Abstract
Most existing expression quantitative trait locus (eQTL) mapping studies have been focused on individuals of European ancestry and are underrepresented in other populations including populations with African ancestry. Lack of large-scale well-powered eQTL mapping studies in populations with African ancestry can both impede the dissemination of eQTL mapping results that would otherwise benefit individuals with African ancestry and hinder the comparable analysis for understanding how gene regulation is shaped through evolution. We fill this critical knowledge gap by performing a large-scale in-depth eQTL mapping study on 1,032 African Americans (AA) and 801 European Americans (EA) in the GENOA cohort. We identified a total of 354,931 eSNPs in AA and 371,309 eSNPs in EA, with 112,316 eSNPs overlapped between the two. We found that eQTL harboring genes (eGenes) are enriched in metabolic pathways and tend to have higher SNP heritability compared to non-eGenes. We found that eGenes that are common in the two populations tend to be less conserved than eGenes that are unique to one population, which are less conserved than non-eGenes. Through conditional analysis, we found that eGenes in AA tend to harbor more independent eQTLs than eGenes in EA, suggesting potentially diverse genetic architecture underlying expression variation in the two populations. Finally, the large sample sizes in GENOA allow us to construct accurate expression prediction models in both AA and EA, facilitating powerful transcriptome-wide association studies. Overall, our results represent an important step toward revealing the genetic architecture underlying expression variation in African Americans.
Keywords: gene expression, lymphoblastoid cell lines, quantitative trait loci, eQTLs, eSNPs, eGenes, European American, African American, GENOA
Introduction
Genome-wide association studies (GWASs) have identified thousands of genetic variants that are associated with various diseases and disease-related complex traits. However, the vast majority of these disease-associated variants reside in non-coding regions and have unknown functions.1, 2, 3, 4 While variants in non-coding regions cannot directly alter the function of a gene through disrupting protein-coding sequencing, they can influence the level of gene expression through impacting the regulatory mechanisms underlying expression. Indeed, in recent years, expression quantitative trait loci (eQTL) mapping studies have successfully identified many cis-acting genetic variants that are associated with gene expression levels.5, 6, 7, 8 These identified eQTLs can help elucidate the molecular mechanisms underlying disease associations and facilitate the identification of biological pathways underlying disease etiology. For example, it has been shown that genetic variants associated with common diseases tend to be eQTLs and vice versa.1,9,10 In addition, identified eQTLs in mapping studies can also provide invaluable information to enhance the power of future GWASs.11
To date, most existing eQTL mapping studies have been performed on individuals with European ancestry. eQTL mapping studies in other populations are noticeably underrepresented, with a particularly noticeable absence of large studies in populations with African ancestry. Indeed, only a few eQTL mapping studies were carried out thus far on individuals with African ancestry and these studies often had small sample sizes that limited the statistical power of eQTL mapping. For example, HapMap3 included only 108 Yoruba (YRI) samples; the Geuvadis study included 89 YRI samples;12 a study on population difference in immune response collected 100 individuals with African ancestry;13 and the Multi-Ethnic Study of Atherosclerosis (MESA) cohort included 233 African Americans.14 Because of differences in allele frequencies and linkage disequilibrium (LD) patterns, eQTL mapping results can vary, sometimes quite substantially, across populations with diverse genetic backgrounds.15 Consequently, eQTLs identified in one population are not necessarily eQTLs in another population, and eQTL mapping results from one population may not necessarily benefit or transfer to another population. In addition, and equally importantly, a lack of eQTL mapping studies in populations with African ancestry also hinders the progress of comparative analysis between Africans and other populations in terms of the genetic architecture differences underlying gene expression variation. Indeed, only a limited number of comparative studies have been performed between populations, and again with small sample sizes.12,16,17 Comparative studies on the genetic regulation of gene expression across populations can provide important insights into the genetic differences among populations that may have been shaped by evolutionary forces. Therefore, well-powered eQTL mapping studies with large sample sizes in populations with African ancestry is critically needed to realize the potential and benefits of eQTL mapping studies across human populations.
To fill the above critical knowledge gap, here, we collected gene expression and genotype data from 1,032 African American (AA) samples and 801 European American (EA) samples from the Genetic Epidemiology Network of Arteriopathy (GENOA) study. We paired genotypes and gene expression data in these samples together to perform a comprehensive cis-eQTL mapping in both populations. By comparing eQTL mapping results in the two populations, our results reveal the genetic architecture differences underlying gene expression variation between African Americans and European Americans.
Material and Methods
Subjects
The Genetic Epidemiology Network of Arteriopathy (GENOA) is a community-based study of hypertensive sibships that was designed to investigate the genetics of hypertension and target organ damage. The study includes both African Americans (AA) from Jackson, Mississippi and European Americans (EA) from Rochester, Minnesota.18 In the initial phase of GENOA (phase I: 1996–2001), all members of sibships containing at least two individuals with essential hypertension clinically diagnosed before age 60 were invited to participate, including both hypertensive and normotensive siblings. Exclusion criteria for GENOA included secondary hypertension, alcoholism or drug abuse, pregnancy, insulin-dependent diabetes mellitus, or active malignancy. Eighty percent of AA (n = 1,482) and 75% of EA (n = 1,213) from the initial study population returned for the second examination (phase II: 2001–2005). Demographic information, medical history, clinical characteristics, lifestyle factors, and blood samples were collected in each phase. Written informed consent was obtained from all subjects and approval was granted by participating institutional review boards (University of Michigan, University of Mississippi Medical Center, and Mayo Clinic).
Genotyping Data and Quality Control
AA and EA blood samples were genotyped using either the Affymetrix Genome-wide Human SNP Array 6.0 platform or the Illumina Human1M-DUO Beadchip. For each platform, participants were excluded if they had an overall SNP call rate < 95% or sex mismatch between genotype and self-report. SNPs were excluded if they had a call rate < 95%. Principal component analysis was performed to identify and remove samples whose genotype profile appeared to be different from all other samples (outliers). After removing outliers, there were 1,599 AA samples and 1,464 EA samples with available genotype data. Imputation was performed using the Segmented HAPlotype Estimation & Imputation Tool (SHAPEIT19), v.2.r and IMPUTE v.220 using the 1000 Genomes project phase I integrated variant set release (v.3) in NCBI build 37 (hg19) coordinates (released on March 2012). Since genotyping was performed on multiple platforms, imputation was performed separately by platform and then the imputed data were combined. After imputation, SNPs with minor allele frequency (MAF) ≤0.01 or imputation quality score (info score) ≤0.4 in any platform-based imputation were removed. The final set of genotype data included 30,022,375 and 26,079,446 markers for AA and EA, respectively, covering both SNPs and SNVs/indels. In our eQTL mapping analysis (more details below), we focused on genotype information for 6,432,684 imputed cis-SNPs on 1,032 AA individuals, and genotype information for 3,818,520 imputed cis-SNPs on 801 European American individuals that also have gene expression data.
In EA and AA separately, the GENESIS package in R was used to infer population structure.21 We used the PC-AiR function to extract the first five genotype PCs and used GEMMA22 to estimate an individual relatedness matrix. Both PCs and the relatedness matrix were included as covariates in the eQTL mapping analysis.
Gene Expression Data and Quality Control
Since eQTL architectures can change dynamically during the development and differentiation of cells, it is essential to map eQTLs in purified cell types.23,24 In our study, gene expression levels were measured using lymphoblastoid cell lines (LCLs) from a subset of AAs (n = 1,233) and EAs (n = 919) in order to minimize environmental influences. The gene expression levels of AA samples were measured using the Affymetrix Human Transcriptome Array 2.0 and those of EA samples were measured using the Affymetrix Human Exon 1.0 ST Array. We used the Affymetrix Expression Console provided by Affymetrix for array quality control and all array images passed visual inspection. In AA, we removed 28 samples due to either low signal-to-noise ratio (n = 1), abnormal polyadenylated RNA spike-in controls (Lys < Phe < Thr < Dap; n = 24), sample mislabeling (n = 2), or low RNA integrity (n = 1), leaving a total of n = 1,205 for analysis. In EA, we removed duplicated samples (n = 31), control samples (n = 11), and sex mismatch samples (n = 2), leaving n = 875 for analysis. We processed data in each population separately. Specifically, raw intensity data were processed using the Affymetrix Power Tool software.25 Affymetrix CEL files were normalized using the Robust Multichip Average (RMA) algorithm which included background correction, quantile normalization, log2-transformation, and probe set summarization.26 The algorithm also includes GC correction (GCCN), signal space transformation (SST), and gain lock (value = 0.75) to maintain linearity. The Brainarray custom CDF27 version 19 (see Web Resources) was used to map the probes to genes. This custom CDF27 uses updated genomic annotations and multiple filtering steps to ensure that the probes used are specific for the intended gene cluster. In particular, it removes probes with non-unique matching cDNA/EST sequences that can be assigned to more than one gene cluster. Consequently, the gene expression data processed through the custom CDF in27 is expected to be largely free of mappability issues. However, we do acknowledge that alignment bias may still exist due to genetic variation, errors in the reference genome and other complications.28 After mapping, we used ComBat software29 to remove batch effects. Finally, the gene expression data were quantile normalized across genes for the following eQTL analysis. A total of 17,616 autosomal protein-coding genes in AA and 17,360 autosomal protein-coding genes in EA were available for analysis. In AA samples 17,572 genes and in EA samples 17,343 genes were mapped to the corresponding imputed SNPs.
eQTL Mapping Analysis
The eQTL mapping analysis was performed in AA and EA samples separately, using individuals with both genotype and gene expression data (n = 1,032 in AA and n = 801 in EA). For each gene, we first extracted cis-SNPs that are within 100 kb of transcription start site or transcription end site of genes, following recommendations by Peters et al.30 A total of 17,572 genes in AA and a total of 17,343 genes in EA had non-zero cis-SNPs. The median number of cis-SNPs per gene is 722 for AA (mean = 825.8; SD = 649.8) and 418 for EA (mean = 491.4, SD = 410.8), with range varying from 1 to 19,808 for AA and from 1 to 13,379 for EA. We focused our analysis on genes with at least one cis-SNP. For each gene, we then applied a linear mixed model implemented in GEMMA22 for eQTL mapping, adjusting for age, gender, the top five genotype PCs from PC-AiR, and the genetic relatedness matrix from GEMMA. Afterward, we selected the SNP with the lowest p value for each gene as the candidate eQTL and used its p value as the gene-level significance measure. We permuted the sample labels ten times and applied the same eQTL mapping procedure to obtain an empirical null distribution of gene-level p values.31, 32, 33 In each population, after each permutation, we kept the most significant p value per gene. With the empirical null distribution, we computed the false discovery rate (FDR) associated with each p value threshold following Barreiro et al.31 and Pickrell et al.32 and selected the p value threshold that provided a 5% FDR control. The p value threshold used is 6.245907e−05 in AA and 1.385504e−4 in EA. We refer to the genes that pass an FDR threshold of 5% as the identified eGenes and refer to the SNPs with the lowest p value in these genes as the identified (primary) eQTLs. We refer to the significant cis-SNPs in the eGenes as eSNPs. We also used Plink34 to calculate Weir and Cockerham’s Fst35 to measure the degree of population differentiation between AA and EA. Negative values of Weir and Cockerham’s Fst were treated as zero. The summary statistics from eQTL mapping analysis along with all analysis scripts are available on our website (see Web Resources).
