Abstract
EIF2AK1 and EIF2AK2 encode members of the eukaryotic translation initiation factor 2 alpha kinase (EIF2AK) family that inhibits protein synthesis in response to physiologic stress conditions. EIF2AK2 is also involved in innate immune response and the regulation of signal transduction, apoptosis, cell proliferation, and differentiation. Despite these findings, human disorders associated with deleterious variants in EIF2AK1 and EIF2AK2 have not been reported. Here, we describe the identification of nine unrelated individuals with heterozygous de novo missense variants in EIF2AK1 (1/9) or EIF2AK2 (8/9). Features seen in these nine individuals include white matter alterations (9/9), developmental delay (9/9), impaired language (9/9), cognitive impairment (8/9), ataxia (6/9), dysarthria in probands with verbal ability (6/9), hypotonia (7/9), hypertonia (6/9), and involuntary movements (3/9). Individuals with EIF2AK2 variants also exhibit neurological regression in the setting of febrile illness or infection. We use mammalian cell lines and proband-derived fibroblasts to further confirm the pathogenicity of variants in these genes and found reduced kinase activity. EIF2AKs phosphorylate eukaryotic translation initiation factor 2 subunit 1 (EIF2S1, also known as EIF2α), which then inhibits EIF2B activity. Deleterious variants in genes encoding EIF2B proteins cause childhood ataxia with central nervous system hypomyelination/vanishing white matter (CACH/VWM), a leukodystrophy characterized by neurologic regression in the setting of febrile illness and other stressors. Our findings indicate that EIF2AK2 missense variants cause a neurodevelopmental syndrome that may share phenotypic and pathogenic mechanisms with CACH/VWM.
Keywords: hypomyelination, abnormal myelination, hypotonia, movement disorders, cognitive impairment, regression, febrile illnesses, integrated stress response, EIF2S1, EIF2α
Main Text
The eukaryotic translation initiation factor 2 alpha kinase (EIF2AK) family is comprised of four mammalian kinases that regulate the cytoprotective integrated stress response (ISR) required for cellular adaptation to stress conditions.1,2 EIF2AK1 (MIM: 613635; HGNC: 24921), also known as Heme-Regulated Inhibitor, responds to heme deprivation and proteasome inhibition and maintains basal endoplasmic reticulum (ER) stress.3, 4, 5, 6, 7 EIF2AK1 contains two protein kinase domains and two heme binding sites. EIF2AK2 (MIM: 176871; HGNC: 9437), also known as Protein Kinase R, is activated by double-stranded RNA (dsRNA) and can block the translation of viral mRNA in response to infection,8, 9, 10 activation also occurs in response to oxidative stress, ER stress,11, 12, 13, 14 cytokines,14,15 and growth factors.16 EIF2AK2 contains two dsRNA binding motifs (DSRM) and a protein kinase domain. In the presence of their respective cellular stressors, both EIF2AK1 or EIF2AK2 activate ISR by phosphorylating Eukaryotic Translation Initiation Factor 2 Subunit 1 (EIF2S1, also known as EIF2α), a major regulator of the initiation of mRNA translation and the rate of protein synthesis. The phosphorylation of EIF2S1 on serine 51 by EIF2AK family members prevents mRNA translation and results in transient suppression of general protein synthesis.17,18 Prior studies have linked missense, nonsense, and splicing variants in EIF2AK3 (MIM: 604032) to autosomal recessive epiphyseal dysplasia with early onset diabetes mellitus (MIM: 226980) and truncating variants in EIF2AK4 (MIM: 609280) to autosomal recessive pulmonary veno-occlusive disease type 2 (MIM: 234810).19,20 However, neither of these disorders present with primary neurologic findings. The phenotypic consequences of rare variants in human EIF2AK1 and EIF2AK2 are currently unknown.
Nine probands were found via trio exome sequencing (ES) with Sanger sequencing confirmation to have rare missense variants in either EIF2AK1 or EIF2AK2. DNA was extracted from peripheral blood mononuclear cells for ES. Maternity and paternity were confirmed by the inheritance of rare SNPs from the parents and sample swap was excluded. There were no pathogenic copy number variants identified by chromosomal microarray. Clinical data were obtained after written informed consent was obtained in accordance with the ethical standards of the participating institutional review boards (IRB) on human research at each respective institution. A summary of the molecular findings and recurrent phenotypes of all nine individuals in our cohort is in Tables 1 and S1–S3. Probands 1 through 3 were identified through the Undiagnosed Diseases Network (UDN)21,22 (Table 1, probands 1–3). Probands 4 through 9 were identified through curation of ∼13,500 clinical ES from Baylor Genetics (BG) and GeneMatcher23,24 (Table 1, probands 4–9).
Table 1.
Summary of Clinical and Molecular Findings in Individuals with Heterozygous De Novo EIF2AK1 and EIF2AK2 Variants
| Proband 1 | Proband 2 | Proband 3 | Proband 4 | Proband 5 | Proband 6 | Proband 7 | Proband 8 | Proband 9 | |
|---|---|---|---|---|---|---|---|---|---|
| Molecular Findings | |||||||||
| Gene | EIF2AK1 | EIF2AK2 | EIF2AK2 | EIF2AK2 | EIF2AK2 | EIF2AK2 | EIF2AK2 | EIF2AK2 | EIF2AK2 |
| cDNA | NM_014413.4; c.1342A>G | NM_002759.3; c.31A>C | NM_002759.3; c.398A>T | NM_002759.3; c.973G>A | NM_002759.3; c.1382C>G | NM_002759.3; c.326C>T | NM_002759.3; c.325G>T | NM_002759.3; c.95A>G | NM_002759.3; c.290C>T |
| Protein | p.Ile448Val | p.Met11Leu | p.Tyr133Phe | p.Gly325Ser | p.Ser461Cys | p.Ala109Val | p.Ala109Ser | p.Asn32Ser | p.Ser97Phe |
| Inheritance | de novo | de novo | de novo | de novo | de novo | de novo | de novo | de novo | de novo |
| AOH | no | no | no | no | no | 47 Mb on chromosome 17 | no | n/a | no |
| CNV | no | no | no | no | no | no | no | n/a | no |
| Background | |||||||||
| Gender | female | male | male | female | male | male | male | male | male |
| Age at most recent assessment | 6 years | 10 years | 13 years | 3 years | 18 months | 19 months | 3 years | 12 years | 4 years |
| Ancestry | Irish, German | German, Mexican, Spanish | Chinese | European | European | Moroccan, Kuwaiti | European | European | German, Irish, Apache, Cherokee |
| Neurology | |||||||||
| Dysarthria or nonverbal | dysarthria | dysarthria | dysarthria | dysarthria | nonverbal | nonverbal | dysarthria | dysarthria | nonverbal |
| Nonambulatory | no | no | no | no | yes | yes | no | no | yes |
| Gait ataxia | no | yes | yes | yes | n/a | n/a | yes | no | no |
| Truncal ataxia | no | yes | no | no | yes | yes | no | no | no |
| Hypotonia | no | yes | yes | yes | yes | yes | yes | no | yes |
| Hypertonia | yes, lower extremities | yes | yes | no | yes | yes | yes | yes | yes |
| Hyperreflexia | yes, lower extremities | no | yes | no | yes | no | yes | yes | yes |
| Spasticity | yes, lower extremities | no | yes | yes | yes | no | yes | yes | yes |
| Dystonia | no | yes | yes | no | yes | yes | no | no | yes |
| Tremor | no | yes | no | yes | yes | no | no | no | no |
| Myoclonus | no | yes | no | no | no | no | no | no | no |
| Choreathetosis | no | yes | no | no | no | no | no | no | no |
| Hemiballismus | no | yes | no | no | no | no | no | no | no |
| Extrapyramidal signs | bradykinesia | parkinsonism, tremor, dystonia | parkinsonism, bradykinesia, bradyphrenia | tremor | parkinsonism, bradykinesia, rigidity, mask-like facies | no | no | parkisonism, hypomimia, abnormal postural reactions | no |
| Seizures | no | yes | no | no | yes | yes | no | no | yes |
| Seizure history | N/A | GTC | N/A | concern for seizure activity, normal EEG | focal complex seizures, focal epileptiform discharges | focal tonic seizures, multifocal epileptiform discharges, seizure onset at 7 months old | N/A | no | focal complex seizures, focal epileptiform discharges, seizure onset at 4 months old |
| OFC at birth | N/A | 31.5 cm (Z = −0.5) | 35.5 cm (0.82) | n/a | 32 cm (Z = −0.5) | N/A | N/A | 34 cm (Z = −0.5) | N/A |
| OFC at latest assessment | 51.5 cm (Z = 1.0) | 53.2 cm (Z = −0.05) | 52.8 cm (Z score = −0.66) | 44.50 (Z = −1.18, 17 months) | 43 cm (Z = −3.0) | 44.5 cm (Z = −2.42) | 48.8 cm (Z = −1.4) | 49 cm (Z = −1.0) | 49 cm (Z = −1.61) |
| Neurologic regression with febrile illness | not reported | yes | yes | yes | yes | yes | yes | yes | yes |
| Features of neurologic regression | n/a | n/a | neurologic decline with febrile illnesses | neurologic decline with febrile illnesses | neurologic decline with febrile illnesses | loss of eye contact, babbling, and motor skills with influenza A illness at 13 months | abruptly nonverbal with neurologic decline following febrile RSV illness at 4 years old | transient but severe worsening of the postural instability during febile illness | loss of crawling and oral skills with human metapneumovirus illness at 4 years old |
| Additional features | urinary and fecal urgency, slow finger tapping movements | urinary and fecal incontinence, silent aspiration of thin liquids | intellectual disability, dysphagia, poor eye contact | abnormal eye movements concerning for seizure | acquired microcephaly, laryngomalacia, gastroparesis, head titubations | exacerbation of epilepsy with febrile illnesses | progressive contractures, walks in a crouched position with elbows flexed, thumbs adducted, bilateral feet pronation | acquired microcephaly | failure to thrive |
| MRI Brain | |||||||||
| Age at assessment | 3 years | 7 years | 10 years | 17 months | 6 months | 18 months | 4 years | 8.5 years | 4 years |
| Cerebral volume loss | no | yes | no | yes | yes | yes | yes | yes | yes |
| T1W signal | isointense | isointense | isointense | isointense | isointense | isointense | isointense | isointense | hyperintensity throughout the supratentorial and infratentorial white matter |
| T2W signal | hyperintensity, posterior lateral ventricles | hyperintensity, dorsal-most upper cervical cord, dorsal medulla, dorsal pons, periaqueductal gray | hyperintensity, confluent signal in subcortical and periventricular white matter, patchy signal in brainstem | isointense | isointense | isointense | hyperintensity, dorsal medulla and periventricular | hyperintensity, posterior part of putamen, periventricual and deep white matter, inferior cerebellar peduncles | hypointensity throughout the supratentorial and infratentorial white matter |
| Contrast enhancement | no | no | no | no | no | no | no | no | no |
| Diffusion restriction | no | no | no | no | no | no | no | no | no |
| Delayed myelination | N/A, age greater than 2 years | N/A, age greater than 2 years | N/A, age greater than 2 years | N/A, age greater than 2 years | yes | yes | N/A, age greater than 2 years | N/A, age greater than 2 years | N/A, age greater than 2 years |
| Hypomyelination/abnormal myelination | no | yes | yes | yes | N/A, age less than 2 years | N/A, age less than 2 years | yes | yes | yes |
| Thinning of the corpus callosum | no | yes | yes | yes | yes | yes | yes | yes | yes |
| Vermis volume loss | no | yes | yes | no | no | yes | yes | no | yes |
| Additional features | periventricular gliosis | progressive enlargement of lateral ventricles, mild prominence of supratentorial sulci | prominent cisterna magna, prominent ventricles, widening of the sylvian fissures, diffuse hypomyelination | prominent sulci and enlargement of the ventricles, generalized cerebral atrophy, hypomyelination | pronounced delayed myelination, diffuse hypomyelination, generalized cerebral atrophy, prominent ventricles | pronounced delayed myelination in cerebral hemispheres, brainstem, and cerebellum; inferior vermian hypoplasia | bifrontal lobe polymicrogyria, arachnoid cyst | atrophy of posterior part of putamina, hyperintense T2 signal of periventricular and deep white matter | pronounced delayed myelination, bifrontal lobe polymicrogyria, numerous areas of T1 hyperintensity throughout the supra and infratentorial white matter |
Abbreviations: T1W, T1-weighted; T2W, T2-weighted; IUGR, intrauterine growth restriction; SGA, small for gestational age; EEG, electroencephalography; GTC, generalized tonic-clonic; OFC, occipital frontal circumference; AOH, absence of heterozygosity; CNV, copy number variant; N/A, not available.
