Abstract
Background
Epigenetic alterations are involved in various aspects of colorectal carcinogenesis. N6-methyladenosine (m6A) modifications of RNAs are emerging as a new layer of epigenetic regulation. As the most abundant chemical modification of eukaryotic mRNA, m6A is essential for the regulation of mRNA stability, splicing, and translation. Alterations of m6A regulatory genes play important roles in the pathogenesis of a variety of human diseases. However, whether this mRNA modification participates in the glucose metabolism of colorectal cancer (CRC) remains uncharacterized.
Methods
Transcriptome-sequencing and liquid chromatography-tandem mass spectrometry (LC-MS) were performed to evaluate the correlation between m6A modifications and glucose metabolism in CRC. Mass spectrometric metabolomics analysis, in vitro and in vivo experiments were conducted to investigate the effects of METTL3 on CRC glycolysis and tumorigenesis. RNA MeRIP-sequencing, immunoprecipitation and RNA stability assay were used to explore the molecular mechanism of METTL3 in CRC.
Results
A strong correlation between METTL3 and 18F-FDG uptake was observed in CRC patients from Xuzhou Central Hospital. METTL3 induced-CRC tumorigenesis depends on cell glycolysis in multiple CRC models. Mechanistically, METTL3 directly interacted with the 5′/3’UTR regions of HK2, and the 3’UTR region of SLC2A1 (GLUT1), then further stabilized these two genes and activated the glycolysis pathway. M6A-mediated HK2 and SLC2A1 (GLUT1) stabilization relied on the m6A reader IGF2BP2 or IGF2BP2/3, respectively.
Conclusions
METTL3 is a functional and clinical oncogene in CRC. METTL3 stabilizes HK2 and SLC2A1 (GLUT1) expression in CRC through an m6A-IGF2BP2/3- dependent mechanism. Targeting METTL3 and its pathway offer alternative rational therapeutic targets in CRC patients with high glucose metabolism.
Keywords: m6A modification, METTL3, Glycolysis, Colorectal cancer, HK2, GLUT1
Background
As the fourth most common malignancies and the third leading cause of cancer deaths worldwide [1], colorectal cancer (CRC) is a major cause of morbidity and mortality in the world. Despite that the substantial diagnostic and therapeutic strategies have been improved, the survival time of CRC patients have been increased in recent years, the mortality rate of colorectal cancer remains high [2]. The lack of effective interventions and precise biomarkers of the cancer urgently need a better understanding of the molecular mechanism of CRC initiation and progression.
An emerging hallmark of invasive cancer cells is energy metabolism, which includes elevated glycolysis activation and higher lactate fermentation known as the Warburg effect [3–5]. Targeting lactate production, which is the final product of glycolysis, is regarded to be a promising therapeutic approach in cancer [6]. Recent studies found that inhibition of EGFR signaling resulted in dramatically lung cancer reduction by reversing of Warburg effect and reactivation of oxidative phosphorylation [7], which highlights the perspective role for the therapeutic target of glycolysis. However, the molecular basis for glycolysis and its role in cancer growth remain unclear.
N6-methyladenosine (m6A) modification is the most abundant posttranscriptional internal mRNA modification, which mediates various biological processes in eukaryotes [8, 9]. m6A RNA modification is usually enriched near stop codons and terminal exons in over 25% of human transcripts [10, 11]. In mammals, m6A, which is dynamic and reversible RNA modification in mammalian cells, is post-transcriptionally installed by a m6A methyltransferase complex that contains an enzymatic subunit METTL3, and its assistant co-factors METTL14 and WTAP within the consensus motif of G(m6A) C (70%) or A(m6A) C (30%) [12–14]. And this mRNA modification can be removed by demethylases FTO and ALKBH5 [15, 16]. Recent studies have shown that m6A modification in mRNAs participates in RNA metabolisms to regulate mRNA stability, splicing, transport, localization, as well as in stem cell self-renewal and differentiation, tissue development in normal cellular physiological and disease status [10, 17–19]. Recent studies have reported that the writers, readers, and erasers of m6A may play important roles in various cancer initiation, progression and response to immunotherapy via different regulation patterns [18, 20–24]. Despite these recent discoveries between m6A modification with malignant cancer development and treatment, the status of m6A modification and the underlying regulatory mechanism in CRC, especially in the glycolytic metabolism of CRC remains little known. In the current study, we investigated the role of m6A modification in CRC and identified the oncogenic role of METTL3, with a biological, mechanistic, and clinical impact on human CRC and glucose metabolism. Our findings on the implication of METTL3-mediated m6A modification in epigenetic stabilizing of HK2 and SLC2A1 (GLUT1) illustrated the critical role of m6A epitranscriptomic change in human colorectal carcinogenesis and glycolysis pathways.
Methods
Patient specimens
We used three Cohorts of patients with colorectal cancer who underwent surgery between 2012 and 2019. Cohort 1 (fresh tissues and paraffin-embedded tissues) were from Xuzhou Central Hospital, Xuzhou Medical University; Cohort 2 (paraffin-embedded tissues) were from the Eastern Campus of Renji Hospital, Shanghai Jiao Tong University School of Medicine; Cohort 3 (fresh tissues) were from the Western Campus of Renji Hospital, Shanghai Jiao Tong University School of Medicine. The study protocol was approved by the ethics committee of Shanghai Jiao Tong University School of Medicine. Written informed consent was obtained from all participants in this study. All the research was carried out in accordance with the provisions of the Declaration of Helsinki of 1975. None of these patients had received radiotherapy or chemotherapy prior to surgery.
M6A dot blot and m6A quantification
Polyadenylated mRNA was purified by GenElute™ mRNA Miniprep Kit (Sigma, St. Louis, MO) from previously isolated total RNA. The m6A dot blot assay was performed as previously described [25]. The global m6A levels in mRNA were measured with EpiQuik m6A RNA Methylation Quantification Kit (Colorimetric) (Epigentek, Farmingdale, NY) following the manufacturer’s protocol. The detailed m6A dot blot and m6A quantification protocols were described in the Supplementary Materials and Methods.
MeRIP and MeRIP-qPCR
The m6A-immunoprecipitation and library preparation was performed according to a published protocol [10]. Real-time PCR was carried out following m6A-IP to quantify the changes to m6A methylation of a certain target gene. The detailed MeRIP and MeRIP-qPCR protocols were described in the Supplementary Materials and Methods.
Glucose uptake, lactate production, hexokinase activity assay, seahorse metabolic analysis
Glucose Uptake, L-Lactate Colorimetric assay, Hexokinase Colorimetric assay, Seahorse XF Glycolysis Stress Test and Seahorse XF Cell Mito Stress Test were detailed described in the Supplementary Materials and Methods.
RNA Immunoprecipitation
RNA Immunoprecipitation (RIP) assays were conducted using the Magna RIP Kit (Millipore, New Bedford, MA) and the detailed protocol was described in the Supplementary Materials and Methods.
Statistical analysis
Statistical analyses were carried out using the program R (www.r-project.org). Recurrence-free survival was evaluated by Kaplan-Meier survival curve and Log-rank tests. Statistical significance was assessed by unpaired two-tailed Student’s-tests. Single-sample gene set enrichment analysis (ssGSEA) was used to assess gene set activation scores in gene expression profiling data. ssGSEA calculates a sample level gene set score by comparing the distribution of gene expression ranks inside and outside the gene set. The ssGSEA score was calculated by Gene Set Variation Analysis (GSVA) R package. Data were examined to determine whether they were normally distributed with the One-Sample Kolmogorov-Smirnov test. If the data were normally distributed, comparisons of measurement data between two groups were performed using independent sample t test and the comparisons among three or more groups were first performed by one-way ANOVA test. If the results showed significant difference, when the data were skewed distribution, comparisons were performed by nonparametric test. Measurement data between two groups were performed using nonparametric Mann-Whitney test.
Data availability
The raw sequencing data have been deposited in the Gene Expression Omnibus database under the accession number GSE130012. All the other data generated in this study are included in the article and the additional files.
More detailed materials and methods are in the Supplementary Methods.
Results
METTL3 is closely correlated with glycolysis in colorectal cancer
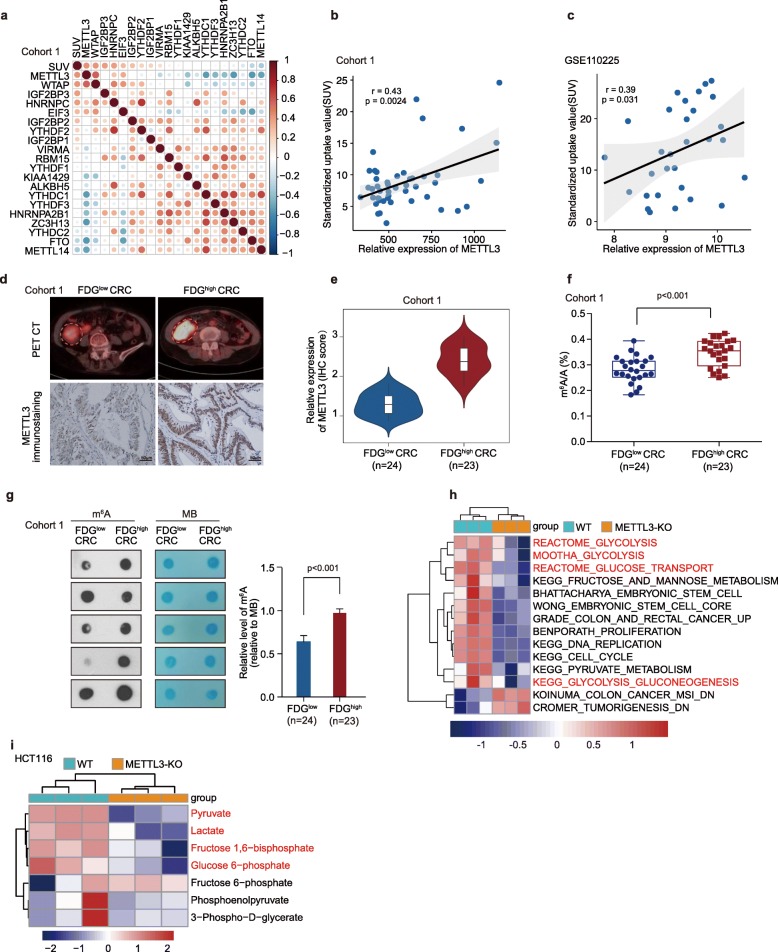
To explore the correlation between m6A modifications with glycolysis metabolism in colorectal cancer (CRC), real-time PCR analysis was performed to compare the regulated-m6A gene expression profiles in 47 CRC patients who had been imaged for 18F-FDG PET (Cohort 1). We observed that there was a most significant correlation between FDG uptake and METTL3 expression in CRC patients (Fig. 1a-b), as measured by FDG maximal standardized uptake value (SUVmax). In addition, we observed that there was a significant correlation between FDG uptake and METTL3 expression in an independent CRC patients’ dataset (GSE110225), as measured by SUVmax (Fig. 1c). Further analysis revealed that a significant correlation between FDG uptake and METTL3 immunohistochemical staining existed in CRC patients of Cohort 1 (Fig. 1d-e). In line with METTL3 higher expression in CRC patients with vigorous glucose metabolism, LC-MS/MS (liquid chromatography-tandem mass spectrometry) (Fig. 1f) and m6A dot blot (Fig. 1g) experiments revealed that the m6A modification level was significantly increased in these CRC patients with higher FDG uptake as well. To elucidate whether METTL3 plays a role in CRC glycolysis and tumorigenesis, we then performed RNA-seq analysis to compare the gene expression profiles of METTL3-knockout and wild type (WT) HCT116 CRC cells, which exhibit higher METTL3 expression (Figure S1a). METTL3 knockout of CRISPR-cas9 generated cell was confirmed by sequencing and Western blot analysis (Figure S1b-c). A total of 2848 downregulated genes and 3046 upregulated genes (p < 0.05) were detected (raw data accessible via GEO number GSE130012) after knockout of METTL3 in HCT116 cells (Table S1). Gene set enrichment analysis (GSEA) of RNA-seq data showed that the gene signatures of “Glucose uptake” were enriched in HCT116 WT cells (Figure S1d). Single-sample GSEA (ssGSEA) [26] revealed that the gene sets related to Reactome_Glycolysis, Mootha_Glycolysis, Reactome_Glucose_Transport and KEGG_Glycolysis_Gluconeogenesis (glycolysis pathway) and Grade_Colon_And_Rectal_Cancer_Up (colorectal-carcinogenesis signature) were negatively correlated with METTL3-knockout CRC cells (Fig. 1h, Table S2). Besides, further mass spectrometric metabolomics analysis [27] was performed in HCT116 WT and METTL3-knockout cells. The high through put metabolic analysis showed that knockout of METTL3 significantly reduced the levels of key components of glycolysis pathway, such as glucose-6-phosphate, fructose 1,6-bisphosphate, pyruvate and lactate in HCT116 cells (Fig. 1i and Table S3). These data indicate that METTL3 may mediate glycolytic metabolism and carcinogenesis in CRC patients.
