Abstract
MERS-CoV is the only lethal human CoV still endemic in the Arabian Peninsula and neither vaccine nor therapeutics against MERS-CoV infection is available. The nsp1 of CoV is thought to be a major virulence factor because it suppresses protein synthesis through the degradation of host mRNA. In contrast, viral RNA circumvents the nsp1-mediated translational shutoff for an efficient propagation. In this study, we identified amino acid residue in MERS-CoV nsp1 that differ from those of SARS-CoV nsp1, and that appear to be crucial for circumventing the translational shutoff. In addition, reverse genetics analysis suggested the presence of a cis-acting element at the 5′-terminus of the nsp1-coding region, which contributes to the specific recognition of viral RNA that is required for an efficient viral replication. Our results suggest the CoVs share a common mechanism for circumventing the nsp1-mediated translational shutoff.
Keywords: Coronavirus, MERS-CoV, Nsp1, RNA recognition, Viral replication
Highlights
-
•
MERS-CoV nsp1 interacts with Stem-loop 1 in 5′ UTR of viral RNA.
-
•
Arginine at position 13 of MERS-CoV nsp1 is critical for viral RNA recognition.
-
•
Nsp1-mediated Viral RNA recognition is important for efficient viral propagation.
-
•
Functional amino acids of nsp1 differ between SARS-CoV and MERS-CoV.
1. Introduction
Coronaviruses (CoVs) are pathogens that infect a large variety of vertebrate animals, resulting in mainly respiratory and enteric diseases (Weiss and Navas-Martin, 2005). Most human CoVs are causative agents of mild illness and common cold (Bradburne et al., 1967, van der Hoek et al., 2006, van der Hoek et al., 2004, Woo et al., 2005). However, an epidemic of severe acute respiratory syndrome (SARS) occurred in China in 2002, and the causative agent was designated as SARS-CoV (Drosten et al., 2003, Ksiazek et al., 2003). Ten years after the SARS outbreak, another highly pathogenic human CoV, designated as Middle East respiratory syndrome (MERS)-CoV, emerged in Saudi Arabia (Zaki et al., 2012). Many patients infected with MERS-CoV have been identified, most of them in the Arabian Peninsula, although MERS-CoV has also spread to several other countries in North Africa, Europe, and Asia (Bermingham et al., 2012, Buchholz et al., 2013, Cowling et al., 2015, Müller et al., 2014, Mailles et al., 2013, Perera et al., 2013, Reusken et al., 2014). In humans, MERS-CoV infection causes high fever, cough, and pneumonia, and is transmitted through close contact with infected dromedary camels (Memish et al., 2014, Reusken et al., 2013).
CoVs are enveloped viruses possessing a large single-stranded and positive-sense RNA genome (~ 32 kb). The 5′ two-thirds of the CoV genome consists of two overlapping open reading frames (ORFs 1a and 1b) that encode non-structural proteins (nsps). The other one-third of the genome consists of ORFs encoding structural proteins, including spike (S), membrane (M), envelope (E) and nucleocapsid (N) proteins, and accessory proteins (Woo et al., 2010). Upon infection of CoV into host cells, the translation of two precursor polyproteins, pp1a and pp1ab, occurs and these polyproteins are cleaved into 15 or 16 nsps by viral proteases, papain-like protease (PLpro: nsp3) and 3C-like protease (3CLpro: nsp5) (Prentice et al., 2004, Thiel et al., 2003). Alpha and beta CoVs possess 16 nsps, while gamma and delta CoVs, lacking nsp1, possess 15 nsps (nsp2 to nsp16) (Neuman et al., 2014). Although the amino acid sequences of nsp3 to nsp16 are predicted to be conserved among CoVs, those of nsp1 are highly divergent (Connor and Roper, 2007). However, the nsp1s of several CoVs, such as porcine transmissible gastroenteritis virus (TGEV), human CoV (HCoV)-229E, mouse hepatitis virus (MHV), and SARS-CoV, exhibit a similar function to induce translational suppression (Huang et al., 2011a, Kamitani et al., 2006, Wang et al., 2010, Züst et al., 2007). The nsp1 of SARS-CoV is the most studied among CoVs and is known to inhibit host gene expression via a two-pronged strategy (Kamitani et al., 2009)—i.e., translational shutoff through interaction with the 40 S ribosomal subunit, and host mRNA degradation through the recruitment of unidentified host nuclease(s) (Huang et al., 2011a, Kamitani et al., 2009, Lokugamage et al., 2012). This two-pronged strategy of nsp1 inhibits expression of the IFN gene (Kamitani et al., 2006, Narayanan et al., 2008, Wathelet et al., 2007). Murine models of SARS-CoV have revealed that the dysregulated type I IFN response is a key factor for inducing lethal pneumonia (Channappanavar et al., 2016, Kindler and Thiel, 2016). In addition, Züst and colleagues showed that a mutant MHV with partial deletion in nsp1 is highly attenuated (Züst et al., 2007). These accumulated data indicate that the nsp1 of CoV is a major virulence factor.
