Abstract
Ebola virus causes severe hemorrhagic fever, often leading to death in humans. The trimeric fusion glycoprotein (GP) is the sole target for neutralizing antibodies and is the major focus of vaccine development. Soluble GP ectodomains are unstable and mostly monomeric when not fused to a heterologous trimerization domain. Here, we report structure-based designs of Ebola and Marburg GP trimers based on a stabilizing mutation in the hinge loop in refolding region 1 and substitution of a partially buried charge at the interface of the GP1 and GP2 subunits. The combined substitutions (T577P and K588F) substantially increased trimer expression for Ebola GP proteins. We determined the crystal structure of stabilized GP from the Makona Zaire ebolavirus strain without a trimerization domain or complexed ligand. The structure reveals that the stabilized GP adopts the same trimeric prefusion conformation, provides insight into triggering of GP conformational changes, and should inform future filovirus vaccine development.
Keywords: filovirus, Ebola, Marburg, glycoprotein, stabilizing substitutions, prefusion trimer, X-ray structure, crystal, instability, size-exclusion chromatography
Graphical Abstract
Highlights
-
•
Filovirus GP expression increases by stabilizing mutations in hinge loop and base helix
-
•
Charged lysine in base helix and GP1 N terminus are trapped in metastable conformation
-
•
Crystal structure of stabilized Makona Δmucin GP confirms successful stabilization
-
•
These findings may be useful for understanding fusion mechanisms and vaccine design
Rutten et al. describe structure-based stabilization of soluble filovirus GP trimers to obtain high yields of trimers. The crystal structure of a stabilized Makona Δmucin GP shows how the introduced substitutions stabilize the trimer and that the N terminus of GP1 plays a crucial role in refolding.
Introduction
Filovirus infections are characterized by high fatality rates, with repeated outbreaks occurring in West Africa. Most are local outbreaks, but in 2014, an epidemic caused by the Zaire ebolavirus spread throughout West Africa, resulting in more than 11,000 deaths (World Health Organization, 2020a). The current epidemic in the Democratic Republic of Congo is the second largest, with more than 2,264 deaths, to date and has been declared a public health emergency of international concern (World Health Organization, 2020b).
The Filoviridae family comprises six genera, with members of the genus Marburgvirus (one species: Marburg marburgvirus) and the genus Ebolavirus (six species: Bombali ebolavirus, Bundibugyo ebolavirus, Reston ebolavirus, Sudan ebolavirus, Tai Forest ebolavirus, and Zaire ebolavirus) being the most significant threats to human health (Kuhn et al., 2019) (International Committee on Taxonomy of Viruses, 2019). Filovirus glycoprotein (GP) is a class I fusion protein that consists of two disulfide-linked subunits, GP1 and GP2, that trimerize to form the active molecule on the virion surface. GP1 consists of a core, a glycan cap, and a mucin-like domain. GP2 is anchored to the membrane and contains the membrane-fusion domains. Like other viral fusion proteins, filovirus GP is a dynamic machine that drives membrane fusion by irreversibly refolding from a metastable prefusion conformation to a stable postfusion conformation. In the case of Ebola virus, the unusually complex entry requirements are (1) binding to a cell-surface receptor (TIM-1, Axl, heparan sulfate, and DC-SIGN) (Alvarez et al., 2002, Brindley et al., 2011, Jemielity et al., 2013, Kondratowicz et al., 2011, Shimojima et al., 2007, Simmons et al., 2003, Takada et al., 2000); (2) macropinocytosis (Mulherkar et al., 2011, Nanbo et al., 2010, Saeed et al., 2010); (3) cleavage of GP at low pH by cathepsins (Chandran et al., 2005, Schornberg et al., 2006); (4) binding to loop C of Niemann-Pick C1 cholesterol transporter in the endosome (Carette et al., 2011, Côté et al., 2011, Miller et al., 2012, Wang et al., 2016); and most likely, (5) a final trigger that drives GP to undergo the conformational changes required for membrane fusion (Fénéant et al., 2019, Wang et al., 2016).
For many class I fusion proteins, stabilizing substitutions have been described for vaccine applications or to facilitate structural analysis (Binley et al., 2000, Fels et al., 2019, Hastie et al., 2017, Krarup et al., 2015, McLellan et al., 2013, Pallesen et al., 2017, Rutten et al., 2018, Sanders et al., 2002). However, no stabilizing substitutions have been described for soluble filovirus GP trimer. Moreover, GP structures that have been obtained previously have included a trimerization domain, complexed antibodies, or a non-native N terminus (Bale et al., 2012, Bornholdt et al., 2016, Dias et al., 2011, Janus et al., 2018, Lee et al., 2008, Murin et al., 2018, Pallesen et al., 2016, Wang et al., 2016, West et al., 2018). Because GP ectodomains form a mixture of species when expressed in HEK293T cells (Figure S8 in Lee et al., 2008), we sought to stabilize the trimeric, prefusion conformation of GP. We mutated elements in refolding region 1 (RR1, from 502 to 584) of GP2 and at the interface between GP1 and GP2 to obtain high yields of near-native soluble filovirus prefusion GP trimers with a native N terminus and without a heterologous trimerization domain. The trimers described in this study allowed structure determination of an unliganded Ebola GP ectodomain with a native N terminus, revealing the importance of the N terminus in the stability of the prefusion GP trimer. The stabilized soluble trimers described here may have applications as superior subunit-based antigens in vaccines or immune assays or as bait for isolation of monoclonal antibodies (mAbs) against filovirus GPs.
Results
Structure-Based Design of Stabilized GP Trimers
The ectodomains of wild-type Zaire ebolavirus GPs from the Yambuku-Mayinga and Makona strains were expressed with or without their mucin-like domains and without an additional C-terminal trimerization domain. Only a small fraction of the total produced protein formed trimers, as judged by analytical size-exclusion chromatography (SEC) or native polyacrylamide gel electrophoresis (PAGE), whereas most of the protein formed dimers and monomers (Figure S1). To increase the trimer yields, we set out to increase the stability of the protein using a structure-based design. Although class I fusion proteins like HIV-1 Env, respiratory syncytial virus (RSV) F, influenza hemagglutinin (HA), and Ebola/Marburg GP have very low sequence conservation, they share structural features in their fusion machinery. Because class I fusion proteins need to transform from a prefusion conformation to a highly stable postfusion conformation, the proteins harbor several regions of instability. The C-terminal end of RR1 just before the base helix is the so-called hinge region (Figure 1 ) that needs to transform from a loop to a coiled-coil structure. Mutations to proline in the hinge loop of RR1 have been successful in arresting this transition and stabilizing other class I fusion proteins (Battles et al., 2017, Hastie et al., 2017, Krarup et al., 2015, Pallesen et al., 2017, Sanders et al., 2002). Other approaches that proved successful were the neutralization of charged repulsions or the substitution of buried charged and polar residues in subdomain interfaces (Krarup et al., 2015, Rutten et al., 2018). Both approaches were explored for Ebola GP.