Fig. 1.
METTL3 is closely correlated with glycolytic metabolism in CRC. (a) The correlation matrix showed the relationship between the expression of m6A genes and SUVmax (FDG maximal standardized uptake value) in 47 CRC patients of Cohort 1. (b) Correlation between FDG uptake (SUVmax) and METTL3 expression in 47 CRC patients of Cohort 1. (c) Correlation between FDG uptake (SUVmax) and METTL3 expression in 30 CRC patients of another independent cohort (GSE110225). (d) Representative PET/CT images and METTL3 immunohistochemical images of CRC patients with FDG high uptake and FDG low uptake in Cohort 1. (e) Statistical analysis of METTL3 expression in CRC patients with FDG high uptake and patients with FDG low uptake, nonparametric Mann-Whitney test. (f) LC-MS/MS quantification of the m6A/A ratio in poly(A) RNA isolated from CRC patients with FDG high uptake or FDG low uptake in Cohort 1, nonparametric Mann-Whitney test. (g) Representative images and relative quantitative information of m6A dot blot assay showed global m6A abundance in CRC patients with FDG high uptake or FDG low uptake in Cohort 1. MB, methylene blue staining (as a loading control). (h) ssGSEA analysis was performed to show the pathways closely correlated with METTL3 expression levels in CRC cells; n = 3. (i) LC-MS/MS-based analysis showed the major metabolites altered in the glycolytic pathway in HCT116 METTL3-KO cells relative to WT cells. The differentially expressed metabolites (p < 0.05) were highlighted in red; n = 3. (WT, wild type; METTL3-KO, METTL3-knockout)
METTL3 drives glycolytic metabolism in colorectal cancer
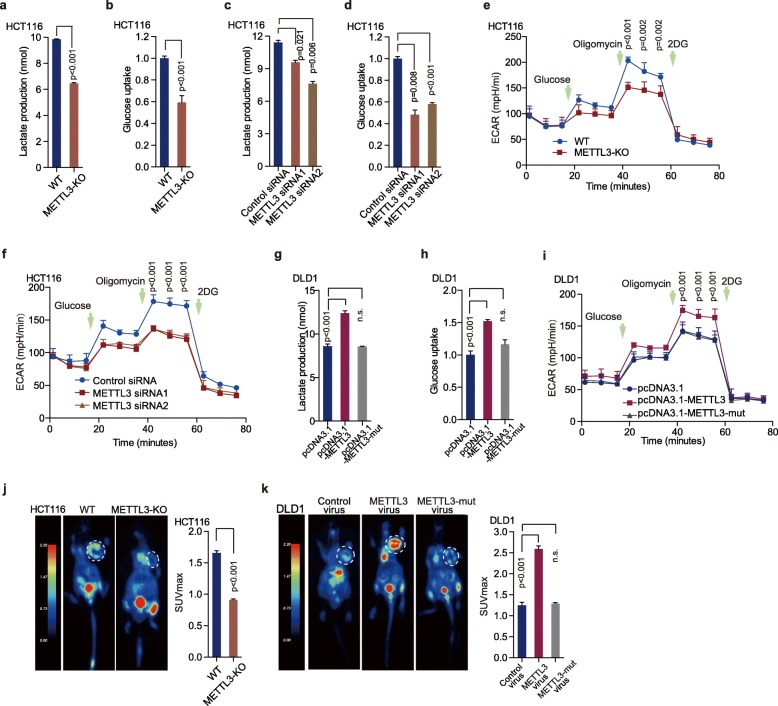
Further functional colorimetric validation showed that lactic acid production (a key metabolite of glycolysis) and glucose uptake were both significantly decreased after METTL3 knockout in HCT116 cells (Fig. 2a-b). Knockdown of METTL3 significantly reduced lactic acid production as well as glucose uptake in HCT116 cells (Fig. 2c-d, Figure S2a) and SW480 cells (Figure S2b-d). To figure out whether alteration of METTL3 directly influence glycolytic metabolism, we measured extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) in CRC cells after manipulating METTL3. Knockout or knockdown of METTL3 significantly reduced ECAR levels in HCT116 (Fig. 2e-f) and SW480 cells (Figure S2e), compared with control cells. However, the deletion of METTL3 had no significant effect on OCR levels in HCT116 cells and SW480 cells (Figure S2f).
Fig. 2.
METTL3 drives glycolytic metabolism in CRC. (a-b) Lactate production (a), glucose uptake (b) were measured in HCT116 WT and METTL3-KO cells by colorimetric analysis, n = 3, nonparametric Mann–Whitney test. (c-d) Lactate production (c) and glucose uptake (d) were measured in HCT116 CRC cells transfected with control siRNA and METTL3 siRNA1/2 by colorimetric analysis; n = 3, nonparametric Mann–Whitney test. (e-f) ECAR was detected in HCT116 WT, METTL3- knockout cells (e) and HCT116 CRC cells transfected with control siRNA or METTL3 siRNA1/2 (f), n = 4, nonparametric Mann–Whitney test. (g-i) Lactate production (g), glucose uptake (h) and ECAR (i) were assessed in DLD1 cells transfected with pcDNA3.1, pcDNA3.1-METTL3 and pcDNA3.1-METTL3-mut, n = 3, nonparametric Mann–Whitney test. (j) Representative images of 18F-FDG uptake by micro-PET imaging in WT and METTL3-KO xenograft mouse models. White circles indicated tumor glucose uptake. Maximum uptake values (SUVmax) for xenografts measured by FDG-PET were presented, nonparametric Mann–Whitney test. (k) Representative images of 18F-FDG uptake by micro-PET imaging in control virus, overexpression of METTL3 or METTL3-mut virus xenograft mouse models. White circles indicated tumor glucose uptake. Maximum uptake values (SUVmax) for xenografts measured by FDG-PET were presented, nonparametric Mann–Whitney test. Fig. 2k and Fig. 6k shared experimental controls and METTL3-overexpression group. (WT, wild type; METTL3-KO, METTL3-knockout; n = 3)
To elucidate whether METTL3-induced CRC glycolysis depends on its methyltransferase function, we constructed METTL3 wild-type (pcDNA3.1-METTL3) and mutant (pcDNA3.1-METTL3-mut, with MTase domain deletion) recombination plasmids (Figure S2g) [28]. Both of pcDNA3.1-METTL3 and pcDNA3.1-METTL3-mut were successfully overexpressed in DLD1 cells (Figure S2h-i). In gain of function assays, overexpression of METTL3 dramatically increased lactic acid production (Fig. 2g), glucose absorption (Fig. 2h) and ECAR levels (Fig. 2i) in DLD1 cells. Deletion of the MTase domain of METTL3 blocked METTL3-induced glycolysis in CRC cells (Fig. 2g-i). To further examine the effects of METTL3 on glycolysis, we used 18F-FDG PET ([18F]-fluoro-2-deoxyglucose positron emission tomography) to measure glucose uptake in vivo. Mouse PET-CT data showed that knockout of METTL3 significantly reduced glucose uptake in xenograft mouse tumor model (Fig. 2j). Overexpression of WT METTL3, but not METTL3-mut, dramatically increased glucose uptake in vivo (Fig. 2k). These data indicate that METTL3 regulates glycolytic metabolism in colorectal cancer via its methyltransferase domain.
METTL3-induced proliferation depends on the activation of glycolysis in colorectal cancer
METTL3 has been reported as a pivotal modulator in tumorigenesis [29]; however, the role of METTL3 in CRC is paradoxical [30, 31]. GSEA of RNA-seq data showed that the gene signatures of CRC and DNA replication were enriched in HCT116 WT cells, compared with METTL3-knockout cells (Figure S3a-b). The top-scoring genes in the gene sets included key carcinogenesis genes, CDK1, PCNA, and CDCA7. Real-time PCR confirmed that knockout or knockdown of METTL3 expression dramatically reduced the expression of the key genes of tumorigenesis (Figure S3c-e). Our functional validation data showed that knockout or knockdown of METTL3 abolished cell proliferation (Fig. 3a, Figure S3f) and colony formation (Fig. 3b, Figure S3g) in HCT116 cells and SW480 cells (Figure S3h-i). Knockout of METTL3 dramatically reduced HCT116 tumor growth (Fig. 3c-d) and tumor weight (Fig. 3e) in xenograft mouse models.
Fig. 3.
METTL3-induced cell survival depends on glycolytic metabolism. (a) Cell proliferation assay was performed by CCK8 assay in HCT116 WT or METTL3-KO cells, n = 6, nonparametric Mann–Whitney test. (b) Colony formation assay was performed in HCT116 WT or METTL3-KO cells, n = 3, nonparametric Mann–Whitney test. (c) Representative images of tumors in nude mice bearing HCT116 WT or METTL3-KO cells, n = 8. (d-e) Tumor volumes(d) and tumor weights (e) were measured in mice bearing HCT116 WT or METTL3-KO cells, n = 8, nonparametric Mann–Whitney test. (f) Cell proliferation of DLD-1 cells was measured by CCK8 assay after transfection of pcDNA3.1, pcDNA3.1-METTL3 and pcDNA3.1-METTL3-mut (METTL3 with MTase domain deletion), n = 6, nonparametric Mann–Whitney test. (g) Colony formation assay was performed in DLD1 cells after transfection with pcDNA3.1, pcDNA3.1-METTL3 and pcDNA3.1-METTL3-mut; n = 3, nonparametric Mann–Whitney test. (h-j) Representative images of tumors (h), statistical analysis of tumor volumes (i) and tumor weights (j) in nude mice bearing DLD1 cells in different groups (n = 8, nonparametric Mann–Whitney test). Fig 3h, Fig. 3m and Figure S6j shared experimental controls and METLL3-overexpression group. (k-l) Cell proliferation (k) and colony formation assay (l) were performed in DLD1 cells after transfection with control plasmids, METTL3 overexpression plasmids, and subsequently with 5 mM 2-DG (2-Deoxyglucose) treatment; nonparametric Mann–Whitney test. (m-o) Representative images of tumors (m), statistical analysis of tumor volumes (n) and tumor weights (o) in nude mice bearing DLD1 cells in different groups (n = 8, nonparametric Mann–Whitney test). Fig. 3h, Fig. 3m and Figure S6j shared experimental controls and METTL3-overexpression group. (WT, wild type; METTL3-KO, METTL3-knockout)
In gain of function assays, overexpression of METTL3 increased cell proliferation (Fig. 3f) and colony formation (Fig. 3g) of DLD1 cells in vitro. Furthermore, overexpression of METTL3 dramatically increased DLD1 tumor growth (Fig. 3h-i) and tumor weight (Fig. 3j) in xenograft mouse models. However, the same phenomenon was not observed both in vitro (Fig. 3f-g) and in vivo (Fig. 3h-j) after overexpression of METTL3-mut. In addition, 2-DG (an inhibitor of glycolysis pathway) treatment significantly blocked METTL3-induced cell proliferation (Fig. 3k), and colony formation (Fig. 3l) in vitro and in vivo (Fig. 3m-o). There was no difference in tumor growth (Figure S3j-k) and tumor weight (Figure S3l) among different control groups. The data strongly suggests that METTL3 may be a tumor-driver gene and promote CRC progression by regulating colorectal cancer glucose metabolism.
