Abstract
Objectives
This study determined associations between respiratory viruses and subsequent illness course in primary care adult patients presenting with acute cough and/or suspected lower respiratory tract infection.
Methods
A prospective European primary care study recruited adults with symptoms of lower respiratory tract infection between November 2007 and April 2010. Real-time in-house polymerase chain reaction (PCR) was performed to test for six common respiratory viruses. In this secondary analysis, symptom severity (scored 1 = no problem, 2 = mild, 3 = moderate, 4 = severe) and symptom duration were compared between groups with different viral aetiologies using regression and Cox proportional hazard models, respectively. Additionally, associations between baseline viral load (cycle threshold (Ct) value) and illness course were assessed.
Results
The PCR tested positive for a common respiratory virus in 1354 of the 2957 (45.8%) included patients. The overall mean symptom score at presentation was 2.09 (95% confidence interval (CI) 2.07–2.11) and the median duration until resolution of moderately bad or severe symptoms was 8.70 days (interquartile range 4.50–11.00). Patients with influenza virus, human metapneumovirus (hMPV), respiratory syncytial virus (RSV), coronavirus (CoV) or rhinovirus had a significantly higher symptom score than patients with no virus isolated (0.07–0.25 points or 2.3–8.3% higher symptom score). Time to symptom resolution was longer in RSV infections (adjusted hazard ratio (AHR) 0.80, 95% CI 0.65–0.96) and hMPV infections (AHR 0.77, 95% CI 0.62–0.94) than in infections with no virus isolated. Overall, baseline viral load was associated with symptom severity (difference 0.11, 95% CI 0.06–0.16 per 10 cycles decrease in Ct value), but not with symptom duration.
Conclusions
In healthy, working adults from the general community presenting at the general practitioner with acute cough and/or suspected lower respiratory tract infection other than influenza impose an illness burden comparable to influenza. Hence, the public health focus for viral respiratory tract infections should be broadened.
Keywords: Disease burden, lower respiratory tract infection, primary healthcare, public health, respiratory tract infection, respiratory virus, symptom duration, symptom severity
Introduction
From the few studies describing the aetiology of acute lower respiratory tract infections (LRTIs) in primary care patients, we know that most LRTIs in the general community are caused by viral pathogens, in particular rhinovirus, influenza virus, coronavirus (CoV), respiratory syncytial virus (RSV), human metapneumovirus (hMPV), and parainfluenza virus (PiV) [1,2]. The illness course of LRTIs in adults presenting in this setting – a relatively healthy, working population – is mostly self-limiting and complications are rare [3]. However, with an average of 3.5 days of sick leave per year, LRTIs cause a substantial socio-economic burden [3,4]. In adults, influenza virus, bacteria and viral–bacterial coinfections are assumed to cause the most severe illnesses, with most systemic symptoms, longest illness durations, and most complications [[5], [6], [7]]. However, evidence on associations between aetiology and severity are mainly derived from hospital care settings with vulnerable patient populations [[8], [9], [10]]. In this setting, a focus on pathogens with the highest complication rates is obvious. Quite often, however, this focus is also applied in the general community, with public health interventions such as the annual influenza vaccinations targeted at the most vulnerable people with the aim of reducing the risk of complications and death [11]. Although data on the impact of respiratory viruses in the primary care setting are limited due to restricted microbial testing and absence of a standardized, validated outcome measure to evaluate illness severity [12], there are studies suggesting that the burden of disease from infections due to respiratory viruses other than influenza – i.e. rhinovirus, coronavirus and RSV – may be greater overall [13]. In this study, we aimed to explore the associations between respiratory viral pathogens, including viral load, and illness course in the adult primary care community, thereby opening up possibilities to base the public health focus on the impact of respiratory viruses in primary care, rather than on extrapolated data from hospital settings. This study was conducted in a large European cohort consisting of prospectively enrolled adult patients with acute cough and/or a clinical suspicion for LRTI.
Methods
Design and study population
This prospective study in primary care is part of the GRACE study (Genomics to combat Resistance against Antibiotics in Community-acquired LRTI in Europe). Participants were recruited between November 2007 and April 2010 by general practitioners (GPs) from 16 primary care networks in 11 European countries (Supplementary Fig. S1). Patients aged ≥18 years presenting with acute cough (duration of ≤28 days) and/or suspected LRTI, were asked to participate in this study, i.e. to fill out study materials and provide written informed consent [14]. Exclusion criteria were pregnancy, breast-feeding, any serious immunocompromised condition and antibiotic use in the previous month [14]. About one-third of these patients agreed to being randomized to either the intervention (amoxicillin) or placebo arm of the original randomized controlled trial [14]. Remaining patients were not randomly assigned but were included in the observational part of the study [1]. In the current study, both trial and observational patients were analysed together, but patients without polymerase chain reaction (PCR) and/or serology results on viral aetiology (all due to practical reasons) were excluded. Ethical approval was obtained for all participating networks.
Clinical measurements
For the collection of clinical data on the day of presentation (baseline), standardized case report forms (CRFs) were used. GPs completed the CRF on the following 12 symptoms rated by the patients using a four-point Likert-scale (1 = no problem, 2 = mild, 3 = moderate, 4 = severe): cough, sputum production, shortness of breath, wheeze, blocked or runny nose, fever, chest pain, muscle aching, headache, disturbed sleep, feeling generally unwell, and interference with normal daily activities. Additionally, the symptoms confusion/disorientation and diarrhoea were rated. Following initial presentation, patients were asked to fill out a symptom diary at home on a daily basis until they had no more symptoms or until the end of follow-up (day 28). Patients were asked to rate the same 12 symptoms by using a seven-point Likert-scale (0 = normal, 1 = very little problem, 2 = slight problem, 3 = moderately bad, 4 = bad, 5 = very bad, 6 = as bad as it could be). This diary was internally reliable, valid, and sensitive to change for acute LRTI [15].