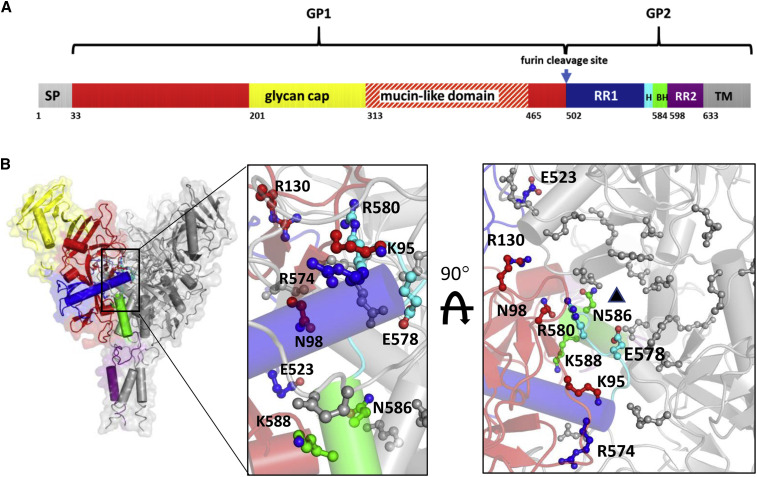
Figure 1.
Structure-Based Design of Stabilizing Mutations
(A) Schematic structure of filovirus GP showing the GP1 head domain (red, yellow) that includes the mucin-like domain (dashed red/white) and the glycan cap (yellow), GP2 that includes RR1 (with the hinge region in cyan), the base helix (BH, between RR1 and RR2) in green, refolding region 2 (RR2, in purple), and the transmembrane domain (TM, in gray).
(B) Cartoon of the 5JQ3 crystal structure used for the structure-based design. Coloring is as in (A). Amino acid residues shown as a ball-and-stick model were selected for mutagenesis.
Hinge Loop Stabilization
Plasmids encoding soluble Makona GP variants with proline substitutions in the hinge loop at positions 575, 576, 577, 579, and 581 were transfected in Expi293F cells. Cell culture supernatants were tested for trimer formation using native PAGE (Figure 2 A). L579P and T577P increased the trimer yield of Makona GP, and the double mutation (T577P/L579P) further increased the yield, as shown by analytical SEC of cell culture supernatants (Figure 2B). T577P increased the melting temperature in which 50% of the protein was unfolded (T m50) by ∼2.5°C, whereas L579P did not increase the T m50 of Makona GP (Figure S2). Introduction of the T577P substitution in Makona GP that lacked the mucin-like region (Δmucin GP) showed a ∼10-fold increase in trimer yield (Figure S3). The hinge loop stabilization was also tested for Mayinga and Sudan (Gulu) (SUDV) GP. Excluding the mucin-like domain, Makona and Mayinga GPs are ∼97% identical and Makona and Sudan (Gulu) GPs are ∼70% identical. Both Mayinga and Sudan (Gulu) GPs showed substantially increased trimer yields when T577P was introduced (Figures 2C and 2D), demonstrating the general applicability of the hinge loop stabilization approach.
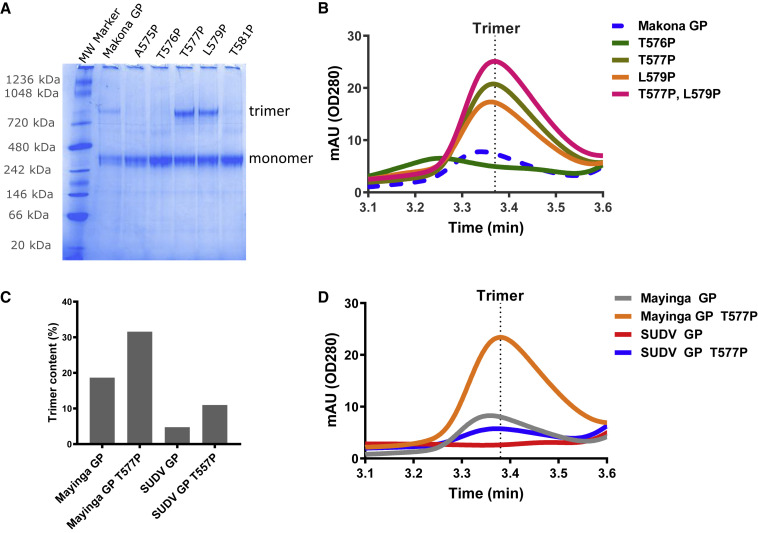
Figure 2.
Increased Trimer Yield by Prolines in RR1 Hinge Loop in Ebola GP
(A and B) Native PAGE (A) and analytical SEC profile (B) of variants with single and double proline mutations in the hinge loop of Makona GP. Analysis was performed on crude cell culture supernatants.
(C) Quantification of expression levels of Mayinga and Sudan (Gulu) (SUDV) GP trimers observed in native PAGE gel (data not shown) with and without the T577P mutation. The y axis shows the trimer content, based on the intensity of the trimer band and monomer bands, as a percentage of trimer content.
(D) Analytical SEC profiles of Mayinga and SUDV GP with and without T577P. Analysis was performed on crude cell culture supernatants.
Stabilization by Optimization of Domain Interfaces
Subdomains in viral fusion proteins need to refold and move relative to each other; therefore, such proteins do not contain the hydrophobic core of a typical protein. We identified nine charged and polar residues that are buried to different extents in the inter-protomer interface or the interface between GP1 and GP2. All of these residues are clustered in the charged center of the structure, close to the hinge loop (Figure 1B). These amino acids were substituted with hydrophobic residues in Makona Δmucin GP. Supernatants of cell cultures transfected with the GP variants with single or double substitutions were analyzed by native PAGE (Figure S4A). The variants that showed increased trimer formation were further analyzed by analytical SEC (Figures 3 A and S4B). The K588W and K588F substitutions showed a substantial increase in trimer expression, and few monomers were detected for the K588F variant (Figure 3A). To investigate the nature of the stabilizing effect of K588F, other substitutions were evaluated at position 588 for Mayinga GP and Mayinga Δmucin GP. Most hydrophobic residues at position 588 increased trimer yield, as shown by analytical SEC (Figures 3B and 3D) or binding to the trimer-specific antibody mAb100 (Figures 3C and 3E). Although the footprint of mAb100 is distributed over two monomers at the trimer interface, it is not known whether mAb100 can also bind to monomeric GP with reduced affinity. However, the association rate of mAb100 with the GP variants, as measured by bio-layer interferometry (BLI), correlated with the amounts of trimer measured using analytical SEC (Figure S5A). The association rate with samples that contain more monomeric GP is low, but at saturation after 300 s, the nanometer shift no longer correlates with the trimer content but with the total amount of GP trimer and monomer in the sample. This indicates that mAb100 may also bind to monomers and could indicate that mAb100 is able to induce trimer formation. Surprisingly, although the K588F mutation increases trimer content, it slightly decreased the T m50 by ∼1.2°C. (Figure S2B).
