Abstract
The identification of functional regions in the noncoding human genome is difficult but critical in order to gain understanding of the role noncoding variation plays in gene regulation in human health and disease. We describe here a co-localization approach that aims to identify constrained sequences that co-localize with tissue- or cell-type-specific regulatory regions, and we show that the resulting score is particularly well suited for the identification of rare regulatory variants. For 127 tissues and cell types in the ENCODE/Roadmap Epigenomics Project, we provide catalogs of putative tissue- or cell-type-specific regulatory regions under sequence constraint. We use the newly developed co-localization score for brain tissues to score de novo mutations in whole genomes from 1,902 individuals affected with autism spectrum disorder (ASD) and their unaffected siblings in the Simons Simplex Collection. We show that noncoding de novo mutations near genes co-expressed in midfetal brain with high confidence ASD risk genes, and near FMRP gene targets are more likely to be in co-localized regions if they occur in ASD probands versus in their unaffected siblings. We also observed a similar enrichment for mutations near lincRNAs, previously shown to co-express with ASD risk genes. Additionally, we provide strong evidence that prioritized de novo mutations in autism probands point to a small set of well-known ASD genes, the disruption of which produces relevant mouse phenotypes such as abnormal social investigation and abnormal discrimination/associative learning, unlike the de novo mutations in unaffected siblings. The genome-wide co-localization results are available online.
Keywords: sequence constraint, epigenomic annotations, colocalization, whole-genome sequencing studies
Introduction
Predicting the functional effects of variants in noncoding regions of the human genome is very challenging but of great interest due to the important role that variants in noncoding regions are likely to play in gene regulation. In particular, most of the variants identified in genome-wide association studies reside in noncoding regions, and comparative genomic studies also suggest that most of the mammalian conserved and recently adapted regions consist of noncoding elements.
Several computational methods have already been proposed to predict functional effects of genetic variants in noncoding regions. One of the traditional approaches is based on assessing the extent of interspecies evolutionary conservation in a region of interest.1,2 This approach has difficulty in identifying a large number of functional elements in noncoding regions of the human genome with rapid (functional and sequence) turnover. With the increasing availability of large numbers of whole-genome sequences from projects such as TOPMed3 and gnomAD,4 it becomes possible to identify regions that show sequence constraint within the human lineage, and several approaches have been proposed to identify such human-specific constraint regions.4, 5, 6 An alternative approach is based on epigenomic annotations from biochemical assays in the ENCODE and Roadmap Epigenomics projects.7,8 The advantage of the epigenomic functional annotations is that they can assess function at the level of tissue or cell type, which is not possible with an approach based solely on sequence constraint metrics. However, measurements from biochemical assays are affected by stochastic fluctuations of biochemical reactions, and hence these epigenomic annotations are not proof of function. Integrative methods to combine various interspecies conservation measures and epigenomic annotations have also been proposed, but the accuracies of the resulting prediction methods tend to be largely driven by one type of annotations (e.g., conservation or epigenomic), depending on how the training was done.9, 10, 11, 12 In the case of common genetic variants, genetic variation and gene expression studies such as GTEx13 can also help identify expression quantitative trait loci (eQTLs).
In this paper, we describe a new tissue-specific functional score that is particularly useful in finding rare regulatory variants that may be difficult to identify using existing approaches. We make use of a newly developed sequence constraint score based on a large number of whole-genome sequences (context-dependent tolerance score or CDTS6) in combination with an integrative epigenomic functional score, GenoNet,12,14 in order to identify regions in the noncoding genome where sequence constraint co-localizes with tissue- or cell-type-specific regulatory regions. The co-localization approach essentially identifies a subset of putative regulatory regions in the noncoding genome that are under sequence constraint, and therefore the regions that are identified have both evidence of sequence constraint and regulatory function based on epigenomic annotations.
Our proposed method is based on co-localization statistics, commonly used in imaging data to quantify co-localization of biomolecules via optical imaging techniques.15 The proposed approach is different from co-localization approaches commonly used in genetics that aim to pinpoint potential causal variants at a locus of interest by co-localizing GWAS results with molecular QTL data.16 It is also different from global co-localization analyses aiming to understand associations between different types of functional annotations.17 Rather, the approach we use here aims to identify individual (tissue- or cell-type-specific) regulatory regions that are under sequence constraint. It also allows us to answer several important questions. For example, for a given region under sequence constraint (within the human lineage), what are the relevant tissues and cell types that are responsible for the observed constraint? Are regulatory regions in specific tissues and cell types more likely to co-localize with sequence constraint regions? Among a set of noncoding variants, can we prioritize a subset of putative functional variants? Our proposed co-localization approach can provide answers to these questions and shows encouraging results in the prioritization of de novo mutations from whole-genome sequencing studies of autism spectrum disorders (ASD [MIM: 209850]).
Material and Methods
Overview of the Co-localization Method
We are interested in detecting co-localization between two sets of signals, i.e., sequence constraint and tissue- or cell-type-specific functional scores in a given region. One natural approach to measure co-localization at the region level is to compute a correlation coefficient. We focus here on Kendall’s τ rank coefficient because it is a non-parametric robust statistic with direct interpretation and fast computation. For a given region of interest R and two sets of signals X and Y, we can define the Kendall’s τ coefficient as
where and . The thresholds tX and tY are pre-specified signal strength thresholds that we discuss in the next section.
Under the null hypothesis that X is independent of Y, and assuming that the observations within X and Y are independent, respectively, the variance of is . Then the test statistic is
The sign of z reflects co-localization or anti-colocalization; in our particular context, the meaningful event is co-localization. The assumption of independence of the observations within X or Y is likely to be violated in our context because observations tend to be positively autocorrelated over small distances, which leads to inflated variance and heavier tails of the distribution compared to N(0,1). When each of X and Y is autocorrelated with a multivariate Gaussian dependence model, the variance of under null hypothesis has been derived by Hamed.18 In classic situations involving a single hypothesis test, one can use the exact variance to compute adjusted z values.
For autocorrelated data, the null distribution of z is still approximately normal, though with a possibly different mean and variance: . We are interested in simultaneously studying co-localization for large numbers of regions genome-wide, and such large-scale hypothesis testing opens up the possibility of empirically estimating the null distribution. The local false discovery rate (local fdr), an empirical Bayes approach that focuses on densities in large-scale simultaneous testing problems,19 assumes a simple Bayes model for a large collection of from N regions, and independence of ’s is not required. Each falls into the null or non-null class, occurring with prior probabilities and , and with the density of z depending on its class, either (null) or (non-null). The mixture density is (see Efron19 for details on estimating the involved densities). Then the posterior probability of being in the null class given z is , and the local fdr is defined as
The local fdr provides a good measure of co-localization since we are primarily interested in identifying the small fraction of regions with small local fdr values, corresponding to interesting regions that are worth further investigation.
