Abstract
Despite next-generation sequencing, which now allows for the accurate detection of segmental aneuploidies from in vitro fertilization embryo biopsies, the origin and characteristics of these aneuploidies are still relatively unknown. Using a multifocal biopsy approach (four trophectoderms [TEs] and one inner cell mass [ICM] analyzed per blastocyst; n = 390), we determine the origin of the aneuploidy and the diagnostic predictive value of segmental aneuploidy detection in TE biopsies toward the ICM’s chromosomal constitution. Contrary to the prevalent meiotic origin of whole-chromosome aneuploidies, we show that sub-chromosomal abnormalities in human blastocysts arise from mitotic errors in around 70% of cases. As a consequence, the positive-predictive value toward ICM configuration was significantly lower for segmental as compared to whole-chromosome aneuploidies (70.8% versus 97.18%, respectively). In order to enhance the clinical utility of reporting segmental findings in clinical TE biopsies, we have developed and clinically verified a risk stratification model based on a second TE biopsy confirmation and segmental length; this model can significantly improve the prediction of aneuploidy risk in the ICM in over 86% of clinical cases enrolled. In conclusion, we provide evidence of the predominant mitotic origin of segmental aneuploidies in preimplantation embryos and develop a risk stratification model that can help post-test genetic counseling and that facilitates the decision-making process on clinical utilization of these embryos.
Keywords: aneuploidies, segmental abnormalities, mosaicism, preimplantation genetic testing, preimplnatation genetic screening, PGT, preimplantation embryo, IVF, meiosis, mitotic aneuploidies
Introduction
The field of preimplantation genetic testing (PGT) of human embryos has been characterized by a continuous technological evolution leading to the introduction of increasingly sensitive, higher-throughput, and more cost-effective platforms for comprehensive chromosome analysis.1, 2, 3 In particular, the implementation of next-generation sequencing (NGS) platforms has provided improved resolution and sensitivity, making it possible to detect a widening dynamic range of chromosomal aberrations that occur de novo.4 These include sub-chromosomal aneuploidies, duplications, and deletions of chromosomal segments affecting regions larger than 5–10 Mb.5,6 Although they are rare in prenatal genetics,7,8 NGS-based studies have revealed that de novo segmental aneuploidies arising from chromosomal structural rearrangements are relatively common in human preimplantation embryos (15.6%).9,10 From a biological standpoint, the occurrence of segmental alterations is primarily generated by meiotic events during gametogenesis, as well as mitotic errors during embryonic development.11 Meiotic events are predicted to give rise to embryos in which all cells inherit the rearranged chromosome, whereas mitotic errors of embryonic origin are predicted to result in a mosaic pattern of segmental aneuploidies across the embryo. However, the nature and prevalence of the two mechanisms at the blastocyst stage of human preimplantation development are still unclear.
Recently, a few studies have assessed karyotype concordance rates between clinical trophectoderm (TE) biopsies carrying a segmental aneuploidy and their correspondent inner cell mass (ICM). These studies showed that concordance rate between TE and ICM for segmental aneuploidies is reduced compared to those involving whole chromosomes, thus suggesting that sub-chromosomal regions’ alterations may be prevalently caused by mitotic events leading to mosaic patterns.6,10,12,13 Nevertheless, either these studies were limited by a small sample size (n = 7)13 or they employed suboptimal techniques (i.e., fluorescence in situ hybridization [FISH]) for confirming the diagnosis on the remaining sections of the embryo.6,10 In particular, the use of the FISH technique prevented the acquisition of informative data on alternative aneuploidy patterns of the affected chromosome, and also prevented the development of a comprehensive view on the entire embryonic karyotype. Furthermore, all studies except one13 failed to report validation data for the ICM biopsy method employed. Indeed, these details are critical for the evaluation of experimental outcomes because the collection of pure ICM fractions, free of TE cell contamination, is an extremely challenging procedure.14, 15, 16 As a result, the potential impact of segmental aneuploidies detection on in vitro fertilization (IVF)/preimplantation genetic testing for aneuploidies (PGT-A) clinical workflow and their real predictive diagnostic value in clinical TE (cTE) biopsies remain an open question deserving further investigation.
In this study, the incidence and type of segmental aneuploidies detected in cTE biopsies has been assessed in a dataset of 8,137 PGT-A analyses, and the meiotic or mitotic nature of sub-chromosomal abnormalities has been investigated via multifocal analysis of 78 disaggregated blastocysts. Furthermore, an ICM risk stratification model able to assist in the interpretation of segmental aneuploidy findings and post-test genetic counselling in PGT-A cycles has been developed and clinically verified.
Material and Methods
Study Design and Objectives
In this study, we have first assessed the incidence and patterns of segmental aneuploidies detected by NGS analysis of 8,137 TE biopsies analyzed in our blastocyst-stage PGT-A program. This evaluation allowed the characterization of the relative contribution of segmental aneuploidies to the overall aneuploidy rate in cTE samples and the assessment of chromosome-specific susceptibility to segmental errors. Next, we performed multifocal biopsies on blastocysts (euploid and with segmental aneuploidies) donated for research purposes to evaluate the positive diagnostic predictive value (PPV) and negative diagnostic predictive value (NPV) for segmental and whole-chromosome aneuploidies. This phase of the study also allowed the evaluation of biological mechanisms responsible for the occurrence of segmental aneuploidies at the blastocyst stage as well as the development of a decisional tree model, based on ICM aneuploidy state, for optimizing clinical management of embryos showing sub-chromosomal abnormalities. The generated decisional tree model has been subsequently verified on an independent cohort of embryos obtained in PGT-A cycles and showing segmental aneuploidies in the cTE as the only abnormality.
Prospective Analysis of NGS-Based PGT-A Results
NGS-based PGT-A results were obtained at Igenomix Italia laboratory between February 2018 and November 2019 (n = 8,137, mean female age = 37.9, 95% confidence interval [CI] = 37.84–38.00). Embryos were defined as normal and/or euploid if no alteration with respect to the reference base line was observed, and embryos were defined as aneuploid if they exhibited uniform single or multiple whole-chromosome and/or segmental (deletion and duplication above 10Mb) abnormalities. Although studies on cell lines have shown the capability of NGS-based protocols to increase the resolution toward chromosome copy number (CN) variations, the diagnostic approach employed here did not consider mosaic classification categories. Because technical and biological variations in cTEs’ NGS profiles cannot be entirely distinguished, this approach was chosen in order to limit the impact of experimental variability on aneuploidy mechanism analysis.17, 18, 19 Therefore, our classification scheme followed a binary approach: disomic or uniform aneuploid.