Although viral mRNAs of CoVs are capped and polyadenylated (Cencic et al., 2011, Chen et al., 2011, Decroly et al., 2011, Yount et al., 2000) like host mRNA, the viral mRNAs are resistant to the translational suppression induced by the expression of nsp1. Like many CoV nsp1s, expression of MERS-CoV nsp1 suppresses host protein synthesis (Lokugamage et al., 2015). The nsp1 of MERS-CoV targets nuclear transcribed host mRNAs for suppression, but mRNAs of cytoplasmic origin are resistant to the nsp1-meditated gene suppression (Lokugamage et al., 2015). However, the mechanisms of the interaction between nsp1 and viral RNA of MERS-CoV and the roles of this interaction on the viral replication are largely unknown.
In this study, we examined the biological significance of the interaction of viral RNA with the nsp1 of MERS-CoV. As we expected, MERS-CoV nsp1 also induced translational suppression and RNA degradation. Like SARS-CoV, MERS-CoV circumvents the nsp1-mediated translational suppression by inducing a specific interaction between SL1 in the 5′ UTR of viral RNA and nsp1. Interestingly, the amino acid residue of MERS-CoV nsp1 required for viral RNA recognition appeared to differ from those of SARS-CoV nsp1. In addition, reverse genetics analysis of MERS-CoV revealed that the specific interaction of nsp1 with SL1 is crucial for the efficient replication of MERS-CoV. These results indicate CoVs share a common mechanism of viral gene expression that is regulated by a specific interaction between SL1 in the 5′ UTR of viral RNA and nsp1.
2. Materials and methods
2.1. Cells
293 T cells (human embryonic kidney) and Huh7 cells (human hepatocellular carcinoma) were maintained in Dulbecco's modified minimum essential medium (DMEM) (Nacalai Tesque, Kyoto, Japan) containing 10% heat-inactivated fetal bovine serum (FBS), 100 U/ml penicillin, and 100 µg/ml streptomycin (Nacalai Tesque). Vero cells (African green monkey kidney) were maintained in DMEM containing 5% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin (Nacalai Tesque). All cells were cultured in a humidified 5% CO2 atmosphere at 37 °C.
2.2. Plasmid constructions
The construction of a firefly luciferase-expressing plasmid, pcD-fLuc, has been described elsewhere (Tanaka et al., 2012). The PCR products of the CAT gene were cloned into pCAGGS-MCS-FLAG, pCAG-CAT. Using the supernatants of Vero cells infected with the MERS-CoV (EMC2012 Strain), first-strand cDNA was prepared by using a SuperScript II First-Strand Synthesis Kit (ThermoFisher Scientific, Waltham, MA) according to the manufacturer's instructions. The cDNA was used for construction of the expression plasmids. The PCR products of the MERS-CoV nsp1 sequence were cloned into pCAGGS-MCS-FLAG, yielding pCAG-nsp1-wt. An inverse PCR procedure using pCAG-nsp1 as the template was employed to generate pCAG-nsp1-Δ1, pCAG-nsp1-Δ2, pCAG-nsp1-Δ3, pCAG-nsp1-G11A, pCAG-nsp1-R13A, pCAG-nsp1-G14A, and pCAG-nsp1-T15A by using a KOD-Plus-Mutagenesis Kit (TOYOBO, Osaka, Japan). The In-fusion HD cloning procedure (Clontech, Mountain View, CA) was employed to generate pCAG-nsp1-Δ4 according to the manufacturer's instructions. The 5′ UTR sequence of MERS-CoV was connected downstream of the cytomegalovirus (CMV) promoter by overlapping PCR using the same method as described previously (Tanaka et al., 2012, Yamshchikov et al., 2001). The fragment was cloned between the CMV promoter and firefly luciferase gene into pcD-fluc, yielding pcD-5′-fluc. For mutational analysis of the nucleotide sequence from position 1–190 in the MERS-CoV 5′ UTR, the inverse PCR procedure using pcD-5′-fluc was employed to generate pcD-ΔSL1-fluc, pcD-ΔSL2-fluc, pcD-ΔSL4-1-fluc, pcD-ΔSL4-2-fluc, and pcD-SL1-fluc by using a KOD Mutagenesis Kit (TOYOBO). All plasmid constructs were confirmed by sequence analysis using a BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA).