We examined the overlap of the detected eGenes and eSNPs in AA or EA in other replication cohorts (AFA, CAU, and HIS in MESA; YRI and EUR in Geuvadis; details of these populations are described below). In particular, we used the qvalue method36,37 to estimate the expected true positive rate between populations. The statistics was estimated by selecting the SNP-gene pairs with FDR ≤ 0.05 in AA or EA population from the GENOA cohort and examining their p value distribution in each replication cohort (YRI and EUR in Geuvadis, AFA, CAU, HIS in MESA). is the proportion of false positives estimated by assuming a uniform distribution of null p values and .37
For each eGene in turn, we performed conditional analysis to identify additional conditional eQTLs following Jansen et al.33 To do so, we refer to the primary eQTLs as E1 SNPs. For each gene in turn, we performed association analysis conditional on the E1 SNP and identified the strongest SNP association among the remaining SNPs. We refer to the identified SNP as an E2 SNP if its conditional p value is below the genome-wide significance threshold established in the above paragraph. Afterward, we performed further association analysis conditional on both E1 and E2 SNPs to identify E3 SNPs. We repeated such process until the smallest p value among the remaining SNPs can no longer exceed the genome-wide significance threshold.
We performed subsampling analysis to check whether gene length is a potential source of bias in eQTL detection. In the first analysis, we focused on half of the genes that have a SNP number greater than or equal to the median value (8,802 genes in AA and 8,694 genes in EA). For each gene in turn, we down-sampled its cis-SNPs to the median value, so that all genes have the same number of cis-SNPs. In the second analysis, we focused on the genes with SNP density higher or equal to the median (8,787 genes in AA and 8,672 genes in EA). The SNP density is defined as the ratio between the number of SNPs in a gene and the length of that gene. Afterward, for each gene in turn, we randomly subsampled a specific number of SNPs to do the analysis, while the specific number is selected as the median SNP density multiple the gene length. For both subsampling analyses, we repeated the down-sampling procedure 10 times and averaged results to account for stochasticity in the down-sampling process.
Controlling for Local Ancestry
We performed local ancestry (LA) inference in AA and EA samples using the software Efficient Local Ancestry Inference (ELAI) v.1.0138 in the FRANC interface (see Web Resources). We used the default settings in ELAI. We downloaded genotype files in plink format for 83 Utah Residents (CEPH) with Northern and Western European Ancestry (CEU) and 88 Yoruba in Ibadan, Nigeria (YRI) populations from the 1000 Genomes Project to serve as reference panels. We focused on the common set of autosomal SNPs that are available both in the 1000 Genomes Project and in the AA or EA samples for ancestry inference. We converted variant base pair positions to centimorgans using the hg19 genetic map. The inferred local ancestry is in the value of the number of African ancestry alleles (0, 1, or 2) for each SNP. We treated these inferred values as an additional covariate in the eQTL mapping using the LAMatrix R package.39 Note that the LAMatrix software is not able to control for family relatedness in GENOA. In the analysis, we also constructed empirical null distributions as described above, computed the false discovery rate (FDR) associated with each p value threshold, and selected the p value threshold that provided a 5% FDR control. Such p value threshold is 5.962974e−05 in AA and 1.376416e−4 in EA. As described above, we refer to genes that pass an FDR threshold of 5% as the identified eGenes and refer to the SNPs with the lowest p value in these genes as the identified (primary) eQTLs. We also refer to the significant cis-SNPs in the eGenes as eSNPs.
Gene Expression Heritability Estimation and Partitioning
For each gene, we estimated the proportion of variance in gene expression level explained by all SNPs using the Bayesian sparse linear mixed model (BSLMM) implemented in GEMMA. Following Mogil et al.,14 we also used BSLMM40 to partition the gene expression variance into a cis-component that is explained by cis-SNPs and a trans-component that is explained by trans-SNPs. To do so, for each gene we fit the following model:
Where y is a n by 1 vector of gene expression levels for n individuals; is the intercept; is the by matrix of genotypes for cis-SNPs of interest and are the corresponding effect sizes; is the by matrix of genotypes for trans-SNPs of interest (i.e., SNPs that are not cis-SNPs); and are the corresponding effect sizes, here was based on all genotyped sites used in our analyses; and is a by vector of residual errors. We used 1,000 burn-in steps and 10,000 sampling steps in the Markov chain Monte Carlo (MCMC) algorithm to fit BSLMM. We used the posterior samples of and to calculate , which represents the proportion of variance in the phenotype explained by cis-SNPs, as well as , which represents the proportion of variance in the phenotype explained by trans-SNPs. Besides the main analysis where we used all trans-SNPs, we also performed sensitivity analysis where we used only trans-SNPs that reside on different chromosomes.
Conservation Scores
We obtained three types of conservation scores: phyloP score,41 phastCons score,42 and dN/dS ratio.43 The phyloP score measures the evolutionary conservation at each individual alignment site and the absolute phyloP score is a −log p value for testing the null hypothesis of neutral evolution. A positive sign of phyloP score indicates conservation and slower evolution than expectation, while a negative sign of phyloP score indicates faster evolution than expectation.44 The phastCons score measures the probability that each nucleotide belongs to a conserved element and aims to compare whether the site is better explained by the conserved model or by the non-conserved model. A higher phastCons score represents more conservation.42 The dN/dS score measures the direction and magnitude of nature selection on the protein-coding genes. A dN/dS ratio greater than 1 implies positive selection; a ratio less than 1 implies negative selection; while a ratio of exactly 1 indicates no selection.45 We obtained the per-site phyloP and phastCons scores from the 100-way vertebrate comparison on the UCSC Genome Browser46 for each base position inside the annotated exons and averaged them to obtain the per-gene phyloP and phastCons scores. We obtained per-gene dN and dS scores using the BioMart R package.47
We compared the conservation scores in eGenes that are identified in both populations, eGenes that are uniquely identified in one population, and non-eGenes. We performed Jonckheere-Terpstra test to test whether there is an observable trend in conservation scores across these three classes of genes.
Functional Enrichment Analysis
We performed GO and KEGG pathway enrichment analyses to investigate the shared biological function among eGenes in the AA and EA populations. We do so by using the g:GOSt tool on the web software g:Profiler and used the expressed genes as background.48 In the analysis, we used the default option g:SCS method in g:Profiler for multiple testing correction. We presented pathways identified with an adjusted p value < 1e−5. To adjust for the potential influence of gene length to the GO analysis, we also carried out GO enrichment using R package GOfuncR.49,50 In the analysis, we computed family-wise error rates (FWER) based on permutations of gene-associated variables and used an FWER threshold of 0.1 to declare enrichment significance.
Comparison of eQTL Results with Previous Studies
We compared our findings (eGenes, eSNPs, and eQTLs) to those from two previous eQTL mapping studies. These two previous studies include the Geuvadis Consortium study8 and the Multi-Ethnic Study of Atherosclerosis (MESA).14 The Geuvadis study was performed on lymphoblastoid cell lines (LCL) of 465 individuals from five different populations: Utah residents (CEPH) with northern and western European ancestry (CEU, n = 92), Finns (FIN, n = 95), British (GBR, n = 96), Toscani (TSI, n = 93), and Yoruba (YRI, n = 89). The MESA study was performed on CD14+ monocytes of individuals from three different populations: African American (AFA, n = 233), Hispanic (HIS, n = 352), and European (CAU, n = 578). In the Geuvadis results, we directly matched their reported Ensembl gene IDs to GENOA and matched SNPs between studies through their positions. In the MESA data, we directly matched their reported Ensembl gene IDs to GENOA and matched SNPs between studies through matching rs IDs.
