Abstract
Glycosylphosphatidylinositol (GPI)-anchored proteins are critical for embryogenesis, neurogenesis, and cell signaling. Variants in several genes participating in GPI biosynthesis and processing lead to decreased cell surface presence of GPI-anchored proteins (GPI-APs) and cause inherited GPI deficiency disorders (IGDs). In this report, we describe 12 individuals from nine unrelated families with 10 different bi-allelic PIGK variants. PIGK encodes a component of the GPI transamidase complex, which attaches the GPI anchor to proteins. Clinical features found in most individuals include global developmental delay and/or intellectual disability, hypotonia, cerebellar ataxia, cerebellar atrophy, and facial dysmorphisms. The majority of the individuals have epilepsy. Two individuals have slightly decreased levels of serum alkaline phosphatase, while eight do not. Flow cytometric analysis of blood and fibroblasts from affected individuals showed decreased cell surface presence of GPI-APs. The overexpression of wild-type (WT) PIGK in fibroblasts rescued the levels of cell surface GPI-APs. In a knockout cell line, transfection with WT PIGK also rescued the GPI-AP levels, but transfection with the two tested mutant variants did not. Our study not only expands the clinical and known genetic spectrum of IGDs, but it also expands the genetic differential diagnosis for cerebellar atrophy. Given the fact that cerebellar atrophy is seen in other IGDs, flow cytometry for GPI-APs should be considered in the work-ups of individuals presenting this feature.
Keywords: glycosylphosphatidylinositol (GPI), PIGK, GPI8, transamidase, inherited GPI deficiency disorders (IGDs)
Introduction
The synthesis and modification of the glycosylphosphatidylinositol (GPI) anchor proteins involves at least 31 enzymes. This process includes multiple steps of synthesis of GPI precursor molecules taking place in the endoplasmic reticulum (ER) membrane, then transfer of the entire precursor GPI to newly synthesized proteins by a multi-protein transamidase complex, which also simultaneously cleaves the C-terminal consensus sequence of the GPI target. Finally, the GPI-bound protein is modified to exit from the ER to the Golgi for further modification in order to be functional.1,2 Gpi8p was first characterized in yeast as a glycosylated transmembrane protein of the ER and was hypothesized to be part of the GPI-transamidase complex.3 Then, in GPI-anchored protein (GPI-AP) class K mutant cells, where endogenous human GPI8 (hGPI8) is mutated, leading to failure of incorporation of GPI anchors into nascent polypeptides, the overexpression of hGPI8 restored the transamidation ability. This protein, later renamed phosphatidylinositol glycan (PIG) anchor biosynthesis class K (PIGK), is coded by a 25-kb gene, PIGK (MIM: 605087), that resides on chromosome 1 in humans.4 It is a cysteine protease and forms a disulfide bridge with PIGT in the GPI-transamidase complex, which is composed of GPI anchor attachment 1 protein (GPAA1), PIGU, PIGT, PIGS, and PIGK, and this bridge is required for normal transamidase activity.5
More than 150 GPI-APs are known, making up about 1% of the human proteome, and they play important roles as hydrolytic enzymes, adhesion molecules, receptors, protease inhibitors, and complement regulatory proteins.1 The disruption of GPI anchor biosynthesis and remodeling has been linked to many inherited GPI deficiency disorders (IGDs). With the development of next-generation sequencing, 19 genes in the GPI-AP biosynthesis pathway have been linked to human diseases.6,7 In this study, we report 10 individuals from eight unrelated families who presented a neurodevelopmental syndrome associated with biallelic variants in PIGK. We further demonstrate GPI-AP deficiency at the cell surface in affected individual cell lines. This deficit could be rescued by overexpression of wild-type (WT) human PIGK. PIGK loss of function therefore shares common phenotypic and biochemical features with PIGT and PIGS deficiencies, as well as with other IGDs.
Material and Methods
Identification of Affected Individuals and Collection of Samples
For Individual 1A, clinical exome sequencing was performed at Baylor Genetics, and the individual was identified after a search for individuals with potentially bi-allelic variants in PIGK and neurological features suggestive of an IGD. The family was enrolled in a protocol approved at the CHU Sainte-Justine after informed consent and samples were obtained. The rest of cohort was then assembled with the help of GeneMatcher.8
For Individuals 2A and 2B, the family was enrolled in an ongoing research program, dedicated to individuals affected by undiagnosed diseases, at the Ospedale Pediatrico Bambino Gesù, Rome. After written informed consent was obtained, clinical data and DNA samples from the participating family were collected, stored, and used following the Institutional Review Board recommendations. Permission was obtained to publish the photographs.
Individual 3 had a research trio exome sequencing performed in a Dijon UHC research project after sequencing of a cerebellar atrophy gene panel and a clinical solo exome, both of which were negative.
Individuals 4 and 5 had exome sequencing performed after informed consent was obtained as part of a study on brain malformations carried out in the Gleeson lab.
Individuals 6 and 7 had clinical exome sequencing performed at GeneDx after informed consent.
Individual 8 had research trio whole-genome sequencing (WGS) performed through the 100,000 Genomes Project, which is a national UK project described in detail elsewhere.9 Clinical data and samples were collected from the participating family after written informed consent was obtained. Permission was obtained to publish the photographs and magnetic resonance imaging (MRI) images.
After informed consent was obtained prior to genetic testing from the family of Individual 9, genomic DNA was extracted from peripheral blood samples (from the proband, parents, one unaffected sister, and one affected brother) according to standard procedures of phenol chloroform extraction.
For GPI studies, blood samples were collected from five probands in three families, Families 1, 2, and 3. Parental blood samples and samples from other family members were used to assess co-segregation between variants and the trait. Skin fibroblasts were established from Individual 1A.
Exome and Genome Sequencing
For Family 1, exome sequencing was performed as described previously.