Multifocal Blastocyst Biopsies to Define Segmental Aneuploidies’ Origin and Predictive Value Analysis
A cohort study blinded to the geneticist was carried out to assess positive predictive values (PPVs) and negative predictive values (NPVs) for segmental and whole aneuploidy detection in TE biopsies. To this aim, 78 blastocysts (53 with a segmental aneuploidy in cTE and 23 otherwise euploid) donated for research at the Bacheci clinic in Cyprus were warmed and disaggregated into four portions and the ICM. Ethical committee approval for the study was obtained from the Institutional Review Board (IRB) at Near East University (project number: YDU/2018/64-685). Approved informed consent forms were signed by all of the individuals donating their embryos to this study. ICM isolation and multiple TE biopsies were performed using a previously described and validated methodology.14 Individual biopsies were blindly analyzed using an NGS platform. In the multifocal analysis, concordance rates were calculated comparing the PGT-A result obtained from the cTE with each of the associated TE and ICM biopsies. In detail, when the same segmental alteration was observed in all biopsies, this outcome was considered to be consistent with a pattern of meiotic origin. When the abnormality was detected in more than one sample, but not in all, the aneuploidy was considered to originate from a mitotic error leading to mosaicism. Furthermore, the aneuploidy was considered to be confined to TE when it was uniformly detected in all TE samples but not in the ICM (confined TE mosaic). Finally, when the alteration was found only in the cTE, the pattern was interpreted as consistent with low-grade mosaicism or as a technical artifact.
Development of a Risk Stratification Model to Enhance Clinical Management and Genetic Counselling on Segmental Aneuploidy Findings in PGT-A Cycles
From the dataset of donated embryos, a risk stratification model was then developed to enhance the predictivity toward ICM ploidy status of a segmental finding in a cTE biopsy, including significantly associated covariates (see Statistical Analysis). A logistic regression model was built in order to identify potential additional variables to enhance segmental aneuploidy predictive values. To this end, a multivariate analysis was conducted in which the independent variable was the confirmation state on ICM and which included as main covariates female age, sperm quality, male age, segmental size, chromosome involved, embryo morphology, and day of biopsy. Recursive partitioning analysis was used to stratify the samples according to predictive variables on confirmation outcome.20 Accordingly, a decision-making model was computed. This model was further verified through an independent population of embryos that were diagnosed with a segmental abnormality in the cTE in clinical PGT-A cycles. These embryos were subjected to a second clinical TE biopsy (scTE) in order to obtain diagnostic confirmation and improve clinical management of the associated PGT-A cycle.
For this purpose, 51 blastocysts from 1,817 consecutive IVF treatments shown to carry segmental aneuploidies during clinical PGT-A cycles were enrolled in the study at the GENERA Center for Reproductive Medicine in Rome, Italy, leading to a total collection of 102 biopsy samples. This analysis served to define the proportion of cases in which the application of the predictive model improved clinical management compared with the information provided by the cTE biopsy alone. IRB approval for the study was also obtained from Clinica Valle Giulia for cTE re-analysis of embryos showing segmental aneuploidies in the original blastocyst biopsy. Informed consent was obtained from individuals donating their embryos to this study.
PGT-A Analysis
Embryo culture and cTE biopsies were performed as previously described.21 NGS-based PGT-A was employed for the analysis of all blastocyst biopsies by performing genomic DNA extraction and whole-genome amplification (WGA) using Ion Reproseq PGS kit (ThermoFisher). In detail, each biopsy was tubed in 2.5μl of 1× PBS, treated with 5μl of Extraction Enzyme master mix and incubated at 75°C for 10 min, followed by incubation at 95°C for 4 min. Extracted genomic DNA was pre-amplified with 5μl of Pre-amplification master mix and incubated according to the following program: 1 cycle at 95°C for 2 min and 12 cycles at 95°C for 15 s, 15°C for 50 s, 25°C for 40 s, 35°C for 30 s, 65°C for 40 s, 75°C for 40 s, and holding at 4°C. Subsequently, 30μl of Amplification master mix and 5μl of Ion SingleSeq Barcode Adaptor were added to each sample. Library amplification was performed with the following program: 1 cycle at 95°C for 3 min, 4 cycles at 95°C for 20 s, 50°C for 25 s, 72°C for 40 s, 12 cycles at 95°C for 20 s, 72°C for 55 s, and holding at 4°C. Libraries were then pooled, purified with AMPure XP beads, quantified using the Qubit dsDNA High Sensitivity Assay kit, and diluted to the final concentration of 80pM. Template preparation and chip loading was performed using Ion Chef system (Thermo Fisher) according to manufacturer instructions. Chip was then loaded and sequenced on Ion S5TM XL SequencerTM (Thermo Fisher).
Interpretation of Sequencing Data and Diagnosis
Sequencing data obtained by the S5TM XL Sequencer were processed and sent to the Ion Reporter software for analysis. Aneuploidies and CN variations were analyzed with the Ion Reporter Software version 5.4 (Thermo Fisher Scientific). This software uses the bioinformatic tool ReproSeq Low-pass Whole-Genome Aneuploidy Workflow v1.0 to detect 24 chromosome aneuploidies from a single whole-genome sample with low coverage (minimum 0.01×). Data obtained from the Ion Reporter files for each embryo were analyzed using the Igenomix proprietary algorithm to release an automated result including detection of segmental aneuploidies.22 Segmental aneuploidies >10Mb were manually identified only if a chromosome fragment deviated from the standard threshold for disomy.
Statistical Analysis
Categorical variables are shown as percentages with 95% CI, and continuous variables are shown as mean ± standard deviation (SD). Statistical analysis was conducted using the two-tailed chi square test for categorical variables and ANOVA with Bonferroni’s correction for continuous variables. In Phase 2 experiments, in order to define sensitivity and specificity of TE result toward ICM ploidy status prediction, we first classified each segmental aneuploidy as true positive (TP, abnormal ICM and abnormal TE), true negative (TN, normal ICM and normal TE), false positive (FP, normal ICM and abnormal TE), or false negative (FN, abnormal ICM and normal TE). Sensitivity was calculated as the percentage of abnormal chromosome correctly predicted as aneuploid, while specificity was defined as the percentage of euploid chromosomes detected for all chromosomes expected to be normal. PPVs and NPVs were calculated as the proportion of positive and negative results that were true positive and true negative [PPV = TP/(TP + FP); NPV = TN/(TN + FN)]. To assess the diagnostic reliability of segmental detection on a single cTE biopsy toward the remaining embryo, concordance measures were calculated as described above but considering as confirmation the presence of at least one additional biopsy showing the same or an alternative aneuploidy pattern for the same chromosomal segment. p value < 0.05 was considered statistically significant.