Gene Expression Prediction
We constructed gene expression prediction models using AA and EA samples in either GENOA or MESA. We then accessed the prediction performance of these models in a separate study, the Geuvadis study. For MESA, we directly downloaded the cis-SNP weights. These weights were produced by fitting the elastic net model for gene expression prediction with PrediXcan in the MESA study.14 For GENOA, we followed the MESA study14 and used the glmnet R package51 to fit the elastic net model for gene expression prediction. Also following the MESA study, we set elastic net regularization penalty α = 0.5. Besides using the elastic net, we also used BSLMM40 for gene expression prediction. After building expression prediction models in either MESA and GENOA, we downloaded individual-level genotype and gene expression data from Geuvadis and examined the prediction performance there. To do so, we processed the Geuvadis gene expression data as described in Lappalainen et al.8 Specifically, we focused our analysis on protein-coding genes that are annotated from GENCODE52 (release 12). We removed lowly expressed genes that have zero counts in at least half of the individuals and retained a total of 15,810 genes. Afterward, we performed PEER normalization to remove confounding effects and unwanted variations.53 In order to remove potential population stratification in Geuvadis, we quantile normalized the gene expression measurements across individuals in each population to a standard normal distribution, and then quantile normalized the gene expression measurements to a standard normal distribution across individuals from all five populations. In addition to the gene expression data, all individuals in Geuvadis also have their genotypes sequenced in the 1000 Genomes project. Among the sequenced genotypes, we retained 7,072,917 SNPs that have a MAF above 0.05. We compared the prediction performance in a set of 2,524 common genes across all seven prediction models (GENOA AA and EA with BSLMM and elastic net; MESA AFA, CAU, and HIS with elastic net). We predicted the expression level of each gene in the Geuvadis data using the cis-SNP weights constructed in either GENOA or MESA, with overlap SNPs between GENOA and Geuvadis and between MESA and Geuvadis. We then measured the prediction performance using the squared Pearson’s correlation coefficient (R2) between the predicted expression level and true expression level as described in Mikhaylova and Thornton.54
TWAS Analysis in WTCCC
The Wellcome Trust Case Control Consortium (WTCCC) study55 data consist of about 14,000 case subjects from seven common diseases and 2,938 shared control subjects. The cases include 1,963 individuals with type 1 diabetes (T1D [MIM: 222100]), 1,748 individuals with Crohn disease (CD [MIM: 266600]), 1,860 individuals with rheumatoid arthritis (RA [MIM: 180300]), 1,868 individuals with bipolar disorder (BD [MIM: 125480]), 1,924 individuals with type 2 diabetes (T2D [MIM: 125853]), 1,926 individuals with coronary artery disease (CAD [MIM: 608320]), and 1,952 individuals with hypertension (HT [MIM: 145500]). We obtained quality-controlled genotypes from WTCCC and imputed missing genotypes using BIMBAM.56 We obtained a total of 458,868 SNPs shared across all individuals. We then imputed SNPs based on the 1000 Genomes project reference panel using SHAPEIT and IMPUTE2.20 For TWAS analysis, we focused on genes and SNPs that are shared between WTCCC and GENOA or shared between WTCCC and MESA. We calculated the predicted gene expression levels in WTCCC using models constructed either in GENOA (BSLMM or elastic net) or MESA (elastic net), with details described in the previous section. We then tested for association between the predicted gene expression level and disease status using logistic regression, with the first ten genetic PCs included as covariates. We considered the association between gene and disease genome-wide significant if its p value is below the Bonferroni corrected genome-wide threshold of 0.05. For results validation, for each prediction model in turn, we counted the number of genes identified in each WTCCC trait (T1D, T2D, RA, HT, CD, CAD, and BD) that is replicated in post-WTCCC studies. In particular, we defined replication as the genes within 100 kb of a previous gene reported to be associated with the same trait in the GeneCards knowledge base.57
Results
eQTL Mapping in AA and EA Samples in the GENOA Study
We performed eQTL mapping in the GENOA study in the AA and EA samples separately. The description of the GENOA study, the gene expression data collection and processing procedure, the genotype data collection and processing procedure, and the eQTL mapping procedure are all provided in Material and Methods. Briefly, the AA data include expression measurements for 17,616 protein-coding genes and genotype information for 30,022,375 imputed SNPs for 1,032 AA individuals. The EA data include 17,360 protein-coding genes and genotype information for 26,079,446 imputed SNPs for 801 EA individuals. We processed gene expression data with Combat29 to remove batch effects or other technical covariates. We extracted cis-SNPs within 100 kb of each gene and used linear mixed models implemented in GEMMA for eQTL mapping.22 In the analysis, we adjusted for age, gender, the top five genetic principal components (PCs), as well as a genetic relatedness matrix to control for familial relationships. Note that, following previous approaches,58 we determined the number of genotype PCs included in the model based on maximizing the number of discoveries (Figure S1). Overall, we examined a total of 17,572 genes and 6,432,684 unique cis-SNPs, with an average of 825.8 cis-SNPs per gene in the AA samples; and 17,343 genes and 3,818,520 unique cis-SNPs, with an average of 491.4 cis-SNPs per gene in the EA samples. Following Tung et al.,59 we refer to a gene that harbors at least one eQTL as an eGene. We used an empirical gene-level FDR threshold of 5% constructed across all genes for identifying eGenes. Following Tung et al.,59 we refer to the lowest p value SNP in each eGene as the (primary) eQTL. Following Barreiro et al.31 and Pickrell et al.,32 we refer to any cis-SNP with a significant association with the eGene as an eSNP. We used the p value threshold corresponding to the same empirical FDR of 5% for eGene detection to declare eSNPs. Besides this primary analysis, we also performed conditional analysis to identify additional eQTLs (more in the following section). The summary statistics from eQTL mapping analysis and all analysis scripts are available on our website (see Web Resources).
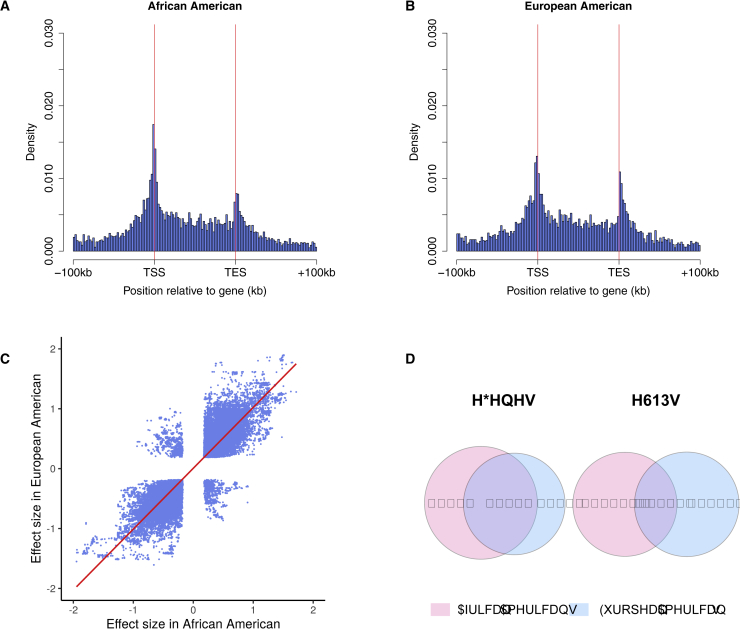
In total, we identified 5,475 eGenes in the AA samples and 4,402 eGenes in the EA samples, with 3,048 overlapping between AA and EA (overlapping Jaccard index = 0.446; Table 1 and Figure 1D). We also identified a total of 354,931 eSNPs in AA and 371,309 eSNPs in EA, with 112,316 eSNPs overlapping between the two populations (overlapping Jaccard index = 0.183). The proportion of overlapped eGenes increases from 53.01% to 59.15% in AA and increases from 66.81% to 69.05% in EA when the FDR threshold is increased from 0.01 to 0.2, though the proportion of overlapped eSNPs remains similar (Table S1). The lack of complete overlap of eGenes or eSNPs between the two populations is in part due to statistical power and in part due to the difference in the genetic architecture underlying gene expression levels between populations. In addition, our results are largely consistent with previous studies, with many eSNPs and eGenes in previous studies replicated in our study. Specifically, compared to the Geuvadis study,8 81.01% of eGenes and 84.01% of eSNPs identified in the Yoruba (YRI) population (n = 89) are also identified in our AA samples. In addition, 65.39% of eGenes and 63.4% of eSNPs identified in the European (EUR) population in Geuvadis (n = 373) are also identified in our EA samples (Table 2). Compared to the MESA study (AFA: n = 233; CAU: n = 578; HIS: n = 352),14 32.21% of eGenes and 24.89% of eSNPs identified in their AFA population are also identified in our AA samples. Also, 31.23% of eGenes and 19.33% of eSNPs identified in the CAU population in the MESA study are also identified in our EA samples (Table 3). However, our analysis also identified many new eGenes and eSNPs that were not identified in these previous studies. For example, compared to the Geuvadis study,8 we identified 4,271 new eGenes and 22,506 new eSNPs in our AA samples than that in the YRI population; 2,116 new eGenes and 55,017 new eSNPs in our EA samples than that in the EUR population. Similarly, compared to the MESA study, we identified 2,345 new eGenes and 146,651 new eSNPs in our AA samples as compared to that in the AFA samples; and 1,220 new eGenes and 99,298 new eSNPs in our EA samples as compared to that in the CAU samples. We also examined the true positive rate of the detected eSNPs in the AA or EA populations that are replicated in the MESA and Geuvadis studies. The results show the true positive rate of eSNPs is in the range of 59.3%–91.9% in AA (with values varying based on which population the comparison is performed on) and that in the 51.4%–90.9% in EA (Table S2). Certainly, a lack of complete overlap of eGenes or eSNPs among different studies is expected, given that statistical power is unlikely achieved fully in any study and that different studies differ in terms of the cis window size, used tissue types (e.g., MESA uses monocytes while the others use LCLs) as well as applied FDR methods (e.g., permutation based versus Benjamini-Hochberg).
Table 1.
Comparison of eQTL Mapping Results for African Americans and European Americans in the GENOA Study
| African American (AA) |
European American (EA) |
Overlapping |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Number | Total | Percentage | Number | Total | Percentage | Number | AA% | EA% | |
| eGenes | 5,475 | 17,572 | 31.16% | 4,402 | 17,343 | 25.38% | 3,048 | 55.67% | 69.24% |
| eSNPs | 354,931 | 14,511,338 | 2.45% | 371,309 | 8,521,801 | 4.36% | 112,316 | 31.64% | 30.25% |
The first row shows the number of eGenes that are identified in AA (first column), the total number of genes analyzed in AA (second column), the percentage of genes that are eGenes in AA (third column), the number of eGenes that are identified in EA (fourth column), the total number of genes analyzed in EA (fifth column), the percentage of genes that are eGenes in EA (sixth column), the number of common eGenes identified in both AA and EA (seventh column), the proportion of eGenes identified in AA that are also identified in EA (eighth column) and the proportion of eGenes identified in EA that are also identified in AA (ninth column), at FDR ≤0.05.
Figure 1.
Overview of eQTL Mapping Results in GENOA
(A) The locations of the eQTLs in the eQTL analysis are shown relative to the most 5′ gene transcription start site (TSS) and the most 3′ gene transcription end site (TES) for each of the 5,475 eGenes in AA.
(B) The locations of the eSNPs are shown relative to the most 5′ TSS and the most 3′ TES for each of the 4,402 eGenes in EA.
In both (A) and (B), identified eQTLs are enriched near TSS, TES, and gene body. For SNPs residing between TSS and TES, we scaled its distance to the transcript starting site by gene length.
(C) The effect size and direction of effect for the shared gene-SNP pairs (112,316) between AA and EA are highly consistent.
(D) Venn diagram plots show the overlap of eGenes and eSNPs identified in AA and EA.