For Family 2, targeted enrichment (SureSelect ClinicalExome V.2, Agilent) and massively parallel sequencing (NexSeq550, Illumina) were performed on genomic DNA extracted from leukocytes through the use of a trio-based approach. About 65 million reads per sample were obtained. Data analysis was performed using an in-house implemented pipeline which mainly takes advantage of the Genome Analysis Toolkit (GATK V.3.7) framework, as previously reported.10 Reads mapping was performed using Burrows-Wheeler Aligner BWA V.0.7.12,11 and GATK tools were used for base quality recalibration and variants calling. SNVs and small insertions, deletions, and deletion-insertions were identified through the use of the GATK’s HaplotypeCaller tool used in gVCF mode, followed by family level joint genotyping and phasing. Finally, variants were quality filtered according to GATK’s 2016 best practices. To retain private and clinically associated variants, we selected annotated variants with unknown frequency or having a minor allele frequency (MAF) <1% (according to dbSNP150 and gnomAD V.2.0) and occurring with a frequency <1% in an in-house database which included frequency data from approximately 1,300 population-matched exomes. SnpEff toolbox (V.4.3) was used to predict the functional impact of variants, which were filtered to retain only splice-site regions (variants located from −3 to +8 with respect to an exon-intron junction) and those located in exons which had any effect on the coding sequence. Moreover, functional annotation of variants was performed using SnpEff and dbNSFP (V.3.5). The functional impact of variants was eventually analyzed using Combined Annotation Dependent Depletion (CADD) V.1.4, M-CAP V.1.0, and InterVar V.2.0 algorithms in order to obtain clinical interpretation according to ACMG/AMP 2015 guidelines. Variant validation and segregation were performed via Sanger sequencing.
For Family 3, genomic DNA extracted from the proband’s blood leukocytes was used for a targeted exon enrichment using the SureSelect Human All Exon V4 kit (Agilent) on a HiSeq 2000 instrument (Illumina) according to the manufacturer’s recommendations for paired-end reads. Raw data were processed as previously described. Files were aligned to the reference human genome (GRCh37/hg19) using BWAv0.6.7, and potential duplicate paired-end reads were removed by Picard v1.109. Indel realignment and base quality score recalibration were conducted with GATK v3.3-0. Variants with a quality score >30 and alignment quality score >20 were annotated with SeattleSeq SNP Annotation. Rare variants present at a frequency above 1% in dbSNP 138 and the NHLBI GO Exome Sequencing Project, Exome Variant Server, or ExAC, or present from local exomes of unaffected individuals were excluded. Variant prioritization focused on variants that were de novo heterozygous, were compound heterozygous, or were hemizygous and affected the coding sequence (missense, nonsense, and splice-site variants and coding indels). Candidate variants were then inspected using the Integrative Genomics Viewer and were validated via Sanger sequencing.
For Families 4 and 5, exome sequencing was performed as described previously.12
For Families 6 and 7, diagnostic trio whole-exome sequencing (WES) was performed on extracted genomic DNA from the peripheral blood of Individual 6 and both of her unaffected parents. The pipeline involved exon targeting with the Agilent SureSelect XT2 All Exon V4 kit, sequencing with Illumina HiSeq 2000 sequencing system with 100bp paired-end reads, and data analysis with XomeAnalyzer in comparison to the published human genome build UCSC hg19 reference sequence. All variants were confirmed through the use of Sanger sequencing.
For Family 8, WGS was performed on extracted genomic DNA from the peripheral blood of Individual 8 and both of her unaffected parents. Sequencing was performed on a HiSeq2500 (Illumina) and alignment was performed using Illumina’s Isaac aligner against the reference human genome GRCh37. The length of paired-end reads was 150bp, and the mean depth of coverage across individuals was 30×. Clinical genome interpretation was performed using Omicia’s Opal platform.
WES on Individual 9 was performed in Macrogen, Korea as described elsewhere.13 Briefly, target enrichment was performed with 2 μg genomic DNA using the SureSelectXT Human All Exon Kit version 6 (Agilent Technologies) to generate barcoded WES libraries. Libraries were sequenced on the HiSeqX platform (Illumina) with 50× coverage. Quality assessment of the sequence reads was performed by generating quality control statistics through the use of FastQC. The bioinformatics filtering strategy included screening for only exonic and donor/acceptor splicing variants. In accordance with the pedigree and phenotype, priority was given to rare variants (<0.01% in public databases, including 1000 Genomes Project; National Heart, Lung, and Blood Institute [NHLBI] Exome Variant Server; Complete Genomics 69; and Exome Aggregation Consortium [ExAC v0.2]) that fit a recessive (homozygous or compound heterozygous) or a de novo model and/or variants in genes previously linked to developmental delay, intellectual disability, and other neurological disorders.
Fluorescence-Activated Cell Sorting (FACS)
Fresh blood samples from the affected children and healthy controls were stained for 1 h on ice with the GPI-AP markers: phycoerythrin (PE)-conjugated anti human CD16 (BioLegend), fluorescein isothiocyanate (FITC)-conjugated mouse anti human CD55 and CD59 (BD PharMingen), or FLAER-Alexa 448 (Cedarlane). Red blood cells were lysed in FACS Lysing Solution (BD Bioscience). For fibroblasts, cells were harvested at 80%–90% confluency; stained with FLAER-Alexa 448, FITC-conjugated mouse anti human CD73, or PE-conjugated mouse anti human CD109 (BioLegend) for 1 h on ice in the incubation buffer containing 0.5% bovine serum albumin (BSA); then fixed in 3.7% formaldehyde. For all assays, non-specific binding was washed off before we analyzed the assays through the use of a BD FACSCanto II system (BD Biosciences) and then analyzed them using Cytobank software.
Rescue Assays of GPI-APs on Fibroblasts
Lentiviruses carrying a WT PIGK-pEZ-Lv105 or an empty-pEZ-Lv105 construct (GeneCopoeia) with the presence of packaging plasmids pMD2.G and psPAX2 (AddGene) were produced in HEK293T cells. Fibroblasts were transduced with the lentiviruses and selected via Puromycin resistance. These cells, untransduced cells, as well as control cells, were subjected to FACS analyses as described above for fibroblasts.