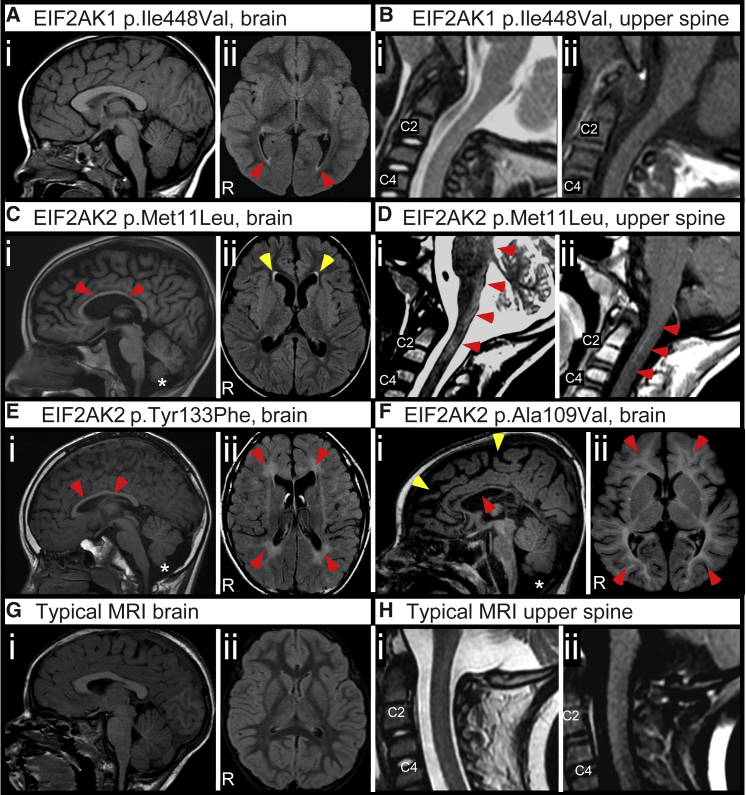
Researchers used Codified Genomics (variation interpretation software) for variant review in probands 1 and 2. Proband 1 is a 6-year-old female of Irish and German descent with developmental delay, progressive lower extremity spasticity, hypertonia, dysarthria, anxiety, and attention deficit and hyperactivity disorder (ADHD). Brain and spinal cord MRI studies revealed non-specific T2-weighted hyperintensities at the posterior lateral ventricles (Figures 1A and 1B). Trio exome sequencing (ES) identified a de novo missense variant in EIF2AK1 (c.1342A>G [p.Ile448Val]) (RefSeq: NM_014413.4). Proband 2 is a 10-year-old male of German, Mexican, and Spanish descent with developmental delay, ataxia, mixed hypotonia and hypertonia, dystonia, hemiballismus, choreoathetosis, myoclonus, dysarthria, parkinsonism, cognitive impairment, epilepsy, and anxiety. At 5 years of age, he abruptly lost developmental milestones including balance and coordination and developed progressively worsening movement disorders due to a febrile illness. Brain and spinal cord MRI studies revealed thinning of the corpus callosum, reduced volume of the cerebellar vermis, and T2-weighted hyperintensities in the dorsal upper cervical cord, dorsal medulla, dorsal pons, and periaqueductal gray (Figures 1C and 1D). Trio ES revealed a de novo missense variant in EIF2AK2 (c.31A>C [p.Met11Leu]; RefSeq: NM_002759.3). Although the EIF2AK2 p.Met11Leu variant is not present in gnomAD,25,26 a different variant affecting the same residue (EIF2AK2 p.Met11Val) was seen in three other heterozygous individuals (mean allele frequency 0.00001061) in gnomAD.25 Proband 3 is a 13-year-old male of Chinese descent with developmental delay, ataxia, mixed hypotonia and hypertonia, spasticity, dystonia, dysarthria, parkinsonism, cognitive impairment, and autism. He exhibited progressive decline in neurologic function and white matter changes with febrile illnesses. Brain and spinal cord MRI studies revealed a prominent cisterna magna, reduced volume of the cerebellar vermis, diffuse hypomyelination, and thinning of the corpus callosum, as well as T2-weighted hyperintensities in the subcortical white matter, periventricular white matter, and patchy signal abnormalities in the brainstem (Figure 1E). Trio ES revealed a de novo missense variant in EIF2AK2 (c.398A>T [p.Tyr133Phe]).
Figure 1.
Delayed Myelination, Cerebral Atrophy, and White Matter Abnormalities Associated with De Novo EIF2AK2 Missense Variants
(A and B) Representative images from proband 1 with EIF2AK1 p.Ile448Val variant at 2 years old acquired on a 1.5 Tesla (1.5T) MRI.
(A) MRI brain without contrast images. (i) Mid-sagittal T1-weighted image with appropriate size of corpus callosum and cerebellar vermis. (ii) Axial FLAIR image showing non-specific T2-weighted hyperintensities at the posterior lateral ventricles (red ars).
(B) MRI upper spinal cord images. Mid-sagittal T2-weighted (i) and T1-weighted (ii) images showing unremarkable upper spinal cord appearance.
(C and D) Representative images from proband 2 with EIF2AK2 p.Met11Leu variant at 7 years old acquired on a 1.5 Tesla (1.5T) MRI.
(C) MRI brain without contrast images. (i) Mid-sagittal T1-weighted image showing thinning of the corpus callosum (red ars) and mild cerebellar vermis hypoplasia (asterisk). (ii) Axial FLAIR image showing mild reduction in cerebral volume with thinning of the gyri and widening of the sulci.
(D) MRI upper spinal cord images. (i) Mid-sagittal T2-weighted image showing hyperintensities in the dorsal-most upper cervical cord, dorsal medulla, and dorsal pons (red ars). (ii) Post-contrast mid-sagittal T1-weighted image showing contrast enhancement in the upper cervical cord.
(E) Representative images from proband 3 with EIF2AK2 p.Tyr133Phe variant at 10 years old acquired on a 1.5T MRI. (i) Mid-sagittal T1-weighted image showing thinning of the corpus callosum (red ars) with cerebellar vermis hypoplasia and prominent cisterna magna (asterisk). (ii) Axial FLAIR image showing diffuse hyperintensities throughout the white matter (red ars).
(F) Representative images from proband 6 with EIF2AK2 p.Ala109Val variant at 18 months old acquired on a 3.0T MRI. (i) Mid-sagittal T1-weighted image showing thinning of the corpus callosum (red ar), cerebral atrophy (yellow ars), and inferior cerebellar vermis hypoplasia (asterisk). (ii) Axial FLAIR image showing pronounced delayed myelination in the cerebral hemispheres.
(G) Representative typical control MRI brain (i) mid-sagittal T1-weighted and (ii) axial FLAIR images from a 3-year-old acquired on a 1.5T MRI.
(H) Representative typical control (i) mid-sagittal T2-weighted and (ii) post-contrast mid-sagittal T1-weighted images from a 2-year-old acquired on a 1.5T MRI.