Table 2.
Comparison of eQTL Mapping Results between GENOA and Geuvadis
| Genes (or Gene-SNP Pairs) Analyzed in Both Studies | Detected in Geuvadis | Detected in GENOA | Overlapped between the Two Studies | |
|---|---|---|---|---|
| GENOA AA (n = 1,205) versus Geuvadis YRI (n = 89) | ||||
| eGenes | 10,539 | 416 | 4,608 | 337 |
| eSNPs | 51,611 | 7,791 | 29,051 | 6,545 |
| GENOA EA (n = 801) versus Geuvadis EUR (n = 373) | ||||
| eGenes | 11,130 | 2,800 | 3,947 | 1,831 |
| eSNPs | 331,784 | 159,262 | 156,027 | 101,010 |
The first row shows the number of eGenes that are identified in GENOA AA and analyzed in Geuvadis YRI (first column), the number of eGenes in AA that are also eGenes in Geuvadis YRI (second column), the percentage of also eGenes in Geuvadis YRI that are also eGenes in AA (third column), the number of eGenes that are identified in GENOA EA and analyzed in Geuvadis EUR (first column), the number of eGenes in EA that are also eGenes in Geuvadis EUR (second column), the percentage of also eGenes in Geuvadis EUR that are also eGenes in EA (third column).
Table 3.
Comparison of eQTL Mapping Results between GENOA and MESA
| Genes (or Gene-SNP Pairs) Analyzed in Both Studies | Detected in MESA | Detected in GENOA | Overlapped between the Two Studies | |
|---|---|---|---|---|
| GENOA AA (n = 1,205) versus MESA AFA (n = 233) | ||||
| eGenes | 9,221 | 4,275 | 3,722 | 1,377 |
| eSNPs | 5,038,835 | 197,277 | 195,747 | 49,096 |
| GENOA EA (n = 801) versus MESA CAU (n = 578) | ||||
| eGenes | 9,151 | 5,559 | 2,956 | 1,736 |
| eSNPs | 3,154,038 | 533,989 | 202,503 | 103,205 |
The first row shows the number of eGenes that are identified in GENOA AA and analyzed in MESA AFA (first column), the number of eGenes in AA that are also eGenes in MESA AFA (second column), the percentage of also eGenes in MESA AFA that are also eGenes in AA (third column), the number of eGenes that are identified in GENOA EA and analyzed in MESA CAU (first column), the number of eGenes in EA that are also eGenes in MESA CAU (second column), the percentage of also eGenes in MESA CAU that are also eGenes in EA (third column).
We used ELAI to infer local ancestry and treated the inferred local ancestry as covariates in the eQTL mapping analysis using the LAMatrix package. We found that the eQTL mapping results controlling for local ancestry are largely consistent with the main results. Specifically, after adjusting for local ancestry, we identified 5,553 eGenes in AA and 4,586 eGenes in EA, with 5,312 and 4,333 genes overlapped with the main results (overlapping Jaccard index = 0.929 in AA and 0.931 in EA; Table S3). We identified a total of 357,072 eSNPs in AA and 372,240 eSNPs in EA, with 325,571 and 347,949 eSNPs overlapped with the main results (Jaccard index = 0.843 in AA and 0.880 in EA). The estimated effect sizes after adjusting for local ancestry are highly correlated with the main results (Pearson’s correlation = 0.991, p value < 2.23e−308 in AA; correlation = 0.994, p value < 2.23e−308 in EA; Figure S2). In addition, the −log10 p values are also highly correlated between these two approaches (Spearman’s correlation = 0.963, p value < 2.23e−308 in AA; correlation = 0.972, p value < 2.23e−308 in EA; Figure S3).
Characteristics of eGenes and eQTLs
We first examined the properties of the identified eQTLs and eSNPs. As expected,60 the eQTLs identified in both AA and EA samples are strongly enriched near gene transcription start sites, inside gene bodies, and near transcription end sites (Figures 1A and 1B), validating the eQTL mapping results. Within each population, the absolute eQTL effect size is negatively correlated with its minor allele frequency (MAF) (Pearson coefficient = −0.47, p value < 2.23e−308 in AA; Pearson coefficient = −0.46, p value < 2.23e−308 in EA; Figures S4A and S4B), likely reflecting either winner’s curse or negative selection.59 In addition, the significance level of the eQTLs in terms of −log10 p value is positively correlated with MAF in each of the two populations (Spearman’s coefficient = 0.414, p value < 2.23e−308 in AA; Spearman’s coefficient = 0.341, p value < 2.23e−308 in EA), likely reflecting the increased power with increasing MAF. Between populations, the −log10(p value) difference between the two populations is positively correlated with MAF difference (Spearman’s correlation = 0.07, p value < 2.23e−308; Figure S5). The average MAF difference between AA and EA is −0.0133 in non-eSNPs (Wilcoxon test p value < 2.23e−308), 0.0396 in AA-specific eSNPs (p value < 2.23e−308), −0.0496 in EA-specific eSNPs (p value < 2.23e−308), and −0.0053 in common eSNPs (p value = 9.15e−27; Figures S6 and S7), suggesting that AA-specific eSNPs tend to have higher MAF in AA than in EA while the EA-specific eSNPs tend to have higher MAF in EA than in AA. Consistent with Glassberg et al.,61 we also found that eSNPs tend to have higher MAF as compared to tested SNPs in both AA and EA (Figure S8). Besides the influence of MAF on power, we found that the effect sizes of the identified common eSNPs shared between EAs and AAs are highly correlated with each other (Pearson coefficient = 0.903; p value < 2.23e−308; Figure 1C), with 97.3% of eSNPs sharing the same effect sign between AA and EA. The significance level in terms of −log10 p value of the corresponding eSNPs in the two populations are also positively correlated (Spearman’s coefficient = 0.42, p value < 2.23e−308), partially reflecting the MAF correlation between the two populations (Pearson coefficient = 0.35, p value < 2.23e−308). In addition, we found that EA unique eSNPs tends to have larger effect size in EA as compared to AA, and vice versa (Figure S9). The primary eQTLs also tend to have the largest effect size across all cis-SNPs for a given gene: in AA, 428 (7.8%) out of the 5,475 primary eQTLs have the largest effect sizes (versus 0.15% by chance alone); in EA, 488 (11.09%) out of 4,402 primary eQTLs also have the largest effect sizes (versus 0.27% by chance alone). In addition, in AA, 1,105 (20.18%) out of the 5,475 primary eQTLs have the largest MAF across all SNPs mapped to a given gene. In EA, 715 (16.24%) out of 4,402 primary eQTLs also have the largest MAF across all SNPs mapped to a given gene.
We next examined the properties of the identified eGenes. Within each population, we first found that eGenes with longer length tended to have a higher number of eSNPs (p value in AA = 7.9e−04; p value in EA = 1.7e−3) but a lower density of eSNPs (p value in AA = 2.51e−13; p value in EA = 1.81e−8). Because we defined eGenes as genes that harbor at least one significant eSNP, longer genes with a larger number of SNPs will tend to be eGenes. Therefore, we performed subsampling-based analyses to avoid such potential confounding and more carefully examine the property of eGenes in terms of their gene length and SNP density (details in Material and Methods). The first subsampling analysis ensures that all analyzed genes have the same number of cis-SNPs. In such analysis, we found that the lowest p value for each gene is negatively correlated with gene length (before subsampling: Spearman’s correlation = −0.146 and −0.141 in AA and EA; p value = 2.31e−43 and 4.77e−40; after subsampling: correlation = −0.107 and −0.109; p value = 6.04e−24 and 1.9e−24) while positively correlated with SNP density (before subsampling: correlation = 0.135 and 0.118; p value = 3.48e−37 and 3.66e−28; after subsampling: correlation = 0.103 and 0.094; p value = 2.45e−22 and 1.912e−18), consistent with our above conclusion (note that a negative correlation between p value and gene length means that an eGene tends to have longer gene length). The second subsampling analysis ensures that all analyzed genes have the same SNP density. In such analysis, we found that the lowest p value for each gene is also negatively correlated with gene length (before subsampling: Spearman’s correlation = −0.267 and −0.269 in AA and EA; p value = 3.99e−143 and 7.13e−144; after subsampling: correlation = −0.458 and −0.425; p value < 2.23e−308 and < 2.23e−308). Overall, these subsampling-based analyses support the conclusion that an eGene tends to have a longer gene length and lower SNP density.
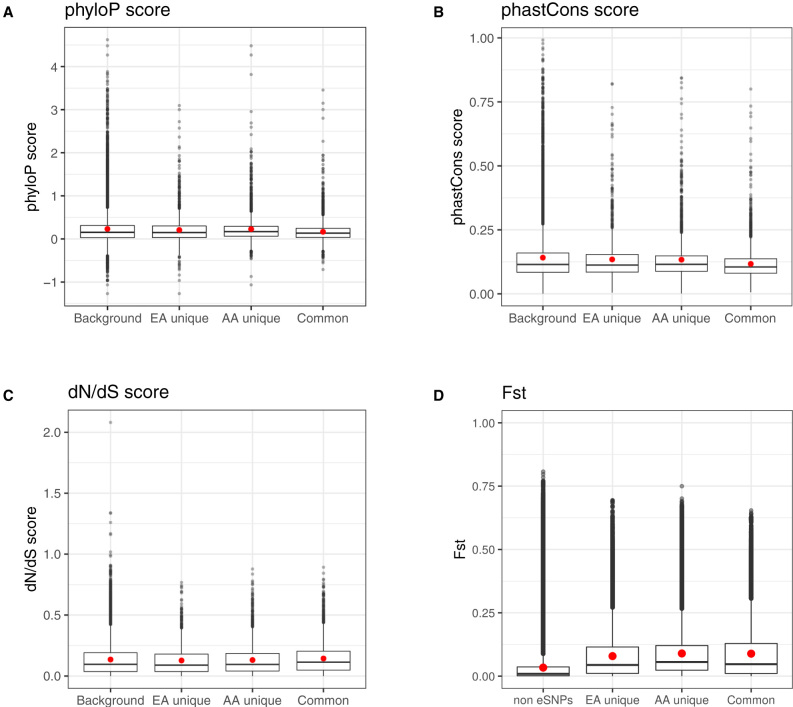
Next, between populations, we found that the common eGenes shared between the two populations are often less evolutionarily conserved than unique eGenes that are identified in a single population, which are also less evolutionarily conserved than non-eGenes. This decreased conservation pattern in non-eGenes versus unique eGenes versus common eGenes can be clearly visualized using each of the three commonly used conservation scores: phyloP (p value = 1.86e−07; Figure 2A), phastCons (p value = 1.6e−35; Figure 2B), and dN/dS ratio (p value = 2.94e−10; Figure 2C). For example, the mean phyloP score is 0.255, 0.207, 0.228, and 0.166, for non-eGenes, eGenes unique to EA, eGenes unique to AA, and eGenes shared between the two populations, respectively. The corresponding mean phastCons scores are 0.151, 0.135, 0.133, and 0.117 and the corresponding mean dN/dS ratios are 0.135, 0.127, 0.131, and 0.144. The decreased conservation in common eGenes shared between populations dovetails an early study in primates.59 In addition, we found that eSNPs in AA and EA tended to have a higher Fst value than non-eSNPs, supporting the previous observation that eSNPs are more variable than non-eSNPs13 (Figures 2D, S10A, and S10B). The mean Fst for the eSNPs shared between EA and AA, eSNPs unique to either EA or AA, and non-eSNPs are 0.0888, 0.079, 0.0895, and 0.0338, respectively (Figure 2D).