In Vitro Functional Assays
PIGK-deficient Chinese hamster ovary (CHO) cells (clone 10.2.2 previously published) were transfected by electroporation with mutant or WT PIGK cDNA expressing strong (signal recognition [SR] alpha) or weak (thymidine kinase) promoter-driven plasmids. After two days, the cells were analyzed using FACS, and immunoblotting was performed.
Results
Clinical Descriptions
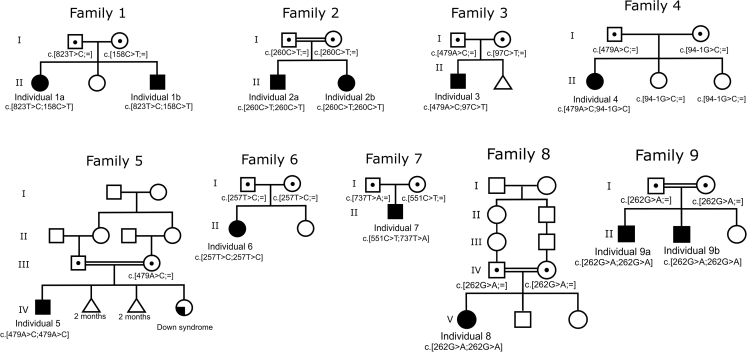
The individuals with bi-allelic pathogenic PIGK variants present several clinical features common to other known IGDs (see (see Table 1 for clinical features, and Figure 1 for pedigrees). Hypotonia was present in all affected individuals, and all affected individuals presented with developmental delay or, when it could be assessed, intellectual disability (from mild to severe, see Table S1 for details). Cerebellar atrophy was noted in eight individuals and was progressive, ataxia was noted in five individuals, and other movement disorders were noted in three. Epilepsy was seen in four out of eight individuals and was well controlled in three of the four. One individual had three episodes of fever-related seizures and no afebrile seizures by the age of 3 years and 5 months. Facial dysmorphisms were noted in seven individuals and varied between individuals (see Figure 2 and Table S1). Other malformations or anomalies were relatively infrequent, and included brachydactyly, hydronephrosis, and teeth anomalies. These issues are also seen in other IGDs.6
Table 1.
Phenotypic Features of Individuals with Biallelic Inactivating Variants in PIGK
| Individual | 1A | 1B | 2A | 2B | 3 | 4 | 5 | 6 | 7 | 8 | 9A | 9B | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hypotonia | + | + | + | + | + | + | + | + | + | + | + | + | 12/12 |
| DD/ID | + | + | + | + | + | + | + | + | + | + | + | + | 12/12 |
| Cerebellar atrophy | + | - | + | + | + | + | + | + | - | + | + | + | 10/12 |
| Ataxia | - | - | + | + | + | + | + | na1 | + | na | na | na | 6/8 |
| Other movement disorder | + | + | - | - | - | - | - | + | - | - | - | - | 3/12 |
| Epilepsy/seizures | + | - | + | + | - | - | - | + | + | +2 | + | + | 8/12 |
| Dysmorphisms | + | + | + | + | + | + | - | + | - | - | + | + | 9/12 |
| Ophthalmological anomalies | + | - | - | + | + | + | - | - | + | + | + | + | 8/12 |
| Genito-urinary malformation | + | + | - | - | - | - | - | - | - | - | - | - | 2/12 |
| Gastrointestinal anomalies including GERD | - | - | - | - | + | - | - | - | + | - | - | - | 2/12 |
| Teeth anomalies | + | - | + | + | - | - | - | + | - | - | + | - | 5/12 |
| Hand/foot anomalies | + | - | + | + | - | - | - | - | - | - | - | - | 3/12 |
| Skeletal findings | - | - | + | + | - | - | - | - | - | - | - | - | 2/12 |
| Low serum alkaline phosphatase | - | - | + | + | - | - | - | - | - | - | na | na | 2/10 |
na = not available. DD/ID = developmental disability and/or intellectual disability. GERD = gastroesophageal reflux disease.
Difficult to assess due to non-ambulatory status and no purposeful hand use. However, cerebellar dysfunction signs include saccadic ocular pursuit and end-gaze nystagmus.
Febrile seizures only.
Figure 1.
Pedigrees of Families with Bi-allelic PIGK Variants
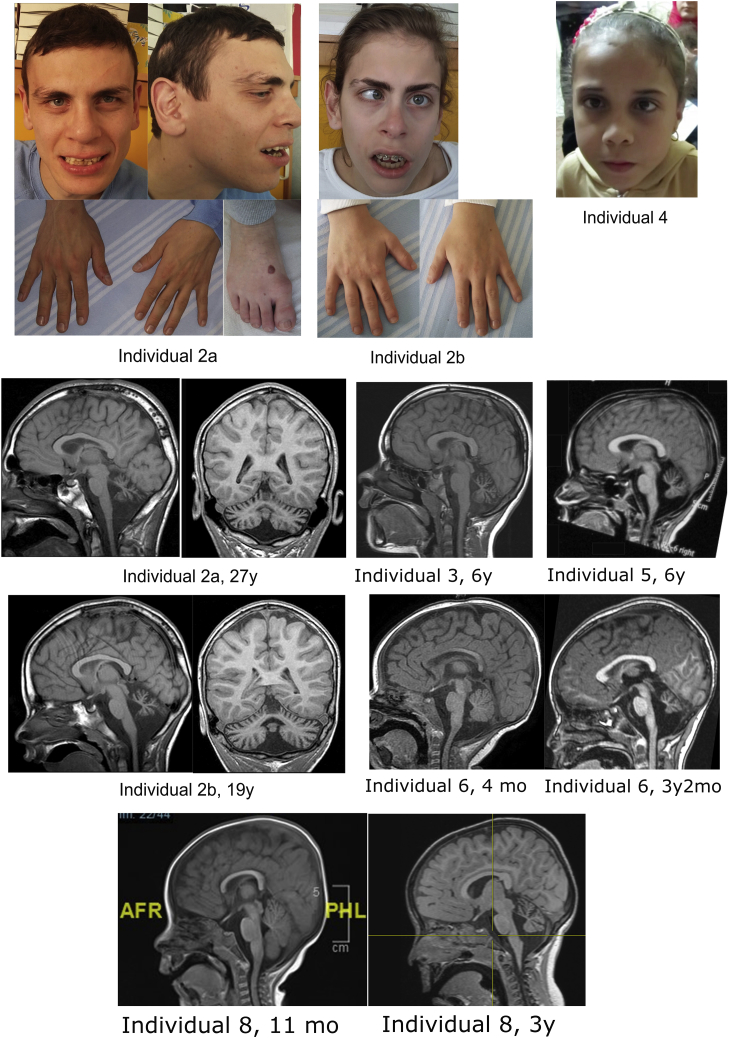
Figure 2.