2.3. Luciferase assay
293T cells were transfected with reporter and expression plasmids by using TransIT LT1 (Mirus, Madison, WI) according to the manufacturer's instructions. At 24 h posttransfection, transfected cells were collected and luciferase activities were determined by using a luciferase assay system (Promega, Madison, WI) and AB-2200 luminometer (Atto, Tokyo, Japan). Luciferase activities were shown after standardization with those in cells expressing CAT.
2.4. Northern blot analysis
Intracellular RNA of transfected 293T cells was extracted using a PureLink RNA Mini Kit (ThermoFisher Scientific) according to the manufacturer's instructions and stored at −80 °C until use. RNA samples were diluted to 1 µg in 5 µl by UltraPure DW (Invitrogen, Waltham, MA) and then mixed with 5 µl of 2× Loading Dye (New England Biolabs, Ipswich, MA). After heating at 65 °C for 5 min, 10 µl RNA samples were electrophoresed through 1.2% denaturing agarose gel and then transferred onto a positively charged nylon membrane (Roche, Basel, Switzerland). Northern blot analysis was performed using a digoxigenin (DIG) Wash and Block Buffer Set and a DIG Luminescence Detection Kit (Roche). The DIG-labeled riboprobe to detect the firefly luciferase gene was generated by using a DIG RNA Labeling Kit (SP6/T7) (Roche) as described previously (Kamitani et al., 2006, Tanaka et al., 2012).
2.5. Western blot analysis
Transfected 293T cells were lysed using RIPA buffer (25 mM Tris-HCl (pH 7.6), 150 mM NaCl, 1% NP40, 1% sodium deoxicholate, 0.1% sodium dodecyl sulfate (SDS) and 2.5U benzonase) and then centrifuged at 16,000×g for 10 min at 4 °C. Supernatants were collected and mixed with 2× sample buffer (0.1 M Tris-HCl (pH6.8), 4% SDS, 20% glycerol, 0.004% bromophenol blue and 10% 2-mercaptoethanol). Ten microliters of boiled samples was electrophoresed by SDS-poly acrylamide gel electrophoresis (PAGE). Electrophoresed gels were transferred onto a polyvinylidene difluoride (PVDF) membrane (Merck Millipore, Billerica, MA). The transferred membranes were blocked by 3% skim milk in phosphate-buffered saline (PBS) containing 0.05% tween 20 (Nacalai Tesque) (PBS-T). Anti-DYKDDDDK (FLAG) mouse antibody (Wako, Osaka, Japan) or Anti-β-Actin mouse antibody (Sigma, St. Louis, MO) was used as a primary antibody, and goat anti-mouse IgG-horseradish peroxidase (HRP) (Sigma) was used as a secondary antibody. ChemiLumi One Ultra (Nacalai Tesque) was used for visualization.
