Abstract
The neuro-oncological ventral antigen 2 (NOVA2) protein is a major factor regulating neuron-specific alternative splicing (AS), previously associated with an acquired neurologic condition, the paraneoplastic opsoclonus-myoclonus ataxia (POMA). We report here six individuals with de novo frameshift variants in NOVA2 affected with a severe neurodevelopmental disorder characterized by intellectual disability (ID), motor and speech delay, autistic features, hypotonia, feeding difficulties, spasticity or ataxic gait, and abnormal brain MRI. The six variants lead to the same reading frame, adding a common proline rich C-terminal part instead of the last KH RNA binding domain. We detected 41 genes differentially spliced after NOVA2 downregulation in human neural cells. The NOVA2 variant protein shows decreased ability to bind target RNA sequences and to regulate target AS events. It also fails to complement the effect on neurite outgrowth induced by NOVA2 downregulation in vitro and to rescue alterations of retinotectal axonal pathfinding induced by loss of NOVA2 ortholog in zebrafish. Our results suggest a partial loss-of-function mechanism rather than a full heterozygous loss-of-function, although a specific contribution of the novel C-terminal extension cannot be excluded.
Keywords: NOVA2, alternative splicing, intellectual disability, de novo mutations, autism, C-terminal part, KH domains
Introduction
Many genes that play important roles in the development of the nervous system undergo alternative splicing (AS) to generate protein variants with different functions. The use of alternative mRNA isoforms is a critical process crucial during neuronal development, since specific proteins are required at different times and places. Such mRNA diversity is coordinated by different RNA-binding proteins (RBPs). NOVA (neuro-oncological ventral antigen) proteins NOVA1 and NOVA2 are two RBPs involved in neuronal-specific alternative splicing.1, 2, 3 They have been first described as antigens in individuals affected by a paraneoplastic neurologic syndrome (POMA), an acquired autoimmune neurological disorder characterized by ataxia with or without opsoclonus-myoclonus, with or without dementia, encephalopathy, and cortical deficits along other symptoms.4,5 The two proteins share three similar KH-domains through which they bind directly to YCAY motifs (where Y stands for a pyrimidine) in the messenger RNA (mRNA) sequence.6, 7, 8 According to the binding location on mRNA, they can induce either exon skipping or exon retention.9
Both NOVA1 and NOVA2 are mainly expressed in the central nervous system. However, they show differential expression in specific brain regions in mouse.4 For instance, Nova2 is primarily expressed in cortex and hippocampus, whereas Nova1 is mainly present in midbrain and spinal cord.4,10 Both the Nova1- and the Nova2-null mice manifest growth retardation, progressive motor dysfunction, and death shortly after birth while corpus callosum agenesis is present only in Nova2−/− mice.2,10 This peculiar defect suggested that NOVA1 and NOVA2 are controlling different set of RNA transcripts. As a matter of fact, NOVA2 seems to be mainly associated with a splicing regulation of genes involved in axonal guidance and projection during the development of a mouse cortex (E18.5), as well as genes implicated in cerebellar function or synapse formation.10,11 These AS events are developmentally regulated between E12.5 and E18.5 in mouse cortex, highlighting an important role of NOVA2 as an axonal pathfinder modifier during cortical development.
Intellectual disability (ID) is a group of neurodevelopmental disorders (NDD) characterized by significant limitations in both intellectual functioning and adaptive behavior reflected by an intellectual quotient below 70 before the age of 18. It is genetic in origin in a majority of the case subjects and these genetic anomalies include chromosomal abnormalities, copy number variants, and point mutations or small insertions/deletions (indels) affecting one of the thousands of genes identified so far as causing ID/NDD when mutated.12 Large-scale sequencing studies combined with international data exchange allow identification of many novel genes. We report here six individuals with frameshift variants in NOVA2 that cluster between sequences encoding the second and the third KH domains. We showed that NOVA2 regulates a series of AS events linked to neurite outgrowth and axonal projections in human neural stem cells (hNSCs), and that variant proteins lose, at least partially, the ability to regulate these AS events. We demonstrated that inactivation of NOVA2 affects neurite formation in neuronal cells in vitro and that inactivation of NOVA2 ortholog impaired axon outgrowth in vivo using zebrafish model. Variant proteins, in contrast to wild-type NOVA2, failed to rescue these phenotypes.
Subjects and Methods
Affected Individuals and Genetic Analyses
Individual 1 and biological parents were followed at the Service for Medical Genetics of Nantes University Hospital and were enrolled for genetic testing in the laboratory of genetic diagnosis of Strasbourg University Hospital. Targeted sequencing of hundred candidate genes for ID did not reveal pathogenic variants.13 A family trio-based WES was performed as previously described14 using 100 bp paired-end sequencing. The NOVA2 de novo pathogenic variant c.782del was confirmed by Sanger sequencing and trio compatibility was checked by using polymorphic microsatellite markers (PowerPlex 16HS system, Promega). Individuals 2 and 6 and biological parents were enrolled for genetic diagnosis in Dijon University Hospital and in Assistance publique – Hôpitaux de Paris (APHP), respectively. Proband-only WES was performed as previously described using 75 bp paired-end sequencing.15 Individuals 3, 4, 5, and their biological parents were enrolled, respectively, at the following centers: Cook Children’s Genetics in Fort Worth; Department of Genetics, University of Illinois College of Medicine in Chicago; and Department of Neurology, Baylor College of Medicine in Houston, Texas, USA. Trio whole-exome sequencing was obtained in a CLIA-approved clinical genetic diagnostic laboratory, GeneDx (https://www.genedx.com) as described previously.16 Sequencing was done on an Illumina system with 100 bp paired-end reads. Reads were aligned to human genome build GRCh37/UCSC hg19 and analyzed for sequence variants using a custom-developed analysis tool. The different genetic studies were approved by the local Ethics Committee and written informed consent for genetic testing and authorization for publication were obtained from their legal representative.
NOVA2 Plasmids and siRNA
Human cells were transfected with a pcDNA3.1 plasmid containing the sequence of human NOVA2 cDNA. The plasmid was optimized to reduce GC content while keeping the amino acid sequence and GFP location at the N-terminal side of NOVA2. Mut1 variant (c.782del [p.Val261Glyfs∗135], ClinVar: VCV000812085.1), identified in individual 1, was introduced by site-directed mutagenesis. We also introduced a substitution leading to a premature truncated protein: c.693C>A (p.Tyr231∗). For experiments in zebrafish, the human wild-type and variant (Mut1) NOVA2 cDNA were introduced into pSC2+ plasmids. The sequences of all constructs were confirmed by Sanger sequencing (GATC Biotech). Pools of four siRNA against human (and mice) NOVA2 gene and of Scramble siRNA were purchased from Dharmacon (GE Healthcare).
Cell Culture and Transfection
Human neuronal stem cells (hNSCs) were derived from human embryonic stem cell line (SA001) and from reprogrammed fibroblasts (GM01869). They were obtained from I-Stem (Cellartis, work supervised by the French Bioethics Agency, and Coriell Institute for Medical Research) as described previously.17 hNSCs were seeded on poly-ornithine- and laminin-coated dishes and maintained in N2B27 medium (DMEM/F12 and Neurobasal medium [1:1] supplemented with N2, B27, 2-mercaptoethanol [all from Invitrogen]), BDNF (20 ng/mL), FGF-2 (10 ng/mL) (both from PeproTech), and EGF (R&D Systems; 10 ng/mL). Culture and quality controls of hNSCs were performed as described in Quartier et al.18 hNSCs were transfected using INTERFER in reverse transfection protocol (Polyplus-transfection) with scramble siRNA, NOVA2 siRNA (pool of siRNA, at 120 nM final concentration), or transfecting agent only. HeLa cells were maintained in DMEM supplemented by 1 g/L glucose with gentamycin and 5% Fetal Calf Serum in a 37°C, 5% CO2 humidified incubator with medium renewed every 2 days. Cells were transfected at 60%–70% of confluence in 6-well plates using Lipofectamine 2000 DNA transfection reagent (Invitrogen) in Opti-MEM according to manufacturer’s instructions with 2 μg of each NOVA2 plasmid. Cells were stopped 24/48 h after transfection for RNA extraction.
Western Blot and Immunofluorescence
Four series of HeLa cells were transfected with empty pcDNA3.1 or pcDNA3.1 containing wild-type or variant (Mut1 and p.Tyr231∗) NOVA2 sequences. To control transfection efficiency, cells were co-transfected using a previously published pcDNA3.1-FLAG-THOC6 constructs.14 HeLa cells were lysed in RIPA buffer with protease inhibitor cocktail (Roche) 24 h after transfection. Proteins were separated after denaturation on a 10% acrylamide gel and transferred onto a PVDF membrane. EGFP-tagged NOVA2 proteins were visualized using an in-house mouse anti-GFP antibody (1:10,000). Their protein levels were normalized using the FLAG staining (FLAG antibody: 1:1,000; F1804, Sigma-Aldrich).