Facial and MRI Features of Subjects with PIGK Variants
Clinical features in two siblings of Family 2. Facial dysmorphisms: long face, sparse hair, high anterior hairline, prominent forehead, broad and laterally sparse eyebrows, thin upper lip, antihelix shelf, prominent antitragus, dental crowding, abnormally shaped teeth, and brachydactyly in hands and feet. Magnetic resonance imaging (MRI) findings: cerebellar atrophy in Individuals 2A, 2B, 3, 5, 6, and 8.
Molecular Analyses
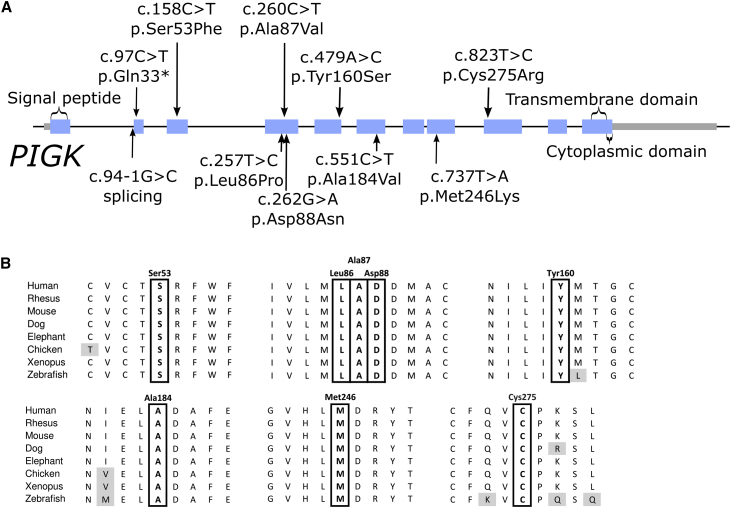
Via WES and WGS, we identified eight different PIGK variants in ten affected individuals in a homozygous or compound heterozygous state (Table 2). Individuals 1A and 1B had two missense changes, c.823T>C (p.Cys275Arg) and c.158C>T (p.Ser53Phe), which had been inherited from their father and mother, respectively (Figure 1, RefSeq accession number NM_005482.3). Note that p.Cys275 is not the cysteine forming the aforementioned disulfide bridge with PIGT (Cys92). The two affected individuals in Family 2 were homozygous for the c.260C>T nucleotide substitution (p.Ala87Val), whereas Individual 3 was compound heterozygous for the c.97C>T (p.Gln33∗) and c.479A>C (p.Tyr160Ser) changes. Notably, the same c.479A>C variant was also found to be heterozygous in Individual 4 and homozygous in Individual 5, and all three families have African Arab origins; thus the variant might represent a founder mutation in these populations. Individual 4 also carries maternal c.94-1G>C, which is an essential splice acceptor site variant (Figure 3). Individual 6 has a homozygous variant, c.257T>C (p.Leu86Pro). Individual 7 is compound heterozygous for c.551C>T (p.Ala184Val) and c.737T>A (p.Met246Lys). Individuals 8 and 9 have a homozygous variant, c.262G>A (p.Asp88Asn), and both families are of Indian origin. As shown in Figure 3, all the variants causing missense mutations affect highly conserved residues.
Table 2.
List of the PIGK Variants Identified in the Subjects Included in the Study
| Family | Genomic Variant (hg19) | DNA Variant (RefSeq: NM_005482.3) | Protein Variant | Inheritance | gnomAD Minor Allele Frequency (no homozygotes found for any variant) |
|---|---|---|---|---|---|
| 1 | chr1:g.77620297A>G | c.823T>C | p.Cys275Arg | compound heterozygous | 0 |
| 1 | chr1:g.77672406G>A | c.158C>T | p.Ser53Phe | compound heterozygous | 0 |
| 2 | chr1:g.77635060G>A | c.260C>T | p.Ala87Val | homozygous | 0.000004129, highest 0.000008978 (European non-Finnish population) rs772948495 |
| 3 | chr1:g.77632412T>G | c.479A>C | p.Tyr160Ser | compound heterozygous | 0.00001504, highest 0.00004152 (African population) |
| 3 | chr1:g.77676171G>A | c.97C>T | p.Gln33∗ | compound heterozygous | 0 |
| 4 | chr1:g.77632412T>G | c.479A>C | p.Tyr160Ser | compound heterozygous | 0.00001504, highest 0.00004152 (African population) |
| 4 | chr1:g.77676175C>G | c.94-1G>C | splice site | compound heterozygous | 0 |
| 5 | chr1:g.77632412T>G | c.479A>C | p.Tyr160Ser | homozygous | 0.00001504, highest 0.00004152 (African population) |
| 6 | chr1:g.77635063A>G | c.257T>C | p.Leu86Pro | homozygous | 0 |
| 7 | chr1:g.77629564G>A | c.551C>T | p.Ala184Val | compound heterozygous | 0.000004011, highest 0.000008837 (European, non-Finnish) |
| 7 | chr1:g.77627056A>T | c.737T>A | p.Met246Lys | compound heterozygous | 0 |
| 8 | chr1:g.77635058C>T | c.262G>A | p.Asp88Asn | homozygous | 0.0000124, highest 0.0001071 (South Asian population) |
| 9 | chr1: g.77635058C>T | c.262G>A | p.Asp88Asn | homozygous | 0.0000124, highest 0.0001071 (South Asian population) |
Figure 3.