Transcriptome-wide m6A-seq and RNA-seq assays identified potential targets of METTL3 in colorectal cancer
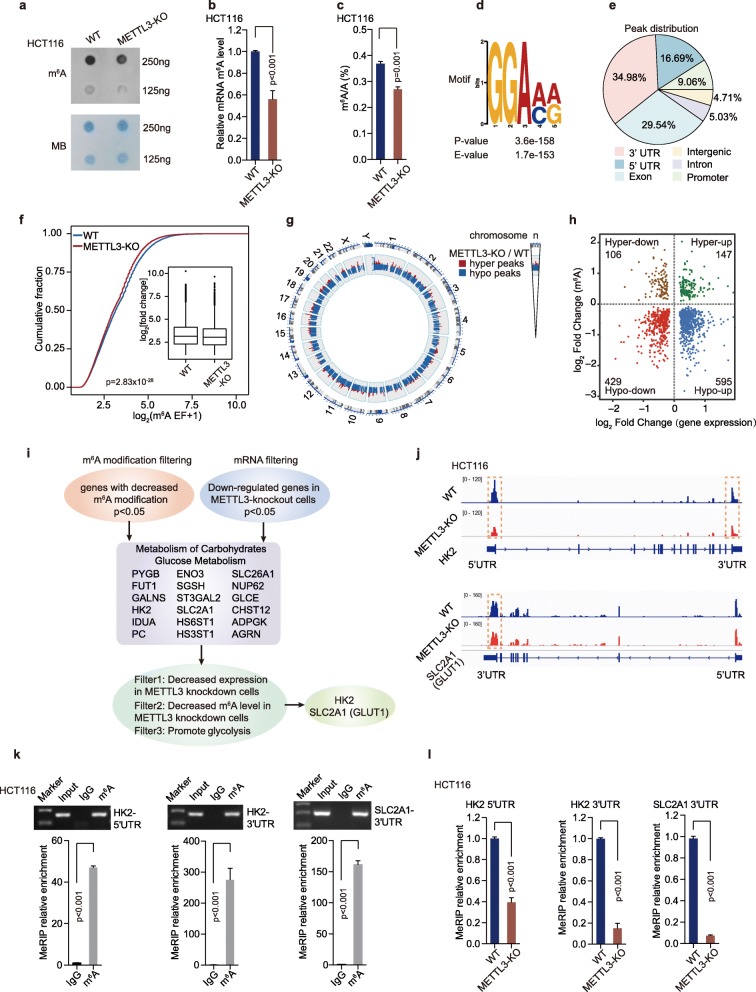
To identify potential mRNA targets of METTL3 whose m6A levels were increased by METTL3 in CRC cells, METTL3-knockout and WT HCT116 cells were selected for transcriptome-wide m6A-sequencing (m6A-seq, MeRIP-seq) and RNA-sequencing (RNA-seq) assays. A noticeable decrease in m6A levels was observed in polyadenylated RNAs (poly(A) RNAs) of the METTL3-knockout cells, compared to the WT HCT116 cells, as measured by m6A dot blot, EpiQuik™ m6A quantification assay and LC-MS/MS analysis (Fig. 4a-c). Consisted with the previous researches, the most common m6A motif ‘GGAC’ is significantly enriched in the m6A peaks (Fig. 4d). Furthermore, most of the METTL3-binding sites (> 80%) were located in protein-coding transcripts (CDS region) and were highly enriched in 5’UTR and 3’UTR, especially enriched in the vicinity of the stop codon, which was coincidence with the m6A distribution (Fig. 4e and Figure S4a).
Fig. 4.
Transcriptome-wide m6A-seq and RNA-seq assays identified potential targets of METTL3 in colorectal cancer. (a) The m6A dot blot assay of global m6A abundance in mRNA of HCT116 WT and METTL3-knockout cells. MB, methylene blue staining (as a loading control). (b) The global m6A levels in mRNA of HCT116 WT and METTL3- knockout cells were measured by the EpiQuik™ m6A RNA Methylation Quantification Kit, n = 3, nonparametric Mann–Whitney test. (c) LC-MS/MS quantification of the m6A/A ratio in poly(A) RNA in HCT116 WT and METTL3-KO cells. (d) HOMER motif analysis revealed the top consensus m6A motif in HCT116 cells. (e) Graphs of m6A peak distribution illustrating the proportion of common m6A peaks in the indicated regions in HCT116 cells. (f) Cumulative curves of m6A abundance (log2(m6A EF + 1)) in HCT116 WT and METTL3-KO cells. The abundance of m6A immunoprecipitation was normalized to input when calculating the enrichment fold. P value was calculated using two-sided Wilcoxon and Mann–Whitney test. EF, enrichment folds. (g) Circos plot showing the distribution of hypermethylated (hyper) and hypomethylated (hypo) m6A peaks in the human transcriptome of HCT116 METTL3-KO cells compared with WT cells. (h) Distribution of genes with a significant change in both m6A level and gene expression level in HCT116 METTL3-KO cells compared with WT cells. (i) The flow chart for selected candidate target genes of METTL3 in HCT116 METTL3-KO cells is shown. (j) The relative abundance of m6A sites along HK2 and SLC2A1 (GLUT1) mRNA in HCT116 WT and METTL3-KO cells, as detected by m6A-seq. The orange rectangles indicated that the m6A peaks had a significantly decreased abundance. (k) Agarose electrophoresis and real-time PCR analysis of MeRIP assays in CRC cells showing the direct binding between the m6A antibody and HK2 5’UTR, HK2 3’UTR and SLC2A1 (GLUT1) 3’UTR. (l) MeRIP-qPCR analysis of HK2 5’UTR, HK2 3’UTR and SLC2A1 (GLUT1) 3’UTR m6A levels in HCT116 WT and METTL3-KO cells, n = 3, nonparametric Mann–Whitney test. (WT, wild type; METTL3-KO, METTL3-knockout)
We next compared the genes with altered-m6A modifications between METTL3-knockout cells and the WT cells. The analysis of m6A-seq revealed a global hypo-methylation of m6A in the transcription level after knockout of METTL3 in HCT116 cells (Fig. 4f-g). A total of 2632 and 584 peaks showed a significant decrease or increase in m6A modifications (p < 0.05, Table S4) in METTL3-knockout cells relative to WT cells respectively, and thereby they were termed as hypo-methylated and hyper-methylated m6A peaks (Figure S4b). Through analysis of the RNA-seq data, we identified 429 hypo-methylated m6A genes whose mRNA transcripts were down-regulated (p < 0.05, Hypo-down) and 595 hypo-methylated m6A genes whose mRNA transcripts were up-regulated (p < 0.05, Hypo-up) in METTL3-knockout cells, compared with WT cells (Fig. 4h). Considering the role of METTL3 in the m6A methyltransferase complex, mRNA transcripts carrying hypo-methylated m6A peaks in HCT116 METTL3 knockout cells were likely potential targets.
We further found that the hypo-down transcripts (p < 0.05) were significantly enriched with genes involved in ‘Metabolism of carbohydrates’ and ‘Glucose metabolism’, which included the glycolysis pathway through gene ontology analysis (Figure S4c). Nevertheless, those hypo-up transcripts were significantly enriched with “Post-translational protein modification”, “Chromatin organization”, which are not associated with glycolytic metabolism (Figure S4c). Therefore, the hypo-methylated m6A genes, with decreased expression in METTL3-knockout CRC cells (p < 0.05) (hypo-down), were selected for further validation (Fig. 4i). Eighteen genes (PYGB, FUT1, GALNS, HK2, IDUA, PC, ENO3, SGSH, ST3GAL2, SLC2A1, HS6ST1, HS3ST1, SLC26A1, NUP62, GLCE, CHST12, ADOGK and AGRN), which involved in ‘Carbohydrates metabolism’ and ‘Glucose metabolism’, were sorted as potential target genes (Fig. 4i). Real-time PCR and MeRIP-qPCR assays were performed to measure the mRNA and m6A levels of eighteen hypo-down candidates in different cells. Real-time PCR data showed that the expression of six target genes was significantly impaired both in METTL3-knockout HCT116 (Figure S4d) and METTL3-knockdown SW480 CRC cells (Figure S4e). METTL3 up-regulation increased the RNA level of these six transcripts in DLD1 cells (Figure S4f-g). The m6A peaks of these six transcripts were remarkably decreased in METTL3-knockout HCT116 cells, compared with WT cells (Fig. 4j and Figure S4h) according to the MeRIP-seq data. MeRIP-PCR data was consistent with the MeRIP-seq data (Fig. 4k and Figure S4i). Further MeRIP-qPCR [18, 32] analyses showed the m6A levels in five of the six target genes were dramatically reduced in HCT116 METTL3-knockout cells (Fig. 4l, Figure S4j). The rest five target genes were further validated by functional glycolysis assay. Lactate production was only significantly reduced in HCT116 cells with Hexokinase 2 (HK2) and SLC2A1 (GLUT1) downregulation, but not in those cells with the other three genes downregulation (Figure S4k). In addition, we performed MeRIP-seq in 5 CRC patients with METTL3 high expression and 5 CRC patients with METTL3 low expression. The Integrative Genomics View (IGV) data revealed that the m6A level of HK2 and SLC2A1 (GLUT1) was higher in 5 CRC patients with METTL3 high expression than those with METTL3 low expression (Figure S4l), indicating that higher m6A level of HK2 and SLC2A1 (GLUT1) depends on METTL3 modification in CRC patients. The MeRIP-seq data in CRC patients is consistent with the data in CRC cell lines. Therefore, we selected HK2 and SLC2A1 (GLUT1) as candidate targets of METTL3 for further investigation.
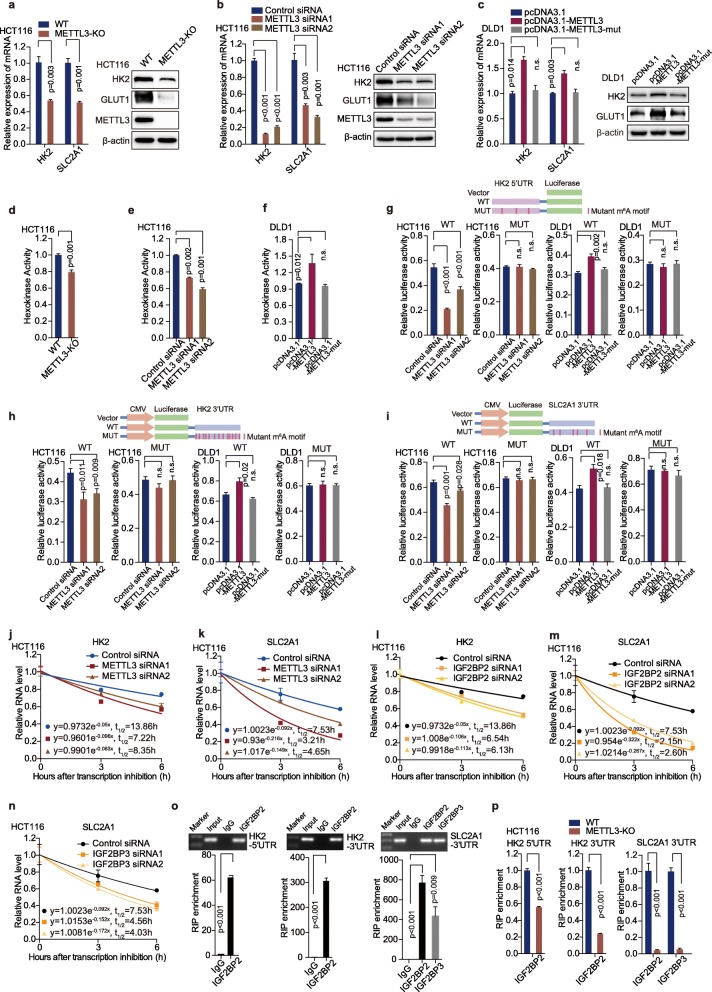
METTL3 regulates HK2 and SLC2A1 (GLUT1) mRNA levels and stability depending on its m6A methyltransferase activity
In biological validation assay, HK2 and SLC2A1 (GLUT1) expression were confirmed to be downregulated in METTL3-knockout cells (Fig. 5a) and METTL3-knockdown cells (Fig. 5b and Figure S5a). In addition, overexpression of METTL3, but not METTL3-mut dramatically increased HK2 and SLC2A1 (GLUT1) mRNA and protein levels (Fig. 5c). We further measured m6A levels of glycolysis genes HK2 and SLC2A1 (GLUT1) by MeRIP-qPCR after transfection of control, METTL3 and METTL3-mut plasmids in DLD1 cells. The MeRIP-qPCR data showed that overexpression of METTL3, but not METTL3-mut, dramatically increased the m6A levels of HK2 and SLC2A1 (GLUT1) in DLD1 cells (Figure S5b). It indicates that m6A modification of HK2 and SLC2A1 (GLUT1) directly affects its mRNA level. We detected the hexokinase activity in CRC cells as well. The hexokinase activity was noticeably decreased in METTL3-knockout cells and METTL3-knockdown cells (Fig. 5d-e and Figure S5c). Moreover, overexpression of METTL3, but not METTL3-mut, significantly up-regulated the hexokinase activity (Fig. 5f). To further address the effect of m6A modification on HK2 and SLC2A1 (GLUT1) gene, we constructed both wild-type and mutant HK2 and SLC2A1 (GLUT1) luciferase reporter plasmids, which contained the wild-type 5′ and 3’UTR of HK2, the 3’UTR of SLC2A1 (GLUT1) or mutations (m6A was replaced by T) in the m6A sites (Fig. 5g-i). As expected, compared with control siRNA, METTL3 siRNA substantially reduced luciferase activity of the individual reporter constructs bearing wild-type HK2 5′ and 3’UTR that have intact m6A sites, and wild-type 3’UTR of SLC2A1 (GLUT1). However, downregulation of METTL3 had no significant effect on the luciferase activity in these reporter plasmids with m6A sites mutations in HCT116 (Fig. 5g-i) and SW480 cells (Figure S5d-f). Furthermore, overexpression of METTL3, but not METTL3 mutant, significantly increased luciferase activity of the individual reporter constructs bearing wild-type HK2 5′ and 3’UTR, and wild-type SLC2A1 (GLUT1) 3’UTR in DLD1 cells (Fig. 5g-i). Overexpression of METTL3 and METTL3-mut had no significant effect on HK2 and SLC2A1 (GLUT1) luciferase reporter plasmids with m6A sites mutation (Fig. 5 g-i).
