human gastrointestinal cancer neoantigens by circulating PD-1+ lymphocytes. J Clin Invest. 2019;129(11):4992–5004. doi: 10.1172/JCI127967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Beatty G.L., Chiorean E.G., Fishman M.P. CD40 agonists alter tumor stroma and show efficacy against pancreatic carcinoma in mice and humans. Science. 2011;331(6024):1612–1616. doi: 10.1126/science.1198443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Byrne K.T., Vonderheide R.H. CD40 stimulation obviates innate sensors and drives T cell immunity in cancer. Cell Rep. 2016;15(12):2719–2732. doi: 10.1016/j.celrep.2016.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.O’Hara M.H., O’Reilly E.M., Rosemarie M. Abstract CT004: a phase Ib study of CD40 agonistic monoclonal antibody APX005M together with gemcitabine (Gem) and nab-paclitaxel (NP) with or without nivolumab (Nivo) in untreated metastatic ductal pancreatic adenocarcinoma (PDAC) patients. Cancer Res. 2019;79(13 Supplement) CT004-CT. [Google Scholar]
- 95.Daley D., Mani V.R., Mohan N. Dectin 1 activation on macrophages by galectin 9 promotes pancreatic carcinoma and peritumoral immune tolerance. Nat Med. 2017;23(5):556–567. doi: 10.1038/nm.4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sanford D.E., Belt B.A., Panni R.Z. Inflammatory monocyte mobilization decreases patient survival in pancreatic cancer: a role for targeting the CCL2/CCR2 axis. Clin Cancer Res. 2013;19(13):3404–3415. doi: 10.1158/1078-0432.CCR-13-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhu Y., Knolhoff B.L., Meyer M.A. CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer Res. 2014;74(18):5057–5069. doi: 10.1158/0008-5472.CAN-13-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kaneda M.M., Cappello P., Nguyen A.V. Macrophage PI3Kgamma drives pancreatic ductal adenocarcinoma progression. Cancer Discov. 2016;6(8):870–885. doi: 10.1158/2159-8290.CD-15-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Klug F., Prakash H., Huber P.E. Low-dose irradiation programs macrophage differentiation to an iNOS(+)/M1 phenotype that orchestrates effective T cell immunotherapy. Cancer Cell. 2013;24(5):589–602. doi: 10.1016/j.ccr.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 100.Blair A.B., Kim V.M., Muth S.T. Dissecting the stromal signaling and regulation of myeloid cells and memory effector T cells in pancreatic cancer. Clin Cancer Res. 2019;25(17):5351–5363. doi: 10.1158/1078-0432.CCR-18-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Provenzano P.P., Cuevas C., Chang A.E., Goel V.K., Von Hoff D.D., Hingorani S.R. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21(3):418–429. doi: 10.1016/j.ccr.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jiang H., Hegde S., Knolhoff B.L. Targeting focal adhesion kinase renders pancreatic cancers responsive to checkpoint immunotherapy. Nat Med. 2016;22(8):851–860. doi: 10.1038/nm.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]