Microbiological measurements
At baseline, two nasopharyngeal flocked swabs were taken by trained staff within 24 h after recruitment and before any antimicrobial treatment had started. Swabs were placed in universal transport medium immediately, frozen locally, and transported on dry ice to the central laboratory (University of Antwerp). Real-time in-house PCR (RT-PCR) testing was performed either as four multiplex RT-PCRs (combining INF-A, INF–B, and RSV; PIV1-4; HRV, hMPV, and the EAV internal control; and finally the human CoV: 229E, OC43, NL63, and HKU1), or as monoplex (all other viruses) [16]. RNA/DNA extractions and amplification methods were described previously [1,16]. Based on the results from our study comparing the prevalence of viral pathogens between symptomatic and asymptomatic matched controls [1], we evaluated rhinoviruses, influenza viruses, coronaviruses, RSV, hMPV and PiV. Because (pan-)adenovirus (1.3% vs 1.1%, p = 0.33), bocavirus (0.6% vs 0.8%, p = 0.43) and WU/KI polyomaviruses (2.2% vs. 2.5%, p = 0.02) were not detected more frequently in symptomatic patients than in controls, they were not considered pathogenic respiratory viruses and therefore excluded from our analyses [1]. A cycle threshold (Ct) value – an inverse, logarithmic, quantitative measurement of viral load – below 45 was chosen as cut-off for a positive result. We adjusted our analyses for bacterial infections, which were defined as having at least one of the following pathogens detected in a sputum or nasopharyngeal sample: Streptococcus species, Gram-negative species, or Aspergillus (fungus). Commensals and Candida species were considered contaminants for which analyses were not adjusted. Microbiologists who determined the results were blinded to clinical information.
Outcome parameters
We focused on two main outcome parameters: symptom severity at presentation and illness duration. Symptom severity was measured as the mean CRF score for all 12 symptoms (scored 1–4) at baseline [14,[17], [18], [19]]. Illness duration was defined as the duration until absence of any symptoms rated moderately bad or severe (score 3 or above) in the symptom diary following initial presentation [14,[17], [18], [19]]. Additionally, the severity of all individual symptoms was analysed, dichotomizing symptom severity at no/mild/moderate vs severe.
Statistical analysis
Baseline characteristics were reported as n (%), means (standard deviation (SD)) or medians (interquartile range (IQR)) as appropriate. Symptom severity at baseline was analysed with linear regression models and expressed as differences in mean symptom severity with a 95% confidence interval (CI). In an additional step, we analysed the presence of individual symptoms with logistic regression, expressed as odds ratios (ORs). Duration until absence of symptoms rated moderately bad or severe were analysed with Cox proportional hazard models. For the latter analysis, patients were censored at the end of follow-up or if fewer than 10 symptoms were filled out in the symptom diary. If patients already met the event criteria at baseline (n = 104), we defined their time to event as 1 day. Results were expressed as hazard ratios (HRs).
For all analyses, we adjusted for the potential confounders defined beforehand (please see Supplementary Material). Statistical analyses were performed using SPSS v.25.0 for Windows and the ‘survival’ and ‘survminer’ packages in R v.4. Details of the statistical analysis are described in the Supplementary Material.
Results
Study population
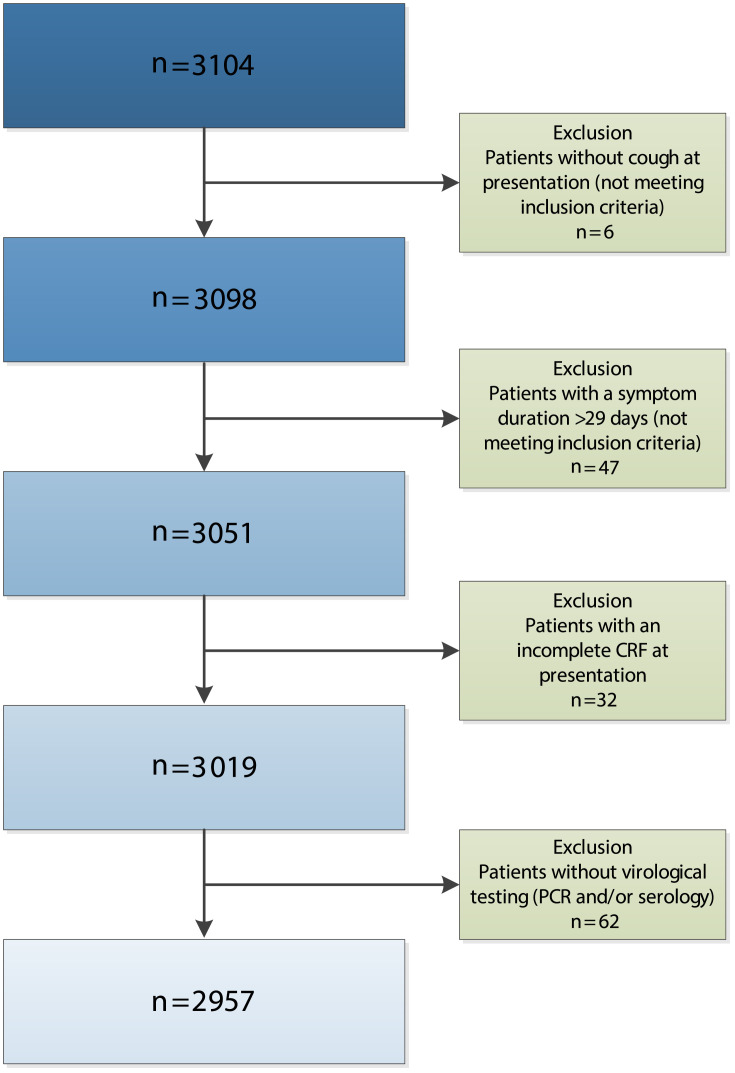
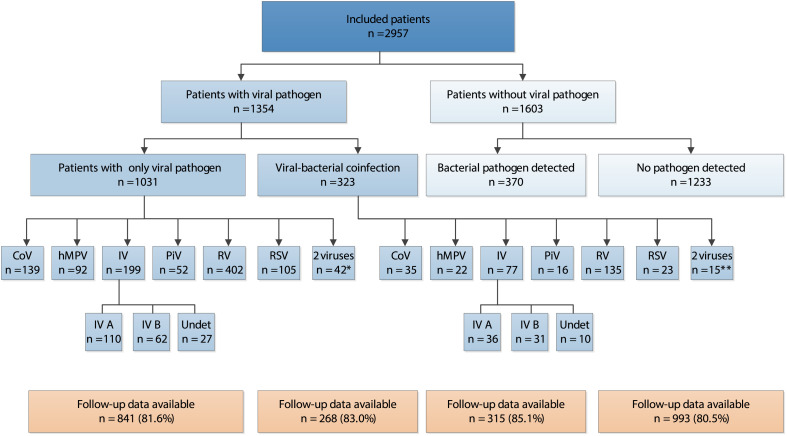