PIGK Variants and Conservation of Affected Residues
(A) Location of the variants on the PIGK gene and corresponding protein. Introns not drawn to scale.
(B) Multiple alignment of phosphatidylinositol glycan (PIG) anchor biosynthesis class K (PIGK) orthologs showing conservation of the affected residues in vertebrates.
Cell Surface Abundance of GPI-AP in Blood Cells
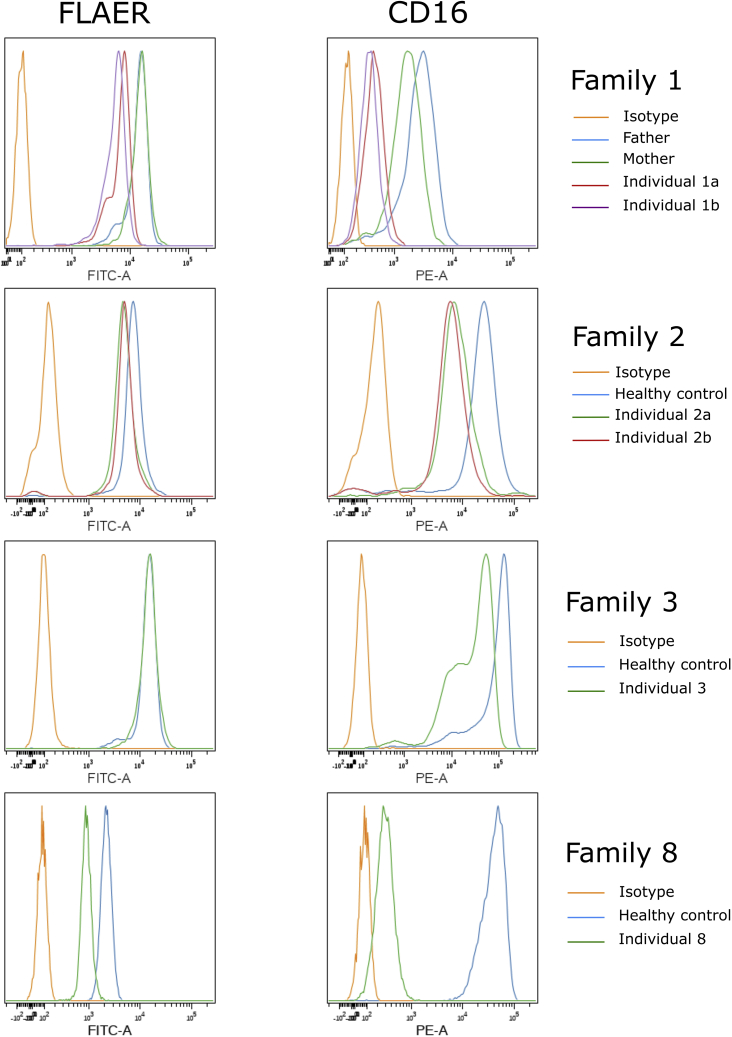
Flow cytometry on blood samples from affected individuals of Families 1, 2, and 3 showed very low levels of cell surface CD16 on granulocytes (the most sensitive marker for inherited GPI deficiency) (Figure 4). The two individuals in Family 1 have 21–26% of CD16 cell surface abundance compared with their parents. These levels were found to be about 35% and 25% in Individuals 2A and 2B, respectively, versus unrelated healthy controls, and a similar diminution was also seen in Individual 3. In Individual 8, significantly less CD16 is found at the cell surface; only 5% of the levels found in the control. For FLAER (marker for all GPI-APs), a low signal was also found in Families 1 and 2, whereas this marker appeared to be normal in Family 3. Individuals 1A and 1B have 50% lower levels compared to their parents, and a more moderate decrease of 30–35% was observed in the individuals of Family 2. Individual 8 has a 60% decrease compared to an unrelated control (Figure 4). There was also a decrease of CD24 in granulocytes and CD14 in monocytes (data not shown). In all tested families, no significant decrease was noted for CD55 or CD59 (Figure S1).
Figure 4.
Cell Surface GPI-AP Levels in Blood Cells of Affected Individuals
Blood samples from the individuals in Families 1, 2, and 3 and control cells were stained with FLAER and CD16. Figure shows representative analysis of cell surface GPI-AP levels of granulocytes from triplicate experiments.