Figure 2.
The Relationship between Conservation Scores and Four Categories of Genes
(A) Boxplot of phyloP scores, together with the mean (red dot), across four groups of genes: eGenes that are shared between EA and AA; eGenes that are unique to either EA or AA; non-eGenes. phyloP scores are based on a 100-way primate genome comparison.
(B) Boxplot of phastCons scores, together with the mean (red dot), across the same four groups of genes. phastCons scores are based on a 100-way primate genome comparison.
(C) Boxplot of dN/dS ratio, together with the mean (red dot), across the same four groups of genes.
(D) Boxplot of Fst, together with the mean (red dot), across the same four groups of genes.
Note that a high phyloP score, a high phastCons score, or a low dN/dS ratio represents the gene is more conserved. The results show that the non-eGenes, which are treated as background genes, are most conserved. The eGenes unique in either European Americans (EA unique) or African Americans (AA unique) are less conserved. The common eGenes between European Americans and African Americans are the least conserved. The Jonckheere-Terpstra tests for testing such trend are significant for all these scores: p value = 2.94e−10 for dN/dS score; p value = 1.86e−07 for phyloP; p value = 1.6e−35 for phastCons; and p value < 2.23e−308 for Fst. Again, the red dots represent the mean values in each boxplot.
We also performed a gene ontology (GO) analysis on eGenes versus non-eGenes to examine whether eGenes are enriched in particular pathways (details in Material and Methods). We found that, in both EA and AA, the eGenes are highly enriched in catalytic activity, protein binding, and transferase activity among the GO molecular functions (Tables S4 and S8); are enriched in metabolic processes among the GO biological processes (Tables S5 and S9); and are enriched in intracellular part, cytoplasmic part, and mitochondrion among the GO cellular components (Tables S6 and S10). In human phenotype ontology analysis, we found that eGenes are enriched in autosomal-recessive inheritance in both AA and EA (Tables S7 and S11). The GO analysis results are consistent with the previous finding that eGenes tend to be less conserved, are enriched for targets of purifying selection, and are more likely to be observed in recessive disorders.62,63 In addition, the GO enrichment using R package GOfuncR49,50 that controls for the influence of gene length also yields consistent results (Tables S20–S22).
Gene Expression Heritability Estimation and Partitioning
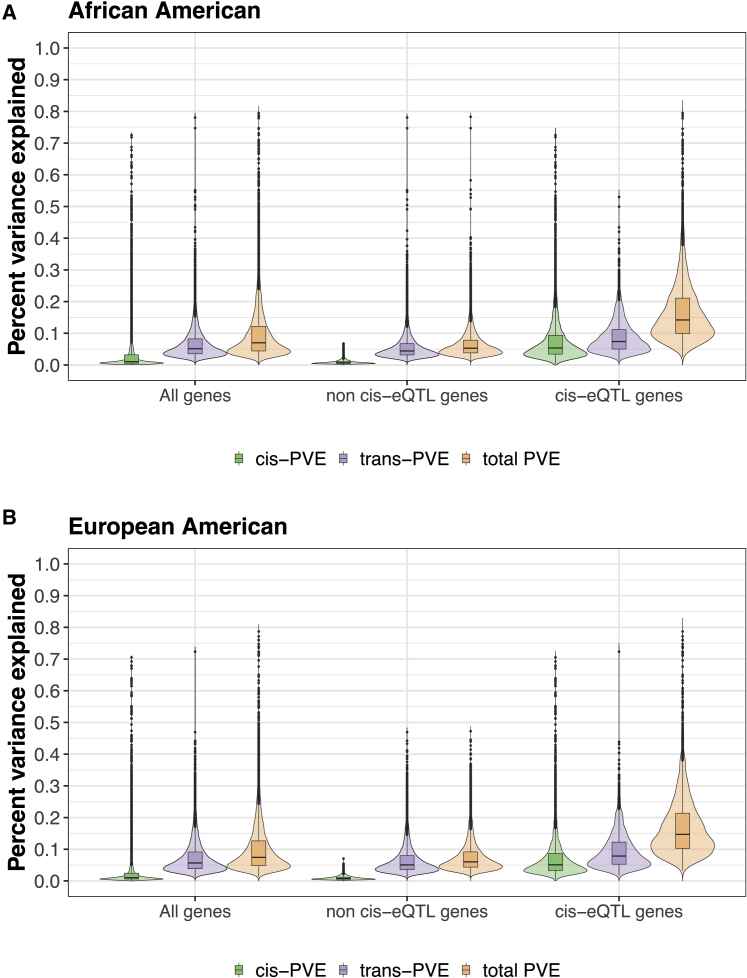
Next, we estimated the genetic architecture underlying gene expression variation through heritability estimation and partitioning. For each gene in turn, we estimated the proportion of variance (PVE) in gene expression levels that are accounted for by all SNPs using the Bayesian sparse linear mixed model (BSLMM). This quantity is commonly referred to as SNP heritability. We used Benjamini-Hochberg false discovery rates (FDR) to correct for multiple testing,64 with an FDR ≤ 0.05 used as the threshold for significant PVE. In the analysis, we found that 11.3% of genes in AA and 8.3% of genes in EA have a PVE that significantly deviates from zero at FDR ≤ 0.05. In AA, the median PVE is 24.6% across these significant genes (1,986 genes, mean = 26.67%; SD = 10.5%), with PVE estimates ranging from 8.49% to 79.46% (Figure S11A). In EA, the median PVE is 26.78% across these significant genes (1,440 genes, mean = 28.25%; SD = 9.88%), with PVE estimates ranging from 9.81% to 78.7% (Figure S11B). The PVE of tested common genes is generally consistent between AA and EA (Pearson’s correlation = 0.57, p value < 2.23e−308), and the PVE of common eGenes is also consistent between AA and EA (Pearson’s correlation = 0.502, p value = 2.69e−194) (Figures S12A and S12B). As one might expect, eGenes tend to have a higher PVE than non-eGenes (p value < 2.23e−308) (Figures 3A and 3B): the median PVE is 14.19% across eGenes and 5.28% across non-eGenes in AA, and is 14.7% across eGenes and 6.03% across non-eGenes in EA.
Figure 3.
Estimation and Partitioning of SNP Heritability Underlying Gene Expression Variation in African Americans and European Americans
Proportion of variance (PVE; aka SNP heritability) in gene expression levels estimated for all genes (left three plots), genes without detectable eQTLs (middle three plots), and genes with detectable eQTLs (right three plots) in African Americans (A) and European Americans (B). PVE explained by all SNPs is referred to as total PVE (orange), which is partitioned into a component that is explained by cis-SNPs (cis-PVE; green) and another component that is explained by trans-SNPs (trans-PVE; purple). As expected, PVE in eGenes tends to be larger than that in non-eGenes or in all genes. The total PVE as well as the cis-PVE in cis-eQTL genes also tend to be larger than that in non cis-eQTL genes or in all genes.
With BSLMM, we partitioned the PVE of each gene into two parts: one that is explained by cis-SNPs and the other that is explained by trans-SNPs. Consistent with previous studies,14 we found that the majority of PVE is explained by trans-SNPs, with only a fraction explained by cis-SNPs: the median proportion of PVE explained by cis-SNPs is only 1.03% (mean = 3.09%; SD = 5.44%) across all genes in AA and is 1.01% (mean = 2.63%; SD = 4.85%) across all genes in EA. As one might expect, cis-SNPs explain a higher proportion of PVE in eGenes than in non-eGenes. Specifically, the median proportion of PVE explained by cis-SNPs is 5.36% (mean = 7.93%; SD = 7.76%) across eGenes in AA and is 5.09% (mean = 7.61%; SD = 7.63%) across eGenes in EA. The proportion of PVE in eGenes explained by cis-SNPs is correlated between AA and EA (Pearson’s correlation coefficient = 0.62, p value < 2.23e−308), and the proportion of PVE in eGenes explained by trans-SNPs is also correlated between AA and EA (Pearson’s correlation coefficient = 0.18, p value = 1.33e−22). For most eGenes, the primary eQTLs is able to explain a large proportion of the total cis-PVE (the PVE explained by cis-SNPs) in both AA and EA populations (Figures S13 and S14): the primary eQTL explains a median of 64.52% cis-PVE in AA and 80.43% in EA. Note that the heritability estimates obtained using trans-SNPs residing on different chromosomes are almost identical to those obtained using all trans-SNPs (Figure S15). In particular, the cis heritability estimates obtained from these two approaches are highly correlated (0.9988 or 0.9937 in AA and EA, respectively) and so are the trans heritability estimates (correlation = 0.9989 or 0.9933 in AA and EA, respectively).