Fig. 5.
METTL3 regulates HK2 and SLC2A1 (GLUT1) mRNA levels and stability depending on its m6A methyltransferase activity. (a-b) Real-time PCR and Western blot assays were performed to analyze the relative HK2 levels and SLC2A1 (GLUT1) in HCT116 cells after knockout (a) or knockdown (b) of METTL3 in HCT116 cells, n = 3, nonparametric Mann–Whitney test. (c) Real-time PCR and Western blot assays were performed to analyze the relative HK2 levels and SLC2A1 (GLUT1) in DLD-1 cells after transfection with pcDNA3.1, pcDNA3.1-METTL3 and pcDNA3.1-METTL3-mut, n = 3, nonparametric Mann–Whitney test. (d-e) Hexokinase activity was measured by colorimetric analysis after knockout (d) or knockdown (e) of METTL3 in HCT116 cells, n = 3, nonparametric Mann–Whitney test. (f) Hexokinase activity was measured by colorimetric analysis after transfection with pcDNA3.1, pcDNA3.1-METTL3 and pcDNA3.1-METTL3-mut in DLD1 cells, n = 3, nonparametric Mann–Whitney test. (g-i) Luciferase activity was measured in HCT116 cells transfected with control siRNA, METTL3 siRNA, pcDNA3.1, pcDNA3.1-METTL3 and pcDNA3.1-METTL3-mut. The luciferase reporters expressing WT or mutant human HK2 5’UTRs (g) and WT or mutant human HK2 3’UTRs (h) and WT or mutant human SLC2A1 (GLUT1) 3’UTRs (i) were used. The results were showed in the form of relative firefly luciferase activity normalized to Renilla luciferase activity. n = 4, nonparametric Mann–Whitney test. (j-k) The HK2 and SLC2A1 (GLUT1) mRNA half-life (t1/2) was detected by real-time PCR in HCT116 cells transfected with control siRNA or METTL3 siRNA1/2, n = 3, nonparametric Mann–Whitney test. (l) The HK2 mRNA half-life (t1/2) was detected by real-time PCR in HCT116 cells transfected with control siRNA or IGF2BP2 siRNA1/2, n = 3, nonparametric Mann–Whitney test. (m-n) The SLC2A1 (GLUT1) mRNA half-life (t1/2) was detected by real-time PCR in HCT116 cells transfected with control siRNA, IGF2BP2 or IGF2BP3 siRNA1/2, n = 3, nonparametric Mann–Whitney test. (o) Agarose electrophoresis and real-time PCR analysis of RIP assays in CRC cells showing the direct binding between the IGF2BP2 protein with HK2 5’UTR and HK2 3’UTR, and as well as the direct binding between IGF2BP2/3 with SLC2A1 (GLUT1) 3’UTR. (p) RIP-qPCR revealed the binding enrichment of IGF2BP2 to HK2 5’UTR and HK2 3’UTR, as well as the binding enrichment of IGF2BP2/3 to SLC2A1 (GLUT1) HK2 3’UTR, were decreased following knockout of METTL3 in HCT116 cells, n = 3, nonparametric Mann–Whitney test
To analyze the effect of m6A modification on the stability of METTL3 target transcripts, we conducted RNA stability assays [8]. The RNA stability curves showed that knockdown of METTL3 reduced the half-life of HK2 and SLC2A1 (GLUT1) mRNA in HCT116 (Fig. 5j-k) and SW480 cells (Figure S5g-h). Thus, METTL3-induced upregulation of HK2 and SLC2A1 (GLUT1) level is at least in part due to the increased stability of HK2 and SLC2A1 mRNA transcript. Insulin-like growth factor 2 mRNA binding proteins (IGF2BP1, IGF2BP2 and IGF2BP3) are three well-established m6A readers in mammalian cells [33], and responsible for stabilizing mRNA. m6A readers YTHDF2, YTHDF3 and YTHDC2 are responsible for mRNA decay [34–36]. Real-time PCR data showed that knockdown of IGF2BP2, but not IGF2BP1/3 significantly reduced the mRNA level of HK2 in HCT116 and SW480 cells (Figure S5i). Downregulation of IGF2BP2/3, but not IGF2BP1 noticeably reduced the mRNA level of SLC2A1 (GLUT1) (Figure S5j). Knockdown of YTHDF2, YTHDF3 or YTHDC2 did not influence the mRNA levels of HK2 or SLC2A1 (GLUT1) (Figure S5k-l). Furthermore, the half-life of HK2 mRNA was significantly decreased in HCT116 cells after transfection with IGF2BP2 siRNA (Fig. 5l), and the half-life of SLC2A1 (GLUT1) mRNA was significantly reduced in IGF2BP2/3 down-regulated HCT116 cells (Fig. 5m-n). RIP assay showed that IGF2BP2 directly bound to the 5′ and 3’UTR of HK2 mRNA (Fig. 5o) and IGF2BP2/3 directly bound to the 3’UTR of SLC2A1 (GLUT1) mRNA (Fig. 5o). Further RIP real-time PCR assay indicated that knockout of METTL3 significantly reduced the binding efficiency of IGF2BP2 to the 5′/3’UTR of HK2 mRNA and the binding efficiency of IGF2BP2/3 to the 3’UTR of SLC2A1 (GLUT1) mRNA (Fig. 5p). Taken together, our data demonstrate that METTL3-mediated m6A modification increases HK2 and SLC2A1 (GLUT1) expression through IGF2BP2-dependent and IGF2BP2/3-dependent mRNA stability regulation, respectively.
HK2 and SLC2A1 (GLUT1) are functionally essential target genes of METTL3 in CRC
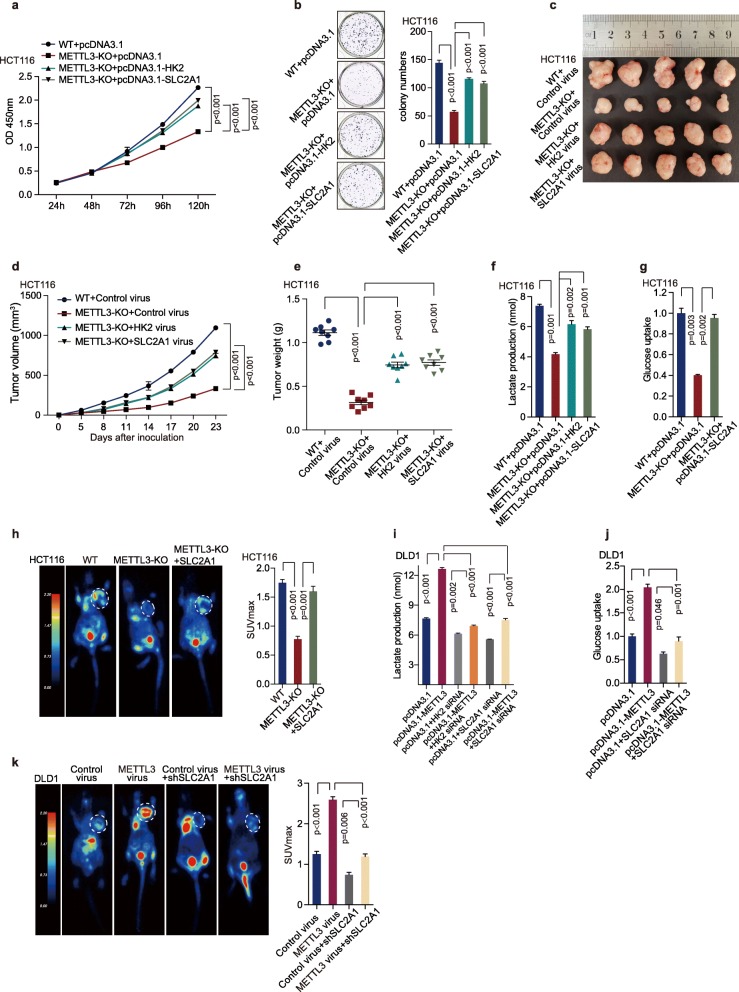
Hexokinase 2 (HK2) is a pivotal kinase in the glycolytic pathway [37]. Previous studies have demonstrated that HK2 activity is remarkably increased in various malignant neoplasms, as well as in CRC [38, 39]. Glucose transporter 1 (GLUT1), encoded by SLC2A1, is the predominant glucose transporter expressed on colonic epithelial cells [40]. Here we first revealed that METTL3 increased HK2 and SLC2A1 (GLUT1) expression via promoting their mRNA stability. Therefore, we next performed rescue experiments to investigate whether HK2 and SLC2A1 (GLUT1) participated in the biological function of METTL3 in CRC. HCT116 WT and METTL3-knockout cells were transfected with control, HK2 or SLC2A1 overexpression plasmids. The overexpression efficiency of HK2 and SLC2A1 were confirmed by real-time PCR and Western blot analysis (Figure S6a-b). Ectopic expression of HK2 or SLC2A1 partially restored the proliferation (Fig. 6a) and colony formation ability (Fig. 6b) of METTL3-knockout cells and tumor growth (Fig. 6c-e, Figure S6c-e). In addition, DLD1 cells were transfected with METTL3 overexpression plasmid and then treated with HK2 or SLC2A1 siRNA. The downregulation efficiency of HK2 and SLC2A1 siRNA were confirmed by real-time PCR and Western blot analysis (Figure S6f-g). Downregulation of HK2 or SLC2A1 dramatically impaired METTL3-induced cell proliferation (Figure S6h) and colony formation (Figure S6i) in vitro and tumor growth in vivo (Figure S6j-l), which further supported HK2 and SLC2A1 (GLUT1) as critical target genes of METTL3 in CRC.
Fig. 6.
