Abstract
We report the dynamic change process of target genes by RT-PCR testing of SARS-Cov-2 during the course of a COVID-19 patient: from successive negative results to successive single positive nucleocapsid gene, to two positive target genes (orf1ab and nucleocapsid) by RT-PCR testing of SARS-Cov-2, and describe the diagnosis, clinical course, and management of the case. In this case, negative results of RT-PCR testing was not excluded to diagnose a suspected COVID-19 patient, clinical signs and symptoms, other laboratory findings, and chest CT images should be taken into account for the absence of enough positive evidence. This case highlights the importance of successive sampling and testing SARS-Cov-2 by RT-PCR as well as the increased value of single positive target gene from pending to positive in two specimens to diagnose laboratory-confirmed COVID-19.
Keywords: Coronavirus, SARS-Cov-2, COVID-19, RT-PCR testing, Nucleocapsid
1. Introduction
In December 2019, a cluster of cases of pneumonia causing by unknown virus were reported in Wuhan, Hubei Province, China, origin of the virus was traced back to Huanan Seafood Wholesale Market in Wuhan. On January 7, 2020, Chinese health authorities confirmed that this cluster was associated with a novel coronavirus, 2019-nCoV [1]. The virus was subsequently renamed SARS-Cov-2 as it is similar to the coronavirus responsible for severe acute respiratory syndrome (SARS-CoV), a member of the subgenus Sarbecovirus (Beta-CoV lineage B), with which it shares more than 79% of its sequence [2]. Coronaviruses (CoVs) are single-stranded RNA viruses which belong to order Nidovirales, family Coronaviridae, subfamily Coronavirinae [3], and have been classified into four major groups: α-CoVs, β-CoVs, γ-CoVs and δ-CoVs [4]. SARS-CoV-2 is the seventh known coronavirus to infect humans, after 229E, NL63, OC43, HKU1, MERS-CoV, and the original SARS-CoV [5]. The whole genome sequence of Wuhan new virus (WH-Human_1) was first released on Jan 10, 2020 [6]. Five typical ORFs were identified on one complete SARS-CoV-2 genome (29870-bp, excluding the poly (A) tail) in GenBank (accession number MN908947), including ORF1ab polyprotein (7096-aa), spike glycoprotein (1273-aa), envelope protein (75-aa), membrane protein (222-aa), and nucleocapsid protein (419-aa) [7]. Coronavirus Disease 2019 (COVID-19) caused epidemic in China, as well as infection cases worldwide. This report describes diagnostic process of a COVID-19 patient from successive negative results to successive single positive nucleocapsid gene, then to two positive target genes (orf1ab and nucleocapsid) by RT-PCR testing. The case provides an example for diagnosing suspected COVID-19, despite successive negative results of reverse transcriptase polymerase chain reaction (RT-PCR) testing in the early stages. Successive sampling and testing SARS-Cov-2 by RT-PCR as well as clinical signs and symptoms, other laboratory findings, and chest CT images, should be taken into account for the absence of enough positive evidence.
2. Case report
On January 23, 2020, a 54-year-old man presented to fever clinic in Ningbo First Hospital, Zhejiang Province, China, with a 6-day history of subjective fever and cough, and was admitted in respiratory ward in this hospital, diagnosed with pneumonia (admission day 0). He disclosed that he had dinner with friends in the restaurant in Ningbo on January 14. He and his friends had no tour history to Wuhan, the epidemic focus of COVID-19 outbreak in China. After dinner, he visited Ningbo Haishu Third Hospital for drunkenness, nausea, and vomiting (details not available). He had a fever three days later, and the highest temperature was up to 38.5 °C. On January 18, he presented to the emergency in Ningbo First Hospital, with cough, chills, headache dizziness, and muscle ache, but without expectoration, stuffy nose and running nose, chest distress and chest pain. Routine blood test revealed lymphocytopenia (Table 1 ). The empiric therapy were administrated with levofloxacin, cefuroxime axetil and aloxicillin (details not available). He still had a fever and visited Ningbo Haishu Third Hospital on January 22. C-reactive protein increased to 8.50 mg/L, antigen test for influenza A and B was negative, chest CT images showed evidence of pneumonia in the lower lobe of the left lung.