Practical Implementation of the Co-localization Approach
For the sequence constraint within the human lineage, we use the recently developed context-dependent tolerance score (CDTS6). For the tissue- and cell-type-specific regulatory score we use GenoNet.14 The co-localization statistic τ depends on several parameters, specifically the thresholds for the signals (tX and tY) and the size of the region. For the analyses described here, we choose for the GenoNet score tX = 90th percentile for each tissue (positions with GenoNet score above tX), and for the CDTS score we choose tY = 0 (in practice we work with −CDTS, so we select positions with CDTS score below tY). The local fdr for each 1 Kb window are calculated using the R package locfdr. After estimating genome-wide local fdr values, we only consider windows with a minimum number of 10 positions with GenoNet and −CDTS scores above the pre-selected thresholds for further analyses, since we have higher confidence in the co-localization results from such windows.
CDTS Imputation by Cubic Smoothing Splines
The GenoNet score is computed every 25 bp, so for CDTS we compute average values for each 25-bp bin. The resulting 25 bp CDTS scores have 13.46% missing rate. In our analyses, we apply a Sequential Divide and Recombine approach20 and impute the missing data by cubic smoothing spline estimates whenever possible, as follows. (1) Divide the range of positions into disjoint intervals so that the CDTS values are either available or missing in each interval. (2) No imputation will be performed for intervals with missing CDTS scores that are longer than 1 kb. (3) For intervals with missing CDTS values and that are shorter than 1 kb, we stretch the interval from center to total length 10 times its length. (4) If the missing rate of stretched interval is less than 40%, which is based on sample size calculation theory of smoothing spline with some assumptions,21 we impute the original interval using cubic smoothing splines fit from the stretched interval. The spline estimates are calculated by the R function ssr in the assist package (see Web Resources).
Results
Genome-wide Scores
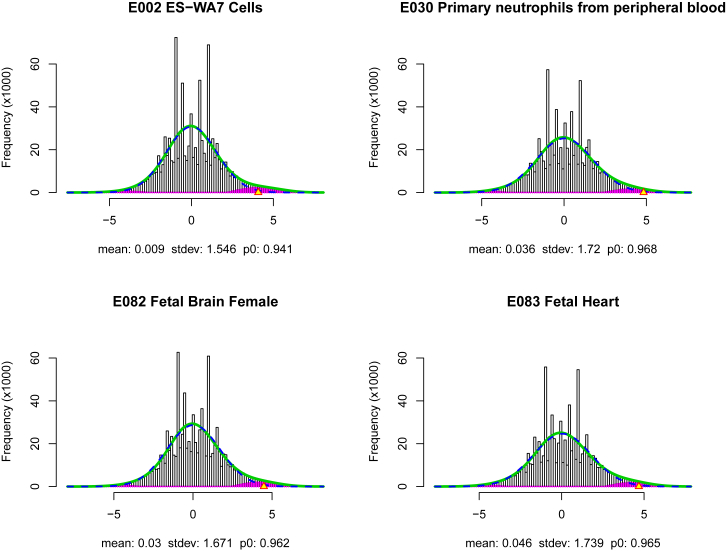
Using the above co-localization local fdr, we compute scores genome-wide. Specifically, we employ a sliding-window type approach with window size 1 Kb, and 500 bps overlap, where for each window a z statistic is computed as described. The collection of z statistics from all windows are used in estimating the empirical null distribution and . An estimated local fdr is computed for each window. See Figure 1 for the fitted mixture densities for selected tissues. Note that the overwhelming majority of windows with local fdr 0.3 show co-localization (i.e., positive z values) rather than anti-colocalization, evidence that we are discovering biologically meaningful regions. Specifically, on average 98.63% (range 88.93%–100%) of windows with local fdr 0.3 across the different tissues show co-localization, with only 1.37% (range 0%–11.07%) showing anti-colocalization.
Figure 1.
Histograms of z Values for Several Roadmap Tissues and Cell Types
Histograms of ’s and fitted mixture density f (green solid curve) and null subdensity (blue dashed cuve) for Roadmap tissues/cell types E002, E030, E082, and E083. Violet colored histogram bars indicate estimated non-null counts. Yellow triangles on the positive horizontal axis mark the threshold values for local fdr(z). Maximum likelihood estimates of mean and standard deviation of the empirical null distribution, and proportion of null cases are listed below each histogram.
In the rest of the paper, we only focus on co-localization statistics for regions with sufficient number of positions with both signals above pre-specified signal thresholds as described in the Material and Methods section.
Genome-wide Co-localization between Tissue- and Cell-Type-Specific Functional Score and Sequence Constraint
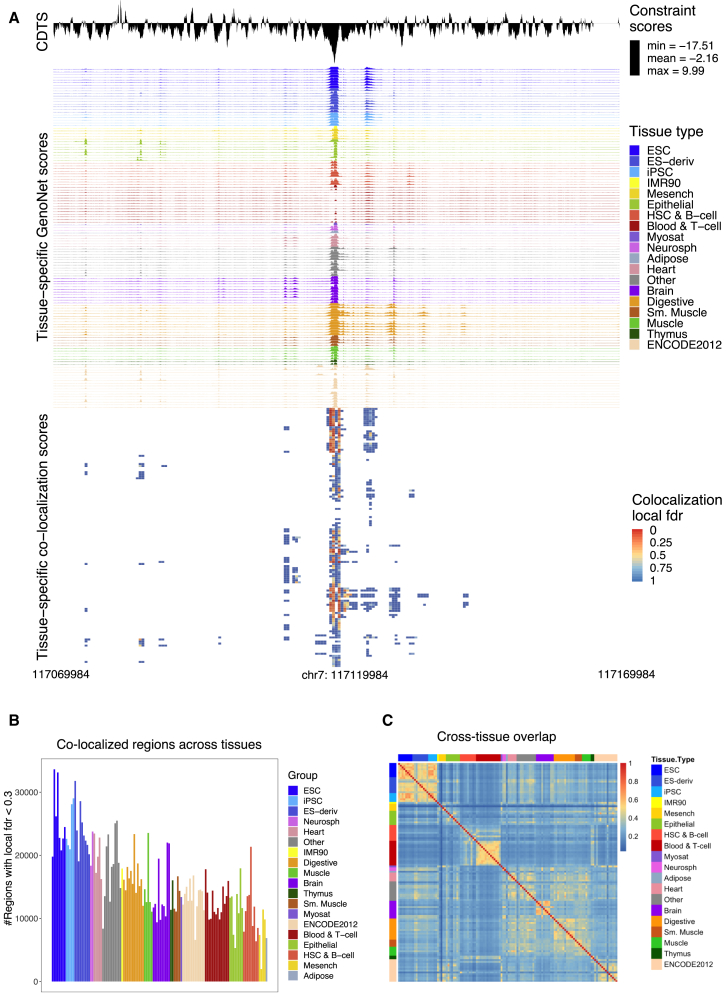
We perform co-localization analyses between regulatory scores (GenoNet scores12,14) for 127 Roadmap tissues and cell types, and sequence constraint scores as defined by CDTS. For each tissue or cell type, we compute co-localization statistics genome-wide. We focus on results from 1 Kb regions that have at least ten positions with CDTS and GenoNet scores above pre-specified thresholds as described in the Material and Methods section. The resulting percentage of the genome with co-localization statistics has a mean of 9% across tissues (range 7.4%–9.6%). A typical result from the co-localization analysis is shown in Figure 2A for a 100 Kb region containing a small window with strong co-localization signal across tissues (see also Figure S1 for several examples of co-localized regions).