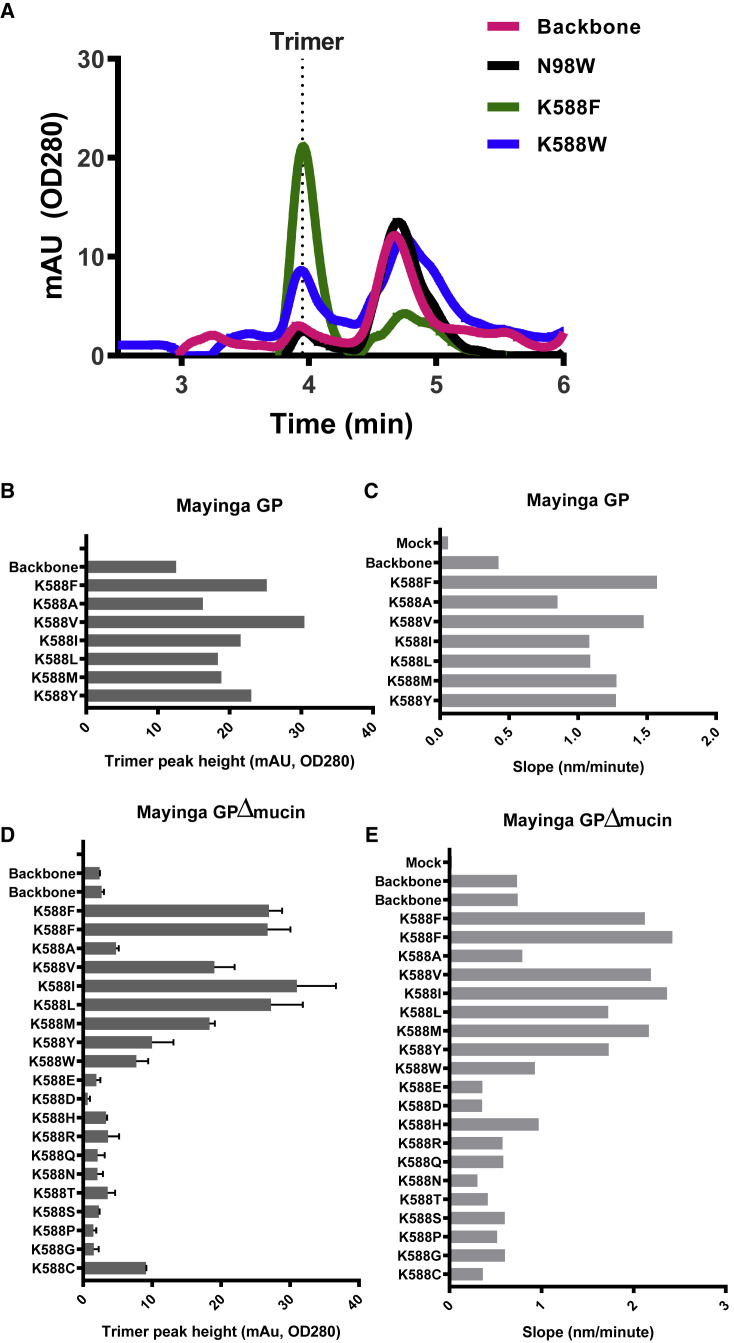
Figure 3.
Substitutions at Position 588 Increase Trimer Expression
(A) Analytical SEC profile of the variants with substitutions that increased the trimer content in the Makona Δmucin GP T577P/T42A backbone. The trimer peak is labeled ∼4 min.
(B) Expression levels of Mayinga GP trimers based on analytical SEC trimer peak heights for variants with hydrophobic substitutions of Lys588.
(C) BLI binding rates after 10 s of binding of mAb100.
(D) Expression levels of Mayinga Δmucin GP trimers based on analytical SEC trimer peak heights for variants with all possible natural substitutions of Lys588. Data are represented as mean ± SEM.
(E) BLI binding rates after 10 s of binding of mAb100.
Analysis was performed on crude cell culture supernatant.
See also Figures S4 and S5 and Table S1.
Combination of Substitutions and General Application of Approach for Other Filovirus GPs
The combined T577P and K588F substitutions were evaluated for their impact on Mayinga, Makona, and SUDV GP trimer stabilization with and without the mucin-like domain. The Mayinga GP with an N-terminal HA tag and T42A/T230V substitutions (Lee et al., 2008) was included as a control. Trimer content was determined using analytical SEC, and association phase analysis with mAb100 was determined using BLI. The T577P and K588F substitutions increased the trimer yield, and the combination of both substitutions resulted in the highest trimer yields (Figures 4 A, 4B, and S5B) and increased the fraction of trimeric GP in the HA-tagged T42A/T230V variant (Lee et al., 2008) (Figure 4A, gray curve in lower left panel). The substitutions at positions 578 (equivalent to 577 in Ebola GP) and 589 (equivalent to 588 in Ebola GP) were also introduced in Marburg GP, which only has ∼32% sequence identity with Ebola GP. Although the T578P substitution did not show a stabilizing effect on the Marburg Δmucin GP trimer (data not shown), the H589F and H589I substitutions increased the levels of the Marburg Δmucin GP trimers compared with the wild-type ectodomain. The wild-type Marburg Δmucin predominantly formed high-molecular-weight aggregates, as shown by analytical SEC and native PAGE (Figures 4C and S5C). Although the retention time of the Marburg Δmucin GP trimers in analytical SEC is shorter than that of the Ebola Δmucin GP trimers, they have similar average molecular weights according to multi-angle light scattering (MALS) (214.0 and ∼213.7 kDa for the H589I and H589F Marburg Δmucin GP variants, respectively) (Figure 4D; Table S1). When only the protein fraction molecular weights (MWs) are calculated, without glycosylation, the MWs are close to the theoretical MW based on the sequence (Table S1). From this, we conclude that the elution peak at 3.6 min contains Marburg Δmucin GP trimers.
Figure 4.
Increase in Trimer Formation after Combination of K588F and T577P in Mayinga, Makona, and SUDV GPs and Trimer Increase in Marburg GP after H589I/F Substitution
(A) Analytical SEC profiles of Ebola GP, Ebola Δmucin GP, and HA-Mayinga Δmucin GP with T577P and/or K588F substitutions. Analysis was performed on crude cell culture supernatant 3 days after transient transfection. The trimer peak is indicated with a dashed line labeled Trimer.
(B) BLI binding rates at 10 s of binding of monoclonal mAb100. Data are represented as mean ± SEM.
(C) Analytical SEC signals of crude cell culture supernatants of the Marburg Δmucin GPs.
(D) SEC-MALS with purified Marburg Δmucin and Mayinga Δmucin GP trimers. The red lines show the molar mass traces (right y axis). The dn/dc values used are 0.185 for all three trimers.
See also Figures S1 and S5.