HK2 and SLC2A1 were functionally important target genes of METTL3 in CRC. (a-b) CCK8 assays (a) and Colony formation assay (b) were measured after transfection with pcDNA3.1, pcDNA3.1-HK2 and pcDNA3.1-SLC2A1 plasmid in HCT116 WT and METTL3-knockout cells. Nonparametric Mann–Whitney test. (c-e) Representative images of tumors (c), statistical analysis of tumor volumes (d) and tumor weights (e) in nude mice bearing HCT116 cells in different groups (n = 8, nonparametric Mann–Whitney test). (f) Lactate production assay was performed after transfection with pcDNA3.1, pcDNA3.1-HK2 and pcDNA3.1-SLC2A1 plasmid in HCT116 WT and METTL3-KO cells, n = 3, Nonparametric Mann–Whitney test. (g) Glucose uptake was measured after transfected with pcDNA3.1, and pcDNA3.1-SLC2A1 plasmid in HCT116 WT and METTL3-knockout cells, n = 3, Nonparametric Mann–Whitney test. (h) Representative images of 18F-FDG uptake by micro-PET imaging in nude mice bearing HCT116 cells in different groups. White circles indicated tumor glucose uptake. Maximum uptake values (SUVmax) for xenografts measured by FDG-PET were presented; nonparametric Mann–Whitney test. (i) Lactate production assay was performed in DLD1 cells with different treatment, n = 3, nonparametric Mann–Whitney test. (j) Glucose uptake was detected in DLD1 cells with different treatment, n = 3, nonparametric Mann–Whitney test. (k) Representative images of 18F-FDG uptake by micro-PET imaging in nude mice bearing DLD1 cells in different groups. White circles indicated tumor glucose uptake. Maximum uptake values (SUVmax) for xenografts measured by FDG-PET were presented; nonparametric Mann–Whitney test. Fig. 2k and Fig. 6k shared experimental controls and METLL3-overexpression group. (WT, wild type; METTL3-KO, METTL3-knockout)
In glycolytic assays, ectopic expression of HK2 or SLC2A1 (GLUT1) restored the decrease of lactate production in HCT116 METTL3-knockout cells (Fig. 6f). Meanwhile, overexpression of SLC2A1 (GLUT1) significantly restored the decrease of glucose uptake in HCT116 METTL3-knockout cells in vitro (Fig. 6g) and in vivo (Fig. 6h). Furthermore, the downregulation of HK2 or SLC2A1 (GLUT1) significantly reduced METTL3-mediated higher lactate production in DLD1 cells (Fig. 6i). Downregulation of SLC2A1 (GLUT1) dramatically impaired METTL3-induced higher glucose uptake in DLD1 cells in vitro (Fig. 6j) and in vivo (Fig. 6k). Thus, HK2 and SLC2A1 (GLUT1) mediate the regulatory function of METLL3 in CRC cells.
The levels of METTL3 and glycolysis components correlate and are clinically relevant in CRC patients
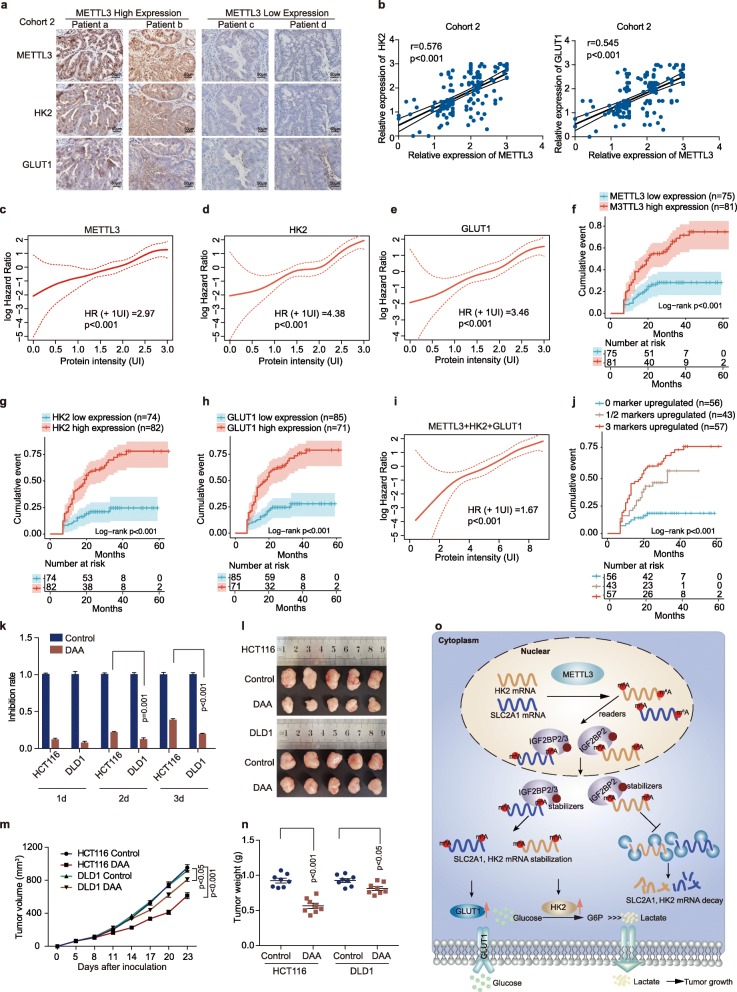
We next performed immunohistochemical staining in CRC patients’ tissues of Cohort 2. Interestingly, the samples with METTL3 higher expression displayed strongly staining for HK2 and GLUT1 (Fig. 7a, left panel). In addition, samples with low expression of METTL3 appeared low levels of HK2 and GLUT1 (Fig. 7a, right panel). Statistically, METTL3 expression was positively correlated with HK2 and GLUT1 expression in CRC tissues (Fig. 7b). There was no significant correlation between METTL3 and IGF2BP2/3 expression in CRC tissues of Cohort 2 (Fig. S7a). We next assessed the association between the intensity of METTL3, glycolysis components, and disease-free survival after tumor resection in this patients’ cohort. This analysis showed that elevated expression of METTL3, HK2 or GLUT1 in CRC tissues predicted robustly shorter disease-free intervals, either as a linear (Fig. 7c-e) or categorized variable (Fig. 7f-h). Furthermore, a linear association was found between the combined expression of METTL3, HK2 and GLUT1 and the risk of relapse after therapy in CRC (Fig. 7i). The shortest disease-free survival times were detected in those patients with three highly expressed markers METTL3/HK2/GLUT1 (Fig. 7j). From these observations, we conclude that elevated expression levels of METTL3 and its target genes may identify CRC patients with poor prognosis. We next analyzed the correlation of METTL3, HK2 or GLUT1 expression and different clinicopathological features in Cohort 2. METTL3 expression was positively correlated with pathological differentiation, AJCC stage, and recurrence in CRC patients (Figure S7b). HK2 and GLUT1 expression were positively correlated with AJCC stage, and recurrence (Figure S7b). In addition, METTL3 expression is gradually increased from normal colorectal epithelial tissues, adenoma to cancer tissues in Cohort 3 (Figure S7c).
Fig. 7.
The levels of METTL3 and glycolysis components correlate and are clinically relevant in CRC patients. (a) Representative immunohistochemical images of METTL3, HK2 and GLUT1 in CRC tissues using IHC analysis in Cohort 2. Scale bars: 50 μm (400X). Fig. 7a and Figure S7a shared representative immunohistochemical images of METTL3 in METTL3 high expression and METTL3 low expression groups. (b) Correlation between METTL3 expression and HK2 IHC scores in CRC tissues of Cohort 2 (left). Correlation between METTL3 expression and GLUT1 IHC scores in CRC tissues of Cohort 2 (Right), n = 156. (c-e) Smooth estimates of HR (+ 1 UI; increase in recurrence risk for every unit of staining intensity) showed a higher risk of relapse for patients with higher expression of METTL3 (c), HK2 (d) or GLUT1 (e) in Cohort 2. Red dashed lines Indicated 95% confidence interval. (f-h) Kaplan-Meier curves showing the disease-free survival of patients with different levels of METTL3 (f), HK2 (g) or GLUT1 (h) protein intensity in Cohort 2. (i) Smooth estimates of HR (+ 1 UI) showed a higher risk of relapse for patients with higher combined scores of molecular markers (METTL3 + HK2 + GLUT1) in Cohort 2. Red dashed lines Indicated 95% confidence interval. (j) Kaplan-Meier analysis of disease-free survival for CRC patients based on the number of upregulated molecular markers (METTL3, HK2 and GLUT1) in Cohort 2. (k) Inhibition rate of DAA (3-Deazaadenosine) (100 μM) in HCT116 and DLD1 cells. (l-n) Representative images of tumors (l), statistical analysis of tumor volume (m), and weights (n) in nude mice bearing HCT116 and DLD1 cells treated with DAA. (o) Schematic diagram of the relationship among METTL3, m6A modification, colorectal cancer cell progression and glycolysis metabolism
So far, we have proved that METTL3 may function as an oncogene by stabilizing HK2 and SLC2A1 mRNA levels in CRC cells. Recently, several RNA methyltransferase inhibitors have been developed as anti-tumor drug candidates. We next hypothesized that those CRC patients, harboring higher METTL3 expression, will be more sensitive to the anti-tumor drug candidates targeting METTL3. To test this prediction, we treated the HCT116 CRC cells, which express a higher METTL3 level, with DAA (3-Deazaadenosine, an inhibitor of the internal N6-Methyladenosine) [41, 42]. DAA treatment resulted in 39% inhibition of cell proliferation in HCT116 cells. Conversely, treatment in DLD1, which has a lower expression of METTL3, with the same dose of DAA, resulted in only 20% inhibition of cellular proliferation (Fig. 7k). These inhibition results could also be recapitulated in vivo. DAA is more efficient to inhibit tumor growth (Fig. 7l-m) and tumor weight (Fig. 7n) in nude mice bearing HCT116 cells than those bearing DLD1 cells. Taken together, these data suggest that targeting the METTL3 may be more effective in CRC cells with higher METTL3 expression than in those CRC cells with lower METTL3 expression.
Discussion
Various epigenetic modifications, including DNA methylation, histone modification, microRNA, and long non-coding RNA regulation, may contribute to colorectal carcinogenesis [43–45]. However, the potential involvement of RNA m6A modification is poorly defined in human colorectal cancer (CRC) and glycolytic metabolism. Through a combination of genomic, biochemical, and cell biological analyses, we have demonstrated that m6A RNA methylation levels and RNA methyltransferase METTL3 expression are highly increased in CRC patients with higher FDG uptake. GSEA analyses have demonstrated that cell proliferation and glycolytic pathways in cancer are significantly enriched in response to METTL3 alteration in the CRC cells. The bioinformatics analyses have been functionally validated in several in vitro and in vivo experimental models. In cultured CRC cells and xenograft mouse models, downregulation of METTL3 markedly suppresses tumor growth and inhibits glycolysis progression in CRC. The data consistently point to the notion that higher METTL3 expression and higher m6A methylation level are decisive factors of controlling human CRC aggressiveness.
RNA Methyltransferase METTL3 may participate in regulating the mRNA stability and modification of target genes [46]. However, the underlying molecular mechanisms of METTL3-regulated genes in CRC remain unknown. Our transcriptome-wide m6A-seq assay, the subsequent validation and functional studies suggest that HK2 and SLC2A1 (GLUT1) are the critical target genes of METTL3 in CRC. As an m6A RNA methyltransferase, METTL3 increases the m6A level of HK2 gene mainly at 3′ and 5′ untranslated regions (UTRs) and the m6A level of SLC2A1 gene mainly at 3′UTR, which in turn leads to the up-regulation of HK2 and SLC2A1 (GLUT1) at the RNA level and protein level. In addition, our luciferase reporter/mutagenesis assays indicate that the m6A sites in the UTRs of METTL3 critical target genes such as HK2 and SLC2A1 are essential for METTL3 to post-transcriptionally regulate their expression. This finding was further supported by the MeRIP assay, RNA stabilization assay, and qRT-PCR in control and METTL3-knockout or METTL3-knockdown CRC cells. Our data and previous studies [38, 39] demonstrate that HK2 and SLC2A1 may act as oncogenes to promote cell glycolysis metabolism. Here, we also firstly reveal that HK2 and GLUT1 participate in METTL3-mediated biological function in CRC. Thus, the METTL3➔HK2/GLUT1 axis likely plays a critical role in the pathogenesis of CRC. A schematic model summarizing our discoveries is shown in Fig. 7o.