Figure 2.
Co-localized Regions across Tissues and Cell Types
(A) A typical result from a region with co-localization. Shown is a 100 Kb region, along with the CDTS score (negative values are indicative of constraint) and tissue-specific functional scores (GenoNet). A heatmap of co-localization local fdr shows the co-localization results in this region.
(B) Number of 1 Kb regions with co-localization local fdr , for each tissue/cell type in Roadmap.
(C) Jaccard index of overlap between different tissues using regions with co-localization local fdr less than 0.3 in each of 127 tissues/cell types in Roadmap.
A summary of the number of 1 Kb regions that have co-localization with local fdr is given in Figure 2B for all 127 tissues and cell types in Roadmap (see Figure S2 for similar results using regions with local fdr ). As shown, human embryonic and induced stem cells have the largest number of co-localized functional regions. This is consistent with our expectation, since stem cells and embryonic tissues have a highly conserved landscape of transcriptional regulation (for reference, in Figure S3 we show the mean GenoNet score per tissue; as shown, stem cells tend to have lower average GenoNet scores relative to other tissues).
We also assess the global sharing of co-localized regions at local fdr across tissues and cell types using the Jaccard index of overlap (more details are in Appendix A), and the results are shown in Figure 2C. Overall, there is substantial overlap among similar tissues and cell types (e.g., within the blood cell types group, or within the stem cell group), with low overlap across different types of tissues. This low overlap likely reflects both tissue and cell type specificity of regulatory regions, but also false negative results.
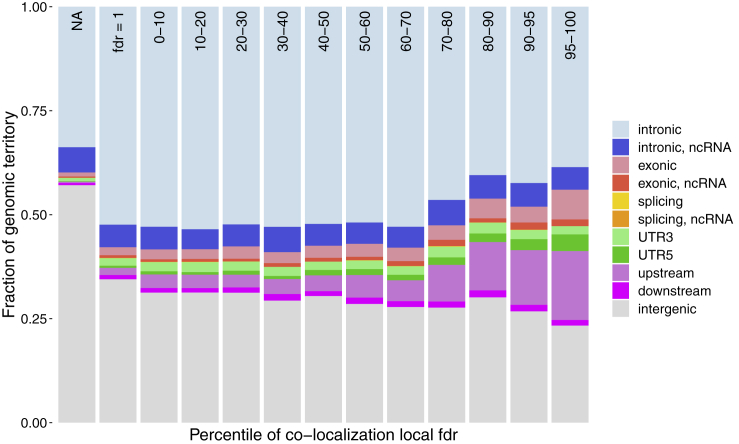
In terms of genomic regions (more details on the genomic annotation are in Appendix A), most positions with low local fdr values fall in intronic and intergenic regions simply due to the large territory occupied by these regions in the genome overall. Note that unlike supervised methods that are limited in their ability to annotate distal cis- or trans-regulatory regions due to a paucity of validated functional noncoding variants outside proximal cis-regulatory regions,11 our co-localization approach does not have this limitation. Results for Roadmap tissue E001 (ES-I3 Cells) are shown in Figure 3, but the rest of the Roadmap tissues show the same patterns. The strongest enrichments are in regions 1 Kb upstream of the transcription start sites (54.86×), 5′ UTR (36×), and exons (7.32×). Additionally, most of the variants in co-localized regions are rare (95.6% of variants with local fdr less than 0.3 have global allele frequency less than 0.001 in gnomAD), with an observed depletion for common variants (Figure S4).
Figure 3.
Genomic Distribution of Co-localized Regions
The barplots show the fraction occupied by different types of genomic regions in the different percentile group, where the percentiles are based on the local fdr values for co-localization in E001 (ES-I3 cells). The NA group corresponds to the approximately 91% of the genome for which we are not able to compute co-localization statistics.
Correlation of Co-localization Scores for Regions Proximal to Transcription Start Sites and the Probability for Genes to Be Intolerant to Loss-of-Function Variation
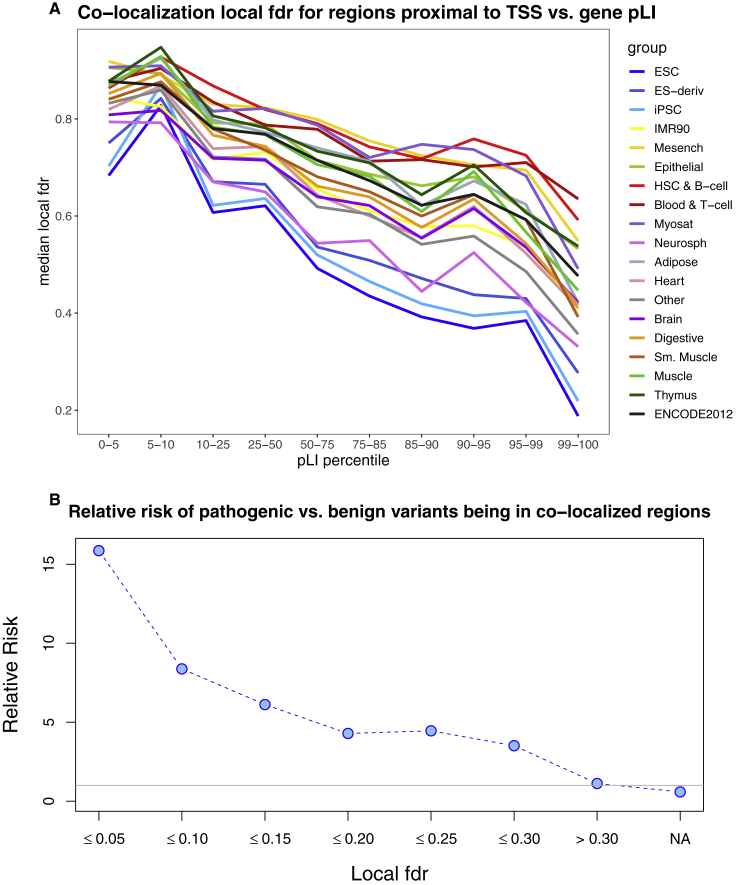
It is interesting to investigate whether genes that are intolerant to loss-of-function variation also have proximal regulatory regions that show co-localization with sequence constraint. We have looked at the correlation between the probability of loss-of-function intolerance (pLI) score for a gene4 and the local fdr values for regions that were less than 3 kb upstream of the annotated transcription start site, for each tissue separately (more details are in Appendix A). As expected, we observed a strong positive correlation, with proximal regulatory regions for genes with high pLI scores showing lower local fdr values compared with proximal regions for genes with low pLI score (Figure 4A). This pattern was consistent across all tissues, and as before we observed higher levels of co-localization for embryonic stem cells.