Crystal Structure of Makona GP with T577P and K588F Mutations
To identify the effect of the stabilizing mutations on the conformation of trimeric prefusion GP, we determined the X-ray crystal structure of apo T577P/K588F Makona GP at pH 5.2 For these studies, the mucin-like region was deleted to facilitate crystallization (Figure 5 A). The 3.5 Å resolution structure (see Table S2 for crystallographic statistics) revealed that stabilized GP is similar to the previously determined Mayinga GP structure (PDB: 5JQ3), with an root-mean-square deviation (RMSD) of 1.87 Å for 353 Cα atoms. Although small differences in the conformation of the fusion loop were evident, the native conformation of the hinge loop and the base helix was preserved (Figures 5B and 5C). Thus, the stabilizing mutations did not disrupt the overall conformation of the trimeric prefusion GP protein. Pro577 is located at the inter-protomer interface (Figure 5B). The proline likely has a stabilizing effect because of prevention of α-helical formation and thus the transition from prefusion to postfusion conformation, but it could also stabilize the packing between two protomers. The proline makes more favorable van der Waals interactions with the neighboring protomer than the original threonine residue, interacting with Asn98, Arg164, and Leu165 of GP1, as well as Phe582 of GP2. As predicted based upon previous GP structures, Phe588 rests within a small hydrophobic pocket that is formed by residues in GP1 (Ile33, Leu35, Leu63, and Phe183) and GP2 (Ile584 and Leu585) (Figures 5B and 6 ). Thus, the crystal structure suggests that Phe588 acts by stabilizing the interface of GP1 and GP2 within one protomer by packing against hydrophobic residues from both subunits.
Figure 5.
Crystal Structure of Stabilized Makona ΔMucin GP
(A) Analytical SEC profile of the crude cell culture supernatant of the T577P/T42A/K588F Makona Δmucin GP that was used for crystallization and a variant lacking K588F.
(B) GP is viewed along the viral membrane, with two protomers shown as gray molecular surfaces and the other protomer shown as ribbons colored according to the schematic in Figure 1A. Insets show the regions surrounding Phe588 (left) and Pro577 (right), with side chains shown as sticks with transparent molecular surfaces. Nitrogen and oxygen atoms are colored blue and red, respectively.
(C) Superposed GP structures of Mayinga Δmucin GP (5JQ3) in blue and stabilized Makona Δmucin GP in red: side view (left) and top view (right).
Figure 6.
Interactions of Lys or Phe at Position 588 with the Ebola GP
(A–C) Wild-type structures determined by cryo-EM (PDB: 5KEL).
(A) Packing of the Lys588 side chain (red) against the hydrophobic pocket composed of the base helix (green) and GP1 (gray).
(B) Residues forming the pocket, including the N terminus of GP1.
(C) Side view of the pocket. Lys588 can either fill the space in the hydrophobic pocket (yellow rotamer) or form a polar interaction with the backbone of the N-terminal Ile33 (red rotamer). Neither solution is energetically favorable (proximity of positive charges versus desolvation of the positive charge).
(D–F) K588F pocket based on the crystal structure.
(D) Phe588 (red) tightly packed in the hydrophobic pocket.
(E) All residues forming the pocket and hydrophobic interactions with Phe588.
(F) Side view on the K588F pocket. Positive charge proximity is removed, and the sidechain of Phe588 is buried in the pocket.
The N Terminus/Lys588 Interaction Network
To better understand the role of amino acid position 588 on trimer stability, we analyzed the environment of Lys588 in existing crystal and electron microscopy (EM) structures (Figure 6). The Lys588 side chain is located next to a hydrophobic cavity and in direct proximity to the N terminus of GP1 (Figure 6). Understanding of the interaction in this area is obfuscated, because most solved EM and X-ray structures contain a non-native N terminus because of fusion with purification tags, and some proteins were crystallized at a pH at which the N terminus may be deprotonated (see Table S3). Of 25 structures, only four contain the native N terminus and do not contain a heterologous trimerization domain. Two of these structures, 5KEL and 5KEN, do not have the flexible β2-β3 loop directly constrained by an antibody and thus are most representative of the native state of the GP. In both structures, Lys588 is close to the N terminus. Because this arrangement seems to be destabilizing, we tested the trimer stability of GP variants with and without an N-terminal extension. The GP variant with a native GP1 N terminus has a lower trimer yield compared with a variant in which the N terminus was elongated by addition of five amino acids ETGRS, used in most crystallized GP proteins (Figure 7 ).
Figure 7.
Stability and Trimer Content of Mayinga ΔMucin GP Variants
(A) Analytical SEC profile of variants with K588F mutation and N-terminal ETGRS extension. Analysis was performed on crude cell culture supernatant.
(B) Analysis of Tm using differential scanning fluorometry (DSF) on variants with N-terminal extension and the K588F variant. Data are represented as mean ± SEM.
(C) Superposed structures with the native N terminus (stabilized Makona Δmucin GP structure in dark green; 5KEL, 5KEN, and 6DZM in green) and structures with extended N termini (6F5U, 6G9I, 5F1B, 5HJ3, 3S88, 3VE0, 3CSY, and 6EAY in pink).
We analyzed the interactions around amino acid 588 in the previously determined cryo-EM structure (PDB: 5KEL). In the cryo-EM structure, the wild-type lysine side chain was built in an orientation that suggests interaction between the lysine’s amino group and the N terminus. However, the density in this region is weak, and an alternative conformation with the lysine packed into the neighboring hydrophobic pocket could be possible (Figure 6C). Interestingly, both arrangements are energetically unfavorable, involving positive charge repulsion or desolvation of the lysine’s amino group, suggesting Lys588 and the N terminus are trapped in a metastable conformation. We believe the N-terminal extension, which does not alter the position of Ile33 in the structure (Figure 7C), effectively decreases the instability arising because of the repulsive interaction of the native N terminus with Lys588 and as a result improves the stability of GP trimers. Mutation of Lys588, as in the K588F variant described here, achieves the same task, but simultaneously fills the neighboring hydrophobic cavity. This seems to result in larger improvement of the protein’s stability and expression than removal of the electrostatic repulsion only (Figure 7) and even stabilizes GP with an extended N terminus (see the last panel of Figure 4A showing GP with an N-terminal HA tag), although to a lesser extent than GP without an extended N terminus. The improvement in both cases stems from eliminating the high energy conformation, which may be involved in the prefusion to postfusion refolding process.
Discussion
Like other class I fusion proteins, Ebola GP refolds from the prefusion to the postfusion state. However, for Ebola GP, the requirements for entry are unusually complex, and most details of the refolding process are unknown. Strategies to stabilize GP in the native soluble prefusion trimeric state have not been described. Production of native trimeric Ebola GP ectodomain is challenging, because most of the protein is monomeric. In recent years, structural knowledge of class I fusion proteins has greatly informed the design of mutations to stabilize these metastable proteins. Introduction of cavity-filling residues and disulfide bridges has been successful in a range of fusion proteins (Hastie et al., 2017, McLellan et al., 2013, Stewart-Jones et al., 2018). Introduction of glycine residues (Guenaga et al., 2017) or a single proline that restricts the movement of the hinge loop in RR1 and prevents the central helix extension and consequential release of the fusion peptide has also been successful, as was shown for RSV F, HIV Env, human metapneumo virus F, Lassa GP, and Middle Eastern respiratory syndrome coronavirus S (Battles et al., 2017, Hastie et al., 2017, Krarup et al., 2015, Pallesen et al., 2017). The substitution of buried or partially buried charged residues with hydrophobic residues resulted in the stabilization of HIV Env (Rutten et al., 2018). Here, the T577P substitution in the hinge loop of RR1 and substitution of the partially buried charged residue Lys588 with a hydrophobic residue in the base helix at the GP1-GP2 interface stabilized the soluble GP trimer and led to a ∼20-fold increase in trimer production. The proline substitution increased stability and expression of several Ebola GPs, and the substitution of the buried charged residue increased stability and expression of Ebola GP and Marburg GP and thus may be universally applied to filoviruses.