We included 2957 adult patients (Fig. 1 ). Demographics and clinical symptoms at presentation are presented in Table 1 . Patients had a median age of 50 years (IQR 36–63), 1195 (40.4%) were male and 1603 (54.2%) were a former or current smoker. The overall mean symptom score at presentation was 2.09 (95% CI 2.07–2.11). Respiratory viruses (1411) were detected in 1354 patient samples (Fig. 2 ). The proportion of influenza virus positive patients was lower among patients who received the annual influenza vaccination during the preceding fall/winter (38/707, 5.4%) than among non-vaccinated patients (259/2250, 11.5%) (p <0.001). Follow-up data were available for 2393 patients (80.9%). Baseline disease characteristics did not differ between patients who did (n = 2393) or did not (n = 564) fill out a symptom diary. Of all 2393 patients included in the symptom duration analysis, 2186 (91.3%) documented resolution of symptoms rated moderately bad or severe before the end of follow-up, with a median duration of 6.00 days (IQR 4.00–11.00 days). At presentation, only two patients were prescribed antiviral medication (oseltamivir).
Fig. 1.
Flow-chart patient exclusion as compared with the total number of patients included in the GRACE cohort [1]. CRF, case report form; PCR, polymerase chain reaction.
Table 1.
Baseline characteristics included patients (n = 2957)
| Demographics | Patients (n = 2957)a |
|---|---|
| Age (years) | 50 (36-63) |
| Gender (male) | 1195 (40.4%) |
| Caucasian ethnicity | 2862 (96.8%) |
| Comorbiditiesb | |
| COPD | 176 (6.0%) |
| Asthma | 307 (10.4%) |
| Other lung disease | 62 (2.1%) |
| Heart failure | 57 (1.9%) |
| Ischemic heart disease | 159 (5.4%) |
| Other hearth disease | 111 (3.8%) |
| Diabetes | 190 (6.4%) |
| Smoking past or current | 1603 (54.2%) |
| Disease related characteristics at presentation | |
| Severe cough | 983 (33.2%) |
| Sputum production | 309 (10.4%) |
| Shortness of breath | 215 (7.3%) |
| Wheeze | 115 (3.9%) |
| Blocked or runny nose | 355 (12.0%) |
| Fever | 122 (4.1%) |
| Chest pain | 155 (5.2%) |
| Muscle aching | 163 (5.5%) |
| Headache | 226 (7.6%) |
| Disturbed sleep | 542 (18.3%) |
| Feeling generally unwell | 349 (11.8%) |
| Interference with normal daily activities | 344 (11.6%) |
| Confusion/disorientation | 6 (0.2%) |
| Diarrhoea | 16 (0.5%) |
| One or more abnormalities at lung auscultation | 1165 (39.4%) |
| Breaths (per min) | 16 (15-18) |
| Heart rate (beats per min) | 76 (70-83) |
| Systolic blood pressure (mmHg) | 127 (117-140) |
| Diastolic blood pressure (mmHg) | 80 (70-85) |
| Oral temperature (°C) | 36.7 (36.4-37) |
| Medication prescribed for illnessc | 2086 (70.5%) |
COPD, chronic obstructive pulmonary disease.
Demographics are given as absolute numbers with % for categorical variables or as median with interquartile range for continuous variables.
Some patients had multiple comorbidities.
Prescribed medication included antibiotics, antitussives, mucolytic drugs, antihistamines, bronchodilators and anti-inflammatory drugs.
Fig. 2.
Detected viral pathogens in included patients (n = 2957) and availability of follow-up data. CoV, coronavirus; hMPV, human metapneumovirus; IV, influenza virus; PiV, Parainfluenza virus; RV, rhinovirus; RSV, respiratory syncytial virus; Undet, influenza virus type undetermined. ∗ The following combinations of viral pathogens were found: CoV + RV (n = 10), IV + RV (n = 8), CoV + hMPV (n = 5), CoV + RSV (n = 4), RV + RSV (n = 4), IV + RSV (n = 3), CoV + IV (n = 2), hMPV + RV (n = 2), IV + PiV (n = 1), CoV + PiV (n = 1), RV + PiV (n = 1), RSV + PiV (n = 1). ∗∗ The following combinations of viral pathogens were found: CoV + RV (n = 5), IV + RV (n = 3), CoV + IV (n = 3), CoV + RSV (n = 1), RV + RSV (n = 1), IV + RSV (n = 1), RV + PiV (n = 1).