Previous studies suggest that mRNA transcripts with m6A modifications tend to be regulated by YTHDFs or IGF2BPs as the direct m6A readers [8–10, 47]. Primarily due to the recognition by different m6A readers, the mRNA transcripts with m6A modifications have a different fate. YTHDF2, YTHDF3, and YTHDC2 tend to reduce the expression of genes by promoting their mRNA decay [34–36], while IGF2BPs tend to regulate the gene expression by promoting mRNA stability [33]. We showed that depleted expression of METTL3 substantially shortened the half-life of its critical targets such as HK2 and SLC2A1, suggesting that METTL3-mediated increase in HK2 and SLC2A1 expression are at least in part due to the increased stability of these two mRNA transcripts. In the further validation study, we demonstrated that METTL3 epigenetically stabilized HK2 and SLC2A1 mRNA levels through an m6A-IGF2BP2 and m6A-IGF2BP2/3-dependent mechanism, and therefore support the malignant state of CRC cells. Since 1) the mRNA levels of HK2 and SLC2A1 were significantly reduced after genetic deficiency of IGF2BP2 or IGF2BP2/3; 2) IGF2BP2 directly bound with the 5’UTR/3’UTR of HK2 mRNA, and IGF2BP2/3 directly bound with the 3’UTR of SLC2A1 mRNA; 3) Knockdown of METTL3 significantly disrupted the binding of IGF2BP2 to the 5’UTR/3’UTR of HK2 mRNA, and the binding of IGF2BP2/3 to the 3’UTR regions of SLC2A1 mRNA. In support of our observation, IGF2BP2 exhibits oncogenic function as m6A readers, which associated with m6A reading processes in tumorigenesis and progression [33, 48]. Dysregulation of IGF2BPs could result in abnormal accumulation of oncogenic products such as MYC in human cancer cells [33].
Therefore, from a therapeutic perspective, the mechanistic understanding of METTL3-induced cancer cell biological function and glucose metabolism in cellular regulation will enable the identification of the novel therapeutic targets. Profiling of m6A modification pattern that is affected by RNA methyltransferase may also allow the development of diagnostic tests of cancer. In short, METTL3 plays an oncogenic role in stabilizing HK2 and SLC2A1 mRNA via IGF2BPs, and further regulate glycolytic metabolism as well as cell proliferation in CRC cells. In addition to METTL3 biological and epigenetic importance, our work may be relevant to the clinical management of CRC patients. As the higher expressions of METTL3, HK2, and GLUT1 are associated with poor outcomes in CRC patients, METTL3 and its target genes may be promising biomarkers to guide early diagnosis and therapy in CRC. We further found that DAA (a chemical inhibitor of the internal N6-Methyladenosine) is more effective to inhibit cell proliferation in CRC cells with higher METTL3 expression than those cells with lower METTL3 expression. Since DAA inhibits other signal pathways [42, 49], a more specific inhibitor of METTL3 or glycolysis pathway needs to be developed for CRC patients, especially for those CRC patients with higher METTL3 expression treatment. Taken together, METTL3 and its associated pathway are crucial for colorectal carcinogenesis as well as glycolysis pathway, and targeting this pathway may be pivotal in the prevention and treatment of colorectal cancer.
Conclusions
Here, for the first time, we found that m6A modification is closely correlated with glycolysis pathway activation in colorectal cancer patients’ tissues. Mechanically, HK2 and GLUT1 were found to be regulated by m6A modification and participate in glycolysis activation in colorectal cancer. The METTL3➔HK2/GLUT1-IGF2BPs axis likely plays a critical role in the pathogenesis of colorectal cancer. Targeting METTL3 and its pathway may be promising for treating colorectal cancer patients with high glucose metabolism.
Supplementary information
Additional file 1. Supplementary Methods
Additional file 2 Table S1. RNA-seq analysis in HCT116 WT and METTL3-knockout cells.
Additional file 3 Figure S1. METTL3 is closely correlated with glycolytic metabolism in CRC. (a) The relative expression of METTL3 was measured by real-time PCR in normal colonic epithelial cell line FHC and CRC cell lines, n = 3, nonparametric Mann–Whitney test. (b) Schematic diagram of METTL3 genomic sequence in HCT116 wild-type (WT) and METTL3-knockout (METTL3-KO) cells. Red label, target sgRNA sequence; Yellow label, A 161 bp DNA fragment was inserted in the cutting site; Red circle, a premature stop site was generated after insertion of 161 bp DNA fragment. (c) The knockout efficiency of METTL3 was confirmed by Western blot. (d) GSEA analysis was conducted to identify the differential gene profiles between HCT116 METTL3-KO and WT cells. (WT, wild type; METTL3-KO, METTL3-knockout). Figure S2. METTL3 drives glycolytic metabolism in CRC. (a-b) The knockdown efficiency of METTL3 siRNA1/2 was evaluated in HCT116 (a) and SW480 (b) cells, n = 3, nonparametric Mann–Whitney test. (c-e) Lactate production (c), glucose uptake (d) and ECAR (e) were measured after transfection of control siRNA and METTL3 siRNA1/2 in SW480 cells, n = 3, nonparametric Mann–Whitney test. (f) OCR was measured in HCT116 WT and METTL3-KO cells (left). OCR was measured in SW480 cells transfected with control siRNA and METTL3 siRNA1/2 (right). (g) Schematic domain structures of METTL3. METTL3 MTase (AA residues 369–580). (h-i) Real-time PCR (h) and Western blot assay (i) were performed to detect METTL3 expression after transfection with pcDNA3.1-METTL3 and pcDNA3.1-METTL3-mut in DLD1 cells. (WT, wild type; METTL3-KO, METTL3-knockout). Figure S3. METTL3 is an oncogenic gene in colorectal cancer. (a-b) GSEA analysis was conducted to identify the differential gene profiles between HCT116 METTL3-KO and WT cells. (c) The expression of CDK1, PCNA and CDCA7 were detected in HCT116 WT and METTL3-KO cells, n = 3, nonparametric Mann–Whitney test. (d-e) The expression of CDK1, PCNA and CDCA7 were detected after transfection with control siRNA and METTL3-siRNA1/2 in HCT116 (d) and SW480 cells (e), n = 3, nonparametric Mann–Whitney test. (f) Cell proliferation of HCT116 cells was measured by CCK8 assay after transfected with control siRNA or METTL3 siRNA, n = 6, nonparametric Mann–Whitney test. (g) Colony formation assay was performed in HCT116 cells after transfection with control siRNA or METTL3 siRNA1/2, n = 3, nonparametric Mann–Whitney test. (h) Cell proliferation assay was performed by CCK8 assay in SW480 cells with METTL3 knockdown, n = 6, nonparametric Mann–Whitney test. (i) Colony formation assay was performed after transfection with control siRNA or METTL3 siRNA1/2 in SW480 cells, n = 3, nonparametric Mann–Whitney test. (j) Representative images of tumors in nude mice bearing DLD1 cells treated with PBS, control adenovirus and shControl adenovirus, n = 8. (k-l) Tumor volumes and (k) tumor weights (l) were measured in mice bearing DLD1 cells treated with PBS, control adenovirus and shControl adenovirus, n = 8, nonparametric Mann–Whitney test. (WT, wild type; METTL3-KO, METTL3-knockout). Figure S4. Transcriptome-wide m6A-seq and RNA-seq assays identified potential targets of METTL3 in colorectal cancer. (a) The distribution of m6A IP signal across the mRNA transcripts. m6A IP signal was enriched around the stop codon of mRNAs. (b) Comparison of the abundance of m6A peaks between HCT116 WT and METTL3-KO cells. A total of 2632 (hypo-methylated) and 584 (hyper-methylated) m6A peaks showed a significant decrease and increase in abundance in HCT116 METTL3-KO cells compared with WT cells. (c) Gene ontology analysis of genes with a significant decrease in m6A modification as well as a significant decrease in mRNA expression level (Hypo-down, p < 0.05) in HCT116 METTL3-KO cells (up). Gene ontology analysis of genes with a significant decrease in m6A modification as well as a significant increase in mRNA expression level (Hypo-up, p < 0.05) in HCT116 METTL3-KO cells (down). (d-e) Relative mRNA expression of METTL3 downstream targets from ‘Metabolism of carbohydrates pathway’ and ‘Glucose metabolism’ of GO analysis in HCT116 WT, HCT116 METTL3-KO (d) and SW480 cells (e) treated with control siRNA or METTL3 siRNA transfection, n = 3, nonparametric Mann–Whitney test. (f) Relative mRNA expression of METTL3 downstream targets in DLD1 cells transfected with METTL3 overexpression plasmids, n = 3, nonparametric Mann–Whitney test. (g) Venn diagram showed 6 genes with consistently decreased levels in both HCT116 and SW480 cells with METTL3 knockout or knockdown and increased expression in DLD1 cells with METTL3 upregulated. (h) The relative abundance of m6A sites along SLC26A1, PC, GLCE and CHST12 mRNA in HCT116 WT and METTL3-KO cells, as detected by m6A-seq. (i) Agarose electrophoresis and MeRIP-qPCR assays in CRC cells showing the direct binding between the m6A antibody and SLC26A1, PC, GLCE and CHST12. (j) MeRIP-qPCR analysis of SLC26A1, PC, GLCE and CHST12 RNA m6A levels in HCT116 cells with METTL3 knockout, n = 3, nonparametric Mann–Whitney test. (k) Lactate production was detected in HCT116 cells after knockdown of HK2, SLC2A1, PC, GLCE and CHST12, respectively, n = 3, nonparametric Mann–Whitney test. (l) The relative abundance of m6A sites along HK2 and SLC2A1 (GLUT1) mRNA in CRC tissues with high or low METTL3 expression as detected by m6A-seq. The orange rectangles indicated that the m6A peaks had a significant decreased abundance. (WT, wild type; METTL3-KO, METTL3-knockout). Figure S5. METTL3 regulates HK2 and SLC2A1 (GLUT1) mRNA levels and stability depending on its m6A methyltransferase activity. (a) Real-time PCR and Western blot assay were performed to detect the levels of HK2 and SLC2A1 (GLUT1) in SW480 cells after transfection of control siRNA and METTL3 siRNA1/2, n = 3, nonparametric Mann–Whitney test. (b) MeRIP-qPCR analysis of HK2 and SLC2A1 m6A levels in DLD1 cells transfected with control, METTL3 overexpression and METTL3-mut plasmids, n = 3, nonparametric Mann–Whitney test. (c) Hexokinase activity was measured in SW480 cells transfected with METTL3 siRNA1/2, n = 3, nonparametric Mann–Whitney test. (d) Relative luciferase activity of WT or mutant HK2 5’UTR firefly luciferase reporter in SW480 cells treated with control siRNA or METTL3 siRNA1/2, n = 4, nonparametric Mann–Whitney test. (e) Relative luciferase activity of WT or mutant HK2 3’UTR firefly luciferase reporter in SW480 cells treated with control siRNA or METTL3 siRNA1/2, n = 4, nonparametric Mann–Whitney test. (f) Relative luciferase activity of WT or mutant SLC2A1 (GLUT1) 3’UTR firefly luciferase reporter in SW480 cells treated with control siRNA or METTL3 siRNA1/2, n = 4, nonparametric Mann–Whitney test. (g-h) The HK2 (g) and SLC2A1 (GLUT1) (h) mRNA half-life (t1/2) were detected by real-time PCR in SW480 cells transfected with control siRNA or METTL3 siRNA1/2, n = 3, nonparametric Mann–Whitney test. (i) The relative expression of HK2 mRNA was measured by real-time PCR in HCT116 and SW480 cells after transfection with control, IGF2BP1, IGF2BP2 or IGF2BP3 siRNA, respectively, n = 3, nonparametric Mann–Whitney test. (j) The relative expression of SLC2A1 (GLUT1) mRNA was measured by real-time PCR in HCT116 and SW480 cells after transfection with control, IGF2BP1, IGF2BP2 or IGF2BP3 siRNA, respectively, n = 3, nonparametric Mann–Whitney test. (k-l) The relative expression of HK2 (k) and SLC2A1 (GLUT1) (l) mRNA were measured by real-time PCR in HCT116 cells or SW480 cells after knockdown of YTHDF2, YTHDF3 or YTHDC2, respectively, n = 3, nonparametric Mann–Whitney test. Figure S6. HK2 and SLC2A1 (GLUT1) are functionally important target genes of METTL3 in CRC (a) The overexpression efficiency of HK2 was confirmed by real-time PCR and Western blot analysis. (b) The overexpression efficiency of GLUT1 was confirmed by real-time PCR and Western blot analysis. (c-e) Representative images (c), tumor volumes (d) and tumor weights (e) in nude mice bearing HCT116 WT cells treated with PBS and control adenovirus, n = 8, nonparametric Mann–Whitney test. (f) The downregulation efficiency of HK2 siRNA was confirmed by real-time PCR and Western blot analysis. (g) The downregulation efficiency of GLUT1 siRNA was confirmed by real-time PCR and Western blot analysis. (h-i) Cell proliferation assay (h) and colony formation assay (i) were performed in DLD1 cells with different treatment, n = 6, nonparametric Mann–Whitney test. (j-l) Representative images of tumors (j), statistical analysis of tumor volumes (k) and tumor weights (l) in nude mice bearing DLD1 cells in different groups (n = 8, nonparametric Mann–Whitney test). Fig. 3h, Fig. 3m and Figure S6j shared experimental controls and METLL3 overexpression group. FigureS7. METTL3 is not correlated with IGF2BP2/3. (a) Representative immunohistochemical images of METTL3, IGF2BP2 and IGF2BP3 in CRC tissues using IHC analysis in Cohort 2. Fig. 7a and Figure S7a shared representative immunohistochemical images of METTL3 in METTL3 high expression and METTL3 low expression groups. (b) Comparing age, gender, histological differentiation, AJCC stage and recurrence between METTL3/HK2/GLUT1 high and low expression tumors of Cohort 2. The heat map illustrated the association of different clinicopathological features with METTL3/HK2/ GLUT1 high and low expression, Chi-square test. (c) Statistical analysis of METTL3 expression in human colorectal cancer (n = 67), colorectal adenoma (n = 75) and normal colorectal tissues (n = 126) of Cohort 3.)