Table 1.
Laboratory test results during the course of a Corona Virus Disease 2019 patient.
| Date | Jan. 18 | Jan. 22 | Jan. 23 | Jan. 25 | Jan. 26 | Jan. 28 | Jan. 30 | Jan. 31 | Feb. 1 | Feb. 2 | Feb. 4 | Feb. 5 | Feb. 7 | Feb. 8 | Feb. 10 | Feb. 11 | Feb. 12 | Feb. 14 | Feb. 15 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Admission day | Day 0 | Day 2 | Day 3 | Day 5 | Day 7 | Day 8 | Day 9 | Day 10 | Day 12 | Day 13 | Day 15 | Day 16 | Day 18 | Day 19 | Day 20 | Day 22 | Discharged | |||
| Fever (oC) | 38.4 | 38.4 | 38.4 | 38.2 | 37.1 | 38.9 | 38.6 | 38.6 | 37.4 | 37.0 | 37.1 | 37.2 | 37.0 | 36.8 | 37.1 | 37.1 | 36.9 | 36.7 | ||
| White cell count (109/L) | 6.71 | 4.02 | 2.85 | 4.22 | 11.54 | 5.22 | 7.28 | 4.05 | ||||||||||||
| Neutrophil (%) | 75.2 | 68.2 | 62.7 | 67 | 92.5 | 64.9 | 70.7 | 65.5 | ||||||||||||
| Lymphocyte (%) | 13.6 | 23.4 | 28.8 | 24.2 | 5.3 | 24.5 | 17.7 | 23.0 | ||||||||||||
| C-reactive protein (mg/L) | 2.9 | 8.5 | 5.66 | 7.04 | 43.4 | 6.73 | 8.77 | 1.58 | ||||||||||||
| RT-PCRa | orf1ab | – | – | – | – | +(Ct, 33.78) | – | – | ||||||||||||
| N | – | – | +(Ct, 32.84) | +(Ct,3 1 0.54) | +(Ct, 34.74) | – | – | |||||||||||||
| RT-PCRb | orf1ab | – | ||||||||||||||||||
| N | – | |||||||||||||||||||
| RT-PCRc | orf1ab | – | ||||||||||||||||||
| N | – | |||||||||||||||||||
Note: RT-PCR: RT-PCR Testing for SARS-CoV-2, a: oropharyngeal swab specimen, b: deep sputum specimen, c: Anal swab specimen, N: nucleocapsid, +: positive, −: negative, Ct: cycle threshold values.
After admission, the physical examination revealed a body temperature of 38.4 °C, blood pressure of 151/80 mm Hg, respiratory rate of 21 breaths per minute, and oxygen saturation of 95% while the patient was breathing ambient air. Lung auscultation revealed rough breath sounds and a few moist rales in both lungs. The patient reported a few dry cough, and received moxifloxacin (0.4 g qd, ivgtt). He has no previous history of hypertension, without antihypertensive treatment, blood pressure reduced to 105/68 mm Hg from the next day and remained stable.
Given the patient’s recurrent fevers, other laboratory testing was performed, but results revealed no abnormalities, including RT-PCR testing for influenza A RNA, blood cultures, sputum cultures, sputum smear for acid fast bacillus, tumor markers (CEA, AFP, PSA, CA125, CA199), blood biochemistry (hepatic function, renal function, myocardial enzyme spectrum, thyroid function, glucose, fat, electrolyte), coagulation function, D-D dimer, routine urine and stool test, erythrocyte sedimentation rate, specific antibody of syphilis, human immunodeficiency virus, hepatitis B virus, hepatitis C virus and rheumatism. Chest CT images on day 2 showed evidence of pneumonia in the bilateral lower lobes. Given high temperature of 38.5 °C persisted, methylprednisolone (40 mg qd, ivgtt) was administrated on day 4, and the patient was afebrile afterwards. After discontinuation of 3 days of methylprednisolone course, the patient was febrile again, the highest temperature was up to 38.9 °C.