Our results point toward Lys588 as an important contributor to the instability of Ebola GP trimers. Not only is Lys588 partially buried, but it is also restrained to interact with a positively charged N terminus (Figures 6A–6C). Such an unstable, high-energy configuration implies the region could act as a structural switch for undergoing conformational change. In the unstable region around Lys588, the N terminus of GP1 can move, as opposed to the lysine, which is fixed on the base helix. Exit of the N terminus may compromise its interaction with the intraprotomeric α3 helix (HR1) of RR1 that contains the internal fusion loop. This HR1 helix is stabilized by Pro34 and by several of the residues that form the hydrophobic pocket (Ile584, Phe183 and the N-terminal GP1 Ile33 residue). Loss of the Ile33-Lys588 interaction would release the N-terminal Ile33, break up the hydrophobic pocket, and thereby disrupt contacts with HR1 in the refolding region. Removal of the instability by mutating the N terminus results in improved expression and trimer stability. An alternative way to remove the instability is by mutating the lysine to a hydrophobic residue, as described in this study. The K588F mutation not only removes the excess positive charge in the region but also adds favorable interactions with a neighboring hydrophobic pocket composed of residues from both GP1 and GP2 subunits (Figures 6D–6F and 7), which also seems to contribute to the stability substantially, given that K588A does not result in substantially increased trimer yields (Figures 3B and 3D).
Interestingly, the K588F mutation described here for Ebola GP has some resemblance to the D589V mutation used to stabilize HIV Env (Rutten et al., 2018). Both substitutions increase trimer yield and are located in the base helix of the fusion domain (GP2 in filovirus GP and gp41 in HIV Env). Both also make an intraprotomeric interaction with the head domain (GP1 and gp120), which forms a hydrophobic pocket. These similarities reported here suggest commonalities in the triggering mechanisms of Ebola GP and HIV Env and indicate that the strategy used here to stabilize GP may be broadly applicable to other class I viral fusion proteins.
In conclusion, stabilization of the hinge loop in RR1 by T577P and stabilization and neutralization of a region of instability in the GP1-GP2 interface by K588F dramatically increases the yield of filovirus prefusion GP trimers. The increasing knowledge of the refolding mechanism of class I fusion proteins is critical for the design of stable proteins that can be used for vaccines, diagnostics, or isolation of antibodies.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| mAb100 | Corti et al., 2016 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| CHT type 1 resin | Bio-Rad | Cat# 158-4000 |
| HisTrap HP 5 mL | GEHC | Cat#17524802 |
| HiLoad Superdex 200 16/600 size-exclusion column | GEHC | Cat# 28989335 |
| mAb Select SuRe 5 mL HiTrap | GEHC | Cat# 11003495 |
| HiPrep 26/10 Desalting column | GEHC | Cat# 17508701 |
| anti-hIgG (AHC) sensors | FortéBio | cat#18-1092 |
| Sypro Orange dye | Thermo Fisher Scientific | Cat# S6650 |
| Critical Commercial Assays | ||
| 50K Amicon Ultra concentrators | Millipore | Cat# UFC905024 |
| NativePage Bis-Tris gradient gels 4-16% | LifeTechnologies | Cat# BN1002BOX |
| Deposited Data | ||
| Stabilized Makona Δ-mucin GP X-ray structure | This paper | PDB: 6VKM |
| Experimental Models: Cell Lines | ||
| Human: Expi293F | Thermo Fischer Scientific | Cat#A14527 |
| Software and Algorithms | ||
| FortéBio Data Analysis 8.1 software | FortéBio | |
| CCP4i interface | Morin et al., 2013, Potterton et al., 2003 | https://www.ccp4.ac.uk/ccp4i_main.php |
| iMOSFLM | Powell et al., 2017 | https://www.mrc-lmb.cam.ac.uk/mosflm/imosflm/ver730/installation.html |
| AIMLESS | Evans and Murshudov, 2013 | http://www.ccp4.ac.uk/dist/html/aimless.html |
| PHASER | McCoy et al., 2007 | https://www.phenix-online.org/documentation/reference/phaser_mr.html |
| Coot | Emsley and Cowtan, 2004 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| PHENIX | Adams et al., 2002 | |
| Astra 7.3 software package | https://www.wyatt.com/products/software/astra.html | |
| Chromeleon 7.2.8.0 software package | https://www.thermofisher.com/order/catalog/product/CHROMELEON7 | |
Lead Contact and Materials Availability
This study did not generate new unique reagents. All requests for reagents and resources should be directed to the lead contact, Johannes P.M. Langedijk (hlangedi@its.jnj.com).
Experimental Model and Subject Details
Cell Lines
Human Expi293F cells (Thermo Fischer Scientific) were maintained in Expi293 Expression medium (Thermo Fisher Scientific).
Method Details
Expression Plasmids and Transient Transfections
The Mayinga and Makona GP proteins contain amino acids 1-647 followed by a His6-tag. For the Mayinga and Sudan (Gulu) GP protein without the mucin-like domain, amino acids 320 until 476 were deleted and for the Makona protein without the mucin-like domain, amino acids 314 until 472 were deleted. DNA encoding the glycoproteins (GPs) were synthesized and codon-optimized for expression in human cells at GenScript (Piscataway, NJ 08854). The codon-optimized sequences were then cloned into the vector pcDNA2004 to generate the GP constructs, which were used as the backbone sequences for introducing further mutations. The genes were expressed in Expi293F cells (Thermo Fischer) according to the manufacturer’s specification. Glucose levels were monitored using the ViCell MetaFlex (Beckmann). Glucose was depleted at day 4 post-transfection and therefore glucose was added at a 15 mM concentration. Supernatants were harvested at day 6 post-transfection by centrifugation and sterile filtration. For small scale 96-well format for analytical SEC experiment, the supernatants were harvested 3 days post-transfection.
Purification of Ebola or Marburg GP Protein
Interfering host-cell proteins (HCPs) were scavenged by applying the supernatant to 13 mL CHT type 1 resin (Biorad) in an XK16/20 column (GEHC) using a flow rate of 300 cm/hr and a running buffer of 5 mM NaPO4, pH 6.8. Bound proteins were eluted by a step elution using 500 mM NaPO4, pH 7.4. The HCP depleted flow through was subsequently applied to a HisTrap HP 5 mL (GEHC) using a flow rate of 300 cm/hr and a running buffer of 20 mM Tris, 500 mM NaCl pH 7.4. Bound proteins were eluted using a step gradient of 15, 30 and 100% elution buffer (20 mM Tris, 500 mM NaCl, 300 mM imidazole pH 7.4) while running the column in upflow with a flow rate of 600 cm/hr. The trimer fractions eluted along with aggregates when 100 mM imidazole was applied. This fraction was concentrated, using 50K Amicon Ultra concentrators (Millipore), and applied to a Superdex 16/600 size-exclusion column (GEHC) using a flow rate of 60 cm/hr to separate the trimer fraction from aggregates and monomers. The fractions containing the trimer peak were pooled, and the identity of the peak confirmed as GP protein using SDS-PAGE, and/or SEC-MALS analysis. The concentration of the purified Ebola or Marburg GP was determined by measuring the optical density at 280 nm, and the purified protein was stored at 4°C until further use.