Association between respiratory viruses and symptom severity
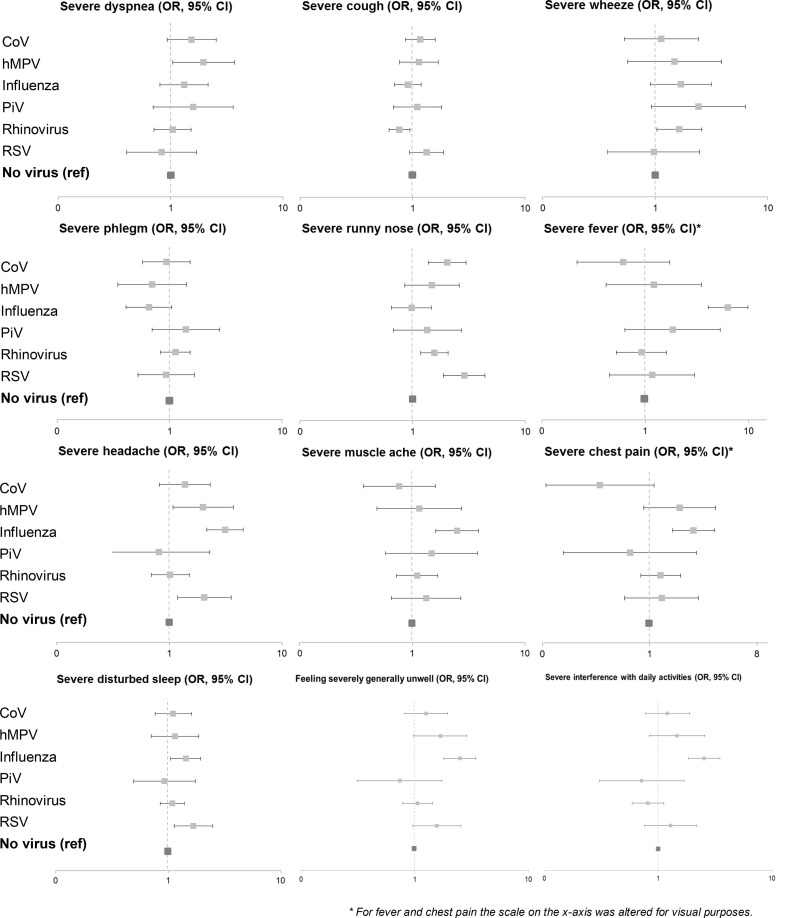
We evaluated the severity of symptoms at presentation for patients with CoV, hMPV, influenza virus, PiV, rhinovirus and RSV, as compared with patients without these viruses, with adjustment for confounders, bacteria and co-viruses. Influenza virus, hMPV, RSV, CoV and rhinovirus were significantly associated with, respectively, 0.25 (95% CI 0.19–0.31), 0.16 (95% CI 0.07–0.26), 0.12 (95% CI 0.04–0.21), 0.09 (95% CI 0.02–0.16) and 0.07 (95% CI 0.02–0.12) points higher symptom scores at presentation as compared with patients without detected virus (Table 2 ). Among patients in whom a virus was detected, a 10 cycles lower Ct value – i.e. a higher viral load – measured at presentation, was associated with a 0.11 (95% CI 0.06–0.16) point higher mean symptom severity as compared with patients without detected virus. After stratification for viral aetiology, we only observed an association between viral load and symptom severity for rhinovirus (increase of 0.12 per 10 cycles reduction in Ct value, 95% CI 0.04–0.20) and for RSV (increase of 0.16 per 10 cycles reduction in Ct value, 95% CI 0.01–0.30). When looking at differences in the severity of individual symptoms of these viruses (Fig. 3 ), influenza virus was independently associated with severe fever (OR 6.3, 95% CI 4.0–9.8), headache (OR 3.1, 95% CI 2.2–4.5), chest pain (OR 2.0, 95% CI 1.3–3.2), muscle pain (OR 2.5, 95% CI 1.6–3.9), disturbed sleep (OR 1.4, 95% CI 1.1–1.9), being generally unwell (OR 2.5, 95% CI 1.8–3.5), and interference with daily activities (OR 2.5, 95% CI 1.8–3.5). RSV was associated with severe headache (OR 2.0, 95% CI 1.2–3.5), disturbed sleep (OR 1.7, 95% CI 1.1–2.5) and a runny nose (OR 2.9, 95% CI 1.9–4.4). hMPV was associated with severe dyspnoea (OR 2.0, 95% CI 1.0–3.7) and headache (OR 2.0, 95% CI 1.1–3.7). Rhinovirus was associated with severe wheeze (OR 1.6, 95% CI 1.0–2.6), a runny nose (OR 1.6, 95% CI 1.2–2.1) and negatively associated with severe cough (OR 0.8, 95% CI 0.6–0.9). CoV was associated with a severe runny nose (OR 2.0, 95% CI 1.4–3.0) and negatively associated with severe chest pain (OR 0.3, 95% CI 0.1–0.9).
Table 2.