RNA Sequencing
Experiments were performed in duplicate from two independent series of hNSCs SA001 treated with the transfecting agent only (INTERFERin) or transfected with scramble siRNA or anti-NOVA2 siRNA for 48 h. Total RNA was extracted using the RNeasy mini kit (QIAGEN) including a DNase treatment. RNA levels and quality were quantified using a Nanodrop spectrophotometer and a 2100 Bioanalyzer (Agilent). cDNA libraries of template molecules suitable for high-throughput sequencing were created using the KAPA mRNA HyperPrep Kit (Roche). Briefly, mRNAs were purified from 300 ng of total RNA using poly-T oligo-attached magnetic beads and fragmented at 85°C with magnesium for 6 min. The cleaved mRNA fragments were reverse transcribed into cDNA using random primers and KAPA Script enzyme. Second strands were synthesized and adapters were added according to manufacturer’s instructions. Libraries were enriched by PCR amplification (12 cycles). PCR products were purified and size selected (400 bp) before sequencing using paired-end 100 bp, Illumina Hiseq 4000 sequencer. Sequencing generated on average 100 to 200 million of reads per sample that were mapped onto the hg19 assembly of the human genome using TopHat 2.0.1419 and the Bowtie 2-2.1.0 aligner.20 Data are available in Gene Expression Omnibus (GEO: GSE138766). Gene expression was quantified using HTSeq-0.6.121 and gene annotations from Ensembl release 75. Only uniquely mapped and non-ambiguously assigned reads were retained for further analyses. Read counts were then normalized across libraries with the median-of-ratios method proposed by Anders and Huber.22 Relative Log Expression (RLE) plots were drawn to check that the distributions were centered around the zero line and as tight as possible to make sure that normalization was performed correctly. Comparisons to untreated cells (receiving transfection agent, INTERFERin, only) were performed using the statistical method proposed by Anders and Huber.22 The Wald test was used to estimate the p values and they were adjusted for multiple testing with the Benjamini and Hochberg method.23 LeafCutter was used to identify alternative splicing (AS) events between NOVA2 inactivation and other conditions.24 Genes with AS events were analyzed using the Database for Annotation, Visualization and Integrated Discovery (DAVID 6.7). Biological processes and molecular functions of Gene Ontology Consortium (GO) were used for the functional annotations.
Confirmation of Alternative Splicing (AS) Events Identified by RNA-Seq
GM01869 hNSCs were used to validate results obtained by RNA-seq in the SA001 line. Cell were treated with transfecting agent, scramble, or NOVA2 siRNA as described for SA001 (each condition in triplicates). For splicing analyses in HeLa cells, 6-well plates of cells were transfected with 2 μg of empty or NOVA2 pcDNA3.1 plasmids alone or combined (three to four series of independent experiments were analyzed). mRNA was extracted 24 h after siRNA transfection using the RNeasy mini kit (QIAGEN). 500 ng of total RNA was reverse transcribed into cDNA using random hexamers and SuperScript IV reverse transcriptase according to manufacturer’s recommendation. PCR was performed (16 cycles of touch-down 70°C–54°C followed by 8 to 21 cycles at 59°C depending of the mRNA), using the following primers NEO1_E25_F 5′-GGAAGGCGAGGAATGAGACCAAAA-3′ and NEO1_E27_R 5′-TGTGCTTGGCAATGCAGGATCA-3′, AKAP13_E11_F 5′-AAGTGCCTGCAAACTGCTCTGT-3′ and AKAP13_E13_R 5′-AAAGAGTCAACCCGTTCCTCACCA-3′, SORBS1_E2_F 5′-TGTGATGAATGGCTTGGCAC-3′ and SORBS1_E4_R 5′-TC™CCTTCCCAGTGCAGATTT-3′, SGCE_E8_F 5′-TGGTGGAGAATACAAACCCC-3′ and SGCE_E10_R 5′-GGACATGTCTCGAAGCTCCT-3′. The PCR products obtained were analyzed by migration on a 2,100 Bioanalyzer instrument (Agilent Technology).
Electrophoretic Mobility Shift Assay
In order to produce recombinant NOVA2 proteins, E. coli BL21 (RIL) pRARE competent cells (Invitrogen) were transformed with wild-type (WT) or variant (Mut1) pet28a-GST-NOVA2. The cells were then incubated at 30°C in 400 mL of LB medium supplemented with Kanamycin until an OD600 of 0.5. Afterward, 0.5 mM IPTG was added and the culture was incubated for additional 4 h at 30°C. Harvested cells were sonicated in 50 mM Tris-Cl (pH 7.5), 300 mM NaCl, 5% glycerol, 1 mM DTT, 5 mM EDTA, centrifuged 20 min at 20,000 × g and recombinant GST-tagged proteins were purified using the GST-Bind™ Kit (Novagen). To synthetize the NOVA2 RNA target, pcDNA3 vector containing the sequence CTAGCGTCATTTCATCTCACCA cloned between the Nhe1 and HinD3 restriction sites was linearized by EcoR1 restriction and 100 ng were transcribed using T7 transcription kit (Ambion) in the presence of 1 μL of [aP32]-UTP (Perkin Elmer). It was analyzed on 8% denaturing polyacrylamide and quantified with LS-6500 counter (Beckman). After transcription, 1 unit of DNase I (Invitrogen) was added, and the sample was incubated for additional 30 min at 37°C. Transcribed RNAs were then purified by micro Bio-Spin 6 chromatography columns (Bio-rad) according to the manufacturer’s instructions. The sizes of RNAs were checked by gel electrophoresis on a denaturing 6% polyacrylamide gel. 10 pM (3000 CPM) of labeled RNA was incubated at 90°C for 5 min in binding buffer (BB, 0.75 mM MgCl2, 50 mM Tris-HCl [pH 7.0], 75 mM NaCl, 37.5 mM KCl, 5.25 mM DTT, 0.1 mg/mL BSA, 0.1 mg/mL Bulk tRNA) and allowed to cool to room temperature. After cooling, RNAsin was added to a final concentration of 0.4 U/μL. Increasing amounts of GST-NOVA2 were then added and the mixture was incubated on ice for 20 min. The solution mixture was loaded onto a non-denaturing 6.0% (w/v) polyacrylamide gel (acrylamide/bisacrylamide, 40:1, w/w) containing 0.5× TBE (1× TBE is 90 mM Tris-base, 89 mM boric acid, and 2 mM EDTA [pH 8.0]), which had been pre-electrophoresed at 110 V for 20 min at 4°C. The gel electrophoresis was run at 110 V at 4°C for 3 h. Gel was then dried, exposed to a phosphorimager screen, and imaged using a Typhoon 9410. The data were fit to the following equation: y = min + ((max − min)/(1 + (x/IC50) − HillSlope)) where y is the percentage of RNA bound, x is the concentration of protein, min and max are the minimum and maximum percentage of RNA bound to NOVA2 (0%–100%) and IC50 is the concentration where 50% of maximum binding is achieved.
Neurite Outgrowth Assay
Mouse Neuroblastoma Neuro2a (N2a) cells provided by the IGBMC cell culture platform (Strasbourg) are mycoplasma free (PCR test Venorgem) and have not been authenticated. N2a cells were cultured in DMEM (GIBCO) supplemented with 1 g/L of glucose, 5% fetal calf serum, and gentamycine. Cells were transfected with different NOVA2 constructs and siRNAs along with the GFP-derived reporter protein Venus. Transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. N2a cells were differentiated 24 h after transfection by replacing the culture media with DMEM without serum and supplemented with 1 μM retinoic acid. After 48 h, cells were fixed in 4% PFA and immunostained by using anti-GFP (Thermo-Fisher, A10262, 1:1,000), anti-tubulin (Abcam, ab6160, 1:1,000) antibodies, and DAPI. Three independent experiments were performed. A total of 80–100 transfected cells were quantified per condition in each experiment.
Zebrafish Experiments
Zebrafish (Danio rerio) were raised and maintained as previously described.25 We injected 6 ng of morpholino targeting nova1 (5′-TCGCCAACTGTCGAGCTTACCTCC-3′) either alone or with 25 pg of mRNA, and 50 pg of mRNA alone into wild-type zebrafish eggs (AB strain) at the 1- to 2-cell stage. At 4 days post fertilization, larvae were scored for defects in the number of the inter tecta axonal tracts. Wild-type (ENST00000263257.6) and variant Mut1 (p.Val261Glyfs∗135) full-length human messages were Sanger sequenced and cloned into the pCS2+ vector and transcribed in vitro using the SP6 Message Machine kit (Ambion). We performed a whole-mount immunostaining with the anti-acetylated tubulin monoclonal antibody in order to examine the integrity of the inter-tecta axonal tracts of the zebrafish larvae. At 4 days post fertilization, the larvae were fixed in Dent’s solution (80% methanol, 20% dimethylsulphoxide [DMSO]) overnight. After rehydration in decreasing concentration of methanol in PBS, larvae were washed with PBS, permeabilized with 10 μg/mL proteinase K, and postfixed with 4% PFA. Larvae were then washed twice with IF buffer (0.1% Tween-20, 1% BSA in 1 × PBS) for 10 min at room temperature. After incubation in blocking solution (10% FBS, 1% BSA in 1 × PBS) for 1 h at room temperature, larvae were incubated with the anti-acetylated tubulin (1:1,000) in blocking solution overnight at 4°C. After two washes in IF buffer for 10 min each, larvae were incubated in the secondary antibody solution, 1:500 Alexa Fluor rabbit anti-mouse IgG (Invitrogen), in blocking solution for 1 h at room temperature and in the dark. After imaging at least 30 larvae per condition from a lateral view on a MacroFluo ORCA Flash (Leica), we scored the number of inter tecta axonal tracts in the brain on each larva for the injected conditions and age-matched controls from the same clutch. All the experiments were repeated three times and a t test was performed to determine significance.