Proband 4 is a 3-year-old female of European descent with developmental delay, ataxia, hypotonia, tremor, dysarthria, cognitive impairment, and concern for seizure activity. She had progressive loss of developmental milestones with febrile illnesses. Brain and spinal cord MRI studies revealed diffuse hypomyelination, cerebral volume loss, and abnormal signal in the central gray matter of the cord. Trio ES identified a de novo missense variant in EIF2AK2 (c.973G>A [p.Gly325Ser]). Proband 5 is an 18-month-old male of European descent with acquired microcephaly, developmental delay, ataxia, mixed hypotonia and hypertonia, dystonia, tremor, parkinsonism, cognitive impairment, and seizures. He exhibited progressive loss of developmental milestones with fevers and illnesses. Brain MRI studies revealed thinning of the corpus callosum, delayed myelination, and cerebral volume loss. He was diagnosed with phenylketonuria (PKU) on newborn screen at 3 days of age and treatment was initiated at diagnosis. Compound heterozygous missense variants were identified in Phenylalanine Hydroxylase (p.Arg158Gln and p.Arg408Trp). Despite consistent medical management, it was difficult to maintain serum phenylalanine levels within normal limits. The Center for Mendelian Genomics and the Broad Institute performed trio ES and analyzed the results with SEQR27 and VExP.28 Trio ES revealed a de novo missense variant in EIF2AK2 (c.1382C>G [p.Ser461Cys]).
Proband 6 is a 19-month-old male of Moroccan and Kuwaiti descent with acquired microcephaly, developmental delay, ataxia, hypotonia, dystonia, and cognitive impairment. At 13 months of age, he abruptly lost developmental milestones including head control, rolling over, eye contact, and vocalizations following a febrile illness due to influenza A. Initial brain MRI study at 7 months of age revealed delayed myelination that was particularly pronounced along the cerebral hemispheres, brainstem, and cerebellum with thinning of the corpus callosum, cerebral volume loss, and inferior cerebellar vermian hypoplasia. Subsequent brain MRI study at 13 months of age following neurologic regression during febrile illness showed progressive global volume loss without substantial progress in myelination (Figure 1F). The family history is significant for consanguinity. Parents are first cousins once removed. A 47-Mb region with absence of heterozygosity on chromosome 17 (17p11.2q24.1) was identified. Chromosome 17 has not been reported with a clinical uniparental disomy (UPD) phenotype and therefore additional UPD testing was not clinically indicated. The region of AOH would be consistent with the family history of consanguinity. Trio ES revealed a de novo missense variant in EIF2AK2 (c.326C>T [p.Ala109Val]).
Proband 7 is a 3-year-old male of European descent with developmental delay, ataxia, mixed hypotonia and hypertonia, progressive lower extremity contractures, dysarthria, and cognitive impairment. He presented at 7–8 months of age with loss of developmental milestones in the setting of a febrile illness. At 4 years of age, he abruptly lost expressive language following a febrile illness due to respiratory syncytial virus. Brain MRI studies revealed thinning of the corpus callosum, progressive cerebral volume loss, reduced volume of the cerebellar vermis, bilateral frontal lobe polymicrogyria, and hypomyelination. Trio ES revealed a de novo missense variant in EIF2AK2 (c.325G>T [p.Ala109Ser]). Proband 8 is a 12-year-old male of European descent. He presented during the first few months of life with developmental delay and acquired microcephaly, and subsequently had progressive lower extremity spasticity with hyperreflexia, parkinsonism, dysarthria, and cognitive impairment. Severe worsening of postural instability was apparent during febrile illnesses. Brain MRI studies revealed abnormal myelination with T2-hyperintensitities in the periventricular and deep white matter, thinning of the corpus callosum, cerebral volume loss, and atrophy of the posterior putamina. Trio ES revealed a de novo missense variant in EIF2AK2 (c.95A>G [p.Asn32Ser]). Proband 9 is a 4-year-old male of German, Irish, Apache, and Cherokee descent with developmental delay, mixed hypotonia and hypertonia, dysarthria, cognitive impairment, and epilepsy. He presented in the first few months of life with bilateral horizontal nystagmus and seizures. At 4 years of age he abruptly lost the ability to crawl with regression in oral skills following a febrile illness due to human metapneumovirus infection. Brain MRI studies revealed thinning of the corpus callosum, cerebral volume loss, reduced volume of the cerebellar vermis, bilateral frontal lobe polymicrogyria, and hypomyelination. Trio ES revealed a de novo missense variant in EIF2AK2 (c.290C>T [p.Ser97Phe]).
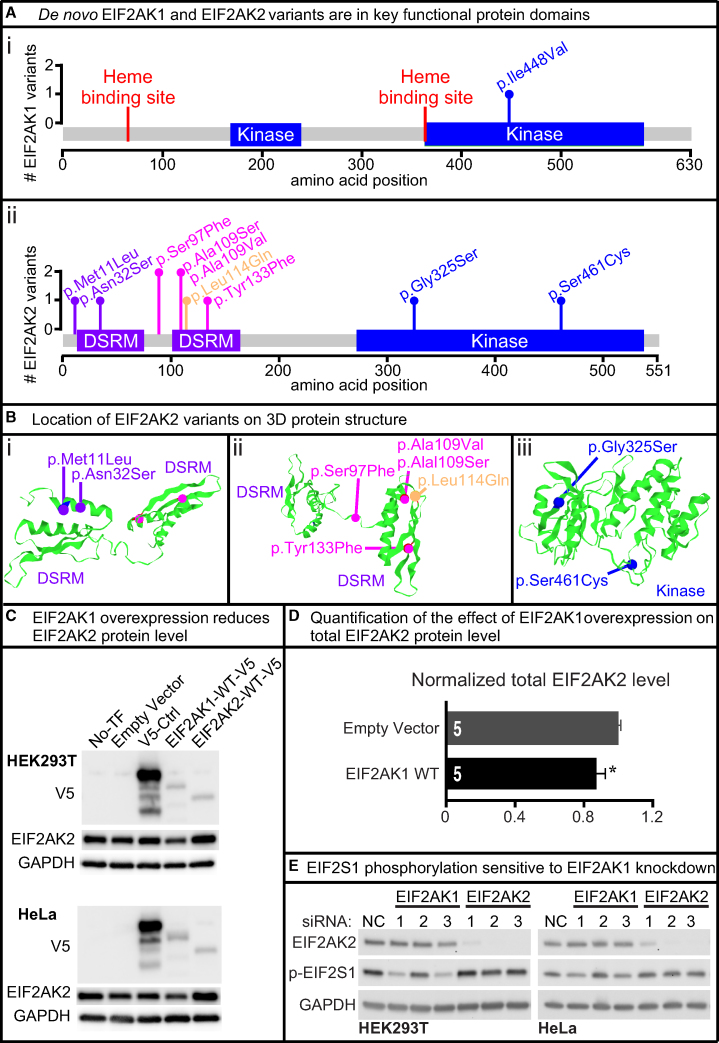
The de novo missense variants identified in these nine individuals predominantly localize to either the protein kinase or DSRM domains of EIF2AK1 and EIF2AK2 (Figures 2A and 2B).9, 10, 11 We also identified a rare missense variant in EIF2AK2 (c.341T>A [p.Leu114Gln]) of unknown inheritance from a proband-only ES in an individual with a discordant phenotype (dysmorphic facies, syndactyly, congenital microcephaly, and global developmental delay). We included the EIF2AK2 p.Leu114Gln variant in the molecular studies as a rare variant control. All of the variants were absent from the Exome Aggregation Consortium Database29 and the Genome Aggregation Database (gnomAD).25
Figure 2.
De novo EIF2AK1 and EIF2AK2 Missense Variants Map to Key Protein Domains and EIF2AK1 Knockdown Impairs EIF2S1 Phosphorylation
(A) Lollipop plots showing variants relative to a schematic representation of the gene adapted from MutationMapper. Heme-binding sites in red, protein kinase domain (Kinase) in blue, and double-stranded RNA-binding motif (DSRM) in purple. EIF2AK1 variant (i) is located in the kinase domain and EIF2AK2 variants (ii) are located in the DSRM and Kinase domains.
(B) 3D structure of EIF2AK2 DSRM and Kinase domains with de novo EIF2AK2 variants in purple, magenta, or blue. The rare variant control, p.Leu114Gln, is in orange. Variants are mapped to the protein 3D structure using Mutation3D.49 PDB: 1QU6, 3UIU.
(C) Full-length human EIF2AK1, EIF2AK2, and unrelated control cDNAs were cloned into pcDNA-DEST40 Vector with a CMV promoter and C terminus V5 tag. Lipofectamine 3000 was used to transfect the cDNA vectors into HEK293T and HeLa cells. Western blots show the protein level of V5-tagged and endogenous EIF2AK2. Increased EIF2AK1 protein level reduces EIF2AK2 protein level in HEK293T and HeLa cell lines. All western blot images in this paper were acquired using the Bio-Rad ChemiDoc Imaging Systems and densitometric analyses of the bands were performed with ImageJ. All images were collected by the imaging system within the linear range.
(D) Quantification of the effect of increased EIF2AK1 in mammalian cell lines on EIF2AK2 protein levels. Statistical significance determined by Student’s t test. Data shown as mean ± SEM; n = 5 independent replicates. ∗p < 0.05.
(E) Lipofectamine RNAiMAX was used to transfect HEK293T and HeLa cells with either control, EIF2AK1, or EIF2AK2 siRNA for 3 days. Three different siRNAs were tested per gene. Western blots show knockdown efficiency for EIF2AK1 and EIF2AK2. Two EIF2AK1 siRNAs show reduced p-EIF2S1 levels in both HEK293T and HeLa cells. Knockdown of EIF2AK2 does not affect p-EIF2S1 levels in either HEK293T or HeLa cells.