Additional file 4 Table S2. Pathways regulated by METTL3 in HCT116 cells.
Additional file 5 Table S3. Metabolites of glycolysis pathway measured by Liquid chromatography-coupled tandem mass spectrometry (LC-MS/MS) in HCT116 WT and METTL3-knockout cells.
Additional file 6 Table S4. m6A-seq analysis in HCT116 WT and METTL3-knockout cells.
Additional file 7 Table S5. Clinical information of 47 cases CRC patients in Cohort 1.
Additional file 8 Table S6. Clinical information of 156 cases CRC patients in Cohort 2.
Additional file 9 Table S7. Clinical information of 30 cases CRC patients in GSE110225.
Additional file 10 Table S8. The sequences of siRNAs and primers used this study.
Abbreviations
- m6A
N6-methyladenosine
- CRC
Colorectal cancer
- 18F-FDG PET
[18F]-Fluoro-2-deoxyglucose positron emission tomography
- SUVmax
Standardized uptake value
- LC-MS/MS
Liquid chromatography-tandem mass spectrometry
- ECAR
Extracellular acidification rate
- OCR
Oxygen consumption rate
- METTL3
Methyltransferase-like 3
- GLUT1
Glucose transporter 1
- HK2
Hexokinase 2
- MeRIP
Methylated RNA immunoprecipitation
- RIP
RNA Immunoprecipitation
- DAA
3-Deazaadenosine
Authors’ contributions
CQ Shen, BQ Xuan, TT Yan, YR Ma, PP Xu, XL Tian, XY Zhang, YY Cao, D Ma and XQ Zhu performed the experiments and analyzed data. YW Zhang provided clinical specimen. J Hong and HY Chen conceived, wrote the manuscript and supervised the study. J Hong, HY Chen and JY Fang designed or/and supervised this project and revised the manuscript. The author (s) read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation (81871901,81790632,81874159,81522008,81421001); “Shu Guang” project supported by Shanghai Municipal Education Commission and Shanghai Education Development Foundation (17SG18); the Program for Professor of Special Appointment (Eastern Scholar No. 201268 and 2015 Youth Eastern Scholar NO. QD2015003) at Shanghai Institutions of Higher Learning; Shanghai Municipal Education Commission—Gaofeng Clinical Medicine Grant Support (No.20152512, 20161309), Program of Shanghai Young Academic/Technology Research Leader; Innovative research team of high-level local universities in Shanghai; Shanghai Sailing Program (No.19YF1428800).
Availability of data and materials
The raw sequencing data have been deposited in the Gene Expression Omnibus database under the accession number GSE130012. All the other data generated in this study are included in the article and the additional files.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Chaoqin Shen, Baoqin Xuan and Tingting Yan contributed equally to this work.
Contributor Information
Jing-Yuan Fang, Email: jingyuanfang@sjtu.edu.cn.
Haoyan Chen, Email: haoyanchen@sjtu.edu.cn.
Jie Hong, Email: jiehong97@sjtu.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12943-020-01190-w.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Bhandari A, Woodhouse M, Gupta S. Colorectal cancer is a leading cause of cancer incidence and mortality among adults younger than 50 years in the USA: a SEER-based analysis with comparison to other young-onset cancers. J Investig Med. 2017;65:311–315. doi: 10.1136/jim-2016-000229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 5.Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 6.Doherty JR, Cleveland JL. Targeting lactate metabolism for cancer therapeutics. J Clin Invest. 2013;123:3685–3692. doi: 10.1172/JCI69741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Rosa V, Iommelli F, Monti M, Fonti R, Votta G, Stoppelli MP, Del Vecchio S. Reversal of Warburg effect and reactivation of oxidative phosphorylation by differential inhibition of EGFR signaling pathways in non-small cell lung Cancer. Clin Cancer Res. 2015;21:5110–5120. doi: 10.1158/1078-0432.CCR-15-0375. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N (6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 11.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, Tang C, et al. Structural basis of N (6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534:575–578. doi: 10.1038/nature18298. [DOI] [PubMed] [Google Scholar]
- 14.Wei CM, Moss B. Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry. 1977;16:1672–1676. doi: 10.1021/bi00627a023. [DOI] [PubMed] [Google Scholar]
- 15.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, He C. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m (6) a mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–594. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, Chen Y, Sulman EP, Xie K, Bogler O, et al. m (6) A Demethylase ALKBH5 maintains Tumorigenicity of Glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millan-Zambrano G, Robson SC, Aspris D, Migliori V, Bannister AJ, Han N, et al. Promoter-bound METTL3 maintains myeloid leukaemia by m (6) A-dependent translation control. Nature. 2017;552:126–131. doi: 10.1038/nature24678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gundert M, Edelmann D, Benner A, Jansen L, Jia M, Walter V, Knebel P, Herpel E, Chang-Claude J, Hoffmeister M, et al. Genome-wide DNA methylation analysis reveals a prognostic classifier for non-metastatic colorectal cancer (ProMCol classifier) Gut. 2019;68:101–110. doi: 10.1136/gutjnl-2017-314711. [DOI] [PubMed] [Google Scholar]
- 21.Han D, Liu J, Chen C, Dong L, Liu Y, Chang R, Huang X, Liu Y, Wang J, Dougherty U, et al. Anti-tumour immunity controlled through mRNA m (6) a methylation and YTHDF1 in dendritic cells. Nature. 2019;566:270–274. doi: 10.1038/s41586-019-0916-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18:176. doi: 10.1186/s12943-019-1109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, Deng X, Wang Y, Weng X, Hu C, et al. R-2HG exhibits anti-tumor activity by targeting FTO/m (6) a/MYC/CEBPA signaling. Cell. 2018;172:90–105. doi: 10.1016/j.cell.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, Jiang Z, Zhang Y, Xu G, Zhang J, et al. METTL3-mediated m (6) a modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2019. [DOI] [PubMed]
- 25.Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, Huang H, Nachtergaele S, Dong L, Hu C, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N (6)-Methyladenosine RNA Demethylase. Cancer Cell. 2017;31:127–141. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shukla SK, Purohit V, Mehla K, Gunda V, Chaika NV, Vernucci E, King RJ, Abrego J, Goode GD, Dasgupta A, et al. MUC1 and HIF-1alpha signaling crosstalk induces anabolic glucose metabolism to impart gemcitabine resistance to pancreatic Cancer. Cancer Cell. 2017;32:71–87. doi: 10.1016/j.ccell.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, Tang C, et al. Corrigendum: structural basis of N (6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2017;542:260. doi: 10.1038/nature21073. [DOI] [PubMed] [Google Scholar]
- 29.Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J, Du P, Kim W, Tang S, Sliz P, et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561:556–560. doi: 10.1038/s41586-018-0538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN, Chen ZH, Zeng ZL, Wang F, Zheng J, et al. METTL3 facilitates tumor progression via an m (6) A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18:112. doi: 10.1186/s12943-019-1038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng R, Cheng Y, Ye S, Zhang J, Huang R, Li P, Liu H, Deng Q, Wu X, Lan P, Deng Y. m6A methyltransferase METTL3 suppresses colorectal cancer proliferation and migration through p38/ERK pathways. Onco Targets Ther. 2019;12:4391–4402. doi: 10.2147/OTT.S201052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Zhao JC. Update: mechanisms underlying N (6)-Methyladenosine modification of eukaryotic mRNA. Trends Genet. 2016;32:763–773. doi: 10.1016/j.tig.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al. Recognition of RNA N (6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang C, Chen Y, Sun B, Wang L, Yang Y, Ma D, Lv J, Heng J, Ding Y, Xue Y, et al. m (6) a modulates haematopoietic stem and progenitor cell specification. Nature. 2017;549:273–276. doi: 10.1038/nature23883. [DOI] [PubMed] [Google Scholar]
- 35.Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, Liu C, He C. YTHDF3 facilitates translation and decay of N (6)-methyladenosine-modified RNA. Cell Res. 2017;27:315–328. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kretschmer J, Rao H, Hackert P, Sloan KE, Hobartner C, Bohnsack MT. The m (6) a reader protein YTHDC2 interacts with the small ribosomal subunit and the 5′-3′ exoribonuclease XRN1. RNA. 2018;24:1339–1350. doi: 10.1261/rna.064238.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu P, Wilhelm K, Dubrac A, Tung JK, Alves TC, Fang JS, Xie Y, Zhu J, Chen Z, De Smet F, et al. FGF-dependent metabolic control of vascular development. Nature. 2017;545:224–228. doi: 10.1038/nature22322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang M, Liu T, Sun H, Weng W, Zhang Q, Liu C, Han Y, Sheng W. Pim1 supports human colorectal cancer growth during glucose deprivation by enhancing the Warburg effect. Cancer Sci. 2018;109:1468–1479. doi: 10.1111/cas.13562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shangguan C, Gan G, Zhang J, Wu J, Miao Y, Zhang M, Li B, Mi J. Cancer-associated fibroblasts enhance tumor (18) F-FDG uptake and contribute to the intratumor heterogeneity of PET-CT. Theranostics. 2018;8:1376–1388. doi: 10.7150/thno.22717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yun J, Mullarky E, Lu C, Bosch KN, Kavalier A, Rivera K, Roper J, Chio GEG, II, Rago C, et al. Vitamin C selectively kills KRAS and BRAF mutant colorectal cancer cells by targeting GAPDH. Science. 2015;350:1391–1396. doi: 10.1126/science.aaa5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Backlund PS, Jr, Carotti D, Cantoni GL. Effects of the S-adenosylhomocysteine hydrolase inhibitors 3-deazaadenosine and 3-deazaaristeromycin on RNA methylation and synthesis. Eur J Biochem. 1986;160:245–251. doi: 10.1111/j.1432-1033.1986.tb09963.x. [DOI] [PubMed] [Google Scholar]
- 42.Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, Okamura H. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 43.Kong X, Chen J, Xie W, Brown SM, Cai Y, Wu K, Fan D, Nie Y, Yegnasubramanian S, Tiedemann RL, et al. Defining UHRF1 domains that support maintenance of human Colon Cancer DNA methylation and oncogenic properties. Cancer Cell. 2019;35:633–648. doi: 10.1016/j.ccell.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valeri N, Braconi C, Gasparini P, Murgia C, Lampis A, Paulus-Hock V, Hart JR, Ueno L, Grivennikov SI, Lovat F, et al. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell. 2014;25:469–483. doi: 10.1016/j.ccr.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun TT, He J, Liang Q, Ren LL, Yan TT, Yu TC, Tang JY, Bao YJ, Hu Y, Lin Y, et al. LncRNA GClnc1 promotes gastric carcinogenesis and may act as a modular scaffold of WDR5 and KAT2A complexes to specify the histone modification pattern. Cancer Discov. 2016;6:784–801. doi: 10.1158/2159-8290.CD-15-0921. [DOI] [PubMed] [Google Scholar]
- 46.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al. Nuclear m (6) a reader YTHDC1 regulates mRNA splicing. Mol Cell. 2016;61:507–519. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 48.Muller S, Bley N, Glass M, Busch B, Rousseau V, Misiak D, Fuchs T, Lederer M, Huttelmaier S. IGF2BP1 enhances an aggressive tumor cell phenotype by impairing miRNA-directed downregulation of oncogenic factors. Nucleic Acids Res. 2018;46:6285–6303. doi: 10.1093/nar/gky229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim JH, Kim JH, Kim SC, Yi YS, Yang WS, Yang Y, Kim HG, Lee JY, Kim KH, Yoo BC, et al. Adenosine dialdehyde suppresses MMP-9-mediated invasion of cancer cells by blocking the Ras/Raf-1/ERK/AP-1 signaling pathway. Biochem Pharmacol. 2013;86:1285–1300. doi: 10.1016/j.bcp.2013.08.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplementary Methods
Additional file 2 Table S1. RNA-seq analysis in HCT116 WT and METTL3-knockout cells.