Independent cis-eQTLs rRevealed through Conditional Analysis
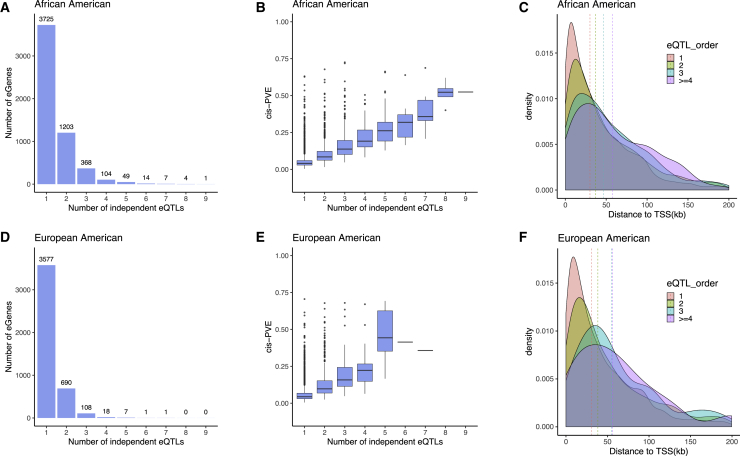
Because the primary eQTL does not fully explain cis-PVE, we performed conditional analysis to identify additional independent eQTLs for eGenes (details in Material and Methods). Through conditional analysis, we identified 8,070 independent eQTLs in AA and 5,401 in EA (Table 4); these include 2,595 conditional eQTLs in AA and 999 conditional eQTLs in EA, in addition to the primary eQTLs identified earlier. We found that most eGenes have only one independent eQTL (i.e., primary eQTL), with the proportion of eGenes with one eQTL lower in AA and higher in EA (68.03% in AA and 81.26% in EA; Fisher’s exact test p value < 2.23e−308; Figures 4A and 4D). A substantial proportion of eGenes have two independent eQTLs, with the proportion higher in AA and lower in EA (21.97% in AA and 15.67% in EA; p value = 1.11e−15). The remaining eGenes have three or more independent eQTLs and those eGenes are more likely to appear in AA than EA (9.99% in AA and 3.07% in EA; p value = 6.94e−45). In addition, the eGenes in AA tend to have a higher number of independent eQTLs: the average number of eQTLs per eGene is 1.47 in AA and 1.23 in EA (Wilcoxon test p value = 2.04e−56). The higher number of independent eQTLs for eGenes in AA may suggest a more complex gene regulatory mechanism in AA, although we also note that the higher number of independent eQTLs in AA may reflect in part the lower linkage disequilibrium among SNPs in AA and hence the higher statistical power in detecting conditional eQTLs in AA. By identifying conditional eQTLs, we can explain a higher proportion of total cis-PVE compared to that explained by primarily eQTLs only (Figures S13 and S14): both primary and conditional eQTL explains a median of 77.83% cis-PVE in AA and 86.28% in EA.
Table 4.
The Number of Independent eQTLs Identified in eGenes through the Conditional Analysis
| Number of Independent eQTLs |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| African American | 3,725 | 1,203 | 368 | 104 | 49 | 14 | 7 | 4 | 1 |
| European American | 3,577 | 690 | 108 | 18 | 7 | 1 | 1 | 0 | 0 |
Table lists the number of eGenes in AA (first row) and EA (second row) that contain different number of independent eQTLs (columns).
Figure 4.
Characterization of the Conditional eQTLs
(A and D) Histogram shows the number of eGenes (y axis) that harbor different numbers of independent eQTLs (x axis) in African Americans (A) or European Americans (D). We displayed eGenes that harbor up to nine independent eQTLs, with the detailed number of eQTLs listed above each bar. A large fraction of eGenes harbor a small number of independent eQTLs.
(B and E) The proportion of variance (PVE) in gene expression levels explained by SNPs are higher for eGenes that harbor a larger number of independent eQTLs in African Americans (B) or European Americans (E).
(C and F) Density plot shows the distance from eQTL to the transcription start site (TSS) of the corresponding eGene. The density plot is stratified by the number of eQTLs: eGenes with one independent eQTL is colored in red; eGenes with two independent eQTL is colored in green; eGenes with three independent eQTL is colored in blue; eGenes with four or more independent eQTL is colored in purple. Dashed lines represent the median distance between eQTL and TSS in the four stratified groups in African Americans (C) or European Americans (F).
As one might expect,65 the conditional eQTLs reside farther away from the TSS compared to the primary eQTLs, though they are still enriched around the TSS when compared with non-eQTLs (Figures 4C and 4F). The number of independent eQTLs across genes in the conditional analysis is positively correlated with the number of eSNPs in the unconditional analysis, more so in AA than in EA (Pearson’s correlation coefficient = 0.41, p value = 3.34e−218 in AA; correlation = 0.18, p value = 1.88e−22 in EA); and positively correlated with the gene length, though to a much lesser extent (Spearman’s correlation between log10 transformed gene length and number of independent eQTLs = 0.08, p value = 0.57 in AA; correlation = 0.034, p value = 0.06 in EA; Figure S16). The number of independent eQTLs across eGenes is also positively correlated with the cis-PVE of each eGene, more so in AA than in EA (Pearson’s correlation coefficient = 0.57, p value < 2.23e−308 in AA; correlation = 0.41, p value = 1.21e−123 in EA; Figures 4B and 4E). The eGenes with more independent eQTLs are less conserved: the number of independent eQTLs across eGenes is positively correlated with the dN/dS scores (Pearson’s correlation = 0.047, p value = 5e−04 in AA; correlation = 0.001, p value = 0.94 in EA); and negatively correlated with the conservation score (phyloP score: Pearson’s correlation coefficient = −0.082, p value = 1.17e−11 in AA; correlation = −0.11, p value = 4.57e−09 in EA; phastCons score: correlation = −0.078, p value = 7.6e−09 in AA; correlation = −0.086, p value = 2.06e−06 in EA).
Large Sample Size in GENOA Enables More Accurate Gene Expression Prediction
Finally, we illustrate how the large sample size in both AA and EA populations in GENOA can allow us to construct accurate gene expression prediction models, thus potentially facilitating powerful transcriptome-wide association analysis (TWAS).66 To do so, we constructed gene expression prediction models for one gene at a time in AA and EA separately. Each prediction model uses all cis-SNPs as covariates and is constructed using BSLMM. Afterward, we evaluated the performance of these prediction models for the same gene in a separate eQTL mapping study, the Geuvadis study. The Geuvadis study consists of five different populations that include CEPH (CEU, n = 92), Finns (FIN, n = 95), British (GBR, n = 96), Toscani (TSI, n = 93), and Yoruba (YRI, n = 89). We evaluated the performance of the prediction models constructed in EA and AA separately in each of the five populations. In each analysis, we calculated the coefficients of determination (R2) between the predicted gene expression and the observed gene expression to measure prediction performance.66 As expected,67 we found that genes with high heritability tend to be predicted with high accuracy. For example, the prediction R2 achieved using GENOA AA samples is positively correlated with PVE across all genes, in each of the five populations in Geuvadis (mean correlation across five populations = 0.36, SD = 0.02; p value < 2.23e−308); similar patterns were observed with the model constructed based on GENOA EA samples (Figure S17). Also as expected,14 we found that the expression prediction models constructed in a population often perform well in the population of the same ancestry. For example, the prediction models based on EA performed better for predicting expression levels in FIN and CEU than in other populations (Tables S26 and S27), while the models constructed based on AA performed better in YRI than in other populations (Figure 5A and Table S23). The better performance of AA models in YRI highlights the need for eQTL mapping studies in the African American population.
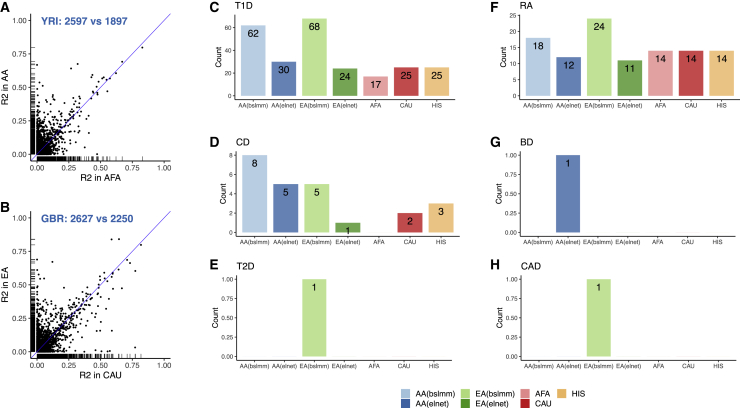
Figure 5.
Application of GENOA eQTL Mapping Results in Gene Expression Prediction and TWAS in WTCCC
(A) Comparison of the prediction performance measured by R2 using GENOA AA (y axis) and MESA AFA (x axis) eQTL mapping results for the Geuvadis YRI data. The panel also lists the number of genes where AA performs better (2,597) and the number of genes where AA performs worse than AFA (1,897). Here we compare the commonly predicted genes between AA and AFA.
(B) Comparison of the prediction performance measured by R2 using GENOA EA (y axis) and MESA CAU (x axis) eQTL mapping results for the Geuvadis GBR data. The panel also lists the number of genes where EA performs better (2,627) and the number of genes where EA performs worse (2,250) than CAU. Here we compare the commonly predicted genes between EA and CAU.
(C–H) Barplots display the number of significant genes from TWAS analysis using gene expression models constructed based on different populations. The significant genes are those passing the genome-wide significance threshold via Bonferroni correction (α = 0.05/number of genes tested) in each of the seven common diseases in WTCCC that include Crohn disease (CD), rheumatoid arthritis (RA), bipolar disorder (BD), type 1 diabetes (T1D), type 2 diabetes (T2D), coronary artery disease (CAD), and hypertension (HT). The result for HT is not shown since no gene was identified in any population. The five populations used to construct the gene expression models include GENOA AA BSLMM (light blue), AA elastic net (deep blue), GENOA EA BSLMM (light green), GENOA EA elastic net (deep green), MESA AFA (pink), MESA CAU (red), and MESA HIS (yellow).
Besides GENOA, we also obtained previously constructed gene expression prediction models using elastic net in the MESA study and evaluated their prediction performance in Geuvadis. The MESA study consists of three populations: African American (AFA, n = 233), Hispanic (HIS, n = 352), and European (CAU, n = 578). For a fair comparison, we also constructed gene expression prediction models using elastic net in GENOA, in addition to using BSLMM above. In the analysis, we found that the gene prediction models constructed based on GENOA AA or EA samples outperform those constructed based on the MESA AFA, CAU, or HIS (Tables S23–S27). For example, for predicting gene expression in YRI, models constructed based on AA in GENOA with elastic net achieve a prediction R2 above 0.1 in 337 genes, which represents a 75.5% gain compared to the prediction models constructed based on the AFA population in MESA with elastic net (Table S23). Similarly, for predicting gene expression in GBR, models constructed based on EA in GENOA with elastic net achieve a prediction R2 above 0.1 in 415 genes, which represents a 30.1% gain compared to the prediction models constructed based on the CAU population in MESA with elastic net (Table S24).