Overexpression of WT PIGK in Fibroblasts from Affected Individuals Can Rescue Its GPI-AP Deficiency
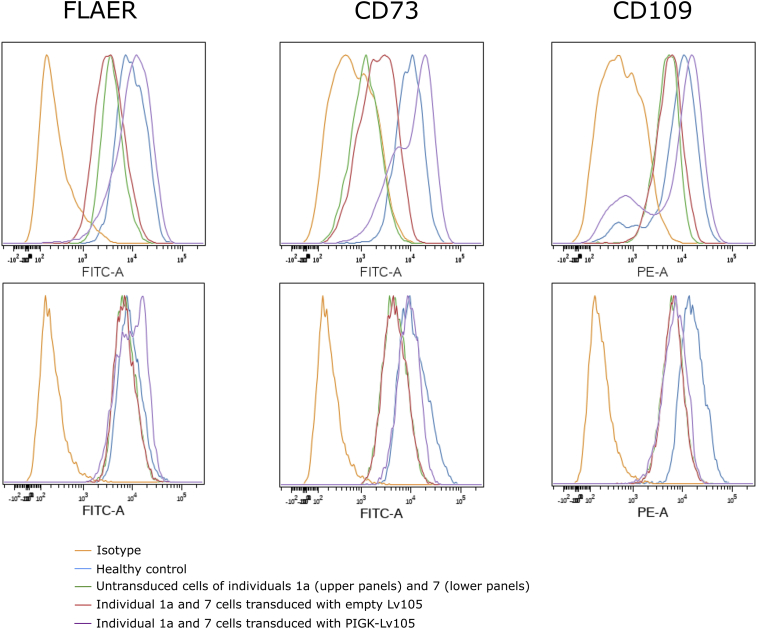
We examined fibroblasts from Individual 1A and Individual 7. For Individual 1A, we found 40% signal for FLAER compared to healthy fibroblasts, 15% for CD73, and 50% for CD109. These marker levels in Individual 7’s cells were 80%, 50%, and 40% for FLAER, CD73, and CD109, respectively (Figure 5). We therefore stably transduced these cells with a lentivirus which expresses WT PIGK. The results indicate that although the empty vector did not change the GPI-AP cell surface levels, the overexpression can increase the cell surface abundance of all of these GPI-APs in Individual 1A’s fibroblasts to similar levels to those seen in healthy control fibroblasts. For Individual 7’s cells, FLAER and CD73 were completely restored, but only partial restoration can be seen for CD109.
Figure 5.
Rescue Assays in Fibroblasts
Skin fibroblasts derived from Individuals 1A (upper panels) and 7 (lower panels) were transduced with PIGK-expressing-Lv105 lentivirus or empty Lv105-lentivirus, and non-transduced cells were stained with FLAER, CD73, and CD109. The figure shows representative results from experiments done in triplicate.
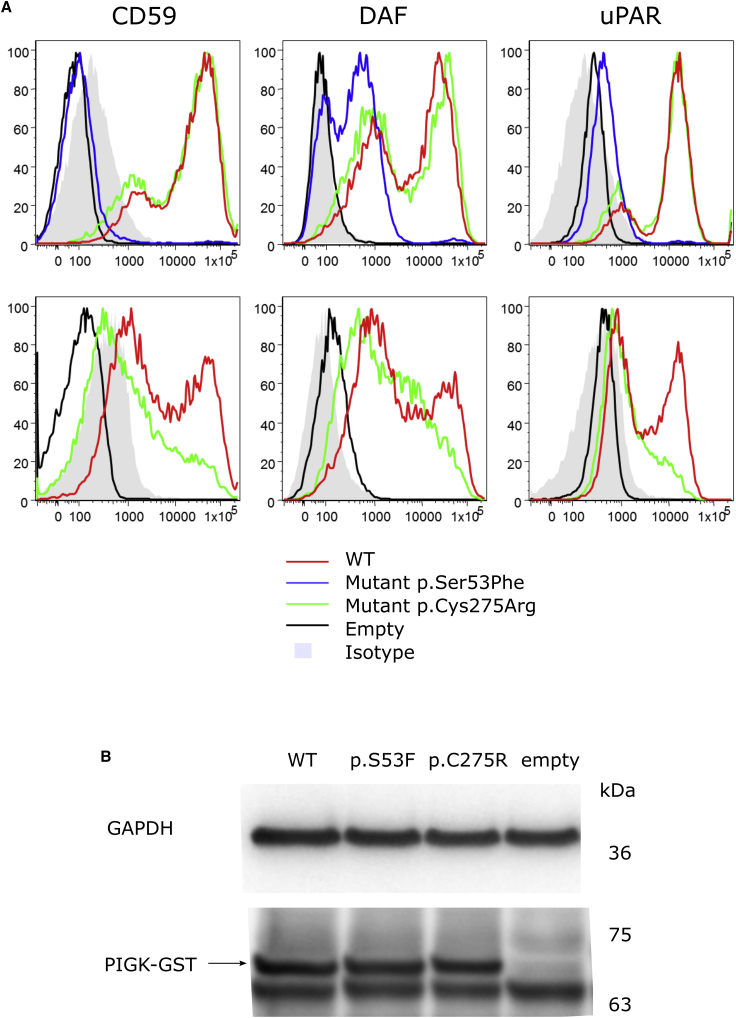
Effects of PIGK Variants in GPI-AP Cell Surface Abundance In Vitro
The effects of the variants found in Family 1 were also studied by using a PIGK-deficient CHO cell model to further demonstrate that they lead to a protein with a loss of function. The cells were transfected with WT or mutant pME-hPIGK GST (pME has a strong SRα promoter) and with pTK-hPIGK GST (pTK has a weaker promoter, thymidine kinase promoter). FACS analysis was performed 2 days post-transfection to check the cell surface abundance of CD59, CD55 (DAF), and CD87 (uPAR). As shown in Figure 6, even using the strong promoter-driven pME vector, PIGK cDNA bearing the Ser53Phe variant could not rescue the surface abundance of GPI-APs such as CD59, DAF, and uPAR, whereas the protein with the p.Cys275Arg variant could. However, using the weak promoter-driven pTK vector, the p.Cys275Arg PIGK protein failed to rescue the surface abundance of GPI-APs completely; this result provides evidence of the hypomorphic behavior of this mutant. These mutant PIGK proteins were expressed at similar levels as was the WT protein, and this suggests that the variant did not affect the protein cell surface abundance (Figure 5B).
Figure 6.
In Vitro Functional Studies for PIGK Variants Identified in Individuals 1A and 1B
(A) Fluorescence-activated cell sorting (FACS) analysis of phosphatidylinositol glycan (PIG) anchor biosynthesis class K (PIGK)-deficient CHO cells transfected with mutant or wild-type (WT) PIGK cDNA driven by a strong promoter, pME vector (upper panels), or a weak promoter, pTK vector (lower panels).
(B) Expression of WT and mutant PIGK-GST protein in PIGK deficient CHO cells transfected with mutant or WT PIGK cDNA driven by a strong promoter, pME vector. GAPDH is a loading control.