Antibody production and purification
The heavy and light chain of mAb100 were cloned into a single IgG1 expression vector to express a fully human IgG1 antibody. mAb100 was made by transfecting the IgG1 expression construct using the ExpiFectamine 293 Transfection Kit (ThermoFisher) in Expi293F (ThermoFisher) cells according to the manufacturer specifications. mAb100 antibodies were purified from serum-free culture supernatants using mAb Select SuRe resin (GE Healthcare) followed by rapid desalting using a HiPrep 26/10 Desalting column (GE Healthcare). The final formulation buffer was 20 mM NaAc, 75 mM NaCl, 5% Sucrose pH 5.5. IgG quality was confirmed to be > 97% monomeric using SEC-MALS.
NativePAGE Analysis
NativePAGE was performed according to manufacturer’s protocol (LifeTechnologies) using 4%–16% NativePage Bis-Tris gradient gels (LifeTechnologies). The GP trimer with mucin-like domain ran at a mass of about 800 kDa, whereas the GP without the mucin-like domain ran at a mass of about 420 kDa.
Analytical SEC and SEC-MALS
The EBOV GP variants were expressed in 96 well format cell cultures. An ultra high-performance liquid chromatography system (Vanquish, Thermo Scientific) and μDAWN TREOS instrument (Wyatt) coupled to an Optilab μT-rEX Refractive Index Detector (Wyatt), in combination with an in-line Nanostar DLS reader (Wyatt), was used for performing the analytical SEC experiment. The cleared crude cell culture supernatants were applied to a TSK-Gel UP-SW3000 4.6x150 mm column with the corresponding guard column (Tosoh Bioscience) equilibrated in running buffer (150 mM sodium phosphate, 50 mM NaCl, pH 7.0) at 0.3 mL/min. When analyzing supernatant samples, μMALS detectors were offline and analytical SEC data was analyzed using Chromeleon 7.2.8.0 software package. The signal of supernatants of non-transfected cells was subtracted from the signal of supernatants of GP transfected cells. When purified proteins were analyzed using SEC-MALS, μMALS detectors were inline and data was analyzed using Astra 7.3 software package. For the protein component, a dn/dc (mL/g) value of 0.1850 was used and for the glycan component a value of 0.1410. Molecular weights were calculated using the RI detector as [C] source and mass recoveries using UV as [C] source.
BioLayer Interferometry (BLI)
A solution of monoclonal antibody mAb10038 at a concentration of 10 ug/mL was used to immobilize the antibody on anti-hIgG (AHC) sensors (FortéBio cat#18-5060) in 1x kinetics buffer (FortéBio cat#18-1092) in 96-half well black flat bottom polypylene microplates (FortéBio cat#3694). The experiment was performed on an Octet HTX instrument (Pall-FortéBio) at 30 °C with a shaking speed of 1,000 rpm. Activation was 60 s, immobilization of antibodies 600 s, followed by washing for 150 s and then binding the GP proteins for 600 s, and a dissociation of 60 s. The data analysis was performed using the FortéBio Data Analysis 8.1 software (FortéBio). The binding was determined by using association phase analysis. The binding slope was determined at 10 s in nm/minute.
Differential scanning fluorometry (DSF)
The purified protein was mixed with SYPRO orange fluorescent dye (Life Technologies S6650) in a 96-well optical qPCR plate. The optimal dye and protein concentration were determined experimentally. All protein dilutions were performed in PBS, and a negative control sample containing the dye only was used for reference subtraction. The measurement was performed in a qPCR instrument (Applied Biosystems ViiA 7) using a temperature ramp from 25–95°C with a rate of 0.015°C per second. Data were collected continuously. The negative first derivative of the Sypro Orange signal was measured at several intervals during a temperature ramp up to 95°C.
For measurement in supernatants, constructs were expressed in a 96-well format and harvested 3-days post transfection. Samples were diluted with PBS pH 7.4 (GIBCO) containing 8-fold diluted supernatant and 500-fold diluted Sypro Orange dye (5000 x stock, Invitrogen). A mock sample was included as background control. The measurement was performed in a qPCR instrument (Applied Biosystems ViiA 7) using a temperature ramp from 25–95°C with a rate of 0.015°C per second. Data were collected continuously. The negative first derivative was plotted as a function of temperature. The melting temperature corresponds to the lowest point in the curve.
Differential scanning calorimetry (DSC)
Melting temperatures for GPs were determined using MicroCal capillary DSC system. 400 μL of 0.5 mg/mL protein sample was used per measurement. The measurement was performed with a start temperature of 20°C and a final temperature of 110°C. The scan rate 100°C/h and the feedback mode; Low ( = signal amplification). The data were analyzed using the Origin J. Software (MicroCal VP-analysis tool).
Crystal structure determination
For crystallization purposes an additional T42A substitution was introduced to remove the glycosylation site at Asn40 (Zhao et al., 2016) and during production a final concentration of 5 μM of kifunensin was used. Although the T577P/T42A variant still has a higher trimer yield than wild-type Makona Δmucin GP, the additional T42A substitution significantly reduced the trimer yield (Figure S3). Deletion of the glycan at position 40 reduced the molecular weight of the monomer as illustrated by the shifted peak in analytical SEC (Figure S3). The trimer retention time, however, was not affected, which may correlate with a relatively compact trimer in the closed prefusion conformation. The stabilized GP T42A/T577P/K588F protein was crystallized by hanging-drop vapor diffusion by mixing 0.5 μL of protein at 8.8 mg/mL with 0.5 μL of water and 1 μL of reservoir solution containing 9.8% polyethylene glycol (PEG) 6000, 0.1 M sodium citrate pH 5.2, and 3% glycerol. Crystals were soaked in reservoir solution supplemented with 25% (v/v) glycerol as a cryoprotectant before being plunge frozen with liquid nitrogen. Data were collected to 3.5 Å resolution at the SBC beamline 19-ID (Advanced Photon Source, Argonne National Laboratory).
X-ray diffraction data were processed using software curated by SBGrid and accessed through the CCP4i interface (Collaborative Computational ProjectNumber 4, 1994, Morin et al., 2013, Potterton et al., 2003). Data indexing and integration were carried out using iMOSFLM (Powell et al., 2017), and merging and scaling were performed with AIMLESS (Evans and Murshudov, 2013). The results of the L-test identified possible twinning, however, since no merohedral or pseudo-merohedral twin laws are possible for space group H32, other possible pathologies were examined. An off-origin peak in the Patterson function with 3.56% the height of the origin peak was identified, suggesting that lattice translocation, which has previously been reported in space group H32 (Wang et al., 2005), could be responsible for the deviation from normal data statistics. Uncorrected data were used to determine the molecular replacement solution in PHASER (McCoy et al., 2007) using the previously determined GP structure (PDB ID: 5JQ3) as a search model. One copy of a single GP protomer was present in the asymmetric unit, although weak density for a second overlapping protomer was also visible and is likely the result of the lattice translocation defect. A single copy of the GP protomer was built manually in Coot (Emsley and Cowtan, 2004) and refined in PHENIX (Adams et al., 2002) to an R work/R free of 28.4%/30.3%.