We utilized statistical models to explore whether EIF2AK1 and EIF2AK2 undergo selective restraint, a process where selection has reduced functional variation. Analysis of the observed to the expected loss-of-function (LoF) variation across the genes for EIF2AK1 and EIF2AK2 revealed observed/expected (o/e) scores of 0.47 and 0.30, respectively.25 These o/e results indicate that there is less LoF variation than predicted.25 Additionally, the Residual Variation Intolerance Score version 4 (RVISv4) is −0.331 for EIF2AK1 and −1.2108 for EIF2AK2, where RIVS < 0 indicates there is less common functional variation in the population than predicted.30 However, both EIF2AK1 and EIF2AK2 have low probability of LoF intolerance scores (pLI = 0 and 0.06, respectively) in gnomAD.25 Based on the gene size and GC content, 34 (EIF2AK1) and 31 (EIF2AK2) LoF variants were expected, and in the gnomAD population 16 (EIF2AK1) and 9 (EIF2AK2) LoF variants were observed.25 Together, these statistical findings indicate that EIF2AK1 and EIF2AK2 likely tolerate the loss of one functional copy of the gene (haplosufficient) but there is less variation in the population than predicted.
To determine the functional consequences of the de novo variants identified in EIF2AK1 and EIF2AK2, we cloned full-length human wild-type (WT) EIF2AK1, EIF2AK2, and unrelated control cDNAs into the mammalian vector pcDNA-DEST40 to generate C-terminal V5 (GKPIPNPLLGLDSD) tagged proteins (EIF2AK1-WT-V5 and EIF2AK2-WT-V5) under the control of a CMV promoter. The pcDNA-DEST40 cDNA constructs were transfected into two human cell lines, HEK293T and HeLa. There were modest increases in EIF2AK1-WT-V5 and EIF2AK2-WT-V5 protein levels compared to an unrelated protein-V5 control (Figure 2C), suggesting that EIF2AK1 and EIF2AK2 protein levels are tightly regulated in these cell lines. Increasing EIF2AK1-WT-V5 protein also reduced the total EIF2AK2 protein level (Figures 2C and 2D). Next, to determine the consequences of EIF2AK1 or EIF2AK2 LoF, we examined the impact of either EIF2AK1 or EIF2AK2 knockdown on EIF2S1 phosphorylation (p-EIF2S1) in HEK293T cells or HeLa cells. We designed three independent siRNAs targeting different regions of EIF2AK1 or EIF2AK2 mRNA and assessed p-EIF2S1 levels. Two of the three EIF2AK1 siRNAs significantly reduced p-EIF2S1 levels in both HEK293T and HeLa cell lines (Figure 2E). However, in all three of the EIF2AK2 siRNAs there were no changes in p-EIF2S1 levels (Figure 2E), suggesting potential redundancy between the EIF2AK family members in HEK293T and HeLa cells.
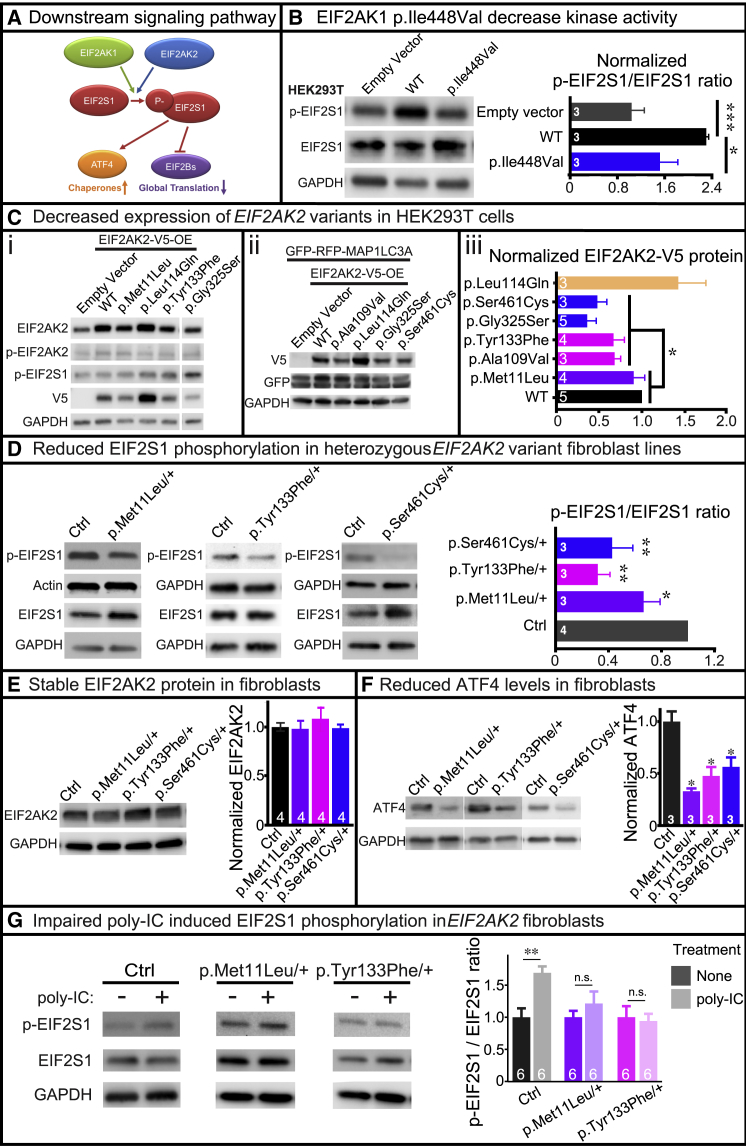
To test whether the EIF2AK1 and EIF2AK2 variants are deleterious, we generated pcDNA-DEST40 cDNA constructs to express the human variants in HEK293T or HeLa cells. The variants were generated via either Agilent QuikChange Lightning or NEB Q5 site-directed mutagenesis and confirmed by Sanger sequencing. We assessed the effects of the EIF2AK1 and EIF2AK2 variants on protein kinase activity and protein stability in both mammalian cell lines and available proband-derived skin fibroblasts. First, we examined whether the EIF2AK1 and EIF2AK2 variants altered protein kinase activity in HEK293T cells, by measuring the phosphorylation of EIF2S1, the substrate of EIF2AK1/2 (Figure 3A). We found that EIF2AK1-WT-V5 protein in HEK293T cells upregulated p-EIF2S1 levels. However, EIF2AK1-Ile448Val-V5 protein in HEK293T cells had no effect on p-EIF2S1 levels, indicating that the EIF2AK1 p.Ile448Val variant impairs protein kinase activity (Figure 3B). Unlike the EIF2AK1 findings, neither EIF2AK2 WT nor the variants tested in this study had an effect on p-EIF2S1 levels in HEK293T (Figure 3C, i) or HeLa (data not shown) cells. This finding is consistent with our previous observation that EIF2AK2 knockdown in HEK293T and HeLa cells had no effect on p-EIF2S1 levels (Figure 2E), suggesting that HEK293T and HeLa cells are insensitive to altered EIF2AK2 protein level.
Figure 3.
De novo EIF2AK1 and EIF2AK2 Missense Variants Impair Kinase Activity
(A) Schematic diagram showing the downstream effectors of the EIF2AK1/EIF2AK2 pathway.
(B) Lipofectamine 3000 was used to transfect HEK293T cells with EIF2AK1-WT-V5 or EIF2AK1-Ile448Val-V5 cDNA vectors. Representative western blot shows that EIF2AK1-Ile448Val-V5 fails to increase EIF2S1 phosphorylation compared to EIF2AK1-WT-V5. Statistical significance determined by confirming the normality of the data (p = 0.05, Shapiro-Wilk test), and then Student’s t test to measure the difference between groups. Data shown as mean + SEM; n = 3 independent replicates. ∗p < 0.05, ∗∗p < 0.01.
(C) Lipofectamine 3000 was used to transfect HEK293T cells with V5-tagged EIF2AK2 WT or variant cDNAs. Western blots show the level of the V5-tagged and endogenous EIF2AK2 protein. (i) EIF2AK2 variants exhibit decreased protein stability compared to WT. No change in EIF2AK2 protein levels were observed with rare variant control, p.Leu114Gln. No change in p-EIF2S1 levels were observed with EIF2AK2 variants. (ii) LipofectamineTM 3000 was used to co-transfect HEK293T cells with GFP-RFP-MAP1LC3A control and either EIF2AK2-WT-V5 or EIF2AK2-variant-V5 cDNAs. The GFP protein level is consistent across all cells, indicating that EIF2AK2 variants do not affect general protein translation. (iii) Statistical significance determined by confirming the normality of the data (p = 0.05, Shapiro-Wilk test), and then Student’s t test to measure the difference between groups. Data shown as mean + SEM; n = 3–5 independent replicates. ∗p < 0.05.
(D) Western blot showing reduced p-EIF2S1 levels in proband-derived skin fibroblasts with heterozygous EIF2AK2 missense variants compared to unrelated control. Statistical significance determined by confirming the normality of the data (p = 0.05, Shapiro-Wilk test), and then Student’s t test to measure the difference between groups. Data shown as mean + SEM; n = 3–4 independent replicates. ∗p < 0.05, ∗∗p < 0.01.
(E) Western blot showing total EIF2AK2 protein level is not affected in proband-derived skin fibroblasts with heterozygous EIF2AK2 missense variants compared to unrelated control. Statistical significance determined by confirming the normality of the data (p = 0.05, Shapiro-Wilk test), and then Student’s t test to measure the difference between groups. n = 4 independent replicates. n.s = not significant.
(F) Western blot showing reduced ATF4 levels in proband-derived skin fibroblasts with heterozygous EIF2AK2 missense variants compared to unrelated control. Statistical significance determined by confirming the normality of the data (p = 0.05, Shapiro-Wilk test), and then Student’s t test to measure the difference between groups. Data shown as mean + SEM; n = 3 independent replicates. ∗p < 0.05.
(G) Control or proband-derived skin fibroblasts were incubated in regular media with or without poly(I:C) (final concentration 10 μg/mL) for 24 h and then protein was collected for western blot analysis. Increased p-EIF2S1 levels observed in control fibroblasts incubated with poly(I:C) but proband-derived skin fibroblasts with heterozygous EIF2AK2 missense variants fail to increase EIF2S1 phosphorylation. Statistical significance determined by Student’s t test. Data shown as mean + SEM; n = 3 independent replicates. ∗∗p < 0.01, n.s = not significant.