Results
Incidence and Type of Segmental Aneuploidies in Human Embryos
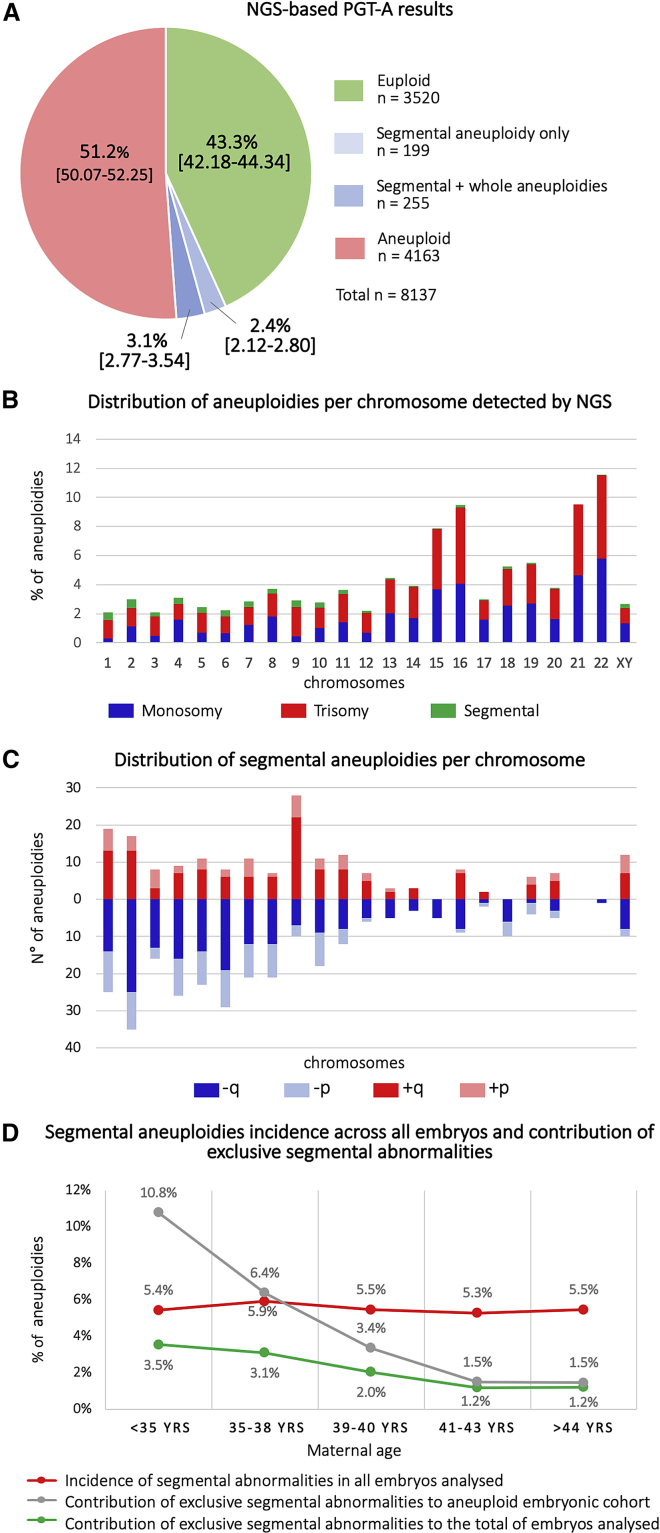
The NGS-based PGT-A protocol used in this study was internally validated for both whole-chromosome and segmental aneuploidies on cell lines with known abnormalities of different sizes and involving different chromosomes (Table S1A, S1B, and S1C). The prospective analysis of 8,137 human NGS-based TE biopsies revealed an overall aneuploidy rate (whole-chromosome and segmental) of 56.7% (n = 4,617/8,137; 95% CI = 55.66–57.82). In particular, the percentage of embryos with at least one segmental aneuploidy was 5.6% (n = 454/8,137; 95% CI = 5.09–6.10), whereas the percentage of embryos carrying a segmental alteration alone was 2.4% (n = 199/8,137; 95% CI = 2.12–2.80) (Figure 1A). The types (i.e., trisomy, monosomy, and segmental) and distributions of abnormalities across chromosomes are shown in Figure 1B. The incidence and distribution of specific segmental aneuploidies (i.e., gain and loss of q and p arms) was further characterized, highlighting an uneven distribution of segmental errors, and larger chromosomes were more frequently involved (Figure 1C). Of note, all detected segmental abnormalities were telomeric. This observation is consistent with the evidence that interstitial aneuploidies commonly detected in pregnancies and/or newborns are of small size (i.e., <10Mb) and therefore fall below the standard resolution limit of the NGS platforms employed for PGT-A analysis.23
Figure 1.
Characterization of Segmental Aneuploidies across All Embryos Analyzed (n = 8,137)
(A) Summary of NGS-based PGT-A results per embryo.
(B) Incidence and distribution of chromosomal abnormalities across the genome. Red = trisomy, blue = monosomy, and green = segmental.
(C) Incidence and distribution of specific segmental aneuploidies across the genome. Dark red = long-arm trisomy, Light red = short-arm trisomy, dark blue = long-arm monosomy, and light blue = short-arm monosomy.
(D) Incidence of segmental aneuploidies according to maternal age. Red line = incidence of all segmental abnormalities over all embryos analyzed, gray line = relative contribution of segmental abnormalities alone to the total of aneuploidies detected, and green line = relative contribution of segmental abnormalities alone to the total of embryos analyzed.
Further analysis showed that the incidence of segmental aneuploidies was not related to female age (Figure 1D, red line), whereas the relative contribution of segmental abnormalities alone to the total of aneuploidies detected (Figure 1D, gray line) and to the total number of embryos analyzed (Figure 1D, green line) decreased with age, reflecting the expected increase in age-related whole chromosomes’ meiotic aberrations.
Multifocal Analysis of the Blastocyst Reveals the Mitotic Origin of Most of Segmental Aneuploidies
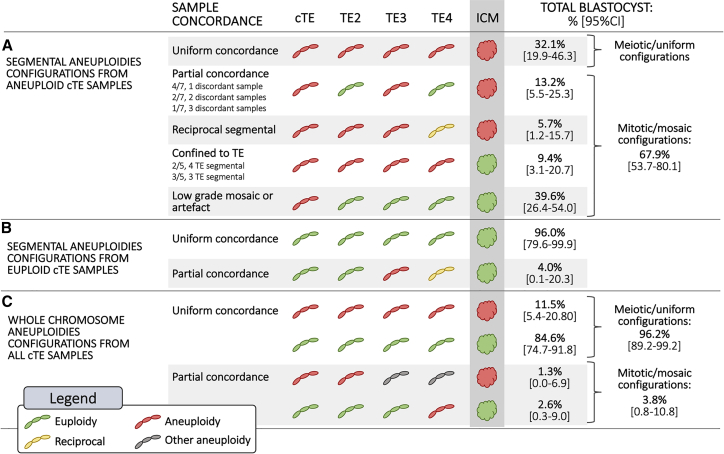
A total of 78 human embryos (25 euploid and 53 aneuploid with a segmental alteration previously detected in the cTE biopsy) were disaggregated into four additional sections and blindly analyzed using NGS-based aneuploidy testing to investigate the mitotic or meiotic origin of the aneuploidy (Table S2). Each embryo provided a total of five diagnostic results, four from the TE (including the biopsy performed for clinical purposes) and 1 from the ICM. In 17 of the 53 aneuploid blastocysts showing a segmental abnormality, all five biopsies, including the ICM, were concordant for the same sub-chromosomal aberration (32.1%; 95% CI = 19.92–46.32, Figure 2A); this result is consistent with an error of meiotic origin. The remaining 68.0% of embryos (n = 36/53; 95% CI = 53.68–80.08) showed a mosaic configuration for the segmental aneuploidy (Figure 2A). In particular, the same segmental aneuploidy was observed in at least one additional biopsy in 13.2% of samples (n = 7/53; 95% CI = 5.48–25.34), whereas reciprocal patterns were observed in 5.7% of embryos (n = 3/53; 95% CI = 1.18–15.66). In a subset of embryos (n = 5/53; 9.4%; 95% CI = 3.13–20.66), a TE-confined mosaic pattern was observed. In contrast, segmental aneuploidies were detected only in the cTE biopsy in 39.6% of cases (n = 21/53; 95% CI = 26.45–54.00); this result suggests an aneuploidy pattern consistent with low-grade mosaicism. However, the presence of a technical artifact in the initial PGT-A analysis for this group of samples cannot be ruled out. Examples of PGT-A profiles of different segmental aneuploidies configurations detected in multifocal TE biopsies are shown in Figure S1. In the 25 euploid blastocysts analyzed for segmental aneuploidies, all ICMs had normal karyotypes and 96.0% (n = 24/25; 95% CI = 79.65–99.90) showed full concordance across all five biopsy specimens (TE + ICM), whereas only one (4.0%; 95% CI = 0.10–20.35) showed partial concordance due to the presence of reciprocal de novo sub-chromosomal errors detected in two different TE biopsies (Figure 2B; sample C068 in Table S2).