2.6. Immunoprecipitation of reporter RNA
At 24 h posttransfection, the 293T cells were lysed with lysis buffer (1% triton X-100, 0.5% sodium deoxycholate, 0.1% SDS in PBS) containing 40U RNase inhibitor (Takara, Shiga, Japan) and protease inhibitor (cOmplete, EDTA-free; Roche). After centrifugation, the supernatants of the cell lysates were collected and mixed with protein A/G Plus-Agarose (Santa Cruz Biotechnology, Santa Cruz, CA), then incubated for 30 min at 4 °C. After centrifugation, the supernatants were collected and incubated with anti-FLAG mouse antibody for 1 h at 4 °C. After incubation, the samples were mixed with protein A/G Plus-Agarose and incubated for 1 h at 4 °C. The bead pellets were washed four times with lysis buffer, and the precipitated RNAs were extracted from the agarose beads by using Sepazol-RNA I Super G reagent (Nacalai Tesque). After ethanol precipitation, the RNA pellets were suspended in 10 µl RNA loading dye (New England Biolabs). The RNAs were subjected to Northern blot analysis as described above.
2.7. BAC constructions
In this study, we employed a reverse genetics system for MERS-CoV by using a bacterial artificial chromosome (BAC) system. The BAC clone carrying a full-length infectious genome of the MERS-CoV EMC2012 strain, pBAC-MERS-wt, was generated according to a previous report (Almazán et al., 2013). The BAC DNA of SARS-CoV-Rep (Almazán et al., 2006), which was kindly provided by Luis Enjuanes, was used as a backbone BAC sequence to generate pBAC-MERS-wt. The BAC infectious clones carrying amino acid substitutions in nsp1 were generated by modification of the pBAC-MERS-wt as a template using a Red/ET Recombination System Counter-Selection BAC Modification Kit (Gene Bridges, Heidelberg, Germany), yielding pBAC-MERS-R13A and pBAC-MERS-A9G/R13A. The sequences of the two introduced mutations were confirmed as described above.
2.8. Recovery of recombinant MERS-CoV from the BAC plasmids
Huh7 cells were grown to approx. 60% confluence on a 6-well plate (VIOLAMO, Osaka, Japan) and then transfected with 4 µg of the indicated BAC DNA using X-tremeGENE 9 DNA Transfection Reagent (Roche). After transfection, the transfected cells were cultured at 37 °C for the indicated durations, and then the culture supernatants and cell pellets were collected. The culture supernatants were used for determination of the infectious viral titers. Intracellular RNAs were extracted by using a PureLink RNA Mini Kit (Thermo Fisher Scientific) and subjected to real-time RT-PCR as described below. Recovered viruses were stored at −80 °C as P0 viruses.
2.9. Virus propagation
Vero cells were seeded onto a 10 cm dish (VIOLAMO) and cultured overnight. After washing with DMEM without FBS, 1.5 ml of P0 viruses was inoculated onto Vero cells with 3.5 ml DMEM without FBS. After incubation at 37 °C for 1 h, the dishes were washed twice with DMEM, and then 10 ml DMEM containing 2% FBS was added. Infected cells were incubated at 37 °C for 3 or 4 days until a cytopathic effect (CPE) was observed. Culture supernatants were collected and then centrifuged at 2500×g for 5 min at 4 °C. The supernatants were collected and stored at −80 °C as P1 viruses.
2.10. Growth kinetics
Vero cells or Huh7 cells were seeded onto 24-well plates (VIOLAMO) and cultured overnight. P1 viruses were diluted using DMEM without FBS to an MOI of 0.001. After washing the cells using DMEM, the diluted viruses were inoculated onto the cells and incubated at 37 °C for 1 h. After incubation, the infected cells were washed twice using DMEM and supplemented with 0.5 ml/well DMEM containing 2% FBS. The cells were then incubated at 37 °C for the indicated amounts of time. Supernatants and cell pellets were collected to investigate the virus titers and levels of subgenomic N mRNA, respectively.