Symptom severitya at presentation in patients consulting in primary care with a detected virus or no detected virus (n = 2957)
| Mean (SD) symptom score at presentation | Unadjusted difference between groups (95% CI) | Adjusted difference between groups (95% CI)b | |
|---|---|---|---|
| No virus(es) (n = 1603) | 2.02 (0.49) | (Ref) | (Ref) |
| ≥1 virus(es) (n = 1354) | 2.18 (0.52) | 0.17 (0.13–0.20) | 0.13 (0.10–0.17) |
| No virus(es) (n = 1603) | 2.02 (0.49) | (Ref) | (Ref) |
| 1 virus (n = 1297) | 2.18 (0.51) | 0.16 (0.13–0.20) | 0.13 (0.09–0.16) |
| 2 viruses (n = 57) | 2.27 (0.54) | 0.13 (0.06–0.19) | 0.22 (0.09–0.35) |
| CoV (n = 205)c | 2.15 (0.48) | 0.10 (0.03–0.18) | 0.09 (0.02–0.16)d |
| hMPV (n = 121)c | 2.18 (0.52) | 0.16 (0.06–0.25) | 0.16 (0.07–0.26)d |
| Influenza virus (n = 297)c | 2.32 (0.55) | 0.30 (0.23–0.36) | 0.25 (0.19–0.31)d |
| PiV (n = 73)c | 2.13 (0.51) | 0.10 (-0.01 to 0.22) | 0.07 (-0.04 to 0.19)d |
| Rhinovirus (n = 572)c | 2.15 (0.50) | 0.12 (0.07–0.16) | 0.07 (0.02–0.12)d |
| RSV (n = 143)c | 2.17 (0.53) | 0.14 (0.05–0.22) | 0.12 (0.04–0.21)d |
CI, confidence interval; CoV, coronavirus; hMPV, human metapneumovirus; Piv, parainfluenza; RSV, respiratory syncytial virus; SD, standard deviation.
Calculated as the mean (SD) symptom severity score for all 12 symptoms at presentation.
Estimates controlled for age, gender, pulmonary comorbidities (asthma, chronic obstructive pulmonary disease and other lung diseases), hearth failure, current smoking, influenza vaccination during the preceding fall or winter, coinfection with at least one respiratory bacterium or with Aspergillus and duration of symptoms before presentation.
Reference group is no CoV, hMPV, influenza virus, PiV, rhinovirus or RSV, respectively.
By including all six viruses in the model, estimates were additionally controlled for coinfection with another respiratory virus.
Fig. 3.
Forest plots showing odds ratios (ORs) with 95% confidence intervals (CIs) on the log scale for coronavirus (CoV), human metapneumovirus (hMPV), influenza virus, parainfluenza virus (PiV), rhinovirus and respiratory syncytial virus (RSV) for a severe burden of individual symptoms at presentation (highest on four-point Likert scale). The reference category is no virus isolated. ORs are derived from logistic regression models (one model per symptom) with adjustment for bacterial and viral coinfections, age, gender, pulmonary comorbidities (asthma, chronic obstructive pulmonary disease and other lung diseases), hearth failure, current smoking, influenza vaccination during the preceding fall or winter and duration of symptoms before presentation. ∗ For fever and chest pain the scale on the x-axis was altered for visual purposes.
Association between respiratory viruses and illness duration
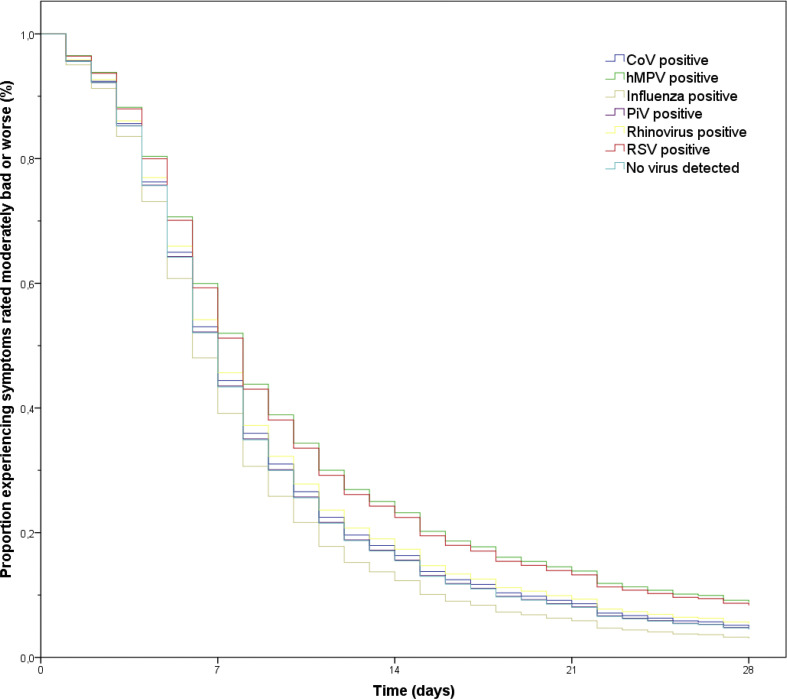
After adjustment for bacterial coinfections, baseline symptom severity and other potential confounders, patients with detected viral pathogen(s) had no significantly different HR (0.93, 95% CI 0.86–1.02) for resolution of moderately bad or severe symptoms compared with patients in which no virus was detected (Table 3 ). We also assessed the duration until resolution of moderately bad or severe symptoms for the six individual viruses as compared with patients without a detected virus (Fig. 4 ). Patients with RSV had an adjusted hazard ratio (AHR) of 0.80 (95% CI 0.65–0.96) and patients with hMPV an AHR of 0.77 (95% CI 0.62–0.94) for symptom resolution, indicating a significantly longer symptom duration as compared with patients without RSV and hMPV, respectively. All other viral pathogens showed no significant differences in AHRs. Among patients in whom a virus was detected, there was no association between baseline viral load and duration of moderately bad or severe symptoms (AHR per unit lower Ct value 1.01, 95% CI 0.99–1.02). After stratification for viral aetiology, no significant associations were found between viral load and symptom duration.
Table 3.