Additional file 3 Figure S1. METTL3 is closely correlated with glycolytic metabolism in CRC. (a) The relative expression of METTL3 was measured by real-time PCR in normal colonic epithelial cell line FHC and CRC cell lines, n = 3, nonparametric Mann–Whitney test. (b) Schematic diagram of METTL3 genomic sequence in HCT116 wild-type (WT) and METTL3-knockout (METTL3-KO) cells. Red label, target sgRNA sequence; Yellow label, A 161 bp DNA fragment was inserted in the cutting site; Red circle, a premature stop site was generated after insertion of 161 bp DNA fragment. (c) The knockout efficiency of METTL3 was confirmed by Western blot. (d) GSEA analysis was conducted to identify the differential gene profiles between HCT116 METTL3-KO and WT cells. (WT, wild type; METTL3-KO, METTL3-knockout). Figure S2. METTL3 drives glycolytic metabolism in CRC. (a-b) The knockdown efficiency of METTL3 siRNA1/2 was evaluated in HCT116 (a) and SW480 (b) cells, n = 3, nonparametric Mann–Whitney test. (c-e) Lactate production (c), glucose uptake (d) and ECAR (e) were measured after transfection of control siRNA and METTL3 siRNA1/2 in SW480 cells, n = 3, nonparametric Mann–Whitney test. (f) OCR was measured in HCT116 WT and METTL3-KO cells (left). OCR was measured in SW480 cells transfected with control siRNA and METTL3 siRNA1/2 (right). (g) Schematic domain structures of METTL3. METTL3 MTase (AA residues 369–580). (h-i) Real-time PCR (h) and Western blot assay (i) were performed to detect METTL3 expression after transfection with pcDNA3.1-METTL3 and pcDNA3.1-METTL3-mut in DLD1 cells. (WT, wild type; METTL3-KO, METTL3-knockout). Figure S3. METTL3 is an oncogenic gene in colorectal cancer. (a-b) GSEA analysis was conducted to identify the differential gene profiles between HCT116 METTL3-KO and WT cells. (c) The expression of CDK1, PCNA and CDCA7 were detected in HCT116 WT and METTL3-KO cells, n = 3, nonparametric Mann–Whitney test. (d-e) The expression of CDK1, PCNA and CDCA7 were detected after transfection with control siRNA and METTL3-siRNA1/2 in HCT116 (d) and SW480 cells (e), n = 3, nonparametric Mann–Whitney test. (f) Cell proliferation of HCT116 cells was measured by CCK8 assay after transfected with control siRNA or METTL3 siRNA, n = 6, nonparametric Mann–Whitney test. (g) Colony formation assay was performed in HCT116 cells after transfection with control siRNA or METTL3 siRNA1/2, n = 3, nonparametric Mann–Whitney test. (h) Cell proliferation assay was performed by CCK8 assay in SW480 cells with METTL3 knockdown, n = 6, nonparametric Mann–Whitney test. (i) Colony formation assay was performed after transfection with control siRNA or METTL3 siRNA1/2 in SW480 cells, n = 3, nonparametric Mann–Whitney test. (j) Representative images of tumors in nude mice bearing DLD1 cells treated with PBS, control adenovirus and shControl adenovirus, n = 8. (k-l) Tumor volumes and (k) tumor weights (l) were measured in mice bearing DLD1 cells treated with PBS, control adenovirus and shControl adenovirus, n = 8, nonparametric Mann–Whitney test. (WT, wild type; METTL3-KO, METTL3-knockout). Figure S4. Transcriptome-wide m6A-seq and RNA-seq assays identified potential targets of METTL3 in colorectal cancer. (a) The distribution of m6A IP signal across the mRNA transcripts. m6A IP signal was enriched around the stop codon of mRNAs. (b) Comparison of the abundance of m6A peaks between HCT116 WT and METTL3-KO cells. A total of 2632 (hypo-methylated) and 584 (hyper-methylated) m6A peaks showed a significant decrease and increase in abundance in HCT116 METTL3-KO cells compared with WT cells. (c) Gene ontology analysis of genes with a significant decrease in m6A modification as well as a significant decrease in mRNA expression level (Hypo-down, p < 0.05) in HCT116 METTL3-KO cells (up). Gene ontology analysis of genes with a significant decrease in m6A modification as well as a significant increase in mRNA expression level (Hypo-up, p < 0.05) in HCT116 METTL3-KO cells (down). (d-e) Relative mRNA expression of METTL3 downstream targets from ‘Metabolism of carbohydrates pathway’ and ‘Glucose metabolism’ of GO analysis in HCT116 WT, HCT116 METTL3-KO (d) and SW480 cells (e) treated with control siRNA or METTL3 siRNA transfection, n = 3, nonparametric Mann–Whitney test. (f) Relative mRNA expression of METTL3 downstream targets in DLD1 cells transfected with METTL3 overexpression plasmids, n = 3, nonparametric Mann–Whitney test. (g) Venn diagram showed 6 genes with consistently decreased levels in both HCT116 and SW480 cells with METTL3 knockout or knockdown and increased expression in DLD1 cells with METTL3 upregulated. (h) The relative abundance of m6A sites along SLC26A1, PC, GLCE and CHST12 mRNA in HCT116 WT and METTL3-KO cells, as detected by m6A-seq. (i) Agarose electrophoresis and MeRIP-qPCR assays in CRC cells showing the direct binding between the m6A antibody and SLC26A1, PC, GLCE and CHST12. (j) MeRIP-qPCR analysis of SLC26A1, PC, GLCE and CHST12 RNA m6A levels in HCT116 cells with METTL3 knockout, n = 3, nonparametric Mann–Whitney test. (k) Lactate production was detected in HCT116 cells after knockdown of HK2, SLC2A1, PC, GLCE and CHST12, respectively, n = 3, nonparametric Mann–Whitney test. (l) The relative abundance of m6A sites along HK2 and SLC2A1 (GLUT1) mRNA in CRC tissues with high or low METTL3 expression as detected by m6A-seq. The orange rectangles indicated that the m6A peaks had a significant decreased abundance. (WT, wild type; METTL3-KO, METTL3-knockout). Figure S5. METTL3 regulates HK2 and SLC2A1 (GLUT1) mRNA levels and stability depending on its m6A methyltransferase activity. (a) Real-time PCR and Western blot assay were performed to detect the levels of HK2 and SLC2A1 (GLUT1) in SW480 cells after transfection of control siRNA and METTL3 siRNA1/2, n = 3, nonparametric Mann–Whitney test. (b) MeRIP-qPCR analysis of HK2 and SLC2A1 m6A levels in DLD1 cells transfected with control, METTL3 overexpression and METTL3-mut plasmids, n = 3, nonparametric Mann–Whitney test. (c) Hexokinase activity was measured in SW480 cells transfected with METTL3 siRNA1/2, n = 3, nonparametric Mann–Whitney test. (d) Relative luciferase activity of WT or mutant HK2 5’UTR firefly luciferase reporter in SW480 cells treated with control siRNA or METTL3 siRNA1/2, n = 4, nonparametric Mann–Whitney test. (e) Relative luciferase activity of WT or mutant HK2 3’UTR firefly luciferase reporter in SW480 cells treated with control siRNA or METTL3 siRNA1/2, n = 4, nonparametric Mann–Whitney test. (f) Relative luciferase activity of WT or mutant SLC2A1 (GLUT1) 3’UTR firefly luciferase reporter in SW480 cells treated with control siRNA or METTL3 siRNA1/2, n = 4, nonparametric Mann–Whitney test. (g-h) The HK2 (g) and SLC2A1 (GLUT1) (h) mRNA half-life (t1/2) were detected by real-time PCR in SW480 cells transfected with control siRNA or METTL3 siRNA1/2, n = 3, nonparametric Mann–Whitney test. (i) The relative expression of HK2 mRNA was measured by real-time PCR in HCT116 and SW480 cells after transfection with control, IGF2BP1, IGF2BP2 or IGF2BP3 siRNA, respectively, n = 3, nonparametric Mann–Whitney test. (j) The relative expression of SLC2A1 (GLUT1) mRNA was measured by real-time PCR in HCT116 and SW480 cells after transfection with control, IGF2BP1, IGF2BP2 or IGF2BP3 siRNA, respectively, n = 3, nonparametric Mann–Whitney test. (k-l) The relative expression of HK2 (k) and SLC2A1 (GLUT1) (l) mRNA were measured by real-time PCR in HCT116 cells or SW480 cells after knockdown of YTHDF2, YTHDF3 or YTHDC2, respectively, n = 3, nonparametric Mann–Whitney test. Figure S6. HK2 and SLC2A1 (GLUT1) are functionally important target genes of METTL3 in CRC (a) The overexpression efficiency of HK2 was confirmed by real-time PCR and Western blot analysis. (b) The overexpression efficiency of GLUT1 was confirmed by real-time PCR and Western blot analysis. (c-e) Representative images (c), tumor volumes (d) and tumor weights (e) in nude mice bearing HCT116 WT cells treated with PBS and control adenovirus, n = 8, nonparametric Mann–Whitney test. (f) The downregulation efficiency of HK2 siRNA was confirmed by real-time PCR and Western blot analysis. (g) The downregulation efficiency of GLUT1 siRNA was confirmed by real-time PCR and Western blot analysis. (h-i) Cell proliferation assay (h) and colony formation assay (i) were performed in DLD1 cells with different treatment, n = 6, nonparametric Mann–Whitney test. (j-l) Representative images of tumors (j), statistical analysis of tumor volumes (k) and tumor weights (l) in nude mice bearing DLD1 cells in different groups (n = 8, nonparametric Mann–Whitney test). Fig. 3h, Fig. 3m and Figure S6j shared experimental controls and METLL3 overexpression group. FigureS7. METTL3 is not correlated with IGF2BP2/3. (a) Representative immunohistochemical images of METTL3, IGF2BP2 and IGF2BP3 in CRC tissues using IHC analysis in Cohort 2. Fig. 7a and Figure S7a shared representative immunohistochemical images of METTL3 in METTL3 high expression and METTL3 low expression groups. (b) Comparing age, gender, histological differentiation, AJCC stage and recurrence between METTL3/HK2/GLUT1 high and low expression tumors of Cohort 2. The heat map illustrated the association of different clinicopathological features with METTL3/HK2/ GLUT1 high and low expression, Chi-square test. (c) Statistical analysis of METTL3 expression in human colorectal cancer (n = 67), colorectal adenoma (n = 75) and normal colorectal tissues (n = 126) of Cohort 3.)
Additional file 4 Table S2. Pathways regulated by METTL3 in HCT116 cells.
Additional file 5 Table S3. Metabolites of glycolysis pathway measured by Liquid chromatography-coupled tandem mass spectrometry (LC-MS/MS) in HCT116 WT and METTL3-knockout cells.
Additional file 6 Table S4. m6A-seq analysis in HCT116 WT and METTL3-knockout cells.
Additional file 7 Table S5. Clinical information of 47 cases CRC patients in Cohort 1.
Additional file 8 Table S6. Clinical information of 156 cases CRC patients in Cohort 2.
Additional file 9 Table S7. Clinical information of 30 cases CRC patients in GSE110225.
Additional file 10 Table S8. The sequences of siRNAs and primers used this study.
Data Availability Statement
The raw sequencing data have been deposited in the Gene Expression Omnibus database under the accession number GSE130012. All the other data generated in this study are included in the article and the additional files.
More detailed materials and methods are in the Supplementary Methods.
The raw sequencing data have been deposited in the Gene Expression Omnibus database under the accession number GSE130012. All the other data generated in this study are included in the article and the additional files.