Figure 2.
Overview of Blastocyst’s Karyotype Configurations and Sample Concordance Rates Detected across Multifocal Analysis of Four Trophectoderm (TE) Sections and the Inner Cell Mass (ICM)
(A) Segmental aneuploidies configurations from embryos showing single segmental alteration in the clinical TE (cTE) biopsy.
(B) Segmental aneuploidies configurations from embryos showing a euploid karyotype in the cTE biopsy.
(C) Karyotype configurations from all embryos in relation to whole-chromosome aneuploidies.
PGT-A analysis across TE biopsies and correspondent ICMs reported 99.3% per chromosome concordance (n = 7,125/7,176; 95% CI = 99.07–99.47) and 83.6% per sample full-karyotype concordance (n = 261/312; 95% CI = 79.07–87.58). Considering that the portion of cells included in a TE biopsy fragment is randomly chosen, we calculated PPV and NPV of all TE biopsies, both from normal and abnormal blastocysts, in relation to their ICM chromosomal status. When considering segmental aneuploidies only, PPV per chromosome and per sample (full-karyotype) was 70.8% (n = 97/137; 95% CI = 62.43–78.25), whereas NPVs were 99.8% (n = 7,028/7,039; 95% CI = 99.72–99.92) and 93.7% (n = 164/175; 95% CI = 89.03–96.82), respectively. In contrast, the analysis of whole-chromosome aneuploidies revealed a remarkably high concordance rate across biopsies from the same embryos. In particular, the concordance rate between individual TE biopsies and ICM was 99.9% when calculated per chromosome (n = 7,171/7,176; 95% CI = 99.84–99.98) and 98.7% when full-karyotype concordance was calculated per sample (n = 308/312; 95% CI = 96.75–99.65) (Figure 2C; Table S1). Only three blastocysts (3.8%; 95% CI = 0.8–10.83) showed partial concordance; this result suggests the presence of a mosaic pattern for whole chromosomes. These data show that whole-chromosome aneuploidies are detected in the blastocyst with very high consistency, and incidence of detectable genuine mosaicism is extremely low.
Development of a Prediction Model for Segmental Aneuploidies Detection in PGT-A
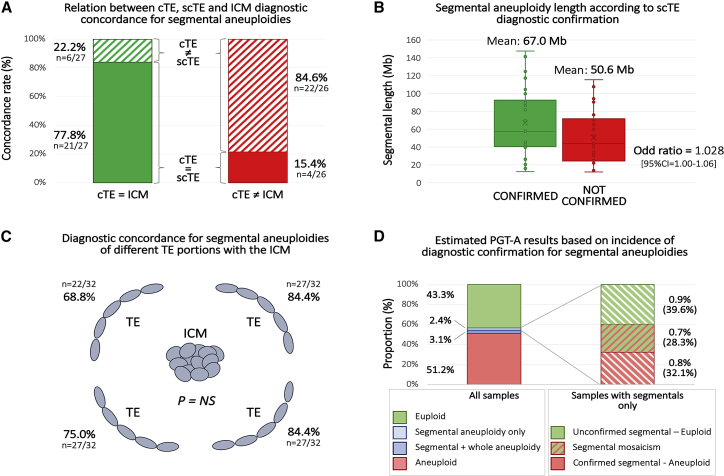
Considering that only one TE biopsy is generally obtained in clinical PGT-A settings, we sought to investigate the predictivity of a segmental finding toward the ICM chromosomal status and whether certain clinical and embryological parameters could enhance it. For this purpose, all segmental alterations were divided into two groups according to whether it was confirmed or not confirmed in ICM (confirmation outcome). Logistic regression analysis revealed that the segment size and the confirmation of the result in the scTE biopsy were the only variables associated with ICM confirmation rate (Table S3). Of these, diagnostic confirmation through detection of the same or an alternative aneuploidy pattern in the scTE was the strongest prognostic factor for ICM confirmation. Indeed, 84.0% (n = 21/25; 95% CI = 63.92–95.46) of cases displaying a positive scTE showed the same or an alternative pattern for that chromosome in the ICM (Figure 3A). Although the statistical effect was weak, the mean segmental length was higher in confirmed diagnoses (67.0 ± 38.5 versus 50.6 ± 30.9, for confirmed and not confirmed, respectively; p = NS; Figure 3B). Additionally, different TE biopsies exhibited similar ICM concordance rates, suggesting an equal representativeness toward the ICM (p = NS; Figure 3C). Based on the evidence brought by multifocal biopsy experiments, we estimated the true incidence of segmental findings in the total study population. According to our data, the occurrence of true segmental aneuploidies (confirmed in additional biopsies as defined in Figure 2) is 1.5%. Around half of these (53%) are expected to be of meiotic origin, while the remaining are expected to follow a mosaic pattern. Interestingly, around 40% of the segmental aneuploidies detected in the first biopsy were not confirmed in additional biopsies; this reveals an origin linked to analytical artifact or to a very-low-level mosaicism. This category involves around 1% of all embryos analyzed for PGT-A purposes (Figure 3D).
Figure 3.
Diagnostic Concordance Rates across Clinical Trophectoderm (cTE), Second Clinical Trophectoderm (scTE), and Inner Cell Mass (ICM)
(A) Relationship between cTE/scTE diagnostic concordance and ICM confirmation. Green bar = concordant diagnosis between cTE and ICM, red bar = non-concordant diagnosis between cTE and ICM, full box = concordant diagnosis between cTE and scTE, and striped box = non-concordant diagnosis between cTE and scTE.
(B) Blox plots of segmental aneuploidy length detected in cTE, according to ICM confirmation. Green plot = cTE diagnosis confirmed in scTE biopsy and red plot = cTE diagnosis not confirmed in scTE biopsy (p = NS).
(C) Diagnostic concordance rates for segmental aneuplodies between different TE portions and the ICM of the same blastocyst (p = NS).
(D) Expected true incidence rate of segmental aneuploidies in preimplantation embryos. Left: proportion of aneuploidy categories as detected in the study total population. Right: expected true diagnostic outcome in blastocysts presenting a segmental aneuploidy in their cTE. Incidence of each subgroup was calculated across all samples and across samples presenting segmental aneuploidies only (in parenthesis).