Figure 4.
Co-localized Regions and Correlations with Genes Intolerant to LoF Variants, and Enrichments in Pathogenic Variants
(A) Correlation between co-localization local fdr for regions proximal to transcription start sites (TSS) of genes and genes’ pLI scores, across 19 tissue groups in Roadmap.
(B) Relative risk of ClinVar pathogenic versus benign variants being in co-localized regions, at different co-localization local fdr thresholds.
We have also looked at the co-localization scores for 3 kb regions upstream of 2,472 human orthologs of mouse essential genes22 and compared to the scores for the remaining genes. We observed significantly lower co-localization local fdr values for the essential genes across all 127 tissues and cell types (with a median p value of 10−20, Figure S5), as expected.
ClinVar Variants
We selected a set of high-confidence noncoding pathogenic variants (n = 446) and benign variants (n = 3,638) in the ClinVar database (more details are in Appendix A), and estimated the relative risk for a pathogenic variant versus a benign variant to fall in co-localized regions (in any tissue) at different thresholds for co-localization local fdr (Figure 4B). We observed that noncoding pathogenic variants are much more likely to fall in co-localized regions than noncoding benign variants (e.g., at co-localization local fdr less than 0.05 the pathogenic variants are 15.9-fold more enriched than benign variants).
Analyses of De Novo Mutations in Autism Spectrum Disorder
The analysis of whole-genome sequencing data has been difficult in part due to the large amount of variation residing in the noncoding part of the genome, and current limitations in interpreting their functional effects. We focus here on the Simons Simplex Collection (SSC) whole-genome sequencing study consisting of 1,902 quartet families including a child affected with ASD, one unaffected sibling control subject, and their parents.23 In total 254,744 de novo mutations were identified, ∼67 de novo mutations per genome. The prioritization of de novo mutations based on the co-localization local fdr can help identify among the large set of observed de novo mutations in ASD probands those more likely to be functional and related to autism. We compute a score for each mutation as the average of the local fdr for the two 1 Kb windows containing the mutation, and focus on ten brain Roadmap tissues and cell types.
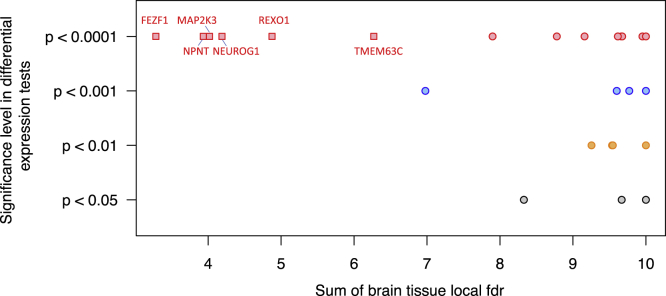
Using experimental data from a dual-luciferase assay on allelic-specific expression in human neuroblastoma cells for proband and sibling allele,24 we show that, among mutations with evidence of allele-specific transcriptional activity, mutations with small co-localization local fdr values in brain tissues (Figures S6 and S7) exhibit the highest level of significance in the differential allelic expression tests (Figures 5 and S8). Therefore, prioritizing mutations with small local fdr values in appropriate tissues can help identify those more likely to validate in experimental studies. In contrast, this is not true if other functional scores are used to prioritize mutations. For example, selecting mutations based on the pLI of nearest gene or CDTS scores or DNA Disease Impact Score24 does not necessarily prioritize the mutations with the highest significance level in differential allelic expression tests (Figure S9).
Figure 5.
Co-localization and Dual-Luciferase Assay Results
Significance level for testing differential expression for proband and sibling allele in a dual-luciferase assay, versus sum of co-localization local fdr of 10 brain tissues for 51 de novo mutations in ASD probands; mutations with sum of co-localization local fdr below 7 are represented as squares, and labeled with their nearest gene; all other mutations are shown as circles. Significance levels for testing differential expression were computed on the basis of a t test and Fisher’s combined probability test (two sided; gray for p < 0.05, orange for p < 0.01, blue for p < 0.001, red for p < 0.0001).