2.11. Titration and Plaque assay
The 50% tissue culture infectious dose (TCID50) method was adopted to determine the infectious titer of each virus. Briefly, Vero cells were seeded onto 96-well plates (VIOLAMO) and cultured overnight at 37 °C. Viruses were diluted by 10-fold serial dilution from 10 times dilution using DMEM without FBS. The diluted viruses were inoculated into Vero cells and incubated at 37 °C for 4 days. After incubation, the infected cells were fixed with buffered formaldehyde (Nacalai Tesque) and stained with crystal violet. The TCID50 was calculated by the Spearman and Karber algorithm. All work with infectious MERS-CoV was performed at a biosafety level 3 facility at Osaka University. We carried out the plaque assay according to the ordinary method. Briefly, 1.5 × 105 cells/ml Vero cells were seeded onto 6-well plate (VIOLAMO) and cultured at 37 °C overnight. Then, Vero cells were washed using DMEM. Recombinant MERS-CoVs were diluted to 1.25 × 102 TCID50/ml with DMEM containing 2% FBS. 400 µl/well diluted viruses were inoculated onto Vero cell monolayer. After adsorption at 37 °C for 1 h, cells were washed twice using DMEM and then overlaid 0.8% agarose (Seaplaque® GTG Agarose; Lonza, Switzerland) in DMEM containing 10% FBS. Infected cells were cultured at 37 °C for 3 days and then fixed by phosphate-buffered formalin. The fixed cells were stained with crystal violet.
2.12. Real-time RT-PCR
Total RNA was prepared from cells by using a PureLink RNA Mini Kit (Thermo Fisher Scientific), and first-strand cDNA was synthesized using a ReverTra Ace qPCR RT kit (TOYOBO) according to the manufacturer's instructions. The level of each cDNA was determined by using Thunderbird Probe qPCR Mix (TOYOBO), and fluorescent signals were analyzed by using a CFX Connect Real-Time PCR Detection System (BioRad, Hercules, CA). For the quantification of subgenomic N mRNA, we used wk1350 (5′- TCGTTCTCTTGCAGAACTTTG -3′) and wk1351 (5′- TTGGATTACGTCCTCTACCTC -3′) as primers and wk1352 (5′- CCTCGTGCTGTTTCCTTTGCCGAT -3′) labeled with FAM as a probe. For quantification of GAPDH mRNA, wk1288 (5′- GAAGGTGAAGGTCGGAGT -3′) and wk1289 (5′- GAAGATGGTGATGGGATTTC -3′) were used as primers and FAM-labeled wk1290 (5′- CAAGCTTCCCGTTCTCAGCC -3′) was used as a probe. Reactions were carried out at 95 °C for 1 min, followed by 40 cycles of 95 °C for 15 s and 58 °C for 1 min.
3. Results
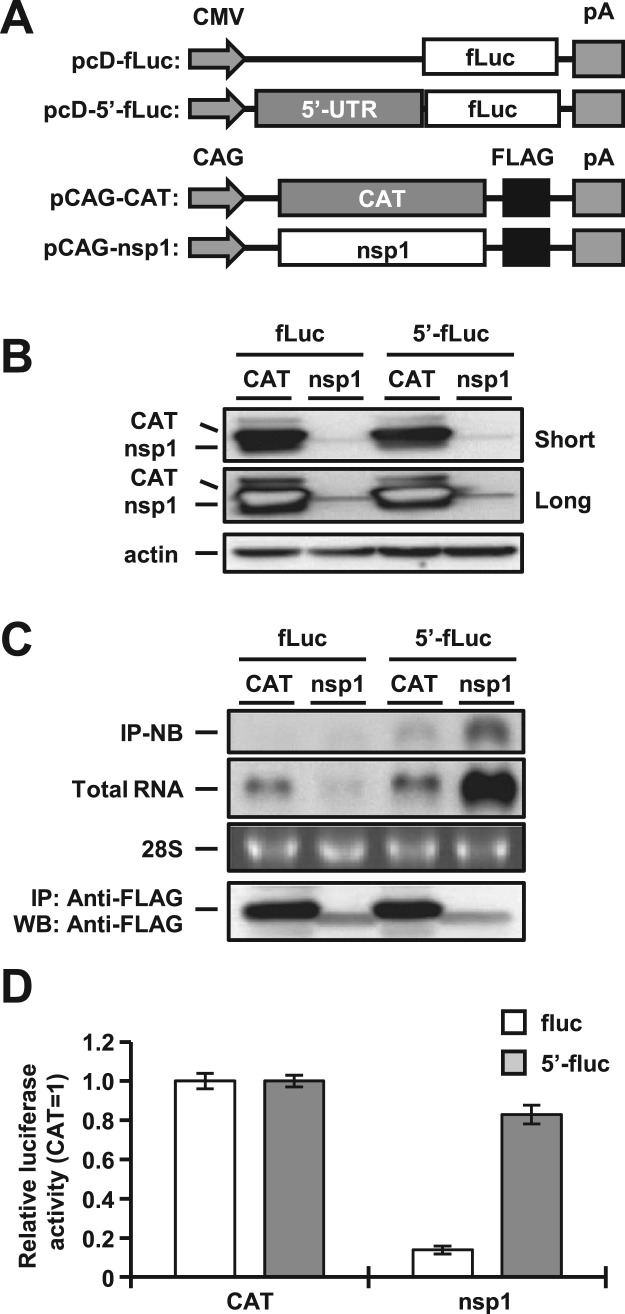
3.1. 5′ UTR of MERS-CoV RNA is required for circumvention of the translational suppression by nsp1
In previous reports, the interaction of nsp1 with the 5′ UTR of SARS-CoV was shown to be important for circumvention of the translational suppression by nsp1 (Tanaka et al., 2012). To determine the role of this interaction in another human pathogenic CoV, MERS-CoV, we constructed two reporter plasmids carrying a firefly luciferase with or without the 5′ UTR of MERS-CoV (pcD-5′-fluc or pcD-fluc) and two expression plasmids, pCAG-nsp1 and pCAG-CAT, carrying a FLAG tag in the C-termini of MERS-CoV nsp1 and chloramphenicol acetyltransferase (CAT), respectively ( Fig. 1A). 