Results
Identification of De Novo Frameshift Variants in NOVA2
We identified a de novo frameshift variant GenBank: NM_002516.3 (c.782del [p.Val261Glyfs∗135]) (Mut1) in NOVA2 (MIM: 601991) in a an individual with severe intellectual disability (ID) (individual 1, S1). We collected five additional unrelated individuals with ID with indel variants in NOVA2, identified by exome sequencing (ES) by different molecular diagnostic laboratories or research centers (Figures 1A and 1B). The compilation of these variants resulted from an international collaborative effort partly facilitated by the web-based tool GeneMatcher.26 All individuals were initially and a posteriori clinically assessed by at least one expert clinical geneticist. Individual 2 (S2) carries a c.710_711dup (p.Leu238Cysfs∗159) found by proband-only WES. Two other deletion/insertion variants causing frameshift in NOVA2, c.701_720dup (p.Ala241Profs∗162) and c.709_748del (p.Val237Profs∗146), were respectively identified in individuals 3 (S3) and 4 (S4), through trio-based WES. Individual 5 (S5) was identified with a deletion of a nucleotide adjacent to the first variant identified: c.781del (p.Val261Trpfs∗135). The variant was absent from mother’s DNA, but father’s DNA was not available for testing. Individual 6 (S6) carries a de novo insertion c.720_721insCCGCGGATGTGCTTCCAGCC, which leads to a truncated protein p.Ala241Profs∗162 with one amino acid difference (Phe245 instead of Cys) with those identified in individual 3. All these variants were unique events never reported in any public variant databases (dbSNP138, 1000 Genomes, NHLBI GO Exome Sequencing Project, ExAC, GnomAD). No truncating variants were reported in the general population (GnomAD) in the canonical transcript GenBank: NM_002516.3, suggesting that NOVA2 is a gene highly intolerant to loss-of-function variants (0 observed versus 12.3 expected, pLI = 0.98). There are no known deletion encompassing NOVA2 in the general population from the Database of Genomic Variants (DGV). However, several large deletions encompassing NOVA2 together with several dozens of other genes are reported in Decipher and ClinVar database in individuals with ID and/or other developmental defects.
Figure 1.
Frameshift Variants in NOVA2 Identified in Individuals with ID
(A) Pictures of individuals 1 (S1), 2 (S2), and 6 (S6).
(B) Schematic representation of NOVA2 protein showing consequences of the frameshift variants identified in the six affected individuals reported here (subjects S1 to S6). p.Tyr231∗ indicates a non-existing truncating variant we made for the need of the study. The four coding exons (E1-4) are represented below the protein.
Individuals Carrying De Novo Frameshift Variants in NOVA2 Present with ID, Autistic Features, and Other Angelman Syndrome-like Clinical Manifestations
The main clinical features of the six individuals are summarized in Table 1. More detailed clinical information for all subjects is provided in the Supplemental Note. All subjects from the case series exhibited developmental delay (DD) including ID, motor and speech delay, autistic features with stereotypic hands movements, and frequent laughter. Other frequent findings included hypotonia (3/4), feeding difficulties (5/6), and spasticity or ataxic gait (4/6). Brain imaging showed Chiari malformation type 1 (n = 1), cortical atrophy (n = 1), and corpus callosum thinning (n = 2). Two individuals presented with seizures. Notable similarities are found between the reported individuals carrying variants in NOVA2 and individuals with Angelman syndrome, including feeding difficulties, hypotonia in childhood, inappropriate bouts of laughter, attraction to water, stereotypic movements, and severe delay or absence of speech. Interestingly, Angelman syndrome (MIM: 105830) diagnosis was clinically evoked for at least four of the individuals, as evident by the prior targeted investigations of the UBE3A region by methylation and Sanger sequencing.
Table 1.
Clinical Features of the Subjects with Frameshift Variants in NOVA2
| Individual | Ind 1 | Ind 2 | Ind 3 | Ind 4 | Ind 5 | Ind 6 |
|---|---|---|---|---|---|---|
| Variant in NOVA2 (NM_002516.3) | c.782del (p.Val261Glyfs∗135) | c.710_711dup (p.Leu238Cysfs∗159) | c.701_720dup (p.Ala241Profs∗162)a | c.709_748del (p.Val237Profs∗146) | c.781del (p.Val261Trpfs∗135) | c.720_721insCCGCGGATGTGC TTCCAGCC (p.Ala241Profs∗162)a |
| Occurence | de novo | de novo | de novo | de novo | not present in the mother | de novo |
| Sex | male | male | female | female | male | female |
| Age at assessment | 5 y 6 m | 2 y 1 m | 2 y 2 m | 2 y 7 m | 21 y | 5 y 5 m |
| Clinical Examination | ||||||
| Birth weight (grams) | 3,515 | 3,640 | 2,327 | 3,459 | N/A | 3,750 |
| Birth length (cm) | 50 | 51 | N/A | 48 | N/A | 51 |
| OFC at birth (cm) | 35 | 36 | N/A | N/A | N/A | 34 |
| Weight (kg/SD) | 14/−2.5 SD | 9.6/−2 SD | 10.38/−1.8 SD | 10.5/−2 SD | N/A | 16/−1 SD |
| Length (cm/SD) | 100/−2 SD | 81/−2 SD | 82.8/−1.5 SD. | 85/−2 SD | N/A | 108/0 SD |
| OFC (cm/SD) | 49/−0.5 SD | 46/−1 SD | 45/−1.8 SD | 46.8/−1 SD | N/A | 48/−2 SD |
| Facial dysmorphy | see Figure 1 | see Figure 1 | N/A | high hairline, brachycephaly, downslanting palpebral fissure, downturned mouth, bilateral ptosis | no | deeply set eyes, anteverted nares, deeply grooved philtrum, see Figure 1 |
| Congenital malformations | none | none | none | limb malformation | none | none |
| Neurological Manifestations | ||||||
| Intellectual disability | + | + | + | + | + | + |
| Motor developmental delay | + | + | + | + | + | + |
| Speech delay | + (no word) | + | + (no word) | + (no word) | + (no word) | + |
| Behavioral disorders | + (autistic traits) | + (anger, autism) | − | + (hypersensitivity when people touch her head and hair) | + (autism) | − |
| Stereotypic movements | + | + (hands) | − | + (hands) | + | + (hand flapping) |
| Hypotonia | N/A | N/A | + | + | − | + |
| Seizures | + | − | − | − | + | − |
| Spasticity/Ataxic gait | + | + | + | − | − | + |
| Frequent laughter | + | − | − | + | − | − |
| Attraction with water | + | − | − | − | − | − |
| Feeding difficulties | + | + | − | + | + | + |
| Magnetic resonance imaging of the brain | cortical atrophy | normal | Chiari malformation, type 1 | corpus callosum thinning and incidental pineal gland cyst | corpus callosum thinning, global white matter volume loss, slight cerebellar volume loss | normal |
| Previous Genetic Testings | ||||||
| Angelman-related genetic investigations | + | + | − | + | − | + |
| Other previous genetic investigations | N/A | X fra, CGH-array, ARX, CDG | SNP array | SNP array, MECP2 sequencing | SNP array | SNP array, MECP2, FMR1, FOXG1, metabolic screening, gene panel of intellectual disability: VUS in CDKL5 |
Abbreviations: +, present; –, absent; N/A, not available; OFC, occipital frontal circumference; SD, standard deviation. hg19 coordinates.
Consequences of variants identified in individuals 3 and 6 differ from only one amino acid (a cysteine residue for individual 3 and a phenylalanine one for individual 6 at amino acid position 245)
Frameshift Variants in NOVA2 Lead to Truncated Proteins Sharing the Same C-Terminal Part
All six variants are located in the last and largest exon of the gene and lead to frameshifts starting from the same region of the protein (between amino acids 237 and 261). They are predicted to remove the third KH domain of the protein (KH3, amino acids 406–473), which binds RNA loops composed of the tetranucleotide YCAY.7 The location of all the variants in the last exon of the gene suggested that the variant transcripts would escape to nonsense-mediated decay (NMD), but the absence of expression in blood prevents confirmation experiment. All the frameshift variants lead to the same alternative frame and the resulting truncated proteins share a common sequence of 133 novel amino acids (Figures S1 and S2). When overexpressed from plasmid in HeLa cells, NOVA2 protein carrying p.Val261Glyfs∗135 variant identified in individual 1 (p.Val261Glyfs∗135, Mut1) is normally localized (Figure S3) with no difference in stability compared to the wild-type protein (cycloheximide treatment, data not shown).