Gene Set Analyses
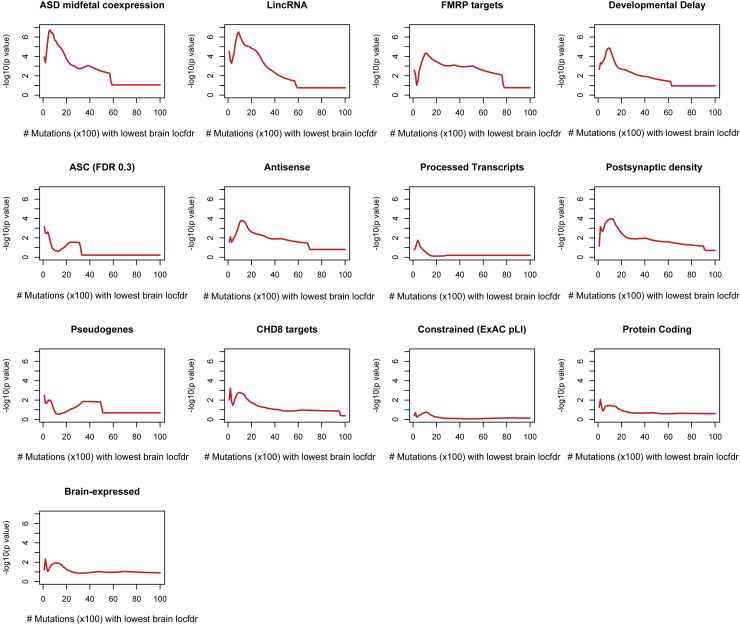
We focus here on 13 gene sets, including ASD risk genes (FDR < 0.3);25 genes coexpressed in midfetal brain with ASD risk genes;26 genes associated with developmental delay from the Development Disorder Genotype-Phenotype Database; CHD8 target genes defined as the union of lists from two ChIP-seq studies;27,28 FMRP target genes;29 human postsynaptic density (PSD) proteins from the Genes2Cognition database; brain expressed genes;30 constrained genes defined as having a probability of loss-of-function intolerance (pLI) score in the ExAC database; and five categories defined by GENCODE (wgEncodeGencodeCompV19): protein coding genes, pseudogenes, lincRNA, antisense genes, and processed transcripts. For each gene set, we assign de novo mutations to genes in the set if they are within / Mb of the transcription start sites of the genes in the set. For each such mutation we compute the sum of locfdr values in ten brain tissues in Roadmap. There are slightly more mutations in ASD probands compared to the unaffected siblings, so we randomly sample a subset of mutations in ASD probands to match the number of mutations in control subjects. We then compare for significant differences in the lower tail distribution of brain locfdr scores for de novo mutations in ASD versus controls (more details are given in Appendix A). Note that we focus on differences in the lower tail of the co-localization score distribution rather than the mean because most de novo variants do not reside in co-localized regions. We find that noncoding de novo mutations near genes co-expressed in midfetal brain with high confidence ASD risk genes are more likely to be in co-localized regions if they occur in ASD probands versus in their unaffected siblings (Figures 6, S10, and S12; p value31 = 0.031). Similarly, we find that de novo mutations near lincRNAs are more likely to reside in co-localized regions if they are in ASD probands versus in their unaffected siblings (p value = 0.030). This is concordant with recent studies showing co-expression of lncRNAs with genes harboring ASD protein coding mutations, suggesting that these lncRNAs are potential ASD risk loci.32 For FMRP target genes, the p value is 0.027. The set of developmental delay genes also shows suggestive enrichment for co-localized de novo mutations in ASD versus unaffected siblings (p value = 0.076). Results for all genesets are reported in Table S1. Note that the gene sets with significant results have been related to ASD risk before, while gene sets defined using GENCODE, with the exception on lincRNA, do not show any significant results in our analyses. Overall, using a conservative choice for the proportion of null p values, we find that the associations with lincRNAs, genes co-expressed in midfetal brain with high confidence ASD risk genes, and FMRP target genes have q-values <0.135 (Table S1).
Figure 6.
Gene Set Analyses
For each gene set, the −log10(p values) from a Wilcoxon rank sum test of difference between brain local fdr for the mutations residing within 2 Mb of TSS of genes in the set, in ASD probands versus unaffected siblings are shown. The test is performed on the 100–10,000 mutations with the lowest brain local fdr in ASD probands and unaffected siblings, respectively, as described in Appendix A.
Mouse Phenotype Enrichment Analyses
We have additionally performed mouse phenotype enrichment analyses as follows. For the ten brain Roadmap tissues and cell types, we identify those de novo mutations with minimum co-localization local fdr less than 0.4 or 0.5 (we have considered more stringent thresholds including 0.3, but the functional enrichment analyses we report below were underpowered at those thresholds). We focus here on describing the details of the analysis using 0.5 as a threshold, but results are also reported at the 0.4 threshold and they are similar. Using local fdr less than 0.5 in at least one brain tissue results in 1,098 de novo mutations in ASD probands and 1,102 in unaffected siblings (Table S2). We then map these to nearby genes using a “nearest gene” approach. We use these selected genes as seed genes in a PPI network analysis in the ToppFun tool33 (all the interactions in the underlying PPI network are derived from large-scale experiments and curated manually) and identify two sets of genes that significantly interact with the set of seed genes, one originating from ASD probands (11 genes) and one from unaffected siblings (8 genes) (Table S2). Note that the set of significantly interacting genes does not contain all the direct neighbors of the seed genes, but only a small number of genes, namely those genes showing significantly more interactions with the set of seed genes than expected by chance, given the interactions in the PPI network (according to a hypergeometric test); in contrast, selecting at random gene sets of same size as the seed gene set leads to empty sets of significantly interacting genes in the vast majority of cases (99%), supporting the idea that the prioritization based on the co-localization local fdr leads to a set of functionally related genes. We focus on the small set of interacting genes and make use of mouse orthologs of human genes in order to interrogate mouse phenotype data, consisting of Mouse Phenotype ontology annotations and gene variants causing these phenotypes in genetically engineered or mutagenesis experiments.
Based on these analyses, we detect clear functional differences between interacting genes in the PPI network for ASD probands versus control subjects (Table 1). In particular, the mouse orthologs of human genes originating from ASD probands point to mouse phenotypes highly relevant to ASD, such as abnormal social investigation, social withdrawal, and abnormal discrimination/associative learning unlike the mouse orthologs of human genes originating from unaffected siblings which point to seemingly unrelated phenotypes, such as delayed hepatic development, abnormal hepatoblast migration, etc. (Table 1). The interacting genes in ASD probands pointing to these mouse phenotypes (Table S2) include some well-known risk genes for ASD (including GRIN1 [MIM: 138249], SHANK3 [MIM: 606230], SYNGAP1 [MIM: 603384], DLGAP1 [MIM: 605445], DLG4 [MIM: 602887], etc.). Four of these proteins (SHANK3, DLGAP1, SYNGAP1, and DLG4) form the core of the postsynaptic density structure, interact with hundreds of other proteins to influence synaptic plasticity, and affect learning and memory.34 Similar results were obtained when using local FDR < 0.4 to prioritize de novo mutations (Table S3). All the significant enrichments (FDR < 5%) are reported in detail in Tables S6, S7, S8, and S9. We note that performing similar analyses but prioritizing de novo mutations based on the DeepSEA disease impact score does not result in a meaningful set of interacting genes and mouse phenotypes (Tables S4 and S5).
Table 1.