The accurate expression prediction performance based on GENOA also translates to a high power for TWAS analysis.66 To illustrate the TWAS power gain brought by GENOA, we applied the prediction models constructed in each of the five populations (GENOA: AA and EA; MESA: AFA, HIS, and CAU) to seven common diseases collected from a GWAS case control study: the WTCCC.55 These seven diseases include Crohn disease (CD), rheumatoid arthritis (RA), bipolar disorder (BD), type 2 diabetes (T2D), coronary artery disease (CAD), and hypertension (HT). For each gene and disease pair in turn, we tested for the association between the predicted gene expression and disease status using logistic regression, with the first ten genetic PCs included as covariates. Overall, we found that models constructed based on GENOA identified more associations with the seven common diseases compared to models constructed based on MESA (Figures 5C–5H and S18 and Table S28). For example, using elastic net for constructing the gene prediction models in GENOA AA, we identified a total of 48 genes in WTCCC, among which 42 are reported to be associated with the same trait in the GeneCards database.57 Using the same elastic net for constructing the gene prediction models in MESA AFA, we identified a total of 40 genes in WTCCC, among which 37 are reported in the GeneCards. Table S29 lists the significant genes identified by TWAS analysis using GENOA AA samples, the majority of which have also been identified in much larger-scale GWASs.68, 69, 70
Discussion
We have presented a comprehensive eQTL mapping analysis in GENOA. Our study is a large eQTL mapping study performed in the African American population. The large AA sample size in GENOA allows us to identify a substantial number of eQTLs and eGenes in AA, many of which were not identified in previous AA eQTL mapping studies.8 In addition, we identified a higher number of eGenes in AA than in EA, likely due to the larger sample size, higher number of cis-SNPs per gene, and/or potentially higher diversity in AA. Importantly, only a small percentage of the significant gene-SNP pairs identified in AA are also identified in EA, highlighting the importance of eQTL mapping with AA samples. The large AA sample size also allows us to construct accurate gene expression prediction models in the African American population, facilitating powerful TWAS analysis there. These analyses and results enhance our understanding of the genetic architecture underlying gene expression variation and facilitate the future investigation of the causal molecular mechanisms underlying common diseases and disease-related complex traits.
The availability of both large-scale AA and EA samples in GENOA allows us to perform comparisons between these two populations. We found that eGenes with multiple independent eQTLs are often less conserved and eGenes shared between AA and EA are also less conserved. Indeed, eGenes are depleted from genes with crucial roles in regulating cell functions.65 The comparison results highlight the importance of negative selection, which constrains biologically important regions, removes large-effect regulatory variants, and reshapes the genetic architecture.36 Through comparison, we found that substantial differences exist between the two populations of AA and EA. Specifically, despite the similar sample sizes between AA and EA, we identified a higher number of independent eQTLs in AA than in EA through conditional analysis. A higher number of independent eQTLs in AA supports the potentially more complex regulatory mechanisms underlying gene expression in AA. While the identified eQTLs vary across populations, the shared eQTLs in the AA and EA populations nevertheless often share similar effect sizes and effect directions. In addition, the gene expression prediction models constructed based on AA apply reasonably accurately for gene expression prediction in EA, and vice versa. Therefore, at least part of the eQTL mapping results from one population can be transferred to the other populations.12 Further integrating the GENOA study with other previous studies for joint eQTL mapping or joint TWAS analysis is an important direction for future exploration.
Finally, while we have identified many primary eQTLs in the main analysis, we acknowledge that the identified primary eQTLs do not explain all cis-SNP heritability in eGenes (median = 64.52% in AA and 80.43% in EA). We can identify many additional eQTLs through conditional analysis. However, these conditional eQTLs in addition to the primary eQTLs again cannot explain all cis-SNP heritability (median = 77.83% AA and 86.28% in EA). In addition, the cis-SNP heritability only represents a small proportion of total SNP heritability, suggesting that a large fraction of SNP heritability remains largely unidentified. The incomplete cis-heritability explained by identified eQTLs support the likely polygenic architecture underlying gene expression variation. Therefore, future studies with larger sample sizes are necessary to fully capture the genetic architecture underlying gene expression variation.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
Support for the Genetic Epidemiology Network of Arteriopathy (GENOA) data collection and analysis was provided by the National Heart, Lung and Blood Institute (HL054457, HL100185, HL119443, and HL133221) and the National Institute of Neurological Disorders and Stroke (NS041558) of the National Institutes of Health. This study was also partially supported by the National Human Genome Research Institutes grant R01HG009124 and National Science Foundation grant DMS1712933. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Published: March 26, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.03.002.
Contributor Information
Sharon L.R. Kardia, Email: skardia@umich.edu.
Xiang Zhou, Email: xzhousph@umich.edu.
Accession Numbers
The accession numbers for the gene expression data used in this analysis are Gene Expression Omnibus (GEO): GSE138914 for AA and GSE49531 for EA. The accession number for the SNP data used in this analysis is Database of Genotypes and Phenotypes (dbGaP): phs001238.v2.p1. Due to IRB restriction, mapping of the sample IDs between genotype data (dbGaP) and gene expression data (GEO) cannot be provided publicly but are available upon written request to JS and SK.
Web Resources
Brainarray, http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF
FRANC interface, http://web.cbio.uct.ac.za/ITGOM/franc/
GEMMA, https://www.xzlab.org/software.html
Geuvadis eQTL mapping results, http://jungle.unige.ch/∼lappalainen/geuvadis/
MESA cis-SNP weights, https://github.com/WheelerLab/DivPop/tree/master/unfiltered_dbs
MESA eQTL mapping results, https://www.dropbox.com/sh/f6un5evevyvvyl9/AAA3sfa1DgqY67tx4q36P341a?dl=0
OMIM, https://www.omim.org/
Zhou lab: summary statistics from eQTL mapping analysis along with all analysis scripts, http://www.xzlab.org/data.html
Supplemental Data
References
- 1.Nica A.C., Montgomery S.B., Dimas A.S., Stranger B.E., Beazley C., Barroso I., Dermitzakis E.T. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 2010;6:e1000895. doi: 10.1371/journal.pgen.1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gusev A., Lee S.H., Trynka G., Finucane H., Vilhjálmsson B.J., Xu H., Zang C., Ripke S., Bulik-Sullivan B., Stahl E., Schizophrenia Working Group of the Psychiatric Genomics Consortium. SWE-SCZ Consortium. Schizophrenia Working Group of the Psychiatric Genomics Consortium. SWE-SCZ Consortium Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am. J. Hum. Genet. 2014;95:535–552. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torres J.M., Gamazon E.R., Parra E.J., Below J.E., Valladares-Salgado A., Wacher N., Cruz M., Hanis C.L., Cox N.J. Cross-tissue and tissue-specific eQTLs: partitioning the heritability of a complex trait. Am. J. Hum. Genet. 2014;95:521–534. doi: 10.1016/j.ajhg.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y.I., van de Geijn B., Raj A., Knowles D.A., Petti A.A., Golan D., Gilad Y., Pritchard J.K. RNA splicing is a primary link between genetic variation and disease. Science. 2016;352:600–604. doi: 10.1126/science.aad9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schadt E.E., Monks S.A., Drake T.A., Lusis A.J., Che N., Colinayo V., Ruff T.G., Milligan S.B., Lamb J.R., Cavet G. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 6.Morley M., Molony C.M., Weber T.M., Devlin J.L., Ewens K.G., Spielman R.S., Cheung V.G. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Battle A., Mostafavi S., Zhu X., Potash J.B., Weissman M.M., McCormick C., Haudenschild C.D., Beckman K.B., Shi J., Mei R. Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res. 2014;24:14–24. doi: 10.1101/gr.155192.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lappalainen T., Sammeth M., Friedländer M.R., ’t Hoen P.A.C., Monlong J., Rivas M.A., Gonzàlez-Porta M., Kurbatova N., Griebel T., Ferreira P.G., Geuvadis Consortium Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emilsson V., Thorleifsson G., Zhang B., Leonardson A.S., Zink F., Zhu J., Carlson S., Helgason A., Walters G.B., Gunnarsdottir S. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 10.Nicolae D.L., Gamazon E., Zhang W., Duan S., Dolan M.E., Cox N.J. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hao X., Zeng P., Zhang S., Zhou X. Identifying and exploiting trait-relevant tissues with multiple functional annotations in genome-wide association studies. PLoS Genet. 2018;14:e1007186. doi: 10.1371/journal.pgen.1007186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stranger B.E., Montgomery S.B., Dimas A.S., Parts L., Stegle O., Ingle C.E., Sekowska M., Smith G.D., Evans D., Gutierrez-Arcelus M. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 2012;8:e1002639. doi: 10.1371/journal.pgen.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quach H., Rotival M., Pothlichet J., Loh Y.E., Dannemann M., Zidane N., Laval G., Patin E., Harmant C., Lopez M. Genetic Adaptation and Neandertal Admixture Shaped the Immune System of Human Populations. Cell. 2016;167:643–656.e17. doi: 10.1016/j.cell.2016.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mogil L.S., Andaleon A., Badalamenti A., Dickinson S.P., Guo X., Rotter J.I., Johnson W.C., Im H.K., Liu Y., Wheeler H.E. Genetic architecture of gene expression traits across diverse populations. PLoS Genet. 2018;14:e1007586. doi: 10.1371/journal.pgen.1007586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelly D.E., Hansen M.E.B., Tishkoff S.A. Global variation in gene expression and the value of diverse sampling. Curr. Opin. Syst. Biol. 2017;1:102–108. doi: 10.1016/j.coisb.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stranger B.E., Nica A.C., Forrest M.S., Dimas A., Bird C.P., Beazley C., Ingle C.E., Dunning M., Flicek P., Koller D. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spielman R.S., Bastone L.A., Burdick J.T., Morley M., Ewens W.J., Cheung V.G. Common genetic variants account for differences in gene expression among ethnic groups. Nat. Genet. 2007;39:226–231. doi: 10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daniels P.R., Kardia S.L.R., Hanis C.L., Brown C.A., Hutchinson R., Boerwinkle E., Turner S.T., Genetic Epidemiology Network of Arteriopathy study Familial aggregation of hypertension treatment and control in the Genetic Epidemiology Network of Arteriopathy (GENOA) study. Am. J. Med. 2004;116:676–681. doi: 10.1016/j.amjmed.2003.12.032. [DOI] [PubMed] [Google Scholar]