Quantification and Statistical Analysis
Bar graphs of analytical SEC data were presented as mean ± standard error from at least two independent transfections when indicated. Data fitting and statistical analysis was performed using GraphPad Prism software (version 7.00).
Data and Code Availability
Atomic coordinates and structure factors for the crystal structure of the stabilized GP trimer have been deposited with the Protein Data Bank with PDB code 6VKM.
Acknowledgments
We thank Mark Bakkers for suggestions and critically reading the manuscript and Ava Sadi and Mark Luinenburg for technical support. These studies were funded by Janssen Vaccines & Prevention.
Author Contributions
Conceptualization, L.R. and J.P.M.L.; Methodology, L.R. and S.B.; Investigation, S.B., J.J., and M.S.A.G.; Writing – Original Draft, L.R., S.B., M.S.A.G., J.J., J.S.M., and J.P.M.L.; Writing – Review & Editing, L.R., S.B., M.S.A.G., J.S.M., and J.P.M.L.; Supervision, L.R., J.S.M., and J.P.M.L.
Declaration of Interests
L.R., S.B., J.J., and J.P.M.L. are employees at Janssen Vaccines & Prevention. L.R., S.B., and J.P.M.L. are inventors on an international patent application describing trimer stabilizing Ebola GP mutations. J.P.M.L. holds stock in Johnson & Johnson. The remaining authors declare no competing financial interests.
Published: March 31, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.03.025.
Supplemental Information
References
- Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Alvarez C.P., Lasala F., Carrillo J., Muñiz O., Corbí A.L., Delgado R. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J. Virol. 2002;76:6841–6844. doi: 10.1128/JVI.76.13.6841-6844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale S., Dias J.M., Fusco M.L., Hashiguchi T., Wong A.C., Liu T., Keuhne A.I., Li S., Woods V.L., Jr., Chandran K. Structural basis for differential neutralization of ebolaviruses. Viruses. 2012;4:447–470. doi: 10.3390/v4040447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battles M.B., Más V., Olmedillas E., Cano O., Vázquez M., Rodríguez L., Melero J.A., McLellan J.S. Structure and immunogenicity of pre-fusion-stabilized human metapneumovirus F glycoprotein. Nat. Commun. 2017;8:1528. doi: 10.1038/s41467-017-01708-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley J.M., Sanders R.W., Clas B., Schuelke N., Master A., Guo Y., Kajumo F., Anselma D.J., Maddon P.J., Olson W.C., Moore J.P. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 2000;74:627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornholdt Z.A., Ndungo E., Fusco M.L., Bale S., Flyak A.I., Crowe J.E., Jr., Chandran K., Saphire E.O. Host-Primed Ebola Virus GP Exposes a Hydrophobic NPC1 Receptor-Binding Pocket, Revealing a Target for Broadly Neutralizing Antibodies. MBio. 2016;7 doi: 10.1128/mBio.02154-15. e02154–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindley M.A., Hunt C.L., Kondratowicz A.S., Bowman J., Sinn P.L., McCray P.B., Jr., Quinn K., Weller M.L., Chiorini J.A., Maury W. Tyrosine kinase receptor Axl enhances entry of Zaire ebolavirus without direct interactions with the viral glycoprotein. Virology. 2011;415:83–94. doi: 10.1016/j.virol.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette J.E., Raaben M., Wong A.C., Herbert A.S., Obernosterer G., Mulherkar N., Kuehne A.I., Kranzusch P.J., Griffin A.M., Ruthel G. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran K., Sullivan N.J., Felbor U., Whelan S.P., Cunningham J.M. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Corti D., Misasi J., Mulangu S., Stanley D.A., Kanekiyo M., Wollen S., Ploquin A., Doria-Rose N.A., Staupe R.P., Bailey M. Protective monotherapy against lethal Ebola virus infection by a potently neutralizing antibody. Science. 2016;351:1126. doi: 10.1126/science.aad5224. [DOI] [PubMed] [Google Scholar]
- Côté M., Misasi J., Ren T., Bruchez A., Lee K., Filone C.M., Hensley L., Li Q., Ory D., Chandran K., Cunningham J. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. 2011;477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias J.M., Kuehne A.I., Abelson D.M., Bale S., Wong A.C., Halfmann P., Muhammad M.A., Fusco M.L., Zak S.E., Kang E. A shared structural solution for neutralizing ebolaviruses. Nat. Struct. Mol. Biol. 2011;18:1424–1427. doi: 10.1038/nsmb.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Evans P.R., Murshudov G.N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fels J.M., Spence J.S., Bortz R.H., 3rd, Bornholdt Z.A., Chandran K. A Hyperstabilizing Mutation in the Base of the Ebola Virus Glycoprotein Acts at Multiple Steps To Abrogate Viral Entry. MBio. 2019;10:e01408–e01419. doi: 10.1128/mBio.01408-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fénéant L., Szymańska-de Wijs K.M., Nelson E.A., White J.M. An exploration of conditions proposed to trigger the Ebola virus glycoprotein for fusion. PLoS ONE. 2019;14:e0219312. doi: 10.1371/journal.pone.0219312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenaga J., Garces F., de Val N., Stanfield R.L., Dubrovskaya V., Higgins B., Carrette B., Ward A.B., Wilson I.A., Wyatt R.T. Glycine Substitution at Helix-to-Coil Transitions Facilitates the Structural Determination of a Stabilized Subtype C HIV Envelope Glycoprotein. Immunity. 2017;46:792–803. doi: 10.1016/j.immuni.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie K.M., Zandonatti M.A., Kleinfelter L.M., Heinrich M.L., Rowland M.M., Chandran K., Branco L.M., Robinson J.E., Garry R.F., Saphire E.O. Structural basis for antibody-mediated neutralization of Lassa virus. Science. 2017;356:923–928. doi: 10.1126/science.aam7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Committee on Taxonomy of Viruses . 2019. Genus: Ebolavirus. talk.ictvonline.org/ictv-reports/ictv_online_report/negative-sense-rna-viruses/mononegavirales/w/filoviridae/1086/genus-ebolavirus. [Google Scholar]