Although patients and their relatives denied the tour history to the outbreak areas, and denied the contact history with persons traveled from the outbreak areas, SARS-CoV-2 nucleic acid were tested by RT-PCR (Novel Coronavirus PCR Fluorescence Diagnostic Kit, Shanghai GeneoDx Biotech). Results of RT-PCR testing using oropharyngeal swab specimens were negative on day 3 and day 8, pending on day 9 and day 10, positive on day 16, and negative on day 19 and day 20.
Given the pending result of RT-PCR testing (single positive nucleocapsid gene) on day 9, combining deteriorating chest CT images on day 7 that infiltrates appeared in the lower lobe of the right lung and bilateral upper lobes, the patient was suspected as a COVID-19 patient, and transferred to the isolation ward to receive antiviral treatment: arbidol tablets (0.2 g tid, p.o), lopinavir and ritonavir tablets (0.5 g bid, p.o), lianhua qingwen capsules (1.4 g tid, p.o). Routine blood test and c-reactive protein revealed secondary bacterial infection, moxifloxacin (0.4 g qd, ivgtt) was administrated.
On day 10, the patient is afebrile, but felt nausea, especially after the medicine. To confirm the pending result, oropharyngeal swab specimen was collected for RT-PCR testing and reported another pending result (single positive nucleocapsid gene) again. Deep sputum specimen was also collected after atomization with hypertonic saline for RT-PCR testing, and reported negative. Treatment plan unchanged.
On day 12, the patient’s clinical condition, routine blood test, and c-reactive protein improved, administration of moxifloxacin was discontinued. Chest CT images on day 13 showed infiltrates in the upper lung lobes increased, the lower lobe of the left lung was consolidated, and dense consolidations in bilateral lower lobes were partially absorbed. On day 16, oropharyngeal swabs for RT-PCR testing was reported positive.
The patient’s clinical condition improved. He reported no fever from day 10, and no cough from day 22. Chest CT images on day 18 and day 22 showed dense consolidations in bilateral lungs absorbed more than before, results of RT-PCR testing of oropharyngeal swab specimens were negative on day 19 and day 20, all symptoms had resolved, the patient was discharged on February 15 (day 23), and sent to Ningbo Haishu Second Hospital for other 14 days. On February 20, 23, 26, the patient was sent to Ningbo First Hospital to undergo evaluation by a provider, and reported no symptom. Oropharyngeal swab specimens were collected for RT-PCR testing and were reported negative every time.
3. Methods
3.1. Specimen collection
Clinical specimens for SARS-CoV-2 diagnostic testing were obtained in accordance with Chinese guidelines [8]. Oropharyngeal swab and anal swab specimens were collected with synthetic fiber swabs, each swab was inserted into a separate sterile tube containing 2 to 3 ml of viral transport medium. Deep sputum specimen was collected after atomization of hypertonic saline, and into a sterile tube containing transport medium. Specimens were stored between 2 °C and 8 °C until ready for shipment to the department of laboratory medicine in Ningbo First Hospital.
3.2. Diagnostic testing for SARS-CoV-2
Nucleic acid extraction was magnetic bead method. Clinical specimens were tested with RT-PCR assay (Novel Coronavirus PCR Fluorescence Diagnostic Kit, Shanghai GeneoDx Biotech). The thermal cycle parameters of RT-PCR amplification were as follows: 42 °C 5 min for reverse transcription, 95 °C 30 s for pre- denaturation, then 40 cycles of PCR (95 °C 10 s, 60 °C 45 s). RT-PCR assay was performed on a Applied Biosystems 7500 Sequence Detection System (ABI, U.S.A). Sequences of primers and fluorescent probe are available from Chinese guidelines [8]. Sample with cycle threshold (Ct) value < 37 was assessed as positive, sample with 37 ≤ Ct value < 40 as gray zone which required retest, sample with no Ct value or Ct value = 40 as negative.
The nucleic acid determination of SARS-CoV-2 is aimed at two target genes: open reading frame 1ab (orf1ab) and nucleocapsid gene. Positive result of RT-PCR testing is determined by two positive target genes in the same specimen. Result of single positive gene is assessed as pending [9], and the result of two negative genes is assessed as negative. If only a single positive gene was tested, it needs to be resampled and retested [9]. However, the criterion of single positive gene was amended in the fifth edition guidelines [8]: if only a single positive gene was tested in two specimens, it can be determined as positive testing of SARS-Cov-2.