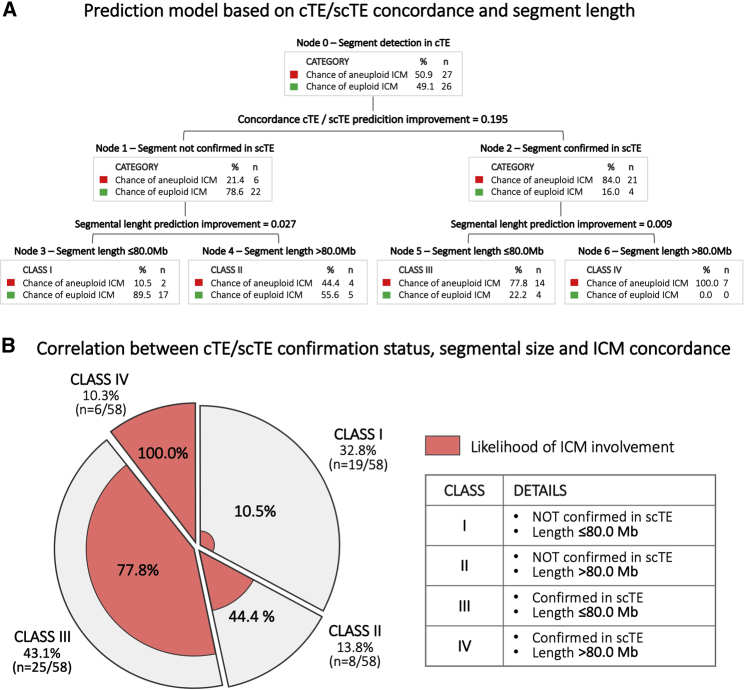
An intuitive risk stratification model for segmental aneuploidies was developed through the use of recursive partitioning analysis. This model can predict the likelihood that the segmental aneuploidy observed in the cTE is also present in the ICM. When an scTE biopsy is available for analysis and confirms the segmental aneuploidy detected in the cTE, the likelihood of diagnostic concordance with the ICM increases from 21.4% to 84% (Figure 4A). Alternatively, when a segmental aneuploidy involving a region smaller than 80Mb is detected in the cTE but not in the scTE, the likelihood of an aneuploid ICM decreases from an a priori 50.9% to 10.5% (Figure 4A).
Figure 4.
Model for Predicting the Inner Cell Mass (ICM) Involvment by the Same Segmental Aneuploidy Detected in Clinical Trophectoderm (cTE)
(A) Decisional tree generated using both confirmation in a second clinical trophectoderm (scTE) biopsy and segmental length.
(B) Clinical application of the risk stratification model to a cohort of PGT-A-derived embryos: segmental aneuploidies detected in cTE biopsy are distributed from Class I to Class IV according to scTE confirmation outcome and aneuploid segment length. For each class, the predicted likelihood of concordance between cTE diagnosis and ICM is shown in red. The clinical verification shows that only around 14% of cases follow in an ICM-predicted risk similar to the a priori risk (Class II).
This risk stratification model was applied clinically to a cohort of PGT-A cycles in which segmental aneuploidies were detected in cTE biopsies and confirmation was undertaken via an scTE. Demographic data are reported in Table S4. Out of 58 segmental aneuploidies identified in 51 cTEs, 46.6% (n = 27/58; 95% CI = 33.34–60.13) were not confirmed in scTEs, while 53.4% (n = 31/58; 95% CI = 39.87–66.66) were confirmed. The contribution of each diagnostic class to the total of segmental aneuploidies detected in this population was 32.8% (n = 19/58; 95% CI = 21.01–46.34), 13.8% (n = 8/58; 95% CI = 6.15–25.38), 43.1% (n = 25/58; 95% CI = 30.16–56.77), and 10.3% (n = 6/58; 95% CI = 3.89–21.17) for Classes I, II, III, and IV, respectively (Figure 4B). In this clinical landscape, only around 13% of cases maintained an ICM aneuploidy risk comparable to the a priori risk (Class II, 44% versus ~50%), whereas in around 87% of cases, risk prediction was significantly improved, thus allowing better post-test genetic counselling and clinical treatment management (Figure 4B).
Discussion
The objective of this study was to further characterize the biological significance of segmental aneuploidies in human blastocysts and, based on these premises, to develop an enhanced diagnostic model for improvement of clinical treatment management.
We report that the general contribution of sub-chromosomal aneuploidies in PGT-A cycles is minimal. Indeed, we showed that only 2.4% of samples analyzed displayed single or multiple segmental aneuploidies as the only alteration, representing around 1% of the embryonic cohort in advanced reproductive age women (>38 years). Moreover, when taking into consideration the proportion of confirmed segmental aneuploidies in multifocal analyses, the overall incidence of sub-chromosomal abnormalities further reduces to 1.5%. It should be noted that our rate of segmental aneuploidies in the clinical biopsy is lower than the rates in several other reports.4,9,24 This discordance likely reflects differences in the technologies used to detected the partial genome changes25 as well as the fact that, in order to minimize the risk for overcalling aneuploidies in embryos, we have considered only segmental abnormalities in the uniform aneuploidy range (non-mosaic) and above 10Mb. Other studies used a less conservative approach which predicted segmental aneuploidies down to 2–5 Mb of chromosome resolution4,24 and also in the mosaicism range.4,9,24
Differently from whole-chromosome aneuploidies, and consistently with other reports,4,10 segmental abnormalities didn’t show a female-age-dependent increase in incidence and were more prevalent in larger chromosomes; this suggests a distinct etiology.26,27 Indeed, our data from multifocal analysis revealed that whole-chromosome aneuploidies were consistently detected across all blastocyst sections, showing minimal evidence of karyotype discordance and mosaicism incidence. In particular, only 1% of ICM and/or TE biopsies (4/390) showed a different aneuploidy pattern compared to the expected analytical profile (Figure 3B). These results corroborate previous studies which showed high concordance rates for whole-chromosome aneuploidies between TE re-biopsies and ICM (as well as embryo outgrowths on day 12) only when uniform aneuploidies were reported13,28 or more reliable criteria for aneuploidy classification was used.12 On the contrary, studies using wider ranges for mosaicism classification have reported lower representativeness of cTE biopsies toward ICM.28, 29, 30 Our results confirm the predominance of meiotic origin for whole-chromosome errors found at the blastocyst stage and highlight the high reliability and accuracy of blastocyst-stage PGT-A analysis when performed with standardized criteria for aneuploidy classification.31
In contrast, multifocal analysis revealed low concordance rates for segmental aneuploidies; this result suggests a true mitotic origin in around three quarters of cases. Indeed, both distinct aneuploidy patterns and reciprocal segmental alterations were detected in multifocal biopsies, providing clear evidence of mitotic non-disjunction events occurring during the early embryonic cell divisions. Mosaic segmental aneuploidies could originate from gross, structural rearrangements of chromosomes that could occur as a result of replication stress, catenenes, and ultrafine anaphase bridges.32, 33, 34 Although DNA damage and markers of replication stress have been reported in human preimplantation embryos,35 repair mechanisms are unclear. Further data will be needed to define time points, mechanisms, and potential susceptibility factors associated with mitotic errors leading to mosaic segmental aneuploidies in blastocyst-stage human embryos. The fact that different TE portions showed discordant PGT-A profiles raises several technical and biological questions regarding the diagnostic accuracy of detecting sub-chromosomal alterations from a single cTE biopsy. To account for this limitation, segmental and whole-chromosome aneuploidies will require separate consideration in future PGT-A predictivity studies. Indeed, because segmental aneuploidies frequently originate as a consequence of mitotic errors during preimplantation development, the observation of discordant intra-blastocyst results should be considered as an expected outcome.12,36 From a clinical standpoint, these data suggest that a diagnosis of segmental aneuploidy from a single cTE biopsy is not sufficient to correctly predict ICM chromosomal constitution or the clinical implications of the aneuploidy observed.