Mouse Phenotypes Affected by Genes Orthologous to the Interacting Genes Derived from De Novo Mutations in ASD Probands and Unaffected Siblings
| Mouse Phenotype | p Value | p Value Bonferroni | Interacting Genes |
|---|---|---|---|
| ASD Proband | |||
| Abnormal social investigation | 9.202e−10 | 7.095e−07 | GRIN1, MAPK1, DLGAP1, DLG4, SHANK3, SYNGAP1 |
| Abnormal glutamate-mediated receptor currents | 2.982e−09 | 2.299e−06 | KALRN, GRIN1, DLG4, SHANK3, SYNGAP1 |
| Social withdrawal | 3.617e−09 | 2.789e−06 | GRIN1, MAPK1, SHANK3, SYNGAP1 |
| Abnormal social/conspecific interaction | 8.313e−09 | 6.410e−06 | KALRN, GRIN1, MAPK1, DLGAP1, DLG4, SHANK3, SYNGAP1 |
| Abnormal discrimination learning | 1.463e−08 | 1.128e−05 | GRIN1, DLG4, SHANK3, SYNGAP1 |
| Abnormal AMPA-mediated synaptic currents | 2.995e−08 | 2.309e−05 | KALRN, GRIN1, DLG4, SYNGAP1 |
| Abnormal associative learning | 1.691e−07 | 1.304e−04 | KALRN, GRIN1, MAPK1, DLG4, SHANK3, SYNGAP1 |
| Increased grooming behavior | 1.706e−07 | 1.315e−04 | GRIN1, DLGAP1, DLG4, SHANK3 |
| Decreased anxiety-related response | 2.603e−07 | 2.007e−04 | KALRN, MAPK1, DLG4, SHANK3, SYNGAP1 |
| Abnormal contextual conditioning behavior | 2.919e−07 | 2.250e−04 | KALRN, GRIN1, MAPK1, DLG4, SHANK3 |
| Control Sibling | |||
| Delayed hepatic development | 1.047e−07 | 1.155e−04 | SMAD2, SMAD3, NF1 |
| Abnormal hepatoblast migration | 1.518e−06 | 1.675e−03 | SMAD2, SMAD3 |
| Abnormal hepatoblast physiology | 3.035e−06 | 3.348e−03 | SMAD2, SMAD3 |
| Abnormal secondary ovarian follicle morphology | 3.038e−06 | 3.350e−03 | SMAD2, SMAD3, FMR1 |
De novo mutations are prioritized based on having co-localization local fdr < 0.5 in at least one brain tissue. Top ten phenotypes with Bonferroni adjusted p value < 0.01 as reported by ToppFun analysis are reported.
In summary, our findings suggest that regulatory de novo mutations in noncoding regions can affect interactions with a small set of highly relevant ASD genes, that can in turn alter the risk to ASD. This is concordant with the omnigenic model, which proposed that a large number of peripheral genes can affect risk to complex diseases through regulatory effects on a much smaller number of core genes with biological relevance to the trait.35
Discussion
We have proposed here a co-localization statistic to detect regions in the noncoding parts of the human genome where tissue- or cell-type-specific regulatory variants co-localize with sequence constraint. Although conceptually simple, the co-localization statistic is a powerful tool in the identification of highly functional regulatory variants and can help prioritize rare regulatory variants implicated in disease, a class of variation that is difficult to identify. Using existing experimental data on allele-specific transcriptional activity of selected ASD mutations, we show that prioritizing mutations based on co-localization score in brain tissues is likely to select those mutations with highest effects on expression.
We demonstrate the usefulness of the co-localization score in identifying regulatory mutations using several applications. Specifically, we have shown that (1) regulatory regions in embryonic and induced stem cells tend to co-localize more often with regions under sequence constraint relative to regulatory regions in other tissues, as expected; (2) the co-localization between potentially regulatory regions and regions under sequence constraint is overwhelmingly in the positive direction, supporting the biological significance of the co-localized regions; (3) we have shown that genes that are intolerant to loss-of-function mutations tend to have proximal regulatory regions that co-localize with constraint; (4) noncoding pathogenic variants in ClinVar are highly enriched in co-localized regions relative to benign variants; (5) noncoding de novo mutations near genes co-expressed in midfetal brain with high confidence ASD risk genes, near FMRP targets, and also near lincRNAs are more likely to be in co-localized regions if they occur in ASD probands versus their unaffected siblings; (6) our ASD findings suggest that regulatory de novo mutations in noncoding regions can affect interactions with a small set of well-known ASD genes, that can in turn alter the risk to ASD. Functional enrichment analyses on the small set of interacting genes points to highly relevant phenotypes in mouse for genes derived from de novo mutations in ASD but not for genes derived from mutations in unaffected siblings. We note that selecting all de novo variants with maximum epigenomic score in brain tissues (i.e., with GenoNet value 1) does not help in prioritizing the relevant mutations and genes due to the large observed number of such mutations. Similarly, prioritizing mutations with high pLI scores (>0.99) for nearest genes does not help (e.g., it uncovers general biological processes, not specific to ASD), due to the large number of genes with high scores and the even larger number of significantly interacting genes. Therefore, the prioritization of mutations based on co-localization scores provides a more effective way to prioritize likely functional mutations with specific relevance to ASD compared with existing methods.
The proposed co-localization score can be useful in prioritizing pathogenic variants in genomes from individuals affected with rare, Mendelian disorders. However, caution is needed because the large number of rare variants per genome makes the confident identification of truly pathogenic variants very challenging. While we can prioritize “outlier” variants in these individuals, other types of data, including transcriptome data in the appropriate tissues, when available, can further help with the prioritization.36 We also note that our method does not provide a means to link putative functional variants to the genes they might regulate.
The co-localization analyses reported here can be improved in several ways. We have shown using allele-specific expression data that the proposed score performs well in prioritizing true regulatory variants, and therefore may perform well in terms of positive predictive value and specificity, but it will naturally miss many true regulatory variants because either they do not reside in co-localized regions or because our co-localization statistic cannot identify them. In particular, common regulatory variants are depleted in these co-localized regions, and hence the proposed score is particularly appropriate for the prioritization of rare regulatory variants. Furthermore, the sequence constraint scores for noncoding regions are still under development but with the rapid increase in whole-genome sequencing datasets and more powerful methods, we should expect substantial improvements. Similarly, tissue- and cell-type-specific regulatory scores can be improved upon by incorporation of additional features and development in experimental functional assays (such as massively parallel reporter assays) that would ultimately lead to improved computational predictions.14 We have computed co-localization statistics for 1 Kb regions genome-wide for 127 tissue and cell types available in ENCODE/Roadmap, and the scores are available online.