Second, we examined whether the EIF2AK2 variants affected the production of EIF2AK2 protein in HEK293T (Figure 3C, i–iii) and HeLa (data not shown) cells by using a V5 antibody to probe for the exogenous protein and an EIF2AK2 antibody to probe for the total EIF2AK2 protein. Interestingly, we found that nearly all EIF2AK2 variants (p.Tyr133Phe, p.Ala109Val, p.Gly325Ser, and p.Ser461Cys) in our cohort had reduced total and V5-tagged EIF2AK2 protein levels. In comparison, the rare variant control EIF2AK2 p.Leu114Gln, which we identified in a proband with discordant phenotypes, had no reduction in protein levels compared to EIF2AK2 WT (Figure 3C, i–iii). To examine whether the reduced EIF2AK2 protein stability associated with the p.Tyr133Phe, p.Ala109Val, p.Gly325Ser, and p.Ser461Cys variants was the result of impaired EIF2S1 signaling on general protein translation, we performed co-transfection with an unrelated protein, MAP1LC3A, tagged with GFP-RFP (GFP-RFP-MAP1LC3A). The co-transfection of EIF2AK2 variants with GFP-RFP-MAP1LC3A had no effect on GFP-RFP-MAP1LC3A levels, indicating that increased levels of EIF2AK2 variants does not affect general protein translation in HEK293T or HeLa cells (Figure 3C, ii).
Based on our findings that HEK293T and HeLa cells are insensitive to changes in EIF2AK2 protein levels, we obtained three independent proband-derived fibroblast lines heterozygous for EIF2AK2 p.Met11Leu, p.Tyr133Phe, and p.Ser461Cys from affected individuals enrolled in the UDN or CMG (probands 2, 3, and 5). First, we examined the levels of EIF2S1 phosphorylation in the fibroblast lines and found a consistent reduction in p-EIF2S1 levels in all three lines (Figure 3D). EIF2AK2 protein levels were stable in the heterozygous proband-derived skin fibroblast lines (Figure 3E), indicating that the reduced p-EIF2S1 levels were likely due to impaired EIF2AK2 kinase activity. Similarly, ATF4 protein level, which is regulated by p-EIF2S1, is significantly decreased in all three fibroblast cell lines (Figure 3F). Next, we examined whether EIF2AK2 kinase activity can be stimulated in the heterozygous fibroblast lines by inducing cellular stress through the addition of polyinosinic:polycytidylic acid (poly(I:C)). Poly(I:C) is structurally similar to dsRNA, which is present in some viruses, and can activate the ISR pathway through EIF2S1 phosphorylation by EIF2AK family members.31 Incubation with poly(I:C) activates Toll-like receptor 3 (TLR3), which recognizes dsRNA,32 and the activated TLR3 recruits TRAF6, TAK1, and TAB2 to form the TAK1-complex.33, 34, 35 EIF2AK2 is present in the poly(I:C)-induced TAK1 complex and a kinase inactive EIF2AK2 mutant protein inhibits poly(I:C) induction of the TLR3-mediated signaling pathway.33 Furthermore, poly(I:C) stimulation of mammalian cells has been shown to upregulate the EIF2AK2-mediated phosphorylation of EIF2S1.10,36,37 Together these findings suggest that poly(I:C) stimulation of mammalian cells triggers both a primary TLR3-mediated signaling event and a secondary EIF2AK2-mediated signaling event following poly(I:C) uptake into cells.10 Therefore, to test the functional consequences of the EIF2AK2 variants, we incubated the proband-derived skin fibroblasts with 10 μg/mL poly(I:C) for 24 h and then assessed p-EIF2S1 levels by western blot. Control (Ctrl) fibroblasts derived from unrelated healthy individuals show an increase in EIF2S1 phosphorylation upon addition of poly(I:C) (Figure 3G). However, the fibroblast lines heterozygous for either EIF2AK2 p.Met11Leu or p.Tyr133Phe failed to upregulate EIF2S1 phosphorylation in the presence of poly(I:C) (Figure 3G). We were unable to test the poly(I:C) induction in the heterozygous EIF2AK2 p.Ser461Cys fibroblasts as the line failed to expand after a few passages. Together, these results demonstrate that EIF2AK2 p.Met11Leu, p.Tyr133Phe, and p.Ser461Cys impair the EIF2AK2 kinase activity required for EIF2S1 phosphorylation in fibroblasts.
The results of our clinical and molecular characterizations in mammalian cell lines and proband-derived fibroblasts show that EIF2AK1 or EIF2AK2 missense variants in key functional domains lead to neurodevelopmental disorders with overlapping symptoms. The EIF2AK1 p.Ile448Val, EIF2AK2 p.Met11Leu, EIF2AK2 p.Tyr133Phe, and EIF2AK2 p.Ser461Cys variants that we tested in either mammalian cell lines or proband-derived skin fibroblasts showed reduced kinase activity with impaired EIF2S1 phosphorylation.
Comparing genotypes and phenotypes within the cohort reveals several findings of interest. First, proband 1 with a de novo EIF2AK1 p.Ile448Val variant has a distinct motor-predominant phenotype compared to the rest of the cohort with de novo EIF2AK2 variants. Proband 1’s phenotype is primarily distinguished by motor developmental delay, speech articulation disorder, progressive spastic hemiplegia with hyper-reflexia, and age-appropriate cognition. The unrelated probands 2–9 have de novo EIF2AK2 missense variants and their phenotypes are relatively more severe compared to proband 1. Common phenotypes in probands 2–9 include motor findings as well as ataxia, movement disorders, cognitive impairment, abnormal white matter findings, cerebral volume loss, and reduced cerebellar vermis volume. The LoF o/e score for EIF2AK1 (0.47) is higher than for EIF2AK2 (0.3), suggesting that EIF2AK1 is more tolerant than EIF2AK2 to LoF mutations. Therefore, the phenotypic spectrum associated with EIF2AK1 variants may be milder than for EIF2AK2 variants or there may be incomplete penetrance of EIF2AK1 pathogenic variants. However, this determination is limited by the small sample size. Second, all eight probands with de novo EIF2AK2 missense variants have a history of neurologic decompensation in the setting of fevers and illnesses. Although an interpretation of genotype to phenotype severity is limited by the small sample size, it is possible that the p.Met11Leu variant is less damaging as it did not reduce EIF2AK2 protein levels in mammalian cell lines.
Our functional data reveal that the de novo missense variants impair EIF2AK1 or EIFAK2 kinase activity and lead to reduced EIF2S1 phosphorylation. This impact on EIF2S1 activity would interfere with downstream molecular pathways critical for responding to cellular stressors. An abnormal stress response may underlie the neurologic decompensation and corresponding white matter alterations associated with fevers and illnesses in our cohort. Potential pathogenic mechanisms for these variants include gain-of-function, haploinsufficiency, and dominant-negative. A gain-of-function mechanism is less likely given the EIF2S1 phosphorylation and protein stability data, as well as the impaired response to poly(I:C) stimulation in fibroblasts. Haploinsufficiency is unlikely to be the primary contributor to the observed phenotypes in our probands given that LoF variants are present in gnomAD,25 a family with thoracic aortic aneurysm syndrome was found to have a heterozygous deletion of chromosome 2p22.3–p22.2 involving EIF2AK2 and ten other genes,38 and mouse models with constitutive loss of either Eif2ak1 or Eif2ak2 are viable and fertile without gross morphological abnormalities or neurologic findings.39,40 These findings are all consistent with EIF2AK1 and EIF2AK2 pLI scores of 0 and 0.06, respectively,25 indicating that a single copy of a functional gene is sufficient to maintain normal function. Therefore, given that EIF2AK1 and EIF2AK2 require dimerization to phosphorylate their downstream target, the most likely pathogenic mechanism of the de novo missense variants are dominant-negative mutations affecting the function of the wild-type protein.
The phosphorylation of EIF2S1 converts EIF2S1 into a competitive inhibitor of EIF2B, which activates the ISR.41 Therefore, the impaired EIF2S1 phosphorylation we observed with the de novo EIF2AK1 and EIF2AK2 missense variants would likely impact the EIF2B-mediated regulation of the ISR. Pathogenic variants in any of the five genes encoding the subunits of the EIF2B protein complex (EIF2B1, EIF2B2, EIF2B3, EIF2B4, and EIF2B5) are associated with autosomal-recessive childhood ataxia with central nervous system hypomyelination/vanishing white matter (CACH/VWM [MIM: 603896]).42, 43, 44, 45, 46 CACH/VWM is a chronic and progressive leukodystrophy characterized by neurologic decompensation in the setting of febrile illness and other stressors. Additional features of CACH/VWM include ataxia, spasticity, optic atrophy, epilepsy, loss of acquired developmental milestones, cognitive impairments, and coma.42,47,48
In conclusion, we show that pathogenic EIF2AK1 and EIF2AK2 missense variants cause a broad phenotypic spectrum including developmental delays, variable cognitive impairments, hypotonia, hypertonia, involuntary movements, ataxia, and white matter alterations. Individuals with EIF2AK2 variants also exhibit sensitivity to febrile illness and commonly experience neurological regression, similar to CACH/VWM. The phenotypic overlap between CACH/VWM and our probands with de novo missense EIF2AK1 and EIF2AK2 variants suggest that deleterious missense variants in EIF2AK1 and EIF2AK2 cause an autosomal-dominant neurodevelopmental syndrome that may share common pathogenic mechanisms with CACH/VWM disease.