293T cells transfected with either pcD-fluc or pcD-5′-fluc together with pCAG-CAT or pCAG-nsp1 were harvested at 24 h posttransfection, and subjected to Western blot analysis. The expression level of MERS-CoV nsp1 was significantly lower than that of CAT protein (Fig. 1B). Cells transfected with the plasmids were harvested at 24 h posttransfection and subjected to immunoprecipitation analysis using anti-FLAG antibody. Total RNAs extracted from the immunoprecipitates were subjected to Northern blot analysis by using a specific probe for the luciferase gene (Fig. 1C, top panel). The luciferase RNAs were co-precipitated with nsp1, but not with CAT, in cells co-transfected with pcD-5′-fluc but not in those co-transfected with pcD-fluc (Fig. 1C, top panel), suggesting that MERS-CoV nsp1 specifically interacts with viral 5′ UTR. Although the expression of luciferase mRNA was impaired in cells co-transfected with pcD-fluc, it was significantly increased in those transfected with pcD-5′-fluc (Fig. 1C, second panel), as reported in SARS-CoV (Tanaka et al., 2012), suggesting that MERS-CoV nsp1 also induces mRNA degradation as reported in SARS-CoV (Tanaka et al., 2012). Although the precise mechanism of the increase of mRNA carrying the 5′ UTR in the presence of nsp1 of either MERS-CoV or SARS-CoV is unclear, the interaction between nsp1 and viral 5′ UTR might contribute to the stabilization of RNA.
Fig. 1.
MERS-CoV nsp1 binds to the 5′ UTR of MERS-CoV to facilitate evasion of nsp1-mediated shutoff and RNA degradation. (A) Schematic diagrams of the reporter plasmids carrying the firefly luciferase (fLuc) gene with or without the 5′ UTR (5′-fLuc) of MERS-CoV under the control of the CMV promoter (CMV) and of the expression plasmids carrying the nsp1 or chloramphenicol acetyltransferase (CAT) gene under the control of the CAG promoter (CAG). pA represents the polyadenylation signal. (B) Lysates of 293T cells transfected with either pCAG-CAT or pCAG-nsp1 together with the indicated reporter plasmids were subjected to Western blot analysis using anti-FLAG and anti-actin antibodies. Short and long represent short exposure and long exposure, respectively. (C) Lysates of 293T cells transfected with either pCAG-CAT or pCAG-nsp1 together with the indicated reporter plasmids were immunoprecipitated with anti-FLAG antibody at 24 h posttransfection. RNAs extracted from the precipitates were subjected to Northern blot analysis using a riboprobe for the luciferase gene (top panel). The second panel represents the amount of intracellular reporter RNAs in the lysate. 28S rRNA was stained with ethidium bromide (third panel). Immunoprecipitated CAT or nsp1 proteins were detected by Western blot analysis using anti-FLAG antibody (bottom panel). (D) Luciferase activities in 293T cells transfected as described for panel B were determined at 24 h posttransfection after standardization with those in cells expressing CAT. The values represent the means ±SD from three independent experiments.