3.3. Diagnostic criterion for COVID-19
A confirmed COVID-19 case is determined by positive result of RT-PCR testing of SARS-Cov-2, or by genome sequencing [10].
4. Results
Results of RT-PCR testing using oropharyngeal swab specimens were negative on day 3 and day 8, pending on day 9 and day 10, positive on day 16 (Fig. 1 ), and negative on day 19 and day 20. After discharge, results of RT-PCR testing using oropharyngeal swab specimens on February 20, 23, 26 were all negative.
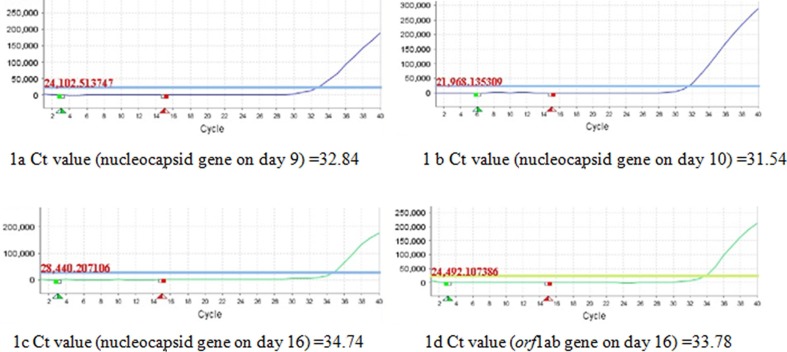
Fig. 1.
RT-PCR amplification curve of positive target genes of SARS-Cov-2, Ct values of positive target of nucleocapsid gene on day 9 (1a), day 10 (1b), day 16 (1c); Ct values of positive target of orf1ab gene (1d) on day 16.
Results using deep sputum specimen on day 10, and anal swab specimen on day 12 were negative (Table 1).
5. Discussion
On January 20, Dr. Zhong Nanshan, an Academician of Chinese Academy of Engineering, made it clear that novel coronavirus was contagious and spread through human-to-human transmission. The policy was initiated that fever patients can only visit fever clinic at first. RT-PCR testing is the gold standard to diagnose SARS-Cov-2. On January 26, first batch of four registration certificate of Novel Coronavirus PCR Fluorescence Diagnostic Kit was issued by the State Food and Drug Administration for the emergency, without a series of clinical trials.
It’s a case in point to explain the amendment for the criterion of single positive gene to diagnose a COVID-19 patient, especially samples with low viral load. Given successive single positive nucleocapsid gene on day9 and day 10, we hypothesize that this fluorescent quantitative PCR kit is more sensitive to amplify nucleocapsid gene than orf1ab gene, samples with low viral load might result in single positive nucleocapsid gene. From Fig. 1, Ct value of nucleocapsid gene on day 10 (31.54) was less than it on day 9 (32.84), which showed a trend toward increasing levels of virus. On day 16, two positive target genes (orf1ab and nucleocapsid) were tested along with higher viral load. If we comply with the pending criterion of single positive gene in old edition guidelines, diagnosis of a COVID-19 patient with low viral load will be delayed or even missed. Therefore, it’s advisable to amend the diagnostic criterion of single positive gene. Fortunately, we treated this case as a COVID-19 patient, 20 days before fifth edition guidelines published on February 21 [9].
The suspected COVID-19 can not be hastily excluded by negative results of RT-PCR testing for SARS-CoV-2. In this case, therapies for recurrent fevers were ineffective in the early stages, we considered the patient as a COVID-19 one, despite negative results of RT-PCR testing on day 3 and day 8. Successive sampling and testing SARS-Cov-2 by RT-PCR caused results of single positive nucleocapsid gene on day 9 and day 10, two positive target genes on day 16. However, false negative factors should be excluded: poor sample quality; sample collection time (too early or too late); incorrect storage, transportation and handling; virus mutation, PCR inhibition and so on. Single positive nucleocapsid gene was reported on day9 and day 10, but deep sputum specimen on day 10 was reported negative, we speculate the negative result might be a false negative for poor sample quality. The sputum specimen was difficult to be collected because of small sputum volume. Although by atomization with hypertonic saline, the sputum specimen was very sticky, we speculate the efficiency of nucleic acid extraction was incomplete, and affected the accuracy of the testing result subsequently. Therefore, collecting a qualified specimen plays a key role in obtaining a correct result.