Discussion
To date, four genes in the transamidase complex, PIGT (MIM: 610272), GPAA1 (MIM: 603048), PIGS (MIM: 610271), and PIGU (MIM: 608528), have been reported to cause IGDs. Phenotype clustering was illustrated by a heatmap in the recent paper on PIGU deficiency.14 The common phenotypes seen in individuals with variants in these genes are brain anomalies (notably cerebellar atrophy), developmental disability and/or intellectual disability (DD/ID), and dysmorphic facial features. These characteristics are also found in individuals with biallelic PIGK variants. Elevated serum alkaline phosphatase (ALP), a feature found in half of other known IGDs, was not seen in individuals with PIGK variants, similarly to what was seen in individuals with variants in PIGS and GPAA1. In fact, two individuals in our cohort actually had low ALP. As for PIGT, seven out of 13 individual with PIGT mutations also had low levels of serum ALP. This was caused by the failure of ALP protein precursor processing by the GPI transamidase complex, a failure which resulted in ER-associated degradation of the protein. Because ALP is critical for the synthesis of hydroxyapatite, this could explain the low bone density found in some individuals with mutations in the transamidase complex (seen with PIGT, PIGU, and GPAA1 mutations). A low bone density was not noted with PIGS deficiency or in the PIGK deficiency we describe here, but bone density studies were not systematically performed in those cohorts. Hypotonia was common in the cohort described here, and ophthalmological anomalies were found in a half of individuals with PIGK variants; these results were comparable to those for individuals with mutations in other components of the transamidase complex. Seizures were a common manifestation in individuals with PIGT, GPAA1, and PIGS variants, and were observed in half of the individuals with PIGK variants.
Flow cytometry analyses showed decreased cell surface presence of CD16 in granulocytes (Figure 4); this is similar to the results for most previously reported IGD cases. Total cell surface GPI-AP, which is measured by the FLAER signal, is also very low in granulocytes. However, CD55 and CD59, two GPI-AP markers that decreased in both granulocytes and lymphocytes of some individuals with PIGS variants, notably were not changed in the individuals with PIGK variants. This is also seen in other IGD syndromes and may be due to cell-type-specific effects. Interestingly, in fibroblasts of Individual 1A, we detected very low levels of all measured GPI markers, especially of CD73 (5′-nucleotidase).
The variants in our cohort affected all regions of PIGK, which lies inside the ER, but there was a clustering of three mutations at amino acids 86, 87, and 88. This clustering lies just upstream of the cysteine residue at amino acid 92, which was previously shown to be important for PIGK to form a disulfide bridge with PIGT. It is possible that these variants affect the interaction of the PIGK cysteine protease with PIGT. Protein structure homology modeling using SWISS-MODEL (based on PDB structure 4FGU15) showed homology with the cysteine protease legumain. PIGK amino acids 86, 87, and 88 in this model lie within the legumain insertion-loop, a well-conserved region thought to perhaps regulate protease activity.15,16 These possibilities could be investigated in future biochemical studies.
A wide variety of genes have been associated with cerebellar ataxia and cerebellar atrophy. When occurrence is in adulthood, a dominant form of spinocerebellar ataxia, often caused by trinucleotide repeat expansion disorders, has to be considered. (Note that repeat expansions can be missed by next-generation sequencing.) With childhood onset, a recessive form is more common, and Friedrich’s ataxia and ataxia telangiectasia are among the most frequent disorders. The initial workup of affected individuals also includes detailed testing for reversible causes of ataxia, including metabolic, toxic, and autoimmune disorders, as well as testing for nutritional disorders (such as measuring vitamin E). Once more common and treatable causes have been screened for, a next-generation sequencing panel or exome sequencing is often the next diagnostic test. Cerebellar atrophy is noted with several IGDs (PIGA, PIGG, PIGL, PIGN, PIGT PIGS,7 GPAA1, and PGAP1) and there does not seem to be a correlation between the location of the protein in the biosynthesis pathway and the presence of cerebellar atrophy. Given that inherited GPI deficiency can be tested for through the use of flow cytometry, such a test could be considered in the initial evaluation of cerebellar hypoplasia, or following negative next-generation sequencing panel if IGD genes were not included, as is often the case, before moving to an exome which is not always clinically available. This test is available in research laboratories, and it could also potentially be made available after specifically requesting CD16 in granulocytes from clinical labs performing high-sensitivity flow cytometry testing for paroxysmal nocturnal hemoglobinuria.
In summary, we present an IGD characterized by hypotonia, DD/ID, cerebellar atrophy, and epilepsy. We show GPI-AP deficiency in affected individual cells, and that this deficiency can be rescued by overexpression of WT PIGK, and we show the effect of individual mutations in PIGK knockout cells. Functional analyses using PIGK-deficient neuronal cells and perhaps a neuron-specific PIGK knockout mouse model could be useful next steps to better characterize which GPI-APs are impaired by PIGK deficiency in various neuronal subpopulations, to better elucidate the downstream effects of GPI-AP abnormalities, and ultimately to understand the neuropathogenesis of IGDs.
Declaration of Interests
The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing conducted at Baylor Genetics. F.M.Z. and R.P. are employees of GeneDx.
Acknowledgments
We thank Kana Miyanagi and Saori Umeshaita for technical assistance. This work is supported by grants from the Ministry of Health, Labour, and Welfare to Y.M.; from the Fondazione Bambino Gesù (Vite Coraggiose) to M.T.; from the Italian Ministry of Health (Ricerca Corrente 2018 and 2019 to A.C. and 2019 M.N.); and from the Canadian Institutes of Health Research (CIHR) and Fonds de Récherche Santé Québec (FRQS) to P.M.C. The identification of the homozygous PIGK variant in Individual 8 was made possible through access to the data and findings generated by the 100,000 Genomes Project. The 100,000 Genomes Project is managed by Genomics England Limited (a wholly owned company of the Department of Health and Social Care UK). The 100,000 Genomes Project is funded by the National Institute for Health Research and National Health Service (NHS) England. The Wellcome Trust, Cancer Research UK, and the Medical Research Council have also funded research infrastructure. The 100,000 Genomes Project uses data provided by individuals and collected by the NHS as part of their care and support. Data from Individual 9 was collected as part of the Synaptopathies (SYNaPS) Study Group collaboration funded by The Wellcome Trust and strategic award (Synaptopathies) funding (WT093205 MA and WT104033AIA). This research was conducted as part of the Queen Square Genomics group at University College London, supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre.