Acknowledgments
We gratefully acknowledge support by National Institutes of Health grants MH106910 (D.X., C.W., I.I.-L.), MH111661 (J.D.B.), and RC2-DK116690 (K.K., C.W., I.I.-L.). We thank Joseph Gogos, Yufeng Shen, Kai Wang, Sam Kim, and Alkes Price for helpful discussions.
Published: April 2, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.03.003.
Appendix A
Jaccard Index of Overlap
Given a threshold α (e.g., 0.2 or 0.3), the window is co-localized for a tissue if the tissue-specific co-localization local fdr is not greater than α and has minimum 10 positions with GenoNet and −CDTS scores above the pre-selected thresholds. The Jaccard index of overlap for a pair of tissues (i,j) is the ratio of the number of windows that are co-localized for both tissues and the number of windows that are co-localized for at least one tissue.
Regions Proximal to Genes
For each region proximal to a transcription start site, defined as less than 3 kb upstream of the annotated transcription start site, we computed the z statistic described in the main text and estimated local fdr values based on z statistics from genome-wide 3 Kb sliding windows.
Annovar Annotation
We selected one million random positions and then used Annovar for their genomic annotation. The genomic annotations include 11 categories: “exonic,” “exonic, ncRNA,” “splicing,” “splicing, ncRNA,” “UTR3,” “UTR5,” “intronic,” “intronic, ncRNA,” “upstream,” “downstream,” and “intergenic.” Definitions of these annotations can be found on the Annovar website.
ClinVar
We selected high-confidence autosomal variants from the ClinVar database for which the review status are “practice guideline” (four gold stars), “reviewed by expert panel” (three gold stars), or “criteria provided, multiple submitters, no conflicts” (two gold stars). The variants were annotated as noncoding variants if the molecular consequences include any term among “3 prime UTR variant,” “5 prime UTR variant,” “500B downstream variant,” “2Kb upstream variant,” and “intron variant.” The resulting set consists of 2,395 pathogenic variants and 3,853 benign variants. We further process this list so that a pathogenic variant will be removed if there is any benign variant located within its neighborhood of radius 500 bps, and the benign variants in the neighborhood will be removed as well. The final dataset consists of 446 pathogenic variants and 3,638 benign variants.
Gene Sets
For a given gene set, we assign de novo mutations to it if the mutations fall within / Mb of the transcription start sites of genes in the set. For each gene set, there are slightly more mutations in ASD probands than in the unaffected siblings, so we randomly sample a subset of mutations in ASD probands to match the number of mutations in control subjects. We focus on ten brain tissues in Roadmap (E067, E068, E069, E070, E071, E072, E073, E074, E081, E082) and compute for each mutation a score as the sum of locfdr values in these brain tissues. For a given gene set, let M be the number of de novo mutations assigned to the gene set in ASD probands and the same number for unaffected siblings. Let be the scores in increasing order for the M mutations in ASD probands and for the unaffected siblings. We compare the two sets of scores, for the lowest scoring mutations, starting with 100 and until 10,000 with a step of 100, using a Wilcoxon rank-sum test. The corresponding p values are denoted as . These are the p values shown in Figures 6 and S10. To evaluate the statistical significance of these observed p values, we compute the following statistic
We evaluate the significance by permutation. Specifically, we permute the ASD proband-unaffected sibling status of the mutations, and recompute the statistic for each permuted dataset. The distributions of the statistic for 1,000 permutations is shown in Figures S11 and S12. The p value is evaluated empirically based on 1,000 permutations. The two p values (using 1 Mb or 2 Mb distance to TSS to assign mutations to gene sets) are combined using the Cauchy combination method.31
Web Resources
ASSIST: A Suite of S-plus functions Implementing Spline smoothing Techniques, http://cran.r-project.org
ClinVar, http://www.clinvar.com/
Co-localization, http://www.funlda.com/colocalization/
FUN-LDA, http://www.funlda.com/
Genes2Cognition database, https://www.genes2cognition.org/
GenoNet, http://www.funlda.com/genonet
Online Mendelian Inheritance in Man, https://www.omim.org/
Toppgene, https://toppgene.cchmc.org/
UCSC Genome Browser, https://genome.ucsc.edu/
Supplemental Data
De novo mutations are prioritized based on having co-localization local fdr < 0.5 in at least one brain tissue. Phenotypes significant at FDR < 5% as reported by ToppFun analysis are shown.
De novo mutations are prioritized based on having co-localization local fdr < 0.4 in at least one brain tissue. Phenotypes significant at FDR < 5% as reported by ToppFun analysis are shown.
De novo mutations are prioritized based on having co-localization local fdr < 0.5 in at least one brain tissue. Phenotypes significant at FDR < 5% as reported by ToppFun analysis are shown.
De novo mutations are prioritized based on having co-localization local fdr < 0.4 in at least one brain tissue. Phenotypes significant at FDR < 5% as reported by ToppFun analysis are shown.