NOVA2 Downregulation Affects Neurite Outgrowth In Vitro
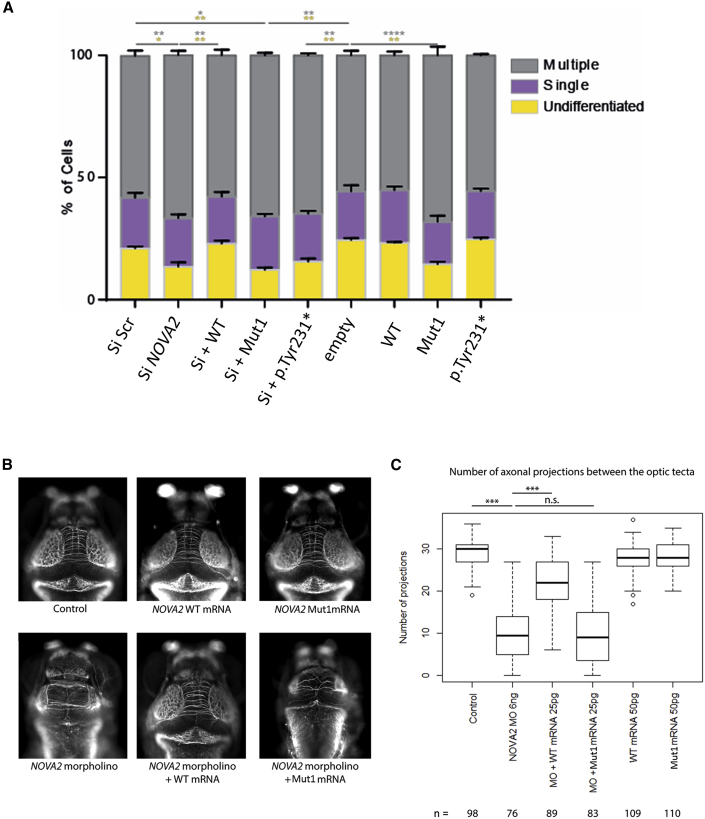
We assessed the effect of NOVA2 inactivation on neurite outgrowth by using N2a cells transfected with NOVA2 siRNA or scramble siRNA, co-transfected with a GFP plasmid. Differentiation was induced by addition of retinoic acid and formation of neurites was observed, revealing three populations of cells: undifferentiated cells (20%), cells with a one neurite (20%), and cells with multiple neurites (60%), consistent with what was previously reported27 (Figure 2A). Inactivation of NOVA2 by siRNA leads to a significant increase in the proportion of cells with multiple neurites and a decrease in the proportion of undifferentiated cells. The cell distribution returns to normal when NOVA2 siRNA are co-transfected with NOVA2 wild-type (WT) cDNA but not with Mut1 NOVA2 nor p.Tyr231∗ NOVA2. Finally, the overexpression of NOVA2 WT alone does not modify the initial cell distribution and that of p.Tyr231∗ NOVA2 either, while overexpression of Mut1 NOVA2 seems to mimic the effect of NOVA2 downregulation.
Figure 2.
NOVA2 Downregulation Affects Neurite Outgrowth In Vitro and Leads to Decreased Number of Inter-tecta Axonal Tracts in Zebrafish In Vivo
(A) NOVA2 downregulation using siRNA affects neurite outgrowth. Neuro2A cells were transfected with different combinations of NOVA2 constructs (wild-type WT, Val261Glyfs∗135 alias Mut1 or p.Tyr231∗) and siRNAs together with the GFP-reporter Venus. 48 h after treatment, cells were stained against GFP and tubulin and counterstained with DAPI. Histograms represent the percentage of transfected cells with either multiple similar sized processes (gray), a single neurite (purple), or round undifferentiated cells without processes (yellow). Data are represented as mean ± SEM. Two-way ANOVA with Tukey’s multiple comparison test. n = 3 independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001, gray asterisks for differences in cells with multiple processes and yellow asterisks for undifferentiated cells.
(B) Top panel: Representative images of dorsal views of control, wild-type, and Mut1 NOVA2 mRNA-injected larva at 4 days post-fertilization stained with anti-acetylated tubulin (AcTub). Bottom panel: In vivo complementation assay. Representative images of dorsal views of morpholino (MO)-injected, MO+WT RNA-injected, and MO+Mut1 RNA-injected larva at 4 days post-fertilization stained with anti-acetylated tubulin (AcTub).
(C) Boxplots of inter-tecta axonal tracts’ count after acetylated Tubulin staining of 4 dpf control larva and larva injected with 6 ng of nova1 morpholino (MO), 6 ng of nova1 MO+25 pg of WT NOVA2 mRNA, 6 ng of nova1 MO+25 pg of Mut1 NOVA2 mRNA, 50 pg of WT NOVA2 mRNA, 50 pg of Mut1 NOVA2 mRNA. A t test was performed between pairs of conditions. p value < 0.001 are indicated by ∗∗∗. n.s.: non-significant. n: number of larvae per condition.
NOVA2 Inactivation Alters Axonal Guidance In Vivo
We inactivated nova1a, the zebrafish orthologous gene for NOVA2 (Uniprot ID: Q1LYC7) sharing 78% of protein sequence identity, using a morpholino (MO). A reduction of the number of axonal tracts formed between optic tecta was observed, as well as a reduction of the tecta size. We counted a total of 29 inter-tecta axonal tracts in control larva whereas the number of inter-tecta axonal tracts was reduced to 10 with an overall altered brain architecture in morphant larva (Figures 2B and 2C). In vivo complementation assay was performed and we observed that 25 pg of human WT NOVA2 mRNA, but not 25 pg of the human Mut1 NOVA2 mRNA, rescued the MO phenotype. The injection of either the WT or Mut1 NOVA2 mRNA alone did not affect significantly the number of inter-tecta axonal tracts compared to controls.
Identification of Splicing Events Regulated by NOVA2 in Human Neuronal Cells
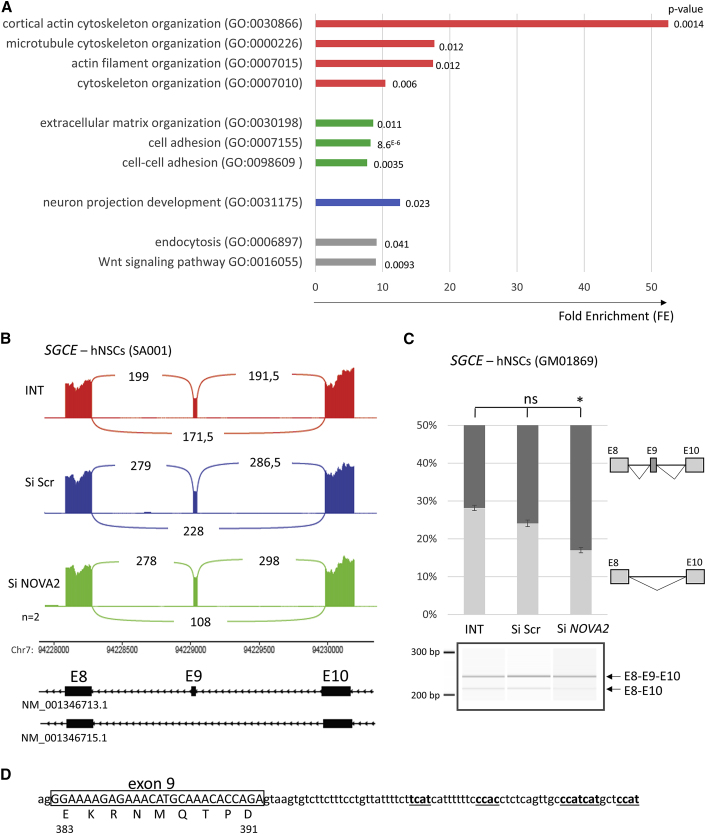
Most of the data concerning splicing target events regulated by NOVA2 have been generated in mice. In order to identify alternative splicing (AS) events regulated by NOVA2 in human neuronal cells, we used human neural stem cells (hNSCs), self-renewal homogeneous precursors of cortical neurons derived from embryonic stem cells (SA001). hNSCs were treated with NOVA2 siRNA, which led to a reduction of 50% in average (±0.09%, n = 6) of NOVA2 mRNA without affecting NOVA1 expression (data not shown). Transcriptomic (RNA-seq) analysis revealed that only few genes (<20) were found to be significantly differentially expressed (DE) in hNSCs treated with NOVA2 siRNA compared to cells treated with the transfection agent only (INTERFERin): 7 upregulated and 11 downregulated, with the most significantly one being NOVA2 (log2 fold change = −0.75, adjusted p value = 3.62e−10) (Table S1, Figure S4A). In comparison, no gene was found significantly DE in hNSCs treated with nonspecific siRNA (scramble) compared to INTERFERin (Figure S4B). Splicing analyses with LeafCutter detected AS events significantly different in hNSCs inactivated for NOVA2 (Table S2) in a total of 41 protein-coding genes (Table 2). Enrichment analysis using DAVID (Database for Annotation, Visualization and Integrated Discovery)28 demonstrated that the set of genes showing difference in AS events was enriched for Gene Ontology terms related to transmembrane proteins and extracellular matrix, cytoskeleton organization, and neuron projection/dendrite development (Figure 3A). We found that two third of these genes (26/41) have been previously reported as differently spliced in cortex of Nova2 KO mice, including SGCE, NEO1, DAB1, SLIT2, SORBS1, and others10 (Table 2). A significant decrease of the skipping of exon 9 from SGCE (sarcoglycan epsilon) transcripts was observed for instance in hNSCs after NOVA2 inactivation (adj. p = 1.02e−5, deltaPsy = 0.11) (Figure 3B). The effect of NOVA2 inactivation on the regulation of this AS event was confirmed in another line of hNSCs (GM01869) (Figure 3C). Consistent with these results, we were able to identify several NOVA2 target sequences YCAY in the beginning of SGCE intron 9 (Figure 3D). The SGCE gene encodes the epsilon-sarcoglycan, a transmembrane protein component of the dystrophin-glycoprotein complex, connecting the actin cytoskeleton to the extracellular matrix, and is known to be submitted to AS. It causes myoclonus dystonia when mutated.29 Among the other AS events significantly affected by NOVA2 inactivation, we can find for instance a decrease of exon 26 skipping in NEO1 transcripts (adj. p = 0.018, deltaPsy = 0.02), which was previously described in mice30 (Figure S5A), and a decrease of exon 3 inclusion in SORBS1 transcripts (adj. p = 5e−3, deltaPsy = −0.28) (Figure S6A) or a decrease of exon 12 skipping for a member of AKAP protein family, AKAP13 (adj. p = 2.6e−4, deltaPsy = 0.19) (Figure S7A). The effects of NOVA2 inactivation on the regulation of these additional AS events were also confirmed in the GM01869 cell line (Figures S5B, S6B, and S7B).