Published: March 26, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.03.001.
Web Resources
1000 Genomes Project, https://www.internationalgenome.org/
100,000 Genomes Project, https://doi.org/10.6084/m9.figshare.4530893.v5
Complete Genomics 69, https://www.completegenomics.com/public-data/69-genomes/
ExAC Browser, http://exac.broadinstitute.org/
FastQC, https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
National Heart, Lung, and Blood Institute (NHLBI) Exome Variant Server, https://evs.gs.washington.edu/EVS/
Online Mendelian Inheritance in Man (OMIM), https://www.omim.org
PIGK mutation database including variants in this manuscript, https://databases.lovd.nl/shared/genes/PIGK
PIGK structure model, https://swissmodel.expasy.org/repository/uniprot/Q92643
Supplemental Data
References
- 1.Kinoshita T., Fujita M. Biosynthesis of GPI-anchored proteins: special emphasis on GPI lipid remodeling. J. Lipid Res. 2016;57:6–24. doi: 10.1194/jlr.R063313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng B.G., Freeze H.H. Human genetic disorders involving glycosylphosphatidylinositol (GPI) anchors and glycosphingolipids (GSL) J. Inherit. Metab. Dis. 2015;38:171–178. doi: 10.1007/s10545-014-9752-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benghezal M., Benachour A., Rusconi S., Aebi M., Conzelmann A. Yeast Gpi8p is essential for GPI anchor attachment onto proteins. EMBO J. 1996;15:6575–6583. [PMC free article] [PubMed] [Google Scholar]
- 4.Yu J., Nagarajan S., Knez J.J., Udenfriend S., Chen R., Medof M.E. The affected gene underlying the class K glycosylphosphatidylinositol (GPI) surface protein defect codes for the GPI transamidase. Proc. Natl. Acad. Sci. USA. 1997;94:12580–12585. doi: 10.1073/pnas.94.23.12580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohishi K., Nagamune K., Maeda Y., Kinoshita T. Two subunits of glycosylphosphatidylinositol transamidase, GPI8 and PIG-T, form a functionally important intermolecular disulfide bridge. J. Biol. Chem. 2003;278:13959–13967. doi: 10.1074/jbc.M300586200. [DOI] [PubMed] [Google Scholar]
- 6.Bellai-Dussault K., Nguyen T.T.M., Baratang N.V., Jimenez-Cruz D.A., Campeau P.M. Clinical variability in inherited glycosylphosphatidylinositol deficiency disorders. Clin. Genet. 2019;95:112–121. doi: 10.1111/cge.13425. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen T.T.M., Murakami Y., Wigby K.M., Baratang N.V., Rousseau J., St-Denis A., Rosenfeld J.A., Laniewski S.C., Jones J., Iglesias A.D. Mutations in PIGS, Encoding a GPI Transamidase, Cause a Neurological Syndrome Ranging from Fetal Akinesia to Epileptic Encephalopathy. Am. J. Hum. Genet. 2018;103:602–611. doi: 10.1016/j.ajhg.2018.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turnbull C., Scott R.H., Thomas E., Jones L., Murugaesu N., Pretty F.B., Halai D., Baple E., Craig C., Hamblin A., 100 000 Genomes Project The 100 000 Genomes Project: bringing whole genome sequencing to the NHS. BMJ. 2018;361:k1687. doi: 10.1136/bmj.k1687. [DOI] [PubMed] [Google Scholar]
- 10.Niceta M., Stellacci E., Gripp K.W., Zampino G., Kousi M., Anselmi M., Traversa A., Ciolfi A., Stabley D., Bruselles A. Mutations Impairing GSK3-Mediated MAF Phosphorylation Cause Cataract, Deafness, Intellectual Disability, Seizures, and a Down Syndrome-like Facies. Am. J. Hum. Genet. 2015;96:816–825. doi: 10.1016/j.ajhg.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dixon-Salazar T.J., Silhavy J.L., Udpa N., Schroth J., Bielas S., Schaffer A.E., Olvera J., Bafna V., Zaki M.S., Abdel-Salam G.H. Exome sequencing can improve diagnosis and alter patient management. Sci. Transl. Med. 2012;4:138ra78. doi: 10.1126/scitranslmed.3003544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mencacci N.E., Kamsteeg E.J., Nakashima K., R’Bibo L., Lynch D.S., Balint B., Willemsen M.A., Adams M.E., Wiethoff S., Suzuki K. De Novo Mutations in PDE10A Cause Childhood-Onset Chorea with Bilateral Striatal Lesions. Am. J. Hum. Genet. 2016;98:763–771. doi: 10.1016/j.ajhg.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knaus A., Kortüm F., Kleefstra T., Stray-Pedersen A., Đukić D., Murakami Y., Gerstner T., van Bokhoven H., Iqbal Z., Horn D. Mutations in PIGU Impair the Function of the GPI Transamidase Complex, Causing Severe Intellectual Disability, Epilepsy, and Brain Anomalies. Am. J. Hum. Genet. 2019;105:395–402. doi: 10.1016/j.ajhg.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dall E., Brandstetter H. Mechanistic and structural studies on legumain explain its zymogenicity, distinct activation pathways, and regulation. Proc. Natl. Acad. Sci. USA. 2013;110:10940–10945. doi: 10.1073/pnas.1300686110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dall E., Brandstetter H. Structure and function of legumain in health and disease. Biochimie. 2016;122:126–150. doi: 10.1016/j.biochi.2015.09.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.