Although there is no clear evidence of the source of infection, we infer that the patient might be infected with SARS-Cov-2 during dinner or emergency period in Ningbo Haishu Third Hospital for drunkenness on January 14. He had a fever three days later, we infer that the incubation period might be three days, which is accorded with the median incubation period of 4 days (interquartile range, 2 to 7) [11]. In this case, fever and cough were the patient’s chief complaints, which is accorded with two most common symptoms described in 1099 patients with laboratory-confirmed COVID-19 in China: fever (43.8% on admission and 88.7% during hospitalization) and cough (67.8%). Lymphocytopenia and abnormalities on chest CT in the case was also presented in 83.2% of the 1099 patients and in 86.2% of the 975 CT scans, respectively [11].
We observed that on day 9, single positive nucleocapsid gene was reported at the first time, white cell count, neutrophils proportion (%), and c-reactive protein, increased significantly, but lymphocytopenia showed (Table 1). Although secondary bacterial infection was considered, and moxifloxacin administration was effective, but we hypothesize that the virus load increased along with the continuous replication of the virus, then the patient’s immunity was consumed to cause secondary bacterial infection. Moreover, secondary bacterial infection accelerated the replication of the virus, even promoted the transfer of the virus from deep part of the lung to the throat. The hypothesis needs further observation and verification, which provides a new way to diagnose highly suspected cases with negative results of RT-PCR testing of SARS-Cov-2.
6. Conclusion
This case highlights the importance of successive sampling and testing SARS-Cov-2 by RT-PCR as well as the increased value of single positive target gene from pending to positive in two specimens to diagnose laboratory-confirmed COVID-19. RT-PCR testing is the gold standard to diagnose SARS-Cov-2, but the suspected COVID-19 patient can not be hastily excluded by negative results of RT-PCR testing. The patient’s clinical condition, results of routine blood test, c-reactive protein, and chest CT images, should be taken into account if short of enough positive evidence. Fever, cough, lymphocytopenia and abnormalities on chest CT were most common features of COVID-19 [11], but these are non-specificity and indistinguishable clinically from other common infectious diseases, particularly during the winter respiratory virus season. Finally, wearing masks is an essential measure to prevent transmission of SARS-Cov-2.
Acknowledgments
Acknowledgements
This work was supported by Ningbo Natural Science Foundation (2019A610381), and Ningbo Public Welfare Foundation (2019C50087), Ningbo Health Branding Subject Fund (PPXK2018–04).
Declaration of Competing Interest
The authors declare that they have no conflict of interest. We obtained the approval letter of Ningbo First Hospital Ethics Committee (Approval No. 2020-R037).
References
- 1.World Health Organization. Novel coronavirus- China. 2020 (https://www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en/).
- 2.Lu R., Zhao X., Li J. Genomic characterisation and epidemiology of novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020 doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D. Schoeman, B.C. Fielding, Coronavirus envelope protein: current knowledge. Virol. J. 16 (1) (2019) 69. http://doi.org/10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed]
- 4.Saminathan M., Chakraborty S., Tiwari R. Coronavirus infection in equines: a review. Asian J. Anim. Vet. Adv. 2014;9:164–176. [Google Scholar]
- 5.Zhu N., Zhang D., Wang W. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. January 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang, Y.Z., 2020. Initial genome release of novel coronavirus. http://virological.org/t/initial-genome-release-of-novel-coronavirus/319?from=groupmessage.
- 7.Wu F., Zhao S., Yu B. A new coronavirus associated with human respiratory disease in China. Nature. 2020 doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.General Office of National Health Commission PRC. Guidelines of prevention and control of COVID-19 [M]. 2020. The fifth edition.
- 9.General Office of National Health Commission PRC. Guidelines of prevention and control of COVID-19 [M]. 2020. The fourth edition.
- 10.General Office of National Health Commission PRC. Guidelines of diagnosis and treatment of COVID-19 [M]. 2020. The sixth edition.
- 11.Guan Wei-jie, Zheng-yi Ni YuHu. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]