Table 2.
List of Genes with Alternative Splicing (AS) Events Altered by NOVA2 Downregulation
| Gene | Description | Mice Cortex10 | Neurodevelopmental or Neurological Disease (OMIM) |
|---|---|---|---|
| AKAP11 | A kinase (PRKA) anchor protein 11 | + | − |
| AKAP13 | A kinase (PRKA) anchor protein 13 | − | − |
| AP1S2 | adaptor-related protein complex 1, sigma 2 subunit | − | mental retardation, X-linked syndromic 5 (304340) |
| APP | amyloid beta (A4) precursor protein | − | cerebral amyloid angiopathy (605714); Alzheimer disease-1 (104300) |
| ARL3 | ADP-ribosylation factor-like 3 | − | Joubert syndrome (618161); retinitis pigmentosa (618173) |
| ATP5C1 | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | − | − |
| CLSTN1 | calsyntenin 1 | + | − |
| COL11A1 | collagen, type XI, alpha 1 | − | − |
| COL4A5 | collagen, type IV, alpha 5 | − | − |
| CSNK1G3 | casein kinase 1, gamma 3 | − | − |
| DAB1 | Dab, reelin signal transducer, homolog 1 (Drosophila) | + | spinocerebellar ataxia 37 (615945) |
| DNM2 | dynamin 2 | − | − |
| DOCK7 | dedicator of cytokinesis 7 | + | epileptic encephalopathy, early infantile (615859) |
| DST | dystonin | − | − |
| EPB41 | erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) | − | − |
| EPB41L2 | erythrocyte membrane protein band 4.1-like 2 | − | − |
| FMNL2 | formin-like 2 | + | − |
| GTF2I | general transcription factor IIi | + | − |
| IMPDH1 | IMP (inosine 5′-monophosphate) dehydrogenase 1 | − | − |
| LRRFIP1 | leucine rich repeat (in FLII) interacting protein 1 | − | − |
| MACF1 | microtubule-actin crosslinking factor 1 | + | lissencephaly 9 with complex brainstem malformation (618325) |
| MAGI1 | membrane associated guanylate kinase, WW and PDZ domain containing 1 | + | − |
| MAP2 | microtubule-associated protein 2 | + | − |
| MPRIP | myosin phosphatase Rho interacting protein | + | − |
| NEO1 | neogenin 1 | + | − |
| OCIAD1 | OCIA domain containing 1 | + | − |
| PBRM1 | polybromo 1 | + | − |
| PPFIBP1 | PTPRF interacting protein, binding protein 1 (liprin beta 1) | + | − |
| PTPRD | protein tyrosine phosphatase, receptor type, D | + | − |
| SGCE | sarcoglycan, epsilon | + | dystonia-11, myoclonic (159900) |
| SLIT2 | slit homolog 2 (Drosophila) | + | |
| SMARCC2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 | + | Coffin-Siris syndrome (618362) |
| SNRPN | small nuclear ribonucleoprotein polypeptide N, SNRPN upstream reading frame | − | Prader-Willi syndrome (176270) |
| SORBS1 | sorbin and SH3 domain containing 1 | + | − |
| SORBS2 | sorbin and SH3 domain containing 2 | + | − |
| SYNE2 | spectrin repeat containing, nuclear envelope 2 | + | Emery-Dreifuss muscular dystrophy 5, autosomal dominant (612999) |
| TPM1 | tropomyosin 1 (alpha) | + | − |
| UAP1 | UDP-N-acteylglucosamine pyrophosphorylase 1 | + | − |
| VCAN | versican | + | |
| VPS29 | vacuolar protein sorting 29 homolog (S. cerevisiae) | + | |
| WNK1 | WNK lysine deficient protein kinase 1 | + | neuropathy, hereditary sensory and autonomic, type II (201300) |
n.d.: not determined
Figure 3.
Transcriptomic Analysis in Human Neural Stem Cells (hNSCs) after NOVA2 Inactivation
(A) Enrichment of GO terms (Biological Process and Molecular Function, analyzed using DAVID) for the genes with differential alternative splicing (AS) events identified in human neuronal precursors (hNSCs – SA001) after NOVA2 inactivation. Fold enrichement (FE) and p value are also indicated.
(B) Sashimi plot established from RNA-seq data representing the AS of SGCE exon 9 (GenBank: NM_001346713.1). The number of reads supporting the existence of each exon-exon junction is indicated as an average between data from the two independent series of hNSCs treated with INTERFERin only (in red), with scramble siRNA (in blue) or with NOVA2 siRNA (in green) during 48 h.
(C) Confirmation of the consequences of NOVA2 inactivation on SGCE splicing in another hNSC cell line (GM01869), treated with the transfecting agent only (INT) or transfected with Scramble (si Scr) or NOVA2 (siNOVA2) siRNA. The RT-PCR products obtained were analyzed by migration on a 2,100 Bioanalyzer instrument (Agilent Technology). Experiments were done in triplicates. The error bars indicate the SEM. Kruskal-Wallis’ ANOVA with Dunn’s multiple comparison test was performed ∗p < 0.05, ns: non-significant.
(D) Sequence of SGCE exon9-intron9 junction with indicated potential NOVA2 binding sequences YCAY (in bold and underlined).