Consortia
Members of the Undiagnosed Diseases Network (UDN): Maria T. Acosta, Margaret Adam, David R. Adams, Pankaj B. Agrawal, Mercedes E. Alejandro, Patrick Allard, Justin Alvey, Laura Amendola, Ashley Andrews, Euan A. Ashley, Mahshid S. Azamian, Carlos A. Bacino, Guney Bademci, Eva Baker, Ashok Balasubramanyam, Dustin Baldridge, Jim Bale, Michael Bamshad, Deborah Barbouth, Gabriel F. Batzli, Pinar Bayrak-Toydemir, Anita Beck, Alan H. Beggs, Gill Bejerano, Hugo J. Bellen, Jimmy Bennet, Beverly Berg-Rood, Raphael Bernier, Jonathan A. Bernstein, Gerard T. Berry, Anna Bican, Stephanie Bivona, Elizabeth Blue, John Bohnsack, Carsten Bonnenmann, Devon Bonner, Lorenzo Botto, Lauren C. Briere, Elly Brokamp, Elizabeth A. Burke, Lindsay C. Burrage, Manish J. Butte, Peter Byers, John Carey, Olveen Carrasquillo, Ta Chen Peter Chang, Sirisak Chanprasert, Hsiao-Tuan Chao, Gary D. Clark, Terra R. Coakley, Laurel A. Cobban, Joy D. Cogan, F. Sessions Cole, Heather A. Colley, Cynthia M. Cooper, Heidi Cope, William J. Craigen, Michael Cunningham, Precilla D’Souza, Hongzheng Dai, Surendra Dasari, Mariska Davids, Jyoti G. Dayal, Esteban C. Dell’Angelica, Shweta U. Dhar, Katrina Dipple, Daniel Doherty, Naghmeh Dorrani, Emilie D. Douine, David D. Draper, Laura Duncan, Dawn Earl, David J. Eckstein, Lisa T. Emrick, Christine M. Eng, Cecilia Esteves, Tyra Estwick, Liliana Fernandez, Carlos Ferreira, Elizabeth L. Fieg, Paul G. Fisher, Brent L. Fogel, Irman Forghani, Laure Fresard, William A. Gahl, Ian Glass, Rena A. Godfrey, Katie Golden-Grant, Alica M. Goldman, David B. Goldstein, Alana Grajewski, Catherine A. Groden, Andrea L. Gropman, Sihoun Hahn, Rizwan Hamid, Neil A. Hanchard, Nichole Hayes, Frances High, Anne Hing, Fuki M. Hisama, Ingrid A. Holm, Jason Hom, Martha Horike-Pyne, Alden Huang, Yong Huang, Rosario Isasi, Fariha Jamal, Gail P. Jarvik, Jeffrey Jarvik, Suman Jayadev, Yong-hui Jiang, Jean M. Johnston, Lefkothea Karaviti, Emily G. Kelley, Dana Kiley, Isaac S. Kohane, Jennefer N. Kohler, Deborah Krakow, Donna M. Krasnewich, Susan Korrick, Mary Koziura, Joel B. Krier, Seema R. Lalani, Byron Lam, Christina Lam, Brendan C. Lanpher, Ian R. Lanza, C. Christopher Lau, Kimberly LeBlanc, Brendan H. Lee, Hane Lee, Roy Levitt, Richard A. Lewis, Sharyn A. Lincoln, Pengfei Liu, Xue Zhong Liu, Nicola Longo, Sandra K. Loo, Joseph Loscalzo, Richard L. Maas, Ellen F. Macnamara, Calum A. MacRae, Valerie V. Maduro, Marta M. Majcherska, May Christine V. Malicdan, Laura A. Mamounas, Teri A. Manolio, Rong Mao, Kenneth Maravilla, Thomas C. Markello, Ronit Marom, Gabor Marth, Beth A. Martin, Martin G. Martin, Julian A. Martínez-Agosto, Shruti Marwaha, Jacob McCauley, Allyn McConkie-Rosell, Colleen E. McCormack, Alexa T. McCray, Heather Mefford, J. Lawrence Merritt, Matthew Might, Ghayda Mirzaa, Eva Morava-Kozicz, Paolo M. Moretti, Marie Morimoto, John J. Mulvihill, David R. Murdock, Avi Nath, Stan F. Nelson, John H. Newman, Sarah K. Nicholas, Deborah Nickerson, Donna Novacic, Devin Oglesbee, James P. Orengo, Laura Pace, Stephen Pak, J. Carl Pallais, Christina G.S. Palmer, Jeanette C. Papp, Neil H. Parker, John A. Phillips III, Jennifer E. Posey, John H. Postlethwait, Lorraine Potocki, Barbara N. Pusey, Aaron Quinlan, Wendy Raskind, Archana N. Raja, Genecee Renteria, Chloe M. Reuter, Lynette Rives, Amy K. Robertson, Lance H. Rodan, Jill A. Rosenfeld, Robb K. Rowley, Maura Ruzhnikov, Ralph Sacco, Jacinda B. Sampson, Susan L. Samson, Mario Saporta, C. Ron Scott, Judy Schaechter, Timothy Schedl, Kelly Schoch, Daryl A. Scott, Lisa Shakachite, Prashant Sharma, Vandana Shashi, Jimann Shin, Rebecca Signer, Catherine H. Sillari, Edwin K. Silverman, Janet S. Sinsheimer, Kathy Sisco, Kevin S. Smith, Lilianna Solnica-Krezel, Rebecca C. Spillmann, Joan M. Stoler, Nicholas Stong, Jennifer A. Sullivan, Angela Sun, Shirley Sutton, David A. Sweetser, Virginia Sybert, Holly K. Tabor, Cecelia P. Tamburro, Queenie K.-G. Tan, Mustafa Tekin, Fred Telischi, Willa Thorson, Cynthia J. Tifft, Camilo Toro, Alyssa A. Tran, Tiina K. Urv, Matt Velinder, Dave Viskochil, Tiphanie P. Vogel, Colleen E. Wahl, Stephanie Wallace, Nicole M. Walley, Chris A. Walsh, Melissa Walker, Jennifer Wambach, Jijun Wan, Lee-kai Wang, Michael F. Wangler, Patricia A. Ward, Daniel Wegner, Mark Wener, Monte Westerfield, Matthew T. Wheeler, Anastasia L. Wise, Lynne A. Wolfe, Jeremy D. Woods, Shinya Yamamoto, John Yang, Amanda J. Yoon, Guoyun Yu, Diane B. Zastrow, Chunli Zhao, Stephan Zuchner.
Declaration of Interests
The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the clinical exome sequencing offered at Baylor Genetics.
Acknowledgments
We thank the families and clinical staff at locations for participation in this study. We thank Seth Masters at the University of Melbourne, Australia for discussions and review of the manuscript. We thank Mingshan Xue, Fairouz Elsaeidi, Darrion Nguyen, Maimuna Sali Paul, and Cole Deisseroth at BCM for critical review and feedback on the manuscript. We thank Dr. Richard A. Lewis and Dr. Karen D. Evankovich at BCM for their input on the clinical findings. Research reported in this manuscript was supported by the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director under Award Number(s) U01HG007709 (BCM clinical site), U01HG007708 (Stanford clinical site), and U01HG007942 (BCM sequencing core). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This was work was supported in part by NIH U54NS093793 to H.J.B., by the Intramural Research Program of the National Human Genome Research Institute, and by the Common Fund, Office of the Director, NIH. H.J.B. is an investigator of the Howard Hughes Medical Institute. L.T.E., J.A.R., L.B., A.T., M.A., and H.-T.C are supported in part by NIH grant U01HG007709. P.L. is supported in part by NIH grant U01HG007942. H.-T.C.’s research effort was also supported in part by the American Academy of Neurology, Child Neurology Foundation, Burroughs Wellcome Fund, NIH grant 1DP1OD026428, and the McNair Medical Institute at The Robert and Janice McNair Foundation. Sequencing and analysis for proband 5 were provided by the Broad Institute of MIT and Harvard Center for Mendelian Genomics (Broad CMG) and was funded by the National Human Genome Research Institute, the National Eye Institute, and the National Heart, Lung, and Blood Institute grant UM1 HG008900 and in part by National Human Genome Research Institute grant R01 HG009141.
Published: March 19, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.02.016.
Contributor Information
Hugo J. Bellen, Email: hbellen@bcm.edu.
Hsiao-Tuan Chao, Email: hc140077@bcm.edu.