- Janus B.M., van Dyk N., Zhao X., Howell K.A., Soto C., Aman M.J., Li Y., Fuerst T.R., Ofek G. Structural basis for broad neutralization of ebolaviruses by an antibody targeting the glycoprotein fusion loop. Nat. Commun. 2018;9:3934. doi: 10.1038/s41467-018-06113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemielity S., Wang J.J., Chan Y.K., Ahmed A.A., Li W., Monahan S., Bu X., Farzan M., Freeman G.J., Umetsu D.T. TIM-family proteins promote infection of multiple enveloped viruses through virion-associated phosphatidylserine. PLoS Pathog. 2013;9:e1003232. doi: 10.1371/journal.ppat.1003232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratowicz A.S., Lennemann N.J., Sinn P.L., Davey R.A., Hunt C.L., Moller-Tank S., Meyerholz D.K., Rennert P., Mullins R.F., Brindley M. T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus. Proc. Natl. Acad. Sci. USA. 2011;108:8426–8431. doi: 10.1073/pnas.1019030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krarup A., Truan D., Furmanova-Hollenstein P., Bogaert L., Bouchier P., Bisschop I.J.M., Widjojoatmodjo M.N., Zahn R., Schuitemaker H., McLellan J.S., Langedijk J.P.M. A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat. Commun. 2015;6:8143. doi: 10.1038/ncomms9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J.H., Adachi T., Adhikari N.K.J., Arribas J.R., Bah I.E., Bausch D.G., Bhadelia N., Borchert M., Brantsæter A.B., Brett-Major D.M. New filovirus disease classification and nomenclature. Nat. Rev. Microbiol. 2019;17:261–263. doi: 10.1038/s41579-019-0187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.E., Fusco M.L., Hessell A.J., Oswald W.B., Burton D.R., Saphire E.O. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan J.S., Chen M., Joyce M.G., Sastry M., Stewart-Jones G.B., Yang Y., Zhang B., Chen L., Srivatsan S., Zheng A. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013;342:592–598. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E.H., Obernosterer G., Raaben M., Herbert A.S., Deffieu M.S., Krishnan A., Ndungo E., Sandesara R.G., Carette J.E., Kuehne A.I. Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J. 2012;31:1947–1960. doi: 10.1038/emboj.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin A., Eisenbraun B., Key J., Sanschagrin P.C., Timony M.A., Ottaviano M., Sliz P. Collaboration gets the most out of software. eLife. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulherkar N., Raaben M., de la Torre J.C., Whelan S.P., Chandran K. The Ebola virus glycoprotein mediates entry via a non-classical dynamin-dependent macropinocytic pathway. Virology. 2011;419:72–83. doi: 10.1016/j.virol.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murin C.D., Bruhn J.F., Bornholdt Z.A., Copps J., Stanfield R., Ward A.B. Structural Basis of Pan-Ebolavirus Neutralization by an Antibody Targeting the Glycoprotein Fusion Loop. Cell Rep. 2018;24:2723–2732. doi: 10.1016/j.celrep.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanbo A., Imai M., Watanabe S., Noda T., Takahashi K., Neumann G., Halfmann P., Kawaoka Y. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 2010;6:e1001121. doi: 10.1371/journal.ppat.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallesen J., Murin C.D., de Val N., Cottrell C.A., Hastie K.M., Turner H.L., Fusco M.L., Flyak A.I., Zeitlin L., Crowe J.E., Jr. Structures of Ebola virus GP and sGP in complex with therapeutic antibodies. Nat. Microbiol. 2016;1:16128. doi: 10.1038/nmicrobiol.2016.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potterton E., Briggs P., Turkenburg M., Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- Powell H.R., Battye T.G.G., Kontogiannis L., Johnson O., Leslie A.G.W. Integrating macromolecular X-ray diffraction data with the graphical user interface iMosflm. Nat. Protoc. 2017;12:1310–1325. doi: 10.1038/nprot.2017.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutten L., Lai Y.T., Blokland S., Truan D., Bisschop I.J.M., Strokappe N.M., Koornneef A., van Manen D., Chuang G.Y., Farney S.K. A Universal Approach to Optimize the Folding and Stability of Prefusion-Closed HIV-1 Envelope Trimers. Cell Rep. 2018;23:584–595. doi: 10.1016/j.celrep.2018.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed M.F., Kolokoltsov A.A., Albrecht T., Davey R.A. Cellular entry of Ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog. 2010;6:e1001110. doi: 10.1371/journal.ppat.1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders R.W., Vesanen M., Schuelke N., Master A., Schiffner L., Kalyanaraman R., Paluch M., Berkhout B., Maddon P.J., Olson W.C. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 2002;76:8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornberg K., Matsuyama S., Kabsch K., Delos S., Bouton A., White J. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 2006;80:4174–4178. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojima M., Ikeda Y., Kawaoka Y. The mechanism of Axl-mediated Ebola virus infection. J. Infect. Dis. 2007;196(Suppl 2):S259–S263. doi: 10.1086/520594. [DOI] [PubMed] [Google Scholar]
- Simmons G., Reeves J.D., Grogan C.C., Vandenberghe L.H., Baribaud F., Whitbeck J.C., Burke E., Buchmeier M.J., Soilleux E.J., Riley J.L. DC-SIGN and DC-SIGNR bind Ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology. 2003;305:115–123. doi: 10.1006/viro.2002.1730. [DOI] [PubMed] [Google Scholar]
- Stewart-Jones G.B.E., Chuang G.Y., Xu K., Zhou T., Acharya P., Tsybovsky Y., Ou L., Zhang B., Fernandez-Rodriguez B., Gilardi V. Structure-based design of a quadrivalent fusion glycoprotein vaccine for human parainfluenza virus types 1–4. Proc. Natl. Acad. Sci. USA. 2018;115:12265–12270. doi: 10.1073/pnas.1811980115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada A., Watanabe S., Ito H., Okazaki K., Kida H., Kawaoka Y. Downregulation of beta1 integrins by Ebola virus glycoprotein: implication for virus entry. Virology. 2000;278:20–26. doi: 10.1006/viro.2000.0601. [DOI] [PubMed] [Google Scholar]
- Wang J., Rho S.H., Park H.H., Eom S.H. Correction of X-ray intensities from an HslV-HslU co-crystal containing lattice-translocation defects. Acta Crystallogr. D Biol. Crystallogr. 2005;61:932–941. doi: 10.1107/S0907444905009546. [DOI] [PubMed] [Google Scholar]
- Wang H., Shi Y., Song J., Qi J., Lu G., Yan J., Gao G.F. Ebola Viral Glycoprotein Bound to Its Endosomal Receptor Niemann-Pick C1. Cell. 2016;164:258–268. doi: 10.1016/j.cell.2015.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West B.R., Moyer C.L., King L.B., Fusco M.L., Milligan J.C., Hui S., Saphire E.O. Structural Basis of Pan-Ebolavirus Neutralization by a Human Antibody against a Conserved, yet Cryptic Epitope. MBio. 2018;9 doi: 10.1128/mBio.01674-18. e01674-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2020. Ebola outbreak 2014–2016.www.who.int/csr/disease/ebola/en/ [Google Scholar]
- World Health Organization . 2020. Ebola in the Democratic Republic of the Congo Health Emergency Update.www.who.int/emergencies/diseases/ebola/drc-2019 [Google Scholar]
- Zhao Y., Ren J., Harlos K., Jones D.M., Zeltina A., Bowden T.A., Padilla-Parra S., Fry E.E., Stuart D.I. Toremifene interacts with and destabilizes the Ebola virus glycoprotein. Nature. 2016;535:169–172. doi: 10.1038/nature18615. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the crystal structure of the stabilized GP trimer have been deposited with the Protein Data Bank with PDB code 6VKM.