Mut1 Variant Alters NOVA2 Ability to Bind Target Sequences and to Regulate Target Alternative Splicing (AS) Events
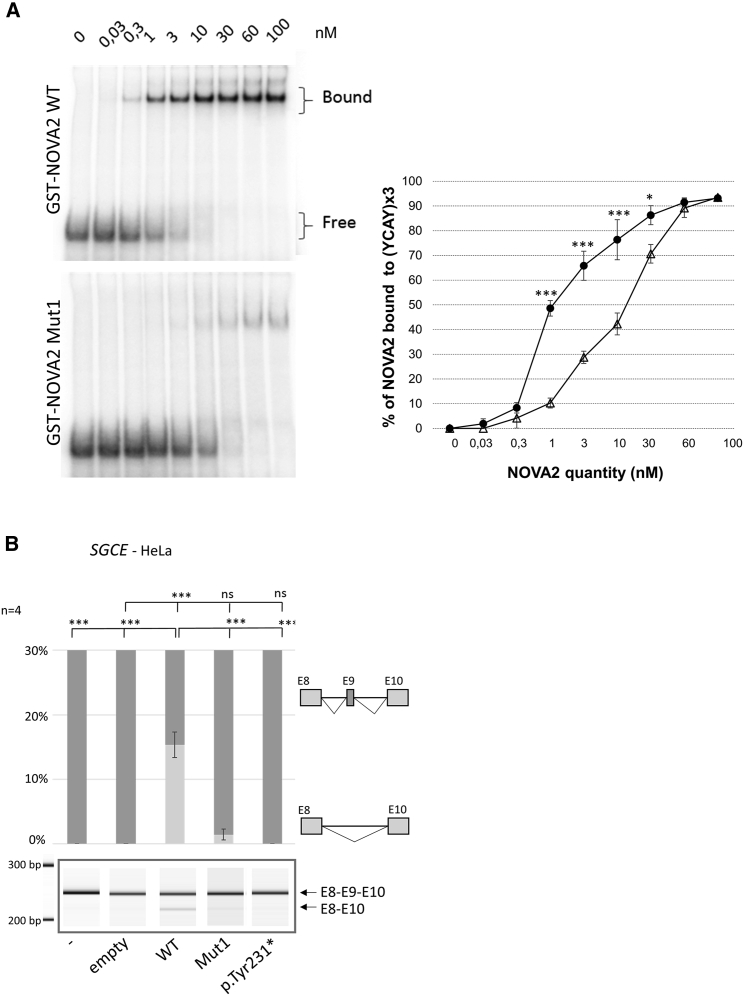
Variant proteins lack the third KH-type domain and we therefore wanted to test their ability to bind RNAs containing the YCAY motif. A gel-shift assays using labeled RNA containing three YCAY sequences showed a significant reduction in RNA binding capacity for NOVA2 Mut1 protein compared to WT NOVA2 (Figure 4A). We then wanted to test the ability of the variant protein to regulate AS events normally regulated by NOVA2. In HeLa cells, which do not express NOVA2, we observed for instance that most of the SGCE transcripts contain exon 9 (Figure 4B). This is consistent with what we observed in hNSCs, where a loss of NOVA2 expression leads to a decrease of exon 9 skipping. The overexpression of WT NOVA2 in HeLa cells leads on the contrary to a significant increase of exon 9 skipping, not observed anymore when Mut1 NOVA2 is overexpressed, demonstrating that the variant protein has lost its ability to regulate this AS event. We also tested ability of Mut1 NOVA2 to regulate the additional AS events identified in NEO1, SORBS1, or AKAP13. The overexpression of WT NOVA2 leads to a significant increase of the skipping of NEO1 exon 26 and AKAP13 exon 12, and the inclusion of SORBS1 exon 3 (Figures S5–S7). Overexpression of Mut1 NOVA2 fails to regulate the skipping of SORBS1 exon 3 but has at least partially preserved activity concerning the regulation of NEO1 exon 26 and AKAP13 exon 12 skipping. In order to test whether the hundred amino acids introduced by the frameshift variants at the C-terminal part of the protein could influence the maintaining of a partial activity of Mut1 NOVA2 protein on splicing regulation, we introduced in NOVA2 cDNA a nonsense variant p.Tyr231∗, located near to the frameshift variants. The resulting protein is a truncated form which does not contain the hundreds of amino acids added by the frameshift variants (Figure S3). In contrast to Mut1 NOVA2, p.Tyr231∗ NOVA2 fails to regulate all the AS events when overexpressed in HeLa cells (Figures 4B, S5C, S6C, and S7C). Cotransfection of the WT and p.Tyr231∗, or WT and Mut1 NOVA2 in HeLa cells have a similar effect on AS than the transfection of WT alone (Figure S8).
Figure 4.
Decreased Ability of the Mut1 Variant NOVA2 Protein to Bind Target RNA Sequences (YCAYx3) and to Regulate Target Alternative Splicing (AS) Events
(A) Binding ability of NOVA2 WT and p.Val261Glyfs∗135 (Mut1) variant proteins to NOVA2-target sequence YCAY 3×. Left panel, gel-shift assays of the indicated amount of purified recombinant WT or Mut1 GST-NOVA2 with 10 pM of uniformly [αP32] internally labeled in vitro transcribed RNAs containing three UCAY binding sites for NOVA proteins. Right panel, gel-shift quantification. Error bars standard error mean (SEM) of three independent experiments. Student’s t test, ∗p < 0.5, ∗∗∗p < 0.001.
(B) Effect of overexpression of wild-type (WT) or variant (Mut1 or p.Tyr231∗) NOVA2 proteins on splicing of SGCE exon 9 in HeLa cells. The RT-PCR products obtained were analyzed by migration on a 2,100 Bioanalyzer instrument (Agilent Technology). Four series of experiments were analyzed. The error bars indicate the SEM. Brown-Forsythe and Welch’s ANOVA with Holm-Sidak’s multiple comparisons ∗∗∗p < 0.001, ∗p < 0.05, ns: non-significant.
Discussion
We report here a case series of six individuals with severe intellectual disability sharing common autistic features and carrying de novo frameshift variants in the same region of NOVA2, between two KH RNA-binding domains. This gene encodes an RNA binding protein which is an alternate splicing regulating factor expressed during brain development. We showed that the variant identified in individual 1 (Mut1 or p.Val261Glyfs∗135) affects NOVA2 ability to regulate alternative splicing (AS) events and to bind RNA target sequences. All the variants identified in affected individuals occur in the last exon of the gene and result in truncated proteins. Therefore, different hypothesis about their molecular consequences can be considered: (1) truncated proteins have the same effect as haploinsufficiency (loss-of-function) and the variants are all located in the last exon just by chance, as it is the larger exon of the gene; (2) truncated proteins might have a more deleterious effect than haploinsufficiency (gain-of-function/dominant-negative effect), as it was described for the distal truncating variants in PPM1D causing NDD (MIM: 617450) for instance,31 and other types of variants leading to haploinsufficiency will not have any pathological consequences; or (3) truncated proteins have less severe effect than haploinsufficiency (partial loss-of-function effect) which is not observed because it would lead to lethality.
Interestingly, in contrast to variants in PPM1D, all the indel variants identified in NOVA2 cause a shift leading to the same reading frame (frame −1) which results in the addition of a common novel C-terminal part of 133 amino acids from Val261, rich in proline (29.3%) and arginine (12.8%). The alternative frame (frame +1) would have led in contrast to a more premature stop codon, 20 amino acids downstream of Val261. To our knowledge, the only case of a specific disorder caused by such specific distal frameshifts not causing nonsense-mediated mRNA decay (NMD) in a single alternate frame resulting in a common long C-terminal extension, is the Robinow developmental syndrome (MIM: 616331 and 616894). This syndrome is characterized by limb and genital anomalies and perturbation of WNT signaling, where in the two orthologous genes implicated, DLV1 and DLV3, common proline-arginine rich C-terminal extensions of 107 aa (DLV1) or 85 aa (DLV3) have been observed in at least 17 and 6 independent affected individuals, respectively, in the absence of any other type of mutations in these two genes.32,33 The authors theorized that the variant proteins would have a dominant-negative or gain-of-function effect, although partial loss-of-function related to the loss of C-terminal functional domains were also observed. In the case of indel frameshift variants in NOVA2, our functional studies presently support more a loss-of-function effect rather than a dominant-negative or gain-of-function effect. Indeed, no effect was observed when we expressed Mut1 NOVA2 mRNA alone in zebrafish. On the contrary, Mut1 NOVA2 mRNA could not rescue the phenotype of loss of axonal projection observed when NOVA2 is inactivated. The Mut1 protein presents also a partial loss of its splicing regulation activity. Its ability to regulate some AS events is strongly affected in some cases (SGCE or SORBS1) while only mildly or not affected in others (NEO1 or AKAP13). However, when co-transfected with the WT protein, the NOVA2 variant does not alter AS, suggesting that the variant protein does not exert a dominant-negative effect on AS regulation.
No other LoF variants (frameshift in +1 or nonsense variants) are reported in Clinvar or in denovo-db database (accessed October 2019). If one takes into account the probability, under a simple heterozygous loss-of-function hypothesis, that nonsense or canonical splice mutations would also lead to the same phenotype, and under the conservative assumption that frameshifts may be twice more frequent than other types of loss-of-function variants (LoF), the chance of observing only six frameshift variants, all leading to the same frame, and no other LoF variants, is low. We therefore speculate that the longer C-terminal part added by variants leading to frame −1 permits maintenance of a residual activity for NOVA2 protein and that indel variants leading to frame +1 are not observed because they would lead to complete loss of NOVA2 function (with more severe consequences). We corroborated this hypothesis by introducing a stop codon p.Tyr231∗ in NOVA2 cDNA: we demonstrated that truncated p.Tyr231∗ NOVA2 protein variant overexpressed in HeLa cells elicits a much stronger loss of AS regulation than Mut1 NOVA2. However, the consequences of NOVA2 haploinsufficiency remain puzzling as three individuals carrying deletions encompassing NOVA2 among several dozens of other genes are reported in the Decipher database, and four large deletions (1 to 4 Mb) encompassing NOVA2 and many other genes are also reported in Clinvar. The individuals do not appear to have a more severe phenotype than those reported here, as they present with ID with or without hypotonia, seizures, or abnormal gait. On the other hand, it is clear from data in gnomAD that NOVA2 is a gene with high intolerance to heterozygous LoF in the general population (none observed, pLI = 0.98) and in fact also to missense variants (observed to expected ratio of 0.26). Thus we propose that the pathomechanism may involve mainly a partial heterozygous loss of function as evidenced by our present results on splicing regulation in vitro and in vivo in the zebrafish model. However, we could not exclude any additional effect linked to the common C-terminal extension that would require longer-term experiments in a mouse model to be manifest. Indeed, overexpression of Mut1 NOVA2 alone seems to have an effect on neurite outgrowth in mouse neuroblastoma N2A cells while no effect was observed with the wild-type or the Tyr261∗ nonsense variant.