Undiagnosed Diseases Network:
Maria T. Acosta, Margaret Adam, David R. Adams, Pankaj B. Agrawal, Mercedes E. Alejandro, Patrick Allard, Justin Alvey, Laura Amendola, Ashley Andrews, Euan A. Ashley, Mahshid S. Azamian, Carlos A. Bacino, Guney Bademci, Eva Baker, Ashok Balasubramanyam, Dustin Baldridge, Jim Bale, Michael Bamshad, Deborah Barbouth, Gabriel F. Batzli, Pinar Bayrak-Toydemir, Anita Beck, Alan H. Beggs, Gill Bejerano, Hugo J. Bellen, Jimmy Bennet, Beverly Berg-Rood, Raphael Bernier, Jonathan A. Bernstein, Gerard T. Berry, Anna Bican, Stephanie Bivona, Elizabeth Blue, John Bohnsack, Carsten Bonnenmann, Devon Bonner, Lorenzo Botto, Lauren C. Briere, Elly Brokamp, Elizabeth A. Burke, Lindsay C. Burrage, Manish J. Butte, Peter Byers, John Carey, Olveen Carrasquillo, Ta Chen Peter Chang, Sirisak Chanprasert, Hsiao-Tuan Chao, Gary D. Clark, Terra R. Coakley, Laurel A. Cobban, Joy D. Cogan, F. Sessions Cole, Heather A. Colley, Cynthia M. Cooper, Heidi Cope, William J. Craigen, Michael Cunningham, Precilla D’Souza, Hongzheng Dai, Surendra Dasari, Mariska Davids, Jyoti G. Dayal, Esteban C. Dell’Angelica, Shweta U. Dhar, Katrina Dipple, Daniel Doherty, Naghmeh Dorrani, Emilie D. Douine, David D. Draper, Laura Duncan, Dawn Earl, David J. Eckstein, Lisa T. Emrick, Christine M. Eng, Cecilia Esteves, Tyra Estwick, Liliana Fernandez, Carlos Ferreira, Elizabeth L. Fieg, Paul G. Fisher, Brent L. Fogel, Irman Forghani, Laure Fresard, William A. Gahl, Ian Glass, Rena A. Godfrey, Katie Golden-Grant, Alica M. Goldman, David B. Goldstein, Alana Grajewski, Catherine A. Groden, Andrea L. Gropman, Sihoun Hahn, Rizwan Hamid, Neil A. Hanchard, Nichole Hayes, Frances High, Anne Hing, Fuki M. Hisama, Ingrid A. Holm, Jason Hom, Martha Horike-Pyne, Alden Huang, Yong Huang, Rosario Isasi, Fariha Jamal, Gail P. Jarvik, Jeffrey Jarvik, Suman Jayadev, Yong-hui Jiang, Jean M. Johnston, Lefkothea Karaviti, Emily G. Kelley, Dana Kiley, Isaac S. Kohane, Jennefer N. Kohler, Deborah Krakow, Donna M. Krasnewich, Susan Korrick, Mary Koziura, Joel B. Krier, Seema R. Lalani, Byron Lam, Christina Lam, Brendan C. Lanpher, Ian R. Lanza, C. Christopher Lau, Kimberly LeBlanc, Brendan H. Lee, Hane Lee, Roy Levitt, Richard A. Lewis, Sharyn A. Lincoln, Pengfei Liu, Xue Zhong Liu, Nicola Longo, Sandra K. Loo, Joseph Loscalzo, Richard L. Maas, Ellen F. Macnamara, Calum A. MacRae, Valerie V. Maduro, Marta M. Majcherska, May Christine V. Malicdan, Laura A. Mamounas, Teri A. Manolio, Rong Mao, Kenneth Maravilla, Thomas C. Markello, Ronit Marom, Gabor Marth, Beth A. Martin, Martin G. Martin, Julian A. Martínez-Agosto, Shruti Marwaha, Jacob McCauley, Allyn McConkie-Rosell, Colleen E. McCormack, Alexa T. McCray, Heather Mefford, J. Lawrence Merritt, Matthew Might, Ghayda Mirzaa, Eva Morava-Kozicz, Paolo M. Moretti, Marie Morimoto, John J. Mulvihill, David R. Murdock, Avi Nath, Stan F. Nelson, John H. Newman, Sarah K. Nicholas, Deborah Nickerson, Donna Novacic, Devin Oglesbee, James P. Orengo, Laura Pace, Stephen Pak, J. Carl Pallais, Christina G.S. Palmer, Jeanette C. Papp, Neil H. Parker, John A. Phillips, III, Jennifer E. Posey, John H. Postlethwait, Lorraine Potocki, Barbara N. Pusey, Aaron Quinlan, Wendy Raskind, Archana N. Raja, Genecee Renteria, Chloe M. Reuter, Lynette Rives, Amy K. Robertson, Lance H. Rodan, Jill A. Rosenfeld, Robb K. Rowley, Maura Ruzhnikov, Ralph Sacco, Jacinda B. Sampson, Susan L. Samson, Mario Saporta, C. Ron Scott, Judy Schaechter, Timothy Schedl, Kelly Schoch, Daryl A. Scott, Lisa Shakachite, Prashant Sharma, Vandana Shashi, Jimann Shin, Rebecca Signer, Catherine H. Sillari, Edwin K. Silverman, Janet S. Sinsheimer, Kathy Sisco, Kevin S. Smith, Lilianna Solnica-Krezel, Rebecca C. Spillmann, Joan M. Stoler, Nicholas Stong, Jennifer A. Sullivan, Angela Sun, Shirley Sutton, David A. Sweetser, Virginia Sybert, Holly K. Tabor, Cecelia P. Tamburro, Queenie K.-G. Tan, Mustafa Tekin, Fred Telischi, Willa Thorson, Cynthia J. Tifft, Camilo Toro, Alyssa A. Tran, Tiina K. Urv, Matt Velinder, Dave Viskochil, Tiphanie P. Vogel, Colleen E. Wahl, Stephanie Wallace, Nicole M. Walley, Chris A. Walsh, Melissa Walker, Jennifer Wambach, Jijun Wan, Lee-kai Wang, Michael F. Wangler, Patricia A. Ward, Daniel Wegner, Mark Wener, Monte Westerfield, Matthew T. Wheeler, Anastasia L. Wise, Lynne A. Wolfe, Jeremy D. Woods, Shinya Yamamoto, John Yang, Amanda J. Yoon, Guoyun Yu, Diane B. Zastrow, Chunli Zhao, and Stephan Zuchner
Accession Numbers
The accession numbers for the variants reported to ClinVar are (1) ClinVar: SCV001142583.1; GenBank: NM_014413.3 (EIF2AK1); c.1342A>G (p.Ile448Val); (2) ClinVar: SCV001142584.1; GenBank: NM_002759.3 (EIF2AK2); c.31A>C (p.Met11Leu); (3) ClinVar: SCV001142597.1; GenBank: NM_002759.3 (EIF2AK2); c.398A>T (p.Tyr133Phe); (4) ClinVar: SCV001161776.1; GenBank: NM_002759.3 (EIF2AK2); c.290C>T (p.Ser97Phe); and (5) ClinVar: SCV001161775.1; GenBank: NM_002759.3 (EIF2AK2); c.326C>T (p.Ala109Val).
Web Resources
ENSEMBL VEP SIFT, http://useast.ensembl.org/info/docs/tools/vep/index.html
ExAC Browser, http://exac.broadinstitute.org/
Genic Intolerance, http://genic-intolerance.org/
gnomAD Browser, https://gnomad.broadinstitute.org/
HUGO Gene Nomenclature Committee, http://www.genenames.org/
MARRVEL, http://marrvel.org/
MutationTaster, http://www.mutationtaster.org/
OMIM, https://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
Supplemental Data
Tables S1–S3 and Supplemental Material and Methods
Article plus Supplemental Information
References
- 1.Wek R.C., Jiang H.Y., Anthony T.G. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 2.Harding H.P., Zhang Y., Zeng H., Novoa I., Lu P.D., Calfon M., Sadri N., Yun C., Popko B., Paules R. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 3.Acharya P., Chen J.J., Correia M.A. Hepatic heme-regulated inhibitor (HRI) eukaryotic initiation factor 2alpha kinase: a protagonist of heme-mediated translational control of CYP2B enzymes and a modulator of basal endoplasmic reticulum stress tone. Mol. Pharmacol. 2010;77:575–592. doi: 10.1124/mol.109.061259. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Liu S., Suragani R.N., Wang F., Han A., Zhao W., Andrews N.C., Chen J.J. The function of heme-regulated eIF2alpha kinase in murine iron homeostasis and macrophage maturation. J. Clin. Invest. 2007;117:3296–3305. doi: 10.1172/JCI32084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J.J. Regulation of protein synthesis by the heme-regulated eIF2alpha kinase: relevance to anemias. Blood. 2007;109:2693–2699. doi: 10.1182/blood-2006-08-041830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yerlikaya A., Kimball S.R., Stanley B.A. Phosphorylation of eIF2alpha in response to 26S proteasome inhibition is mediated by the haem-regulated inhibitor (HRI) kinase. Biochem. J. 2008;412:579–588. doi: 10.1042/BJ20080324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu L., Han A.P., Chen J.J. Translation initiation control by heme-regulated eukaryotic initiation factor 2alpha kinase in erythroid cells under cytoplasmic stresses. Mol. Cell. Biol. 2001;21:7971–7980. doi: 10.1128/MCB.21.23.7971-7980.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin Z., Haynie J., Williams B.R., Yang Y.C. C114 is a novel IL-11-inducible nuclear double-stranded RNA-binding protein that inhibits protein kinase R. J. Biol. Chem. 2003;278:22838–22845. doi: 10.1074/jbc.M212969200. [DOI] [PubMed] [Google Scholar]
- 9.Galluzzi L., Brenner C., Morselli E., Touat Z., Kroemer G. Viral control of mitochondrial apoptosis. PLoS Pathog. 2008;4:e1000018. doi: 10.1371/journal.ppat.1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silva A.M., Whitmore M., Xu Z., Jiang Z., Li X., Williams B.R. Protein kinase R (PKR) interacts with and activates mitogen-activated protein kinase kinase 6 (MKK6) in response to double-stranded RNA stimulation. J. Biol. Chem. 2004;279:37670–37676. doi: 10.1074/jbc.M406554200. [DOI] [PubMed] [Google Scholar]
- 11.Ito T., Yang M., May W.S. RAX, a cellular activator for double-stranded RNA-dependent protein kinase during stress signaling. J. Biol. Chem. 1999;274:15427–15432. doi: 10.1074/jbc.274.22.15427. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura T., Furuhashi M., Li P., Cao H., Tuncman G., Sonenberg N., Gorgun C.Z., Hotamisligil G.S. Double-stranded RNA-dependent protein kinase links pathogen sensing with stress and metabolic homeostasis. Cell. 2010;140:338–348. doi: 10.1016/j.cell.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Onuki R., Bando Y., Suyama E., Katayama T., Kawasaki H., Baba T., Tohyama M., Taira K. An RNA-dependent protein kinase is involved in tunicamycin-induced apoptosis and Alzheimer’s disease. EMBO J. 2004;23:959–968. doi: 10.1038/sj.emboj.7600049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams B.R. PKR; a sentinel kinase for cellular stress. Oncogene. 1999;18:6112–6120. doi: 10.1038/sj.onc.1203127. [DOI] [PubMed] [Google Scholar]