- 19.Delaneau O., Zagury J.F., Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 20.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conomos M.P., Reiner A.P., Weir B.S., Thornton T.A. Model-free Estimation of Recent Genetic Relatedness. Am. J. Hum. Genet. 2016;98:127–148. doi: 10.1016/j.ajhg.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou X., Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012;44:821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ackermann M., Sikora-Wohlfeld W., Beyer A. Impact of natural genetic variation on gene expression dynamics. PLoS Genet. 2013;9:e1003514. doi: 10.1371/journal.pgen.1003514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerrits A., Li Y., Tesson B.M., Bystrykh L.V., Weersing E., Ausema A., Dontje B., Wang X., Breitling R., Jansen R.C., de Haan G. Expression quantitative trait loci are highly sensitive to cellular differentiation state. PLoS Genet. 2009;5:e1000692. doi: 10.1371/journal.pgen.1000692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lockstone H.E. Exon array data analysis using Affymetrix power tools and R statistical software. Brief. Bioinform. 2011;12:634–644. doi: 10.1093/bib/bbq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai M., Wang P., Boyd A.D., Kostov G., Athey B., Jones E.G., Bunney W.E., Myers R.M., Speed T.P., Akil H. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33 doi: 10.1093/nar/gni179. e175–e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saha A., Battle A. False positives in trans-eQTL and co-expression analyses arising from RNA-sequencing alignment errors. F1000Res. 2018;7:1860. doi: 10.12688/f1000research.17145.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 30.Peters J.E., Lyons P.A., Lee J.C., Richard A.C., Fortune M.D., Newcombe P.J., Richardson S., Smith K.G.C. Insight into Genotype-Phenotype Associations through eQTL Mapping in Multiple Cell Types in Health and Immune-Mediated Disease. PLoS Genet. 2016;12:e1005908. doi: 10.1371/journal.pgen.1005908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barreiro L.B., Tailleux L., Pai A.A., Gicquel B., Marioni J.C., Gilad Y. Deciphering the genetic architecture of variation in the immune response to Mycobacterium tuberculosis infection. Proc. Natl. Acad. Sci. USA. 2012;109:1204–1209. doi: 10.1073/pnas.1115761109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pickrell J.K., Marioni J.C., Pai A.A., Degner J.F., Engelhardt B.E., Nkadori E., Veyrieras J.B., Stephens M., Gilad Y., Pritchard J.K. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jansen R., Hottenga J.J., Nivard M.G., Abdellaoui A., Laport B., de Geus E.J., Wright F.A., Penninx B.W.J.H., Boomsma D.I. Conditional eQTL analysis reveals allelic heterogeneity of gene expression. Hum. Mol. Genet. 2017;26:1444–1451. doi: 10.1093/hmg/ddx043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holsinger K.E., Weir B.S. Genetics in geographically structured populations: defining, estimating and interpreting F(ST) Nat. Rev. Genet. 2009;10:639–650. doi: 10.1038/nrg2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Battle A., Brown C.D., Engelhardt B.E., Montgomery S.B., GTEx Consortium. Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group. Statistical Methods groups—Analysis Working Group. Enhancing GTEx (eGTEx) groups. NIH Common Fund. NIH/NCI. NIH/NHGRI. NIH/NIMH. NIH/NIDA. Biospecimen Collection Source Site—NDRI. Biospecimen Collection Source Site—RPCI. Biospecimen Core Resource—VARI. Brain Bank Repository—University of Miami Brain Endowment Bank. Leidos Biomedical—Project Management. ELSI Study. Genome Browser Data Integration &Visualization—EBI. Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz. Lead analysts. Laboratory, Data Analysis &Coordinating Center (LDACC) NIH program management. Biospecimen collection. Pathology. eQTL manuscript working group Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. [Google Scholar]
- 37.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guan Y. Detecting structure of haplotypes and local ancestry. Genetics. 2014;196:625–642. doi: 10.1534/genetics.113.160697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong Y., Perera M.A., Gamazon E.R. On Using Local Ancestry to Characterize the Genetic Architecture of Human Traits: Genetic Regulation of Gene Expression in Multiethnic or Admixed Populations. Am. J. Hum. Genet. 2019;104:1097–1115. doi: 10.1016/j.ajhg.2019.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou X., Carbonetto P., Stephens M. Polygenic modeling with bayesian sparse linear mixed models. PLoS Genet. 2013;9:e1003264. doi: 10.1371/journal.pgen.1003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kryazhimskiy S., Plotkin J.B. The population genetics of dN/dS. PLoS Genet. 2008;4:e1000304. doi: 10.1371/journal.pgen.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siepel A., Pollard K.S., Haussler D. New methods for detecting lineage-specific selection. Lect N Bioinformat. 2006;3909:190–205. [Google Scholar]
- 45.Li W.H., Wu C.I., Luo C.C. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol. Biol. Evol. 1985;2:150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- 46.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Durinck S., Moreau Y., Kasprzyk A., Davis S., De Moor B., Brazma A., Huber W. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 48.Raudvere U., Kolberg L., Kuzmin I., Arak T., Adler P., Peterson H., Vilo J. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47(W1):W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prüfer K., Muetzel B., Do H.H., Weiss G., Khaitovich P., Rahm E., Pääbo S., Lachmann M., Enard W. FUNC: a package for detecting significant associations between gene sets and ontological annotations. BMC Bioinformatics. 2007;8:41. doi: 10.1186/1471-2105-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Friedman J., Hastie T., Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 52.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stegle O., Parts L., Piipari M., Winn J., Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat. Protoc. 2012;7:500–507. doi: 10.1038/nprot.2011.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mikhaylova A.V., Thornton T.A. Accuracy of Gene Expression Prediction From Genotype Data With PrediXcan Varies Across and Within Continental Populations. Front. Genet. 2019;10:261. doi: 10.3389/fgene.2019.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wellcome Trust Case Control C., Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guan Y., Stephens M. Practical issues in imputation-based association mapping. PLoS Genet. 2008;4:e1000279. doi: 10.1371/journal.pgen.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Safran M., Chalifa-Caspi V., Shmueli O., Olender T., Lapidot M., Rosen N., Shmoish M., Peter Y., Glusman G., Feldmesser E. Human Gene-Centric Databases at the Weizmann Institute of Science: GeneCards, UDB, CroW 21 and HORDE. Nucleic Acids Res. 2003;31:142–146. doi: 10.1093/nar/gkg050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aguiar V.R.C., César J., Delaneau O., Dermitzakis E.T., Meyer D. Expression estimation and eQTL mapping for HLA genes with a personalized pipeline. PLoS Genet. 2019;15:e1008091. doi: 10.1371/journal.pgen.1008091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tung J., Zhou X., Alberts S.C., Stephens M., Gilad Y. The genetic architecture of gene expression levels in wild baboons. eLife. 2015;4:e04729. doi: 10.7554/eLife.04729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Veyrieras J.B., Kudaravalli S., Kim S.Y., Dermitzakis E.T., Gilad Y., Stephens M., Pritchard J.K. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Glassberg E.C., Gao Z., Harpak A., Lan X., Pritchard J.K. Evidence for Weak Selective Constraint on Human Gene Expression. Genetics. 2019;211:757–772. doi: 10.1534/genetics.118.301833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blekhman R., Man O., Herrmann L., Boyko A.R., Indap A., Kosiol C., Bustamante C.D., Teshima K.M., Przeworski M. Natural selection on genes that underlie human disease susceptibility. Curr. Biol. 2008;18:883–889. doi: 10.1016/j.cub.2008.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gershoni M., Pietrokovski S. Reduced selection and accumulation of deleterious mutations in genes exclusively expressed in men. Nat. Commun. 2014;5:4438. doi: 10.1038/ncomms5438. [DOI] [PubMed] [Google Scholar]
- 64.Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 65.Dobbyn A., Huckins L.M., Boocock J., Sloofman L.G., Glicksberg B.S., Giambartolomei C., Hoffman G.E., Perumal T.M., Girdhar K., Jiang Y., CommonMind Consortium Landscape of Conditional eQTL in Dorsolateral Prefrontal Cortex and Co-localization with Schizophrenia GWAS. Am. J. Hum. Genet. 2018;102:1169–1184. doi: 10.1016/j.ajhg.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zeng P., Zhou X. Non-parametric genetic prediction of complex traits with latent Dirichlet process regression models. Nat. Commun. 2017;8:456. doi: 10.1038/s41467-017-00470-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wheeler H.E., Shah K.P., Brenner J., Garcia T., Aquino-Michaels K., Cox N.J., Nicolae D.L., Im H.K., GTEx Consortium Survey of the Heritability and Sparse Architecture of Gene Expression Traits across Human Tissues. PLoS Genet. 2016;12:e1006423. doi: 10.1371/journal.pgen.1006423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gonzalez S., Gupta J., Villa E., Mallawaarachchi I., Rodriguez M., Ramirez M., Zavala J., Armas R., Dassori A., Contreras J. Replication of genome-wide association study (GWAS) susceptibility loci in a Latino bipolar disorder cohort. Bipolar Disord. 2016;18:520–527. doi: 10.1111/bdi.12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sanjak J.S., Long A.D., Thornton K.R. Efficient Software for Multi-marker, Region-Based Analysis of GWAS Data. G3 (Bethesda) 2016;6:1023–1030. doi: 10.1534/g3.115.026013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zanetti D., Via M., Carreras-Torres R., Esteban E., Chaabani H., Anaibar F., Harich N., Habbal R., Ghalim N., Moral P. Analysis of Genomic Regions Associated With Coronary Artery Disease Reveals Continent-Specific Single Nucleotide Polymorphisms in North African Populations. J. Epidemiol. 2016;26:264–271. doi: 10.2188/jea.JE20150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.