Individuals carrying truncating variants in NOVA2 exhibited autistic features (stereotypic movements, major speech defect or delay) also overlapping Angelman syndrome (hypotonia, ataxia, seizures, and frequent laughter in two individuals each), and indeed some of them were prior tested for Angelman syndrome or for Rett syndrome. Four individuals presented with anomalies in brain MRI, including corpus callosum thinning in two cases and cortical atrophy. Mouse model inactivated for Nova2 recapitulate some of these phenotypic observations. Heterozygous KO mouse models present cortical hyperexcitability and epilepsy34 and the homozygous knock-out (KO) mice manifest progressive weakness, motor dysfunction, corpus callosum agenesis, and death shortly after birth.10 It has recently been reported that conditional mice model with specific inactivation of Nova2 in Purkinje cells show progressive motor discoordination and cerebellar atrophy.11
We demonstrated that NOVA2, like its rodent counterpart, regulates alternative splicing (AS) of different genes encoding proteins involved in signal transduction and cytoskeleton organization, playing a role in neuronal differentiation and migration. In particular, partial inactivation of NOVA2 leads to mis-regulation of splicing of several transcripts encoding proteins essential for axon outgrowth and pathfinding (NEO1, SLIT2, etc.). In vivo, the consequences of axonal projection impairments could be illustrated for instance in mammals by corpus callosum hypoplasia (observed in individuals 4 and 5) or agenesis (previously identified in mice model10), but also in zebrafish by the decrease in number of inter-tecta axonal tracts, whose onset involves molecular mechanisms conserved with those of mammals. AS is an essential mechanism aiming at increasing protein diversity and contributes among others to the different critical steps of brain development such as neuronal differentiation, migration, axon guidance, and synaptogenesis. A quarter of the genes identified with AS events mis-regulated when NOVA2 is partially inactivated are known to cause neurodevelopmental or neurological disorders when mutated, highlighting their crucial role during brain development (Table 2). In addition to NOVA2, another gene encoding a protein playing a pivotal role in the regulation of alternative splicing in brain, RBFOX1, also named Ataxin-2-binding protein 1 (A2BP1) or FOX1, is known to be involved in neurodevelopmental disorders. Its expression was found to be reduced in brains collected postmortem from individuals with ASD leading to alteration of splicing of its predicted targets such as GRIN1 or MEF2C.35 Moreover, structural variants affecting RBFOX1 have been reported in several individuals with ASD, epilepsy, and ID.36, 37, 38
Mis-regulation of splicing of genes important for brain functioning have also been reported in neurodegenerative diseases, such as Alzheimer disease, fronto-temporal dementia, amyotrophic lateral sclerosis, etc.39 Interestingly, the targeting of NOVA proteins by an abnormal immune response causes POMA disease, a neurological condition characterized by ataxia with or without opsoclonus-myoclonus, dementia, encephalopathy, and cortical deficits.4,5 Other genes encoding RBP have already been implicated in both child neurodevelopmental conditions and adult-onset neurological and/or neurodegenerative disorders. For instance, deletions and missense variants in PUM1 have been reported to cause a global developmental delay syndrome characterized by speech delay, ID, ataxia, and seizure, while a rare missense variant with milder functional effect was identified in a family with several members affected by an adult-onset ataxia with incomplete penetrance.40 FMR1, the gene responsible for the most frequent cause of X-linked ID, fragile X syndrome (FXS), has been also implicated in an adult-onset neurodegenerative disease: while large CGG expansion in the 5′ UTR of FMR1 cause FXS, expansions with an intermediate number of repeats cause the fragile-X tremor ataxia syndrome (FXTAS).41 Overall, defects in RBPs causing both neurodevelopment and adult-onset neurological/neurodegenerative disorders highlight the importance of the regulation of gene expression at the post-transcriptional level in neuronal cells for both brain development and brain functioning throughout life.
In conclusion, through a multi-center collaboration, we identified truncating variants in NOVA2 affecting its activity on AS regulation and responsible for a severe syndromic form of NDD/ID with clinical manifestations overlapping Angelman syndrome. The frequency of this syndrome is difficult to evaluate. No frameshift variant has been observed in about 10,000 probands with NDD subjected to exome sequencing in the DDD project (M. Hurles, personal communication) or in 27,000 diagnostic exomes in the Netherlands (H. Brunner and R. Pfundt, personal communication). However, the frameshift variants we report in this study are located in a GC-rich internally repetitive region of NOVA2 that is very poorly represented in most exome-sequencing data (Figure S9). Moreover, half of the frameshifts we identified are caused by indels of 20 to 40 bp that are difficult to detect in standard exome protocols, especially when relatively short read lengths are used (75 to 100 bp). This study also highlights the importance of the regulation of AS events during brain development and how alterations of this regulation might lead to brain dysfunction.
Declaration of Interests
A.T., G.D., and Y.C.S. are employees of GeneDx.
Acknowledgments
The authors thank the families for their participation to the study. The authors also thank the Fondation Jerome Lejeune, Fondation Maladies Rares, and Fondattion APLM for their financial support. This study was also supported by the grant ANR-10-LABX-0030-INRT, a French State fund managed by the Agence Nationale de la Recherche under the frame program Investissements d’Avenir ANR-10-IDEX-0002-02. The authors want also to thank all the people from the IGBMC sequencing platform (Céline Keime, Serge Vicaire, Bernard Jost, Stéphanie Le Gras, Mathieu Jung, etc.) and especially Damien Plassard for their technical and bioinformatics supports. They thank Jean Muller and Véronique Geoffroy for developing and running Varank on whole-exome sequencing data, as well as Paola Rossadillo and Karim Essabri for their help for the cloning and mutagenesis. They thank Alexandra Benchoua and Istem for providing cells and technical support for hNSCs culture. They also thank people for the Molecular Genetic Unit of Strasbourg Hospital (Claire Feger) for Sanger sequencing.
Published: March 19, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.02.013.