Symptom durationa (days) in patients consulting in primary care with detected virus or no detected virus (n = 2393)
| Median (IQR) time to resolution of symptoms rated moderately bad or worse | Unadjusted hazard ratio (95% CI) | Adjusted hazard ratio (95% CI)b | |
|---|---|---|---|
| No virus(es) (n = 1288) | 6 (4–10) | (Ref) | (Ref) |
| ≥1 of six viruses (n = 1105) | 7 (5–11) | 0.93 (0.86–1.01) | 0.93 (0.86–1.02) |
| No virus(es) (n = 1288) | 6 (4–10) | (Ref) | (Ref) |
| 1 of six viruses (n = 1056) | 7 (5–11) | 0.94 (0.87–1.03) | 0.94 (0.86–1.03) |
| 2 of six viruses (n = 49) | 8 (5–15) | 0.74 (0.55–1.00) | 0.76 (0.56–1.03) |
| CoV (n = 177)c | 7 (4–11) | 0.92 (0.78–1.09) | 0.95 (0.80–1.12)d |
| hMPV (n = 108)c | 8 (6–12) | 0.80 (0.65–0.98) | 0.77 (0.62–0.94)d |
| Influenza (n = 243)c | 7 (5–10) | 1.12 (0.97–1.28) | 1.08 (0.93–1.24)d |
| PiV (n = 60)c | 8 (5–11) | 0.98 (0.75–1.28) | 0.97 (0.74–1.26)d |
| Rhinovirus (n = 445)c | 7 (5–11) | 0.90 (0.81–1.01) | 0.93 (0.83–1.04)d |
| RSV (n = 121)c | 8 (5–14) | 0.79 (0.65–0.96) | 0.80 (0.65–0.96)d |
A hazard ratio <1 indicates a disadvantageous effect on symptom resolution. CI, confidence interval; CoV, coronavirus; hMPV, human metapneumovirus; IQR, interquartile range; Piv, parainfluenza; RSV, respiratory syncytial virus.
Calculated as the median (IQR) number of days with symptoms rated moderately bad or worse by the patient following initial presentation.
Estimates controlled for age, gender, pulmonary comorbidities (asthma, chronic obstructive pulmonary disease and other lung diseases), heart failure, current smoking, influenza vaccination during the preceding fall or winter, coinfection with at least one respiratory bacterium or with Aspergillus and duration of symptoms before presentation.
Reference group is no CoV, hMPV, influenza virus, PiV, rhinovirus or RSV, respectively.
By including all six viruses in the model, estimates were additionally controlled for coinfection with another respiratory virus.
Fig. 4.
Cox regression survival curves for the duration of symptoms rated moderately bad or worse in patients with lower respiratory tract infections (LRTIs) and a viral mono-infection (n = 2344), stratified by detected virus. The reference category is no virus detected. Survival curves are derived from multivariate Cox regression models with adjustment for bacterial coinfections, age, gender, pulmonary comorbidities (asthma, chronic obstructive pulmonary disease and other lung diseases), heart failure, current smoking, influenza vaccination during the preceding fall or winter and duration of symptoms before presentation. CoV, coronavirus; hMPV, human metapneumovirus; PiV, parainfluenza virus; RSV, respiratory syncytial virus.
Discussion
Adult patients visiting the GP with acute cough or suspected LRTI due to influenza virus, hMPV, RSV, CoV or rhinovirus had a 0.07–0.25 points (or 2–8%) higher mean symptom severity score (range 1–4) at presentation as compared with patients presenting with acute cough or suspected LRTI without detection of one of these respiratory viruses. In translation, patients with RSV – who have a 0.12-point (4%) higher symptom score at presentation than patients in whom no virus is detected – rate one or two symptoms severe instead of moderate, moderate instead of mild, or mild instead of absent. Additionally, RSV and hMPV were associated with a longer duration of moderately bad or severe symptoms, which might be linked to the pattern of immune response to these viruses [21]. For all respiratory viruses together, a higher viral load measured at presentation, was significantly associated with a higher symptom severity. This was caused by significant associations between viral load and symptom severity for rhinovirus and RSV. There was no association between viral load and the duration of moderately bad or severe symptoms.
Clinical implications
This study does not provide direct clinically actionable insight. However, although we do not provide recommendations on clinical management or treatment, we do think that the large number of patients included in this study provides important information which can be used to prioritize different respiratory viruses in the primary care setting. Currently, public health resources in the general community are guided by the aim to prevent complications in the most vulnerable people, and are focused almost exclusively on influenza [11,22,23]. From a socio-economic perspective, however, targeting public health resources only at influenza virus neglects the substantial illness course in the community caused by other respiratory viruses. From our results we conclude that RSV and hMPV impose a disease burden that compares well to that of influenza virus and should therefore receive more attention in the primary care setting, e.g., by supporting the development and implementation of prevention approaches such as vaccines [[24], [25], [26]].