Currently, the clinical management of embryos which show a segmental aneuploidy as the only abnormality is extremely challenging because their transfer can potentially lead to serious adverse outcomes. In our study, 32% of all segmental aneuploidies detected were of meiotic origin, whereas an additional 28% of cases displayed the aberration in mosaic constitution but involved the ICM. Considering the potentially harmful consequences of transferring embryos with segmental aneuploidies,17 and the limited clinical data available to assess their reproductive potential,37,38 we have developed a risk stratification model that can facilitate the clinical decision-making process. As reported above, segmental length and diagnostic concordance with an independent scTE biopsy are valuable parameters for determining the validity of the finding and tailoring post-test genetic counselling. In particular, the confirmation of the same segmental finding in an scTE rebiopsy was shown to enhance the predictivity for an abnormal ICM from 50.9% to 84.0% (Figure 4A). On the contrary, the use of the segmental size alone, although statistically significant, showed to be a weaker predictor. None of the other potentially useful paramenters investigated, such as type and position on the chromosome involved and demographic data, aided the interpretation and management of segmental aneuplodies findings in a cTE. Therefore, in cases in which a segmental aneuploidy is identified as the only abnormality, the assessment of a second TE biopsy is the most effective approach for enhancing predictivity on ICM constitution and empowering the decision-making process. This model was verified clinically in an independent dataset of cases in which an scTE biopsy was collected following the original identification of a segmental aneuploidy in the cTE. This analysis aimed to investigate the validity of the model by identifying the proportion of cases in which the cTE-based diagnosis would be confirmed or contradicted by the scTE result. In the applied clinical verification phase, only 14% of cases remained with an ICM involvement risk similar to the a priori risk (Class II, segmentals > 80Mb and unconfirmed in scTE; Figure 4B). All other cases showed either a significantly higher risk (78%–100%, Classes III and IV, Figure 4B) or a reduced risk (Class I); this suggests the possibility of respectively using or excluding the embryo from clinical use with increased confidence. Concerning the clinical feasibility of this model, we have recently provided evidence that a second round of TE biopsy and cryopreservation is not expected to reduce implantation outcome or increase pregnancy complications.39 By improving the diagnostic outlook of single segmental aneuploidies, this predictive model could particularly benefit individuals with poor prognoses and few or no euploid embryos available for transfer following PGT-A.
The main limitation of this study lies in the inability to retrieve and analyze individual cells. Single-cell analysis would allow higher resolution to determine mosaicism configuration in the blastocyst. At present, efficient single cell isolation from human blastocysts remains a technical limitation for this type of studies, which will require development and validation. Moreover, in this study, we have only addressed the diagnostic predictive values of uniform segmental aneuploidies without including putative mosaic segmental configurations. This was due to our observations of the poor diagnostic performance of NGS when segmental aneuploid and euploid cell mixtures from cell lines were employed to mimic mosaicism.22 However, it can be speculated that chromosome CN values below the uniform aneuploid thresholds would provide much lower confirmation rates compared to uniform sub-chromosomal aneuploidies. Furthermore, it is possible that reciprocal segmental aneuploidy incidence is underestimated due to the multicellular nature of blastocyst biopsies.
A potential source of error, as described in single blastomeres by Van Der Aa and colleagues, involves S-phase artifacts, in which single-cell DNA replication domains can result in CN changes that may appear like segmental aneuploidy.40 Hence, the cell cycle phase of the analyzed cell should be taken into account when analyzing the NGS CN profile of single S-phase cells. These cells show CN variations across early and late replication domains, leading to a significantly increased detection of DNA imbalances compared with a cell in the G1- or G2/M-phase. These DNA imbalances may be falsely interpreted as genuine structural aberrations, thus leading to aneuploidy overcalling. However, in multi-cell populations like TE biopsies, G0- or G1-cells are generally the predominant class, and S-phase cells will usually not interfere with CN calling.40 Furthermore, even if a few cells collected in a TE biopsy are in the replication phase, this will likely appear as a mosaic, rather than a uniform segmental aneuploidy. As described, our study focused on the analysis of uniform segmental aneuploidies only, thus reducing the possibility of S-phase effect influence. Furthermore, replication domains for EBV-transformed lymphoblastoid cells based on the data from Ryba and colleagues showed an average size of 1.8 Mb,41 far below our chromosome resolution limit of 10 Mb.
The potential impact of biopsy on segmental aneuploidy detection, although theoretically unlikely, cannot be formally ruled out. Reduced proficiency in biopsy technique cannot explain why the same segmental abnormality could be detected throughout several biopsies. Additionally, because segmental abnormalities were detected in each of the chromosomes, poor biopsy technique could not specifically affect certain chromosomes. Furthermore, we have previously shown high inter- and intra-laboratory reproducibility in terms of cellularity and amplification efficiency in our embryo biopsy program,42 and these minimize the impact of the biopsy procedure on the final genetic result.
We have brought evidence that segmental aneuploidies are primarily of mitotic origin occurring during early embryo cleavage divisions. Consequently, the ensuing mosaic configuration of affected embryos poses challenges in their clinical management. Based on our experimental data, we have developed a risk stratification model for segmental aneuploidies. The resulting decisional diagram was applied clinically to assist diagnostic interpretation and clinical management of embryos exclusively showing sub-chromosomal alterations. Although their relative contribution to PGT-A cycles is low, future non-selection studies will be required to investigate the clinical predictive values of segmental abnormalities detected in single or double cTE biopsy, as well as their impact on embryonic reproductive potential and gestational risks.
Declaration of Interests
L.G., C.P., M.P., M.F., S.C., and A.C. are employed by Igenomix Italy S.R.L. L.R. and F.U. are shareholders of GENERA Health Care S.R.L. A.C. and C.R. are employed by Igenomix S.L. C.S. is the head of the Scientific Advisory Board of Igenomix. The other authors declare no competing interests.
Acknowledgments
E.H. is the recipient of a Novo Nordisk Foundation grant (16662).
Published: March 26, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.03.005.
Web Resources
None yet. Add here as needed.