Next, to determine the effect of the 5′ UTR of MERS-CoV on nsp1-mediated translational shutoff, cells transfected with the plasmids were harvested at 24 h posttransfection and subjected to luciferase assay (Fig. 1D). Consistent with previous reports (Lokugamage et al., 2015), the expression of nsp1 of MERS-CoV, but not of CAT, suppressed luciferase expression in the cells transfected with pcD-fluc (Fig. 1D). However, the expression of nsp1 of MERS-CoV failed to suppress the luciferase expression in cells transfected with pcD-5′-fluc (Fig. 1D), suggesting that MERS-CoV nsp1 also participates in circumvention of the nsp1-mediated translational shutoff through the interaction with viral 5′ UTR, as seen in SARS-CoV. In addition, pCAG-nsp1 possesses no nucleotide sequence of viral 5′ UTR upstream of nsp1 gene (Fig. 1A). Therefore, it was thought that low level of nsp1 expression was due to its own translational shutoff (Fig. 1B).
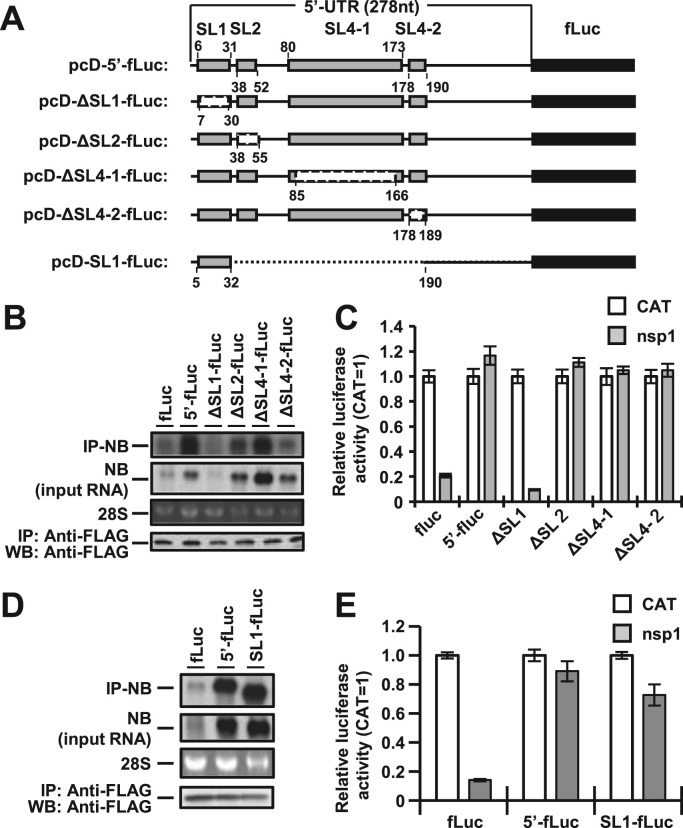
3.2. Interaction between SL1 in the 5′ UTR of MERS-CoV and nsp1 participates in circumvention of the nsp1-mediated shutoff
The 5′ UTR of MERS-CoV genomic RNA possesses four stem-loop (SL) structures, SL1, SL2, SL4-1 and SL4-2 (Yang and Leibowitz, 2015). To determine the region(s) in the 5′ UTR responsible for the interaction with nsp1, we constructed a series of SL deletion mutants based on pcD-5′-fluc as shown in Fig. 2A. Cells transfected with pCAG-nsp1 together with each of the SL deletion mutants were harvested at 24 h posttransfection and subjected to immunoprecipitation assay using anti-FLAG antibody. Total RNAs extracted from the precipitates were subjected to Northern blot analysis. The ΔSL2-fluc, ΔSL4-1-fluc, and ΔSL4-2-fluc RNAs, but not ΔSL1-fluc RNA, were co-immunoprecipitated with nsp1 (Fig. 2B, top panel), suggesting that the SL1 in the 5′ UTR is responsible for the interaction with nsp1. In addition, Northern blot analysis of the total RNA showed that the amounts of ΔSL1-fluc RNA and fluc RNA were low in cells expressing nsp1, suggesting that the specific interaction of nsp1 with SL1 in the 5′ UTR is critical for escape from the nsp1-induced RNA degradation (Fig. 2B, second panel). To confirm this specific interaction between nsp1 and SL1 in the 5′ UTR of MERS-CoV, we constructed a pcD-SL1-fluc plasmid, containing only SL1 in the region upstream of the luciferase gene (Fig. 2A). As we expected, the SL1-fluc RNA was co-immunoprecipitated with nsp1 in cells co-transfected with pcD-SL1-fluc and pCAG-nsp1 (Fig. 2D, top panel). In addition, no RNA degradation of the SL1-fluc was observed in cells transfected with pcD-SL1-fluc and pCAG-nsp1 (Fig. 2D, second panel), indicating that the specific interaction of nsp1 with SL1 in the 5′ UTR of MERS-CoV is critical for evading the nsp1-induced RNA degradation.