- 15.Goh K.C., deVeer M.J., Williams B.R. The protein kinase PKR is required for p38 MAPK activation and the innate immune response to bacterial endotoxin. EMBO J. 2000;19:4292–4297. doi: 10.1093/emboj/19.16.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheshire J.L., Williams B.R., Baldwin A.S., Jr. Involvement of double-stranded RNA-activated protein kinase in the synergistic activation of nuclear factor-kappaB by tumor necrosis factor-alpha and gamma-interferon in preneuronal cells. J. Biol. Chem. 1999;274:4801–4806. doi: 10.1074/jbc.274.8.4801. [DOI] [PubMed] [Google Scholar]
- 17.Ernst H., Duncan R.F., Hershey J.W. Cloning and sequencing of complementary DNAs encoding the alpha-subunit of translational initiation factor eIF-2. Characterization of the protein and its messenger RNA. J. Biol. Chem. 1987;262:1206–1212. [PubMed] [Google Scholar]
- 18.Pathak V.K., Schindler D., Hershey J.W. Generation of a mutant form of protein synthesis initiation factor eIF-2 lacking the site of phosphorylation by eIF-2 kinases. Mol. Cell. Biol. 1988;8:993–995. doi: 10.1128/mcb.8.2.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delépine M., Nicolino M., Barrett T., Golamaully M., Lathrop G.M., Julier C. EIF2AK3, encoding translation initiation factor 2-alpha kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nat. Genet. 2000;25:406–409. doi: 10.1038/78085. [DOI] [PubMed] [Google Scholar]
- 20.Eyries M., Montani D., Girerd B., Perret C., Leroy A., Lonjou C., Chelghoum N., Coulet F., Bonnet D., Dorfmüller P. EIF2AK4 mutations cause pulmonary veno-occlusive disease, a recessive form of pulmonary hypertension. Nat. Genet. 2014;46:65–69. doi: 10.1038/ng.2844. [DOI] [PubMed] [Google Scholar]
- 21.Ramoni R.B., Mulvihill J.J., Adams D.R., Allard P., Ashley E.A., Bernstein J.A., Gahl W.A., Hamid R., Loscalzo J., McCray A.T., Undiagnosed Diseases Network The Undiagnosed Diseases Network: Accelerating Discovery about Health and Disease. Am. J. Hum. Genet. 2017;100:185–192. doi: 10.1016/j.ajhg.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gahl W.A., Wise A.L., Ashley E.A. The Undiagnosed Diseases Network of the National Institutes of Health: A National Extension. JAMA. 2015;314:1797–1798. doi: 10.1001/jama.2015.12249. [DOI] [PubMed] [Google Scholar]
- 23.Sobreira N., Schiettecatte F., Boehm C., Valle D., Hamosh A. New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum. Mutat. 2015;36:425–431. doi: 10.1002/humu.22769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alfoldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv. 2019 [Google Scholar]
- 26.Wang J., Al-Ouran R., Hu Y., Kim S.Y., Wan Y.W., Wangler M.F., Yamamoto S., Chao H.T., Comjean A., Mohr S.E., UDN MARRVEL: Integration of Human and Model Organism Genetic Resources to Facilitate Functional Annotation of the Human Genome. Am. J. Hum. Genet. 2017;100:843–853. doi: 10.1016/j.ajhg.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perić S., Glumac J.N., Töpf A., Savić-Pavićević D., Phillips L., Johnson K., Cassop-Thompson M., Xu L., Bertoli M., Lek M. A novel recessive TTN founder variant is a common cause of distal myopathy in the Serbian population. Eur. J. Hum. Genet. 2017;25:572–581. doi: 10.1038/ejhg.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmitz-Abe K., Li Q., Rosen S.M., Nori N., Madden J.A., Genetti C.A., Wojcik M.H., Ponnaluri S., Gubbels C.S., Picker J.D. Unique bioinformatic approach and comprehensive reanalysis improve diagnostic yield of clinical exomes. Eur. J. Hum. Genet. 2019;27:1398–1405. doi: 10.1038/s41431-019-0401-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petrovski S., Wang Q., Heinzen E.L., Allen A.S., Goldstein D.B. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y., Xu X.L., Zhao D., Pan L.N., Huang C.W., Guo L.J., Lu Q., Wang J. TLR3 ligand Poly IC Attenuates Reactive Astrogliosis and Improves Recovery of Rats after Focal Cerebral Ischemia. CNS Neurosci. Ther. 2015;21:905–913. doi: 10.1111/cns.12469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alexopoulou L., Holt A.C., Medzhitov R., Flavell R.A. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 33.Jiang Z., Zamanian-Daryoush M., Nie H., Silva A.M., Williams B.R., Li X. Poly(I-C)-induced Toll-like receptor 3 (TLR3)-mediated activation of NFkappa B and MAP kinase is through an interleukin-1 receptor-associated kinase (IRAK)-independent pathway employing the signaling components TLR3-TRAF6-TAK1-TAB2-PKR. J. Biol. Chem. 2003;278:16713–16719. doi: 10.1074/jbc.M300562200. [DOI] [PubMed] [Google Scholar]
- 34.Jiang Z., Ninomiya-Tsuji J., Qian Y., Matsumoto K., Li X. Interleukin-1 (IL-1) receptor-associated kinase-dependent IL-1-induced signaling complexes phosphorylate TAK1 and TAB2 at the plasma membrane and activate TAK1 in the cytosol. Mol. Cell. Biol. 2002;22:7158–7167. doi: 10.1128/MCB.22.20.7158-7167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ninomiya-Tsuji J., Kishimoto K., Hiyama A., Inoue J., Cao Z., Matsumoto K. The kinase TAK1 can activate the NIK-I kappaB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature. 1999;398:252–256. doi: 10.1038/18465. [DOI] [PubMed] [Google Scholar]
- 36.Zhang S., Sun Y., Chen H., Dai Y., Zhan Y., Yu S., Qiu X., Tan L., Song C., Ding C. Activation of the PKR/eIF2α signaling cascade inhibits replication of Newcastle disease virus. Virol. J. 2014;11:62. doi: 10.1186/1743-422X-11-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Y., Conway T.W. 2-Aminopurine inhibits the double-stranded RNA-dependent protein kinase both in vitro and in vivo. J. Interferon Res. 1993;13:323–328. doi: 10.1089/jir.1993.13.323. [DOI] [PubMed] [Google Scholar]
- 38.Quiñones-Pérez B., VanNoy G.E., Towne M.C., Shen Y., Singh M.N., Agrawal P.B., Smith S.E. Three-generation family with novel contiguous gene deletion on chromosome 2p22 associated with thoracic aortic aneurysm syndrome. Am. J. Med. Genet. A. 2018;176:560–569. doi: 10.1002/ajmg.a.38590. [DOI] [PubMed] [Google Scholar]
- 39.Han A.P., Yu C., Lu L., Fujiwara Y., Browne C., Chin G., Fleming M., Leboulch P., Orkin S.H., Chen J.J. Heme-regulated eIF2alpha kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J. 2001;20:6909–6918. doi: 10.1093/emboj/20.23.6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Y.L., Reis L.F., Pavlovic J., Aguzzi A., Schäfer R., Kumar A., Williams B.R., Aguet M., Weissmann C. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 1995;14:6095–6106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bogorad A.M., Lin K.Y., Marintchev A. Novel mechanisms of eIF2B action and regulation by eIF2α phosphorylation. Nucleic Acids Res. 2017;45:11962–11979. doi: 10.1093/nar/gkx845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Knaap M.S., Pronk J.C., Scheper G.C. Vanishing white matter disease. Lancet Neurol. 2006;5:413–423. doi: 10.1016/S1474-4422(06)70440-9. [DOI] [PubMed] [Google Scholar]
- 43.Leegwater P.A., Vermeulen G., Könst A.A., Naidu S., Mulders J., Visser A., Kersbergen P., Mobach D., Fonds D., van Berkel C.G. Subunits of the translation initiation factor eIF2B are mutant in leukoencephalopathy with vanishing white matter. Nat. Genet. 2001;29:383–388. doi: 10.1038/ng764. [DOI] [PubMed] [Google Scholar]
- 44.Maletkovic J., Schiffmann R., Gorospe J.R., Gordon E.S., Mintz M., Hoffman E.P., Alper G., Lynch D.R., Singhal B.S., Harding C. Genetic and clinical heterogeneity in eIF2B-related disorder. J. Child Neurol. 2008;23:205–215. doi: 10.1177/0883073807308705. [DOI] [PubMed] [Google Scholar]
- 45.van der Knaap M.S., Leegwater P.A., Könst A.A., Visser A., Naidu S., Oudejans C.B., Schutgens R.B., Pronk J.C. Mutations in each of the five subunits of translation initiation factor eIF2B can cause leukoencephalopathy with vanishing white matter. Ann. Neurol. 2002;51:264–270. doi: 10.1002/ana.10112. [DOI] [PubMed] [Google Scholar]
- 46.van der Knaap M.S., van Berkel C.G., Herms J., van Coster R., Baethmann M., Naidu S., Boltshauser E., Willemsen M.A., Plecko B., Hoffmann G.F. eIF2B-related disorders: antenatal onset and involvement of multiple organs. Am. J. Hum. Genet. 2003;73:1199–1207. doi: 10.1086/379524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schiffmann R., Moller J.R., Trapp B.D., Shih H.H., Farrer R.G., Katz D.A., Alger J.R., Parker C.C., Hauer P.E., Kaneski C.R. Childhood ataxia with diffuse central nervous system hypomyelination. Ann. Neurol. 1994;35:331–340. doi: 10.1002/ana.410350314. [DOI] [PubMed] [Google Scholar]
- 48.Hanefeld F., Holzbach U., Kruse B., Wilichowski E., Christen H.J., Frahm J. Diffuse white matter disease in three children: an encephalopathy with unique features on magnetic resonance imaging and proton magnetic resonance spectroscopy. Neuropediatrics. 1993;24:244–248. doi: 10.1055/s-2008-1071551. [DOI] [PubMed] [Google Scholar]
- 49.Meyer M.J., Lapcevic R., Romero A.E., Yoon M., Das J., Beltrán J.F., Mort M., Stenson P.D., Cooper D.N., Paccanaro A., Yu H. mutation3D: Cancer Gene Prediction Through Atomic Clustering of Coding Variants in the Structural Proteome. Hum. Mutat. 2016;37:447–456. doi: 10.1002/humu.22963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1–S3 and Supplemental Material and Methods
Article plus Supplemental Information