Supplemental Data
References
- 1.Somigliana E., Busnelli A., Paffoni A., Vigano P., Riccaboni A., Rubio C., Capalbo A. Cost-effectiveness of preimplantation genetic testing for aneuploidies. Fertil. Steril. 2019;111:1169–1176. doi: 10.1016/j.fertnstert.2019.01.025. [DOI] [PubMed] [Google Scholar]
- 2.Forman E.J., Hong K.H., Ferry K.M., Tao X., Taylor D., Levy B., Treff N.R., Scott R.T., Jr. In vitro fertilization with single euploid blastocyst transfer: a randomized controlled trial. Fertil. Steril. 2013;100:100–107.e1. doi: 10.1016/j.fertnstert.2013.02.056. [DOI] [PubMed] [Google Scholar]
- 3.Scott R.T., Jr., Upham K.M., Forman E.J., Hong K.H., Scott K.L., Taylor D., Tao X., Treff N.R. Blastocyst biopsy with comprehensive chromosome screening and fresh embryo transfer significantly increases in vitro fertilization implantation and delivery rates: a randomized controlled trial. Fertil. Steril. 2013;100:697–703. doi: 10.1016/j.fertnstert.2013.04.035. [DOI] [PubMed] [Google Scholar]
- 4.Escribà M.J., Vendrell X., Peinado V. Segmental aneuploidy in human blastocysts: a qualitative and quantitative overview. Reprod. Biol. Endocrinol. 2019;17:76. doi: 10.1186/s12958-019-0515-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai H.H., Chuang T.H., Wong L.K., Lee M.J., Hsieh C.L., Wang H.L., Chen S.U. Identification of mosaic and segmental aneuploidies by next-generation sequencing in preimplantation genetic screening can improve clinical outcomes compared to array-comparative genomic hybridization. Mol. Cytogenet. 2017;10:14. doi: 10.1186/s13039-017-0315-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vera-Rodríguez M., Michel C.E., Mercader A., Bladon A.J., Rodrigo L., Kokocinski F., Mateu E., Al-Asmar N., Blesa D., Simón C., Rubio C. Distribution patterns of segmental aneuploidies in human blastocysts identified by next-generation sequencing. Fertil. Steril. 2016;105:1047–1055.e2. doi: 10.1016/j.fertnstert.2015.12.022. [DOI] [PubMed] [Google Scholar]
- 7.Malvestiti F., Agrati C., Grimi B., Pompilii E., Izzi C., Martinoni L., Gaetani E., Liuti M.R., Trotta A., Maggi F. Interpreting mosaicism in chorionic villi: results of a monocentric series of 1001 mosaics in chorionic villi with follow-up amniocentesis. Prenat. Diagn. 2015;35:1117–1127. doi: 10.1002/pd.4656. [DOI] [PubMed] [Google Scholar]
- 8.Ferreira J.C.P., Grati F.R., Bajaj K., Malvestiti F., Grimi M.B., Trotta A., Liuti R., Milani S., Branca L., Hartman J. Frequency of fetal karyotype abnormalities in women undergoing invasive testing in the absence of ultrasound and other high-risk indications. Prenat. Diagn. 2016;36:1146–1155. doi: 10.1002/pd.4951. [DOI] [PubMed] [Google Scholar]
- 9.Babariya D., Fragouli E., Alfarawati S., Spath K., Wells D. The incidence and origin of segmental aneuploidy in human oocytes and preimplantation embryos. Hum. Reprod. 2017;32:2549–2560. doi: 10.1093/humrep/dex324. [DOI] [PubMed] [Google Scholar]
- 10.Zhou S., Cheng D., Ouyang Q., Xie P., Lu C., Gong F., Hu L., Tan Y., Lu G., Lin G. Prevalence and authenticity of de-novo segmental aneuploidy (>16 Mb) in human blastocysts as detected by next-generation sequencing. Reprod. Biomed. Online. 2018;37:511–520. doi: 10.1016/j.rbmo.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 11.Kubicek D., Hornak M., Horak J., Navratil R., Tauwinklova G., Rubes J., Vesela K. Incidence and origin of meiotic whole and segmental chromosomal aneuploidies detected by karyomapping. Reprod. Biomed. Online. 2019;38:330–339. doi: 10.1016/j.rbmo.2018.11.023. [DOI] [PubMed] [Google Scholar]
- 12.Lawrenz B., El Khatib I., Liñán A., Bayram A., Arnanz A., Chopra R., De Munck N., Fatemi H.M. The clinicianś dilemma with mosaicism-an insight from inner cell mass biopsies. Hum. Reprod. 2019;34:998–1010. doi: 10.1093/humrep/dez055. [DOI] [PubMed] [Google Scholar]
- 13.Victor A.R., Griffin D.K., Brake A.J., Tyndall J.C., Murphy A.E., Lepkowsky L.T., Lal A., Zouves C.G., Barnes F.L., McCoy R.C., Viotti M. Assessment of aneuploidy concordance between clinical trophectoderm biopsy and blastocyst. Hum. Reprod. 2019;34:181–192. doi: 10.1093/humrep/dey327. [DOI] [PubMed] [Google Scholar]
- 14.Capalbo A., Wright G., Elliott T., Ubaldi F.M., Rienzi L., Nagy Z.P. FISH reanalysis of inner cell mass and trophectoderm samples of previously array-CGH screened blastocysts shows high accuracy of diagnosis and no major diagnostic impact of mosaicism at the blastocyst stage. Hum. Reprod. 2013;28:2298–2307. doi: 10.1093/humrep/det245. [DOI] [PubMed] [Google Scholar]
- 15.Ström S., Inzunza J., Grinnemo K.H., Holmberg K., Matilainen E., Strömberg A.M., Blennow E., Hovatta O. Mechanical isolation of the inner cell mass is effective in derivation of new human embryonic stem cell lines. Hum. Reprod. 2007;22:3051–3058. doi: 10.1093/humrep/dem335. [DOI] [PubMed] [Google Scholar]
- 16.Hovatta O. Derivation of human embryonic stem cell lines, towards clinical quality. Reprod. Fertil. Dev. 2006;18:823–828. doi: 10.1071/rd06075. [DOI] [PubMed] [Google Scholar]
- 17.Capalbo A., Rienzi L., Ubaldi F.M. Diagnosis and clinical management of duplications and deletions. Fertil. Steril. 2017;107:12–18. doi: 10.1016/j.fertnstert.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Capalbo A., Rienzi L. Mosaicism between trophectoderm and inner cell mass. Fertil. Steril. 2017;107:1098–1106. doi: 10.1016/j.fertnstert.2017.03.023. [DOI] [PubMed] [Google Scholar]
- 19.Goodrich D., Tao X., Bohrer C., Lonczak A., Xing T., Zimmerman R., Zhan Y., Scott R.T.J., Jr., Treff N.R. A randomized and blinded comparison of qPCR and NGS-based detection of aneuploidy in a cell line mixture model of blastocyst biopsy mosaicism. J. Assist. Reprod. Genet. 2016;33:1473–1480. doi: 10.1007/s10815-016-0784-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sox H.C., Blatt M.A., Higgins M.C., Marton K.I. Medical Decision Making. Clinical Chemistry. 1989;35:2020–2021. [Google Scholar]
- 21.Capalbo A., Rienzi L., Cimadomo D., Maggiulli R., Elliott T., Wright G. Correlation between standard blastocyst morphology, euploidy and implantation: An observational study in two centers involving 956 screened blastocysts. Hum. Reprod. 2014;29:1173–1181. doi: 10.1093/humrep/deu033. [DOI] [PubMed] [Google Scholar]
- 22.Rubio C., Rodrigo L., Garcia-Pascual C., Peinado V., Campos-Galindo I., Garcia-Herrero S., Simón C. Clinical application of embryo aneuploidy testing by next-generation sequencing. Biol. Reprod. 2019;101:1083–1090. doi: 10.1093/biolre/ioz019. [DOI] [PubMed] [Google Scholar]