Web Resources
Database of Genomic Variants (DGV), http://dgv.tcag.ca/dgv/app/home
Decipher, https://decipher.sanger.ac.uk/
ExAC Browser, http://exac.broadinstitute.org/
GeneMatcher, https://genematcher.org/
gnomAD Browser, https://gnomad.broadinstitute.org/
Human Genome Variation Society, http://www.hgvs.org/mutnomen/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, https://www.omim.org/
UCSC Genome Browser, https://genome.ucsc.edu
Supplemental Data
References
- 1.Ule J., Ule A., Spencer J., Williams A., Hu J.-S., Cline M., Wang H., Clark T., Fraser C., Ruggiu M. Nova regulates brain-specific splicing to shape the synapse. Nat. Genet. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- 2.Jensen K.B., Dredge B.K., Stefani G., Zhong R., Buckanovich R.J., Okano H.J., Yang Y.Y., Darnell R.B. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron. 2000;25:359–371. doi: 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 3.Ule J., Jensen K.B., Ruggiu M., Mele A., Ule A., Darnell R.B. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 4.Yang Y.Y., Yin G.L., Darnell R.B. The neuronal RNA-binding protein Nova-2 is implicated as the autoantigen targeted in POMA patients with dementia. Proc. Natl. Acad. Sci. USA. 1998;95:13254–13259. doi: 10.1073/pnas.95.22.13254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buckanovich R.J., Yang Y.Y., Darnell R.B. The onconeural antigen Nova-1 is a neuron-specific RNA-binding protein, the activity of which is inhibited by paraneoplastic antibodies. J. Neurosci. 1996;16:1114–1122. doi: 10.1523/JNEUROSCI.16-03-01114.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buckanovich R.J., Darnell R.B. The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol. Cell. Biol. 1997;17:3194–3201. doi: 10.1128/mcb.17.6.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis H.A., Musunuru K., Jensen K.B., Edo C., Chen H., Darnell R.B., Burley S.K. Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell. 2000;100:323–332. doi: 10.1016/s0092-8674(00)80668-6. [DOI] [PubMed] [Google Scholar]
- 8.Jensen K.B., Musunuru K., Lewis H.A., Burley S.K., Darnell R.B. The tetranucleotide UCAY directs the specific recognition of RNA by the Nova K-homology 3 domain. Proc. Natl. Acad. Sci. USA. 2000;97:5740–5745. doi: 10.1073/pnas.090553997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ule J., Stefani G., Mele A., Ruggiu M., Wang X., Taneri B., Gaasterland T., Blencowe B.J., Darnell R.B. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 10.Saito Y., Miranda-Rottmann S., Ruggiu M., Park C.Y., Fak J.J., Zhong R., Duncan J.S., Fabella B.A., Junge H.J., Chen Z. NOVA2-mediated RNA regulation is required for axonal pathfinding during development. eLife. 2016;5:5. doi: 10.7554/eLife.14371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saito Y., Yuan Y., Zucker-Scharff I., Fak J.J., Jereb S., Tajima Y., Licatalosi D.D., Darnell R.B. Differential NOVA2-Mediated Splicing in Excitatory and Inhibitory Neurons Regulates Cortical Development and Cerebellar Function. Neuron. 2019;101:707–720.e5. doi: 10.1016/j.neuron.2018.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vissers L.E.L.M., Gilissen C., Veltman J.A. Genetic studies in intellectual disability and related disorders. Nat. Rev. Genet. 2016;17:9–18. doi: 10.1038/nrg3999. [DOI] [PubMed] [Google Scholar]
- 13.Redin C., Gérard B., Lauer J., Herenger Y., Muller J., Quartier A., Masurel-Paulet A., Willems M., Lesca G., El-Chehadeh S. Efficient strategy for the molecular diagnosis of intellectual disability using targeted high-throughput sequencing. J. Med. Genet. 2014;51:724–736. doi: 10.1136/jmedgenet-2014-102554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattioli F., Isidor B., Abdul-Rahman O., Gunter A., Huang L., Kumar R., Beaulieu C., Gecz J., Innes M., Mandel J.-L., Piton A. Clinical and functional characterization of recurrent missense variants implicated in THOC6-related intellectual disability. Hum. Mol. Genet. 2019;28:952–960. doi: 10.1093/hmg/ddy391. [DOI] [PubMed] [Google Scholar]
- 15.Thevenon J., Duffourd Y., Masurel-Paulet A., Lefebvre M., Feillet F., El Chehadeh-Djebbar S., St-Onge J., Steinmetz A., Huet F., Chouchane M. Diagnostic odyssey in severe neurodevelopmental disorders: toward clinical whole-exome sequencing as a first-line diagnostic test. Clin. Genet. 2016;89:700–707. doi: 10.1111/cge.12732. [DOI] [PubMed] [Google Scholar]
- 16.Retterer K., Juusola J., Cho M.T., Vitazka P., Millan F., Gibellini F., Vertino-Bell A., Smaoui N., Neidich J., Monaghan K.G. Clinical application of whole-exome sequencing across clinical indications. Genet. Med. 2016;18:696–704. doi: 10.1038/gim.2015.148. [DOI] [PubMed] [Google Scholar]
- 17.Boissart C., Nissan X., Giraud-Triboult K., Peschanski M., Benchoua A. miR-125 potentiates early neural specification of human embryonic stem cells. Development. 2012;139:1247–1257. doi: 10.1242/dev.073627. [DOI] [PubMed] [Google Scholar]
- 18.Quartier A., Chatrousse L., Redin C., Keime C., Haumesser N., Maglott-Roth A., Brino L., Le Gras S., Benchoua A., Mandel J.-L., Piton A. Genes and Pathways Regulated by Androgens in Human Neural Cells, Potential Candidates for the Male Excess in Autism Spectrum Disorder. Biol. Psychiatry. 2018;84:239–252. doi: 10.1016/j.biopsych.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 19.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benjamini Y., Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. 1995;57:289–300. [Google Scholar]
- 24.Li Y.I., Knowles D.A., Humphrey J., Barbeira A.N., Dickinson S.P., Im H.K., Pritchard J.K. Annotation-free quantification of RNA splicing using LeafCutter. Nat. Genet. 2018;50:151–158. doi: 10.1038/s41588-017-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolar N.A., Golzio C., Živná M., Hayot G., Van Hemelrijk C., Schepers D., Vandeweyer G., Hoischen A., Huyghe J.R., Raes A. Heterozygous Loss-of-Function SEC61A1 Mutations Cause Autosomal-Dominant Tubulo-Interstitial and Glomerulocystic Kidney Disease with Anemia. Am. J. Hum. Genet. 2016;99:174–187. doi: 10.1016/j.ajhg.2016.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ivanova E.L., Mau-Them F.T., Riazuddin S., Kahrizi K., Laugel V., Schaefer E., de Saint Martin A., Runge K., Iqbal Z., Spitz M.-A. Homozygous Truncating Variants in TBC1D23 Cause Pontocerebellar Hypoplasia and Alter Cortical Development. Am. J. Hum. Genet. 2017;101:428–440. doi: 10.1016/j.ajhg.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang D.W., Sherman B.T., Tan Q., Kir J., Liu D., Bryant D., Guo Y., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169-75. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zimprich A., Grabowski M., Asmus F., Naumann M., Berg D., Bertram M., Scheidtmann K., Kern P., Winkelmann J., Müller-Myhsok B. Mutations in the gene encoding epsilon-sarcoglycan cause myoclonus-dystonia syndrome. Nat. Genet. 2001;29:66–69. doi: 10.1038/ng709. [DOI] [PubMed] [Google Scholar]
- 30.Jelen N., Ule J., Zivin M., Darnell R.B. Evolution of Nova-dependent splicing regulation in the brain. PLoS Genet. 2007;3:1838–1847. doi: 10.1371/journal.pgen.0030173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jansen S., Geuer S., Pfundt R., Brough R., Ghongane P., Herkert J.C., Marco E.J., Willemsen M.H., Kleefstra T., Hannibal M., Deciphering Developmental Disorders Study De Novo Truncating Mutations in the Last and Penultimate Exons of PPM1D Cause an Intellectual Disability Syndrome. Am. J. Hum. Genet. 2017;100:650–658. doi: 10.1016/j.ajhg.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bunn K.J., Daniel P., Rösken H.S., O’Neill A.C., Cameron-Christie S.R., Morgan T., Brunner H.G., Lai A., Kunst H.P.M., Markie D.M., Robertson S.P. Mutations in DVL1 cause an osteosclerotic form of Robinow syndrome. Am. J. Hum. Genet. 2015;96:623–630. doi: 10.1016/j.ajhg.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White J., Mazzeu J.F., Hoischen A., Jhangiani S.N., Gambin T., Alcino M.C., Penney S., Saraiva J.M., Hove H., Skovby F., Baylor-Hopkins Center for Mendelian Genomics DVL1 frameshift mutations clustering in the penultimate exon cause autosomal-dominant Robinow syndrome. Am. J. Hum. Genet. 2015;96:612–622. doi: 10.1016/j.ajhg.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eom T., Zhang C., Wang H., Lay K., Fak J., Noebels J.L., Darnell R.B. NOVA-dependent regulation of cryptic NMD exons controls synaptic protein levels after seizure. eLife. 2013;2:e00178. doi: 10.7554/eLife.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voineagu I., Wang X., Johnston P., Lowe J.K., Tian Y., Horvath S., Mill J., Cantor R.M., Blencowe B.J., Geschwind D.H. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhalla K., Phillips H.A., Crawford J., McKenzie O.L.D., Mulley J.C., Eyre H., Gardner A.E., Kremmidiotis G., Callen D.F. The de novo chromosome 16 translocations of two patients with abnormal phenotypes (mental retardation and epilepsy) disrupt the A2BP1 gene. J. Hum. Genet. 2004;49:308–311. doi: 10.1007/s10038-004-0145-4. [DOI] [PubMed] [Google Scholar]
- 37.Davis L.K., Maltman N., Mosconi M.W., Macmillan C., Schmitt L., Moore K., Francis S.M., Jacob S., Sweeney J.A., Cook E.H. Rare inherited A2BP1 deletion in a proband with autism and developmental hemiparesis. Am. J. Med. Genet. A. 2012;158A:1654–1661. doi: 10.1002/ajmg.a.35396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin C.L., Duvall J.A., Ilkin Y., Simon J.S., Arreaza M.G., Wilkes K., Alvarez-Retuerto A., Whichello A., Powell C.M., Rao K. Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2007;144B:869–876. doi: 10.1002/ajmg.b.30530. [DOI] [PubMed] [Google Scholar]
- 39.Cookson M.R. RNA-binding proteins implicated in neurodegenerative diseases. Wiley Interdiscip. Rev. RNA. 2017;8:8. doi: 10.1002/wrna.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gennarino V.A., Palmer E.E., McDonell L.M., Wang L., Adamski C.J., Koire A., See L., Chen C.-A., Schaaf C.P., Rosenfeld J.A. A Mild PUM1 Mutation Is Associated with Adult-Onset Ataxia, whereas Haploinsufficiency Causes Developmental Delay and Seizures. Cell. 2018;172:924–936.e11. doi: 10.1016/j.cell.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hagerman R.J., Leehey M., Heinrichs W., Tassone F., Wilson R., Hills J., Grigsby J., Gage B., Hagerman P.J. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology. 2001;57:127–130. doi: 10.1212/wnl.57.1.127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.