Fig. 2.
Binding to SL1 in the 5′ UTR of MERS-CoV by nsp1 is important for escape from nsp1-mediated shutoff. (A) Schematic diagrams of deletion mutants derived from the pcD-5′-fluc plasmid. Gray boxes, white boxes and dashed lines, and black boxes represent stem-loops (SL1, SL2, SL4-1, and SL4-2), deletions, and the firefly luciferase gene (fLuc), respectively. The numbers indicate nucleotide positions. (B) Lysates of 293T cells transfected with pCAG-nsp1 together with the indicated reporter plasmids were immunoprecipitated with anti-FLAG antibody at 24 h posttransfection. RNAs extracted from the precipitates were subjected to Northern blot analysis using a riboprobe for the luciferase gene (IP-NB). The second panel represent the amounts of intracellular reporter RNAs in the lysates. 28S rRNA was stained with ethidium bromide (28S). Immunoprecipitated nsp1 was detected by Western blot analysis using anti-FLAG antibody (bottom panel). (C) Luciferase activities in 293T cells transfected with either pCAG-CAT or pCAG-nsp1 together with the indicated reporter plasmids were determined at 24 h posttransfection after standardization with those in cells expressing CAT. The values represent the means ± SD from three independent experiments. (D) Lysates of 293T cells transfected with pCAG-nsp1 together with the indicated reporter plasmids were immunoprecipitated with anti-FLAG antibody at 24 h posttransfection. RNAs extracted from the precipitates were subjected to Northern blot analysis using a riboprobe for the luciferase gene (IP-NB). The second panel represent the amounts of intracellular reporter RNAs in the lysates. 28S rRNA was stained with ethidium bromide (28S). Immunoprecipitated nsp1 was detected by Western blot analysis using anti-FLAG antibody (bottom panel). (E) Luciferase activities in 293T cells transfected with either pCAG-CAT or pCAG-nsp1 together with the indicated reporter plasmids were determined at 24 h posttransfection after standardization with those in cells expressing CAT. The values represent the means ± SD from three independent experiments.
Next, to determine the effect of the SL region(s) on the nsp1-mediated translational suppression, luciferase expression in cells transfected with pCAG-nsp1 together with each of the SL deletion mutant plasmids was determined at 24 h posttransfection. The expression of nsp1 suppressed the luciferase activities in cells transfected with pcD-ΔSL1-fluc or pcD-fluc but not in those transfected with pcD-ΔSL2-fluc, pcD-ΔSL4-1-fluc or pcD-ΔSL4-2-fluc (Fig. 2C). In addition, the expression of nsp1 failed to suppress the luciferase activities in cells transfected with either pcD-5′-fluc or pcD-SL1-fluc (Fig. 2E). These results suggest that the specific interaction between SL1 in the 5′ UTR of MERS-CoV and nsp1 