References
- 1.Cooper G.M., Stone E.A., Asimenos G., Green E.D., Batzoglou S., Sidow A., NISC Comparative Sequencing Program Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taliun D., Harris D.N., Kessler M.D., Carlson J., Szpiech J.A., Torres R., Gagliano Taliun S.A., Corvelo A., Gogarten S.M., Kang H.M. Sequencing of 53,831 diverse genomes from the nhlbi topmed program. bioRxiv. 2019 doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv. 2019 [Google Scholar]
- 5.Petrovski S., Gussow A.B., Wang Q., Halvorsen M., Han Y., Weir W.H., Allen A.S., Goldstein D.B. The intolerance of regulatory sequence to genetic variation predicts gene dosage sensitivity. PLoS Genet. 2015;11:e1005492. doi: 10.1371/journal.pgen.1005492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.di Iulio J., Bartha I., Wong E.H.M., Yu H.-C., Lavrenko V., Yang D., Jung I., Hicks M.A., Shah N., Kirkness E.F. The human noncoding genome defined by genetic diversity. Nat. Genet. 2018;50:333–337. doi: 10.1038/s41588-018-0062-7. [DOI] [PubMed] [Google Scholar]
- 7.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernstein B.E., Stamatoyannopoulos J.A., Costello J.F., Ren B., Milosavljevic A., Meissner A., Kellis M., Marra M.A., Beaudet A.L., Ecker J.R. The nih roadmap epigenomics mapping consortium. Nat. Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ionita-Laza I., McCallum K., Xu B., Buxbaum J.D. A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat. Genet. 2016;48:214–220. doi: 10.1038/ng.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caron B., Luo Y., Rausell A. NCBoost classifies pathogenic non-coding variants in Mendelian diseases through supervised learning on purifying selection signals in humans. Genome Biol. 2019;20:32. doi: 10.1186/s13059-019-1634-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Backenroth D., He Z., Kiryluk K., Boeva V., Pethukova L., Khurana E., Christiano A., Buxbaum J.D., Ionita-Laza I. Fun-lda: a latent dirichlet allocation model for predicting tissue-specific functional effects of noncoding variation: methods and applications. Am. J. Hum. Genet. 2018;102:920–942. doi: 10.1016/j.ajhg.2018.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.GTEx Consortium The genotype-tissue expression (gtex) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He Z., Liu L., Wang K., Ionita-Laza I. A semi-supervised approach for predicting cell-type specific functional consequences of non-coding variation using MPRAs. Nat. Commun. 2018;9:5199. doi: 10.1038/s41467-018-07349-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang S., Arena E.T., Becker J.T., Bement W.M., Sherer N.M., Eliceiri K.W., Yuan M. Spatially adaptive colocalization analysis in dual-color fluorescence microscopy. arXiv. 2017 doi: 10.1109/TIP.2019.2909194. arXiv:1711.00069. [DOI] [PubMed] [Google Scholar]
- 16.Hormozdiari F., van de Bunt M., Segrè A.V., Li X., Joo J.W.J., Bilow M., Sul J.H., Sankararaman S., Pasaniuc B., Eskin E. Colocalization of GWAS and eQTL Signals Detects Target Genes. Am. J. Hum. Genet. 2016;99:1245–1260. doi: 10.1016/j.ajhg.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanduri C., Bock C., Gundersen S., Hovig E., Sandve G.K. Colocalization analyses of genomic elements: approaches, recommendations and challenges. Bioinformatics. 2018;35:1615–1624. doi: 10.1093/bioinformatics/bty835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamed K.H. The distribution of kendall’s tau for testing the significance of cross-correlation in persistent data. Hydrol. Sci. J. 2011;56:841–853. [Google Scholar]
- 19.Efron B. Large-scale simultaneous hypothesis testing: the choice of a null hypothesis. J. Am. Stat. Assoc. 2004;99:96–104. [Google Scholar]
- 20.Xu D., Wang Y. Divide and recombine approaches for fitting smoothing spline models with large datasets. J. Comput. Graph. Stat. 2018;27:677–683. [Google Scholar]
- 21.Wang Y. Sample size calculations for smoothing splines based on bayesian confidence intervals. Stat. Probab. Lett. 1998;38:161–166. [Google Scholar]
- 22.Georgi B., Voight B.F., Bućan M. From mouse to human: evolutionary genomics analysis of human orthologs of essential genes. PLoS Genet. 2013;9:e1003484. doi: 10.1371/journal.pgen.1003484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.An J.Y., Lin K., Zhu L., Werling D.M., Dong S., Brand H., Wang H.Z., Zhao X., Schwartz G.B., Collins R.L. Genome-wide de novo risk score implicates promoter variation in autism spectrum disorder. Science. 2018;362:eaat6576. doi: 10.1126/science.aat6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou J., Park C.Y., Theesfeld C.L., Wong A.K., Yuan Y., Scheckel C., Fak J.J., Funk J., Yao K., Tajima Y. Whole-genome deep-learning analysis identifies contribution of noncoding mutations to autism risk. Nat. Genet. 2019;51:973–980. doi: 10.1038/s41588-019-0420-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanders S.J., He X., Willsey A.J., Ercan-Sencicek A.G., Samocha K.E., Cicek A.E., Murtha M.T., Bal V.H., Bishop S.L., Dong S., Autism Sequencing Consortium Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willsey A.J., Sanders S.J., Li M., Dong S., Tebbenkamp A.T., Muhle R.A., Reilly S.K., Lin L., Fertuzinhos S., Miller J.A. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155:997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cotney J., Muhle R.A., Sanders S.J., Liu L., Willsey A.J., Niu W., Liu W., Klei L., Lei J., Yin J. The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat. Commun. 2015;6:6404. doi: 10.1038/ncomms7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sugathan A., Biagioli M., Golzio C., Erdin S., Blumenthal I., Manavalan P., Ragavendran A., Brand H., Lucente D., Miles J. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc. Natl. Acad. Sci. USA. 2014;111:E4468–E4477. doi: 10.1073/pnas.1405266111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darnell J.C., Van Driesche S.J., Zhang C., Hung K.Y., Mele A., Fraser C.E., Stone E.F., Chen C., Fak J.J., Chi S.W. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang H.J., Kawasawa Y.I., Cheng F., Zhu Y., Xu X., Li M., Sousa A.M., Pletikos M., Meyer K.A., Sedmak G. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y., Xie J. Cauchy combination test: a powerful test with analytic p-value calculation under arbitrary dependency structures. J. Am. Stat. Assoc. 2019 doi: 10.1080/01621459.2018.1554485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parikshak N.N., Swarup V., Belgard T.G., Irimia M., Ramaswami G., Gandal M.J., Hartl C., Leppa V., Ubieta L.T., Huang J. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature. 2016;540:423–427. doi: 10.1038/nature20612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J., Bardes E.E., Aronow B.J., Jegga A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305-11. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J., Wilkinson B., Clementel V.A., Hou J., O’Dell T.J., Coba M.P. Long-term potentiation modulates synaptic phosphorylation networks and reshapes the structure of the postsynaptic interactome. Sci. Signal. 2016;9 doi: 10.1126/scisignal.aaf6716. rs8–rs8. [DOI] [PubMed] [Google Scholar]
- 35.Boyle E.A., Li Y.I., Pritchard J.K. An expanded view of complex traits: From polygenic to omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohammadi P., Castel S.E., Cummings B.B., Einson J., Sousa C., Hoffman P., Donkervoort S., Jiang Z., Mohassel P., Foley A.R. Genetic regulatory variation in populations informs transcriptome analysis in rare disease. Science. 2019;366:351–356. doi: 10.1126/science.aay0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
De novo mutations are prioritized based on having co-localization local fdr < 0.5 in at least one brain tissue. Phenotypes significant at FDR < 5% as reported by ToppFun analysis are shown.
De novo mutations are prioritized based on having co-localization local fdr < 0.4 in at least one brain tissue. Phenotypes significant at FDR < 5% as reported by ToppFun analysis are shown.
De novo mutations are prioritized based on having co-localization local fdr < 0.5 in at least one brain tissue. Phenotypes significant at FDR < 5% as reported by ToppFun analysis are shown.
De novo mutations are prioritized based on having co-localization local fdr < 0.4 in at least one brain tissue. Phenotypes significant at FDR < 5% as reported by ToppFun analysis are shown.