Strengths and limitations
Despite the fact that we had a large cohort in which data were collected in a standardized manner, and outcome measures were in line with previous studies [14,[17], [18], [19]], there are several potential sources of bias that might limit the validity of our results. Firstly, it is possible that non-agreement of patients to participate in this observational study was not random. The extent to which this selection might be present is uncertain because we have no information on the number and characteristics of patients who declined participation. Secondly, the use of medication, such as antibiotic treatment, antiviral treatment, (over-the-counter) symptomatic treatment, and prophylactic antibiotics with antiviral effects (as azithromycin) might have influenced outcomes. We consider it unlikely that receiving antibiotics caused biased results, because the in-study amoxicillin trial showed no differences in outcomes between the intervention and placebo groups [14]. Because only two patients in our cohort were prescribed antivirals (oseltamivir), we consider the effect of antiviral treatment also negligible. Unavailability of data on the use of prophylactic antibiotics and symptomatic medication made adjustment for these factors impossible. Thirdly, there might be bias in the self-reporting of symptoms by patients. However, previous studies showed a high internal reliability, validity and sensitivity of the symptom diary we used [15]. Also, because the 95/207 (46%) patients who did not meet the event criteria and who did not fill out their symptom diary completely were censored for the analysis, we do not expect selection bias due to loss of follow-up. Fourthly, the required sample size for the prospective observational cohort was not determined on the specific requirements of the current study. Hence, inconclusive or non-significant results can therefore not be considered definite to prove the absence of associations. We specifically chose not to correct for multiple testing, as this correction may have further hampered statistical power, especially for viruses only detected in a limited number of patients. Fifthly, the relatively low overall percentage of detected viruses might have been caused by the inclusion of patients with quite long duration of symptoms. Because respiratory fluids are renewed quickly in the patient, viral pathogens in patients with longer duration of symptoms might therefore not have been detectable anymore. Finally, a higher viral load was associated with a higher symptom severity at presentation. Looking at specific viruses we only found this association for RSV and rhinovirus, which confirms previous studies [[27], [28], [29]]. However, the interpretation of single viral load measurements is difficult. Not only are viral loads of respiratory viruses highly dependent on variation in sampling location and technique, they also rise and fall rapidly and it is known that symptoms mostly follow the viral load [30,31].
In conclusion, in this study among relatively healthy adult patients presenting in a primary care setting with acute cough and/or a suspected LRTI, influenza virus, hMPV, RSV, CoV and rhinovirus were associated with an increased symptom severity at presentation as compared with patients without a detected virus. In this general community population, RSV and hMPV were associated with a longer duration of moderately bad or severe symptoms. This study emphasizes that public health policies as vaccinations and awareness among GPs should not remain focused on influenza virus exclusively, but should also include other common respiratory viruses such as RSV and hMPV that pose a high socio-economic burden to the general adult community.
Transparency declaration
The authors declare that they have no conflicts of interest. The study was part of the European Commission Framework Programme 6 (FP6) funded Network of Excellence GRACE, grant number 518226. Orion Diagnostics provided the QuikRead instruments and kits for this study. The study sponsors played no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the paper for publication.
Author contributions
The larger GRACE observational study was designed by C.C.B., T.J.M.V., P.L., D.C. and H.G., and sampling protocols by M.I., C.L., K.L. and H.G. The day-to-day management at study sites was supervised by M.I., C.L., P.L., T.V. and H.G. PCR and serological analyses were performed by K.L., A.M.V.L., C.L., K.Z., E.C.J.C., M.V. and F.C. Data were analysed by L.M.V., R.B., N.P.A.Z., B.D.L.B., J.J.O. and F.C. The manuscript was designed and drafted by L.M.V., N.P.A.Z. and F.C., and was reviewed by all authors.
Editor: L. Leibovici
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2020.03.023.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Ieven M., Coenen S., Loens K., Lammens C., Coenjaerts F., Vanderstraeten A. Aetiology of lower respiratory tract infection in adults in primary care: a prospective study in 11 European countries. Clin Microbiol Infect. 2018;24:1158–1163. doi: 10.1016/j.cmi.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woodhead M., Blasi F., Ewig S., Garau J., Huchon G., Ieven M. Guidelines for the management of adult lower respiratory tract infections – Summary. Clin Microbiol Infect. 2011;17:1–24. doi: 10.1111/j.1469-0691.2011.03602.x. [DOI] [PubMed] [Google Scholar]
- 3.Verheij T., Hermans J., Kaptein A., Mulder J. Acute bronchitis: course of symptoms and restrictions in patients’ daily activities. Scand J Prim Health Care. 1995;13:8–12. doi: 10.3109/02813439508996728. [DOI] [PubMed] [Google Scholar]
- 4.Fragaszy E.B., Warren-Gash C., White P.J., Zambon M., Edmunds W.J., Nguyen-Van-Tam J.S. Effects of seasonal and pandemic influenza on health-related quality of life, work and school absence in England: results from the Flu Watch cohort study. Influenza Other Respir Virus. 2018;12:171–182. doi: 10.1111/irv.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pavia A.T. What is the role of respiratory viruses in community acquired pneumonia; what is the best therapy for influenza and other viral causes of CAP? Infect Dis Clin North Am. 2014;27:157–175. doi: 10.1016/j.idc.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voiriot G., Visseaux B., Cohen J., Nguyen L.B.L., Neuville M., Morbieu C. Viral-bacterial coinfection affects the presentation and alters the prognosis of severe community-acquired pneumonia. Crit Care. 2016;20:1–9. doi: 10.1186/s13054-016-1517-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qu J.X., Gu L., Pu Z.H., Yu X.M., Liu Y.M., Li R. Viral etiology of community-acquired pneumonia among adolescents and adults with mild or moderate severity and its relation to age and severity. BMC Infect Dis. 2015;15:1–8. doi: 10.1186/s12879-015-0808-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sokolow L.Z., Naleway A.L., Li D.K., Shifflett P., Reynolds S., Henninger M.L. Severity of influenza and noninfluenza acute respiratory illness among pregnant women, 2010-2012. Am J Obstet Gynecol. 2015;212 doi: 10.1016/j.ajog.2014.08.004. 202.e1-11. [DOI] [PubMed] [Google Scholar]