- 23.Nevado J., Mergener R., Palomares-Bralo M., Souza K.R., Vallespín E., Mena R., Martínez-Glez V., Mori M.Á., Santos F., García-Miñaur S. New microdeletion and microduplication syndromes: A comprehensive review. Genet. Mol. Biol. 2014;37(1, Suppl):210–219. doi: 10.1590/s1415-47572014000200007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munné S., Blazek J., Large M., Martinez-Ortiz P.A., Nisson H., Liu E., Tarozzi N., Borini A., Becker A., Zhang J. Detailed investigation into the cytogenetic constitution and pregnancy outcome of replacing mosaic blastocysts detected with the use of high-resolution next-generation sequencing. Fertil. Steril. 2017;108:62–71.e8. doi: 10.1016/j.fertnstert.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 25.Vogel I., Blanshard R.C., Hoffmann E.R. SureTypeSC-a Random Forest and Gaussian mixture predictor of high confidence genotypes in single-cell data. Bioinformatics. 2019;35:5055–5062. doi: 10.1093/bioinformatics/btz412. [DOI] [PubMed] [Google Scholar]
- 26.Capalbo A., Hoffmann E.R., Cimadomo D., Ubaldi F.M., Rienzi L. Human female meiosis revised: new insights into the mechanisms of chromosome segregation and aneuploidies from advanced genomics and time-lapse imaging. Hum. Reprod. Update. 2017;23:706–722. doi: 10.1093/humupd/dmx026. [DOI] [PubMed] [Google Scholar]
- 27.Dumont M., Gamba R., Gestraud P., Klaasen S., Worrall J.T., De Vries S.G., Boudreau V., Salinas-Luypaert C., Maddox P.S., Lens S.M. Human chromosome-specific aneuploidy is influenced by DNA-dependent centromeric features. EMBO J. 2020;39:e102924. doi: 10.15252/embj.2019102924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Popovic M., Dheedene A., Christodoulou C., Taelman J., Dhaenens L., Van Nieuwerburgh F., Deforce D., Van den Abbeel E., De Sutter P., Menten B., Heindryckx B. Chromosomal mosaicism in human blastocysts: the ultimate challenge of preimplantation genetic testing? Hum. Reprod. 2018;33:1342–1354. doi: 10.1093/humrep/dey106. [DOI] [PubMed] [Google Scholar]
- 29.Chuang T.H., Hsieh J.Y., Lee M.J., Lai H.H., Hsieh C.L., Wang H.L., Chang Y.J., Chen S.U. Concordance between different trophectoderm biopsy sites and the inner cell mass of chromosomal composition measured with a next-generation sequencing platform. Mol. Hum. Reprod. 2018;24:593–601. doi: 10.1093/molehr/gay043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fragouli E., Munne S., Wells D. The cytogenetic constitution of human blastocysts: insights from comprehensive chromosome screening strategies. Hum. Reprod. Update. 2019;25:15–33. doi: 10.1093/humupd/dmy036. [DOI] [PubMed] [Google Scholar]
- 31.Capalbo A., Treff N.R., Cimadomo D., Tao X., Upham K., Ubaldi F.M., Rienzi L., Scott R.T., Jr. Comparison of array comparative genomic hybridization and quantitative real-time PCR-based aneuploidy screening of blastocyst biopsies. Eur. J. Hum. Genet. 2015;23:901–906. doi: 10.1038/ejhg.2014.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizuno K., Miyabe I., Schalbetter S.A., Carr A.M., Murray J.M. Recombination-restarted replication makes inverted chromosome fusions at inverted repeats. Nature. 2013;493:246–249. doi: 10.1038/nature11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan K.L., North P.S., Hickson I.D. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007;26:3397–3409. doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Putnam C.D., Srivatsan A., Nene R.V., Martinez S.L., Clotfelter S.P., Bell S.N., Somach S.B., de Souza J.E.S., Fonseca A.F., de Souza S.J., Kolodner R.D. A genetic network that suppresses genome rearrangements in Saccharomyces cerevisiae and contains defects in cancers. Nat. Commun. 2016;7:11256. doi: 10.1038/ncomms11256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kort D.H., Chia G., Treff N.R., Tanaka A.J., Xing T., Vensand L.B., Micucci S., Prosser R., Lobo R.A., Sauer M.V., Egli D. Human embryos commonly form abnormal nuclei during development: a mechanism of DNA damage, embryonic aneuploidy, and developmental arrest. Hum. Reprod. 2016;31:312–323. doi: 10.1093/humrep/dev281. [DOI] [PubMed] [Google Scholar]
- 36.Capalbo A. Careful and expert interpretation of PGT-A data can resolve the mosaicism dilemma. Hum. Reprod. 2019;34:2311–2312. doi: 10.1093/humrep/dez175. [DOI] [PubMed] [Google Scholar]
- 37.Fragouli E., Alfarawati S., Spath K., Babariya D., Tarozzi N., Borini A., Wells D. Analysis of implantation and ongoing pregnancy rates following the transfer of mosaic diploid-aneuploid blastocysts. Hum. Genet. 2017;136:805–819. doi: 10.1007/s00439-017-1797-4. [DOI] [PubMed] [Google Scholar]
- 38.Munné S., Spinella F., Grifo J., Zhang J., Beltran M.P., Fragouli E., Fiorentino F. Clinical outcomes after the transfer of blastocysts characterized as mosaic by high resolution Next Generation Sequencing- further insights. Eur. J. Med. Genet. 2020;63:103741. doi: 10.1016/j.ejmg.2019.103741. [DOI] [PubMed] [Google Scholar]
- 39.Cimadomo D., Rienzi L., Romanelli V., Alviggi E., Levi-Setti P.E., Albani E., Dusi L., Papini L., Livi C., Benini F. Inconclusive chromosomal assessment after blastocyst biopsy: prevalence, causative factors and outcomes after re-biopsy and re-vitrification. A multicenter experience. Hum. Reprod. 2018;33:1839–1846. doi: 10.1093/humrep/dey282. [DOI] [PubMed] [Google Scholar]
- 40.Van der Aa N., Cheng J., Mateiu L., Zamani Esteki M., Kumar P., Dimitriadou E., Vanneste E., Moreau Y., Vermeesch J.R., Voet T. Genome-wide copy number profiling of single cells in S-phase reveals DNA-replication domains. Nucleic Acids Res. 2013;41:e66. doi: 10.1093/nar/gks1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryba T., Hiratani I., Lu J., Itoh M., Kulik M., Zhang J., Schulz T.C., Robins A.J., Dalton S., Gilbert D.M. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010;20:761–770. doi: 10.1101/gr.099655.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Capalbo A., Ubaldi F.M., Cimadomo D., Maggiulli R., Patassini C., Dusi L., Sanges F., Buffo L., Venturella R., Rienzi L. Consistent and reproducible outcomes of blastocyst biopsy and aneuploidy screening across different biopsy practitioners: a multicentre study involving 2586 embryo biopsies. Hum. Reprod. 2016;31:199–208. doi: 10.1093/humrep/dev294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.