- 9.Treanor J., Falsey A. Respiratory viral infections in the elderly. Antivir Res. 1999;44:79–102. doi: 10.1016/S0166-3542(99)00062-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaunt E.R., Harvala H., McIntyre C., Templeton K.E., Simmonds P. Disease burden of the most commonly detected respiratory viruses in hospitalized patients calculated using the disability adjusted life year (DALY) model. J Clin Virol. 2011;52:215–221. doi: 10.1016/j.jcv.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demicheli V., Jefferson T., Di Pietrantonj C., Ferroni E., Thorning S., Thomas R.E. Vaccines for preventing influenza in the elderly (Review) Cochrane Database Syst Rev. 2018;2 doi: 10.1002/14651858.CD004876.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rath B., Conrad T., Myles P., Alchikh M., Ma X., Hoppe C. Influenza and other respiratory viruses: standardizing disease severity in surveillance and clinical trials. Expert Rev Anti Infect Ther. 2017;15:545–568. doi: 10.1080/14787210.2017.1295847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicholson K.G., Kent J., Hammersley V., Cancio E. Acute viral infections of upper respiratory tract in elderly people living in the community: comparative, prospective, population based study of disease burden. BMJ. 2011;315:1060–1064. doi: 10.1136/bmj.315.7115.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Little P., Stuart B., Moore M., Coenen S., Butler C.C., Godycki-Cwirko M. Amoxicillin for acute lower-respiratory-tract infection in primary care when pneumonia is not suspected: a 12-country, randomised, placebo-controlled trial. Lancet Infect Dis. 2013;13:123–129. doi: 10.1016/S1473-3099(12)70300-6. [DOI] [PubMed] [Google Scholar]
- 15.Watson L., Little P., Moore M., Warner G., Williamson I. Validation study of a diary for use in acute lower respiratory tract infection. Fam Pract. 2001;18:553–554. doi: 10.1093/fampra/18.5.553. [DOI] [PubMed] [Google Scholar]
- 16.Loens K., Van Loon A.M., Coenjaerts F., Van Aarle Y., Goossens H., Wallace P. Performance of different mono- and multiplex nucleic acid amplification tests on a multipathogen external quality assessment panel. J Clin Microbiol. 2012;50:977–987. doi: 10.1128/JCM.00200-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruyndonckx R., Stuart B., Little P., Hens N., Ieven M., Butler C.C. Amoxicillin for acute lower respiratory tract infection in primary care: subgroup analysis by bacterial and viral aetiology. Clin Microbiol Infect. 2018;24:871–876. doi: 10.1016/j.cmi.2017.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teepe J., Little P., Elshof N., Broekhuizen B.D.L., Moore M., Stuart B. Amoxicillin for clinically unsuspected pneumonia in primary care: subgroup analysis. Eur Respir J. 2016;47:327–330. doi: 10.1183/13993003.00611-2015. [DOI] [PubMed] [Google Scholar]
- 19.Moore M., Stuart B., Coenen S., Butler C.C., Goossens H., Verheij T.J.M. Amoxicillin for acute lower respiratory tract infection in primary care: subgroup analysis of potential high-risk groups. Br J Gen Pract. 2014;64:75–80. doi: 10.3399/bjgp14X677121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ascough S., Paterson S., Chiu C. Induction and subversion of human protective immunity: contrasting influenza and respiratory syncytial virus. Front Immunol. 2018;9:323. doi: 10.3389/fimmu.2018.00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moncion K., Young K., Tunis M., Rempel S., Stirling R., Zhao L. Effectiveness of hand hygiene practices in preventing influenza virus infection in the community setting: a systematic review. Can Commun Dis Rep. 2019;45:12–23. doi: 10.14745/ccdr.v45i01a02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doyon-Plourde P., Fakih I., Tadount F., Fortin É., Quach C. Impact of influenza vaccination on healthcare utilization – a systematic review. Vaccine. 2019;37:3179–3189. doi: 10.1016/j.vaccine.2019.04.051. [DOI] [PubMed] [Google Scholar]
- 24.Zhu T., Zhang C., Yu L., Chen J., Qiu H., Lyu W. The preventive effect of vaccine prophylaxis on severe respiratory syncytial virus infection: a meta-analysis. Virol Sin. 2015;30:371–378. doi: 10.1007/s12250-015-3630-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren J., Phan T., Bao X. Recent vaccine development for human metapneumovirus. J Gen Virol. 2015;96:1515–1520. doi: 10.1099/vir.0.000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papadopoulos N.G., Megremis S., Kitsioulis N.A., Vangelatou O., West P., Xepapadaki P. Promising approaches for the treatment and prevention of viral respiratory illnesses. J Allergy Clin Immunol. 2017;140:921–932. doi: 10.1016/j.jaci.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuller J.A., Njenga M.K., Bigogo G., Aura B., Ope M.O., Nderitu L. Association of the CT values of real-time PCR of viral upper respiratory tract infection with clinical severity, Kenya. J Med Virol. 2013;85:924–932. doi: 10.1002/jmv.23455. [DOI] [PubMed] [Google Scholar]
- 28.Feikin D.R., Fu W., Park D.E., Shi Q., Higdon M.M., Baggett H.C. Is higher viral load in the upper respiratory tract associated with severe pneumonia? Findings from the PERCH Study. Clin Infect Dis. 2017;64(Suppl. l_3):S337–S346. doi: 10.1093/cid/cix148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waghmare A., Kuypers J.M., Xie H., Leisenring W., Campbell A.P., Jerome K.R. Viral load in hematopoietic cell transplant recipients infected with human rhinovirus correlates with burden of symptoms. Biol Blood Marrow Transplant. 2015;21:S317–S318. [Google Scholar]
- 30.Bagga B., Woods C.W., Veldman T.H., Gilbert A., Mann A., Balaratnam G. Comparing influenza and RSV viral and disease dynamics in experimentally infected adults predicts clinical effectiveness of RSV antivirals. Antivir Ther. 2013;18:785–791. doi: 10.3851/IMP2629. [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Mauriño C., Moore-Clingenpeel M., Thomas J., Mertz S., Cohen D.M., Ramilo O. Viral load dynamics and clinical disease severity in infants with respiratory syncytial virus infection. J Infect Dis. 2018;219:1207–1215. doi: 10.1093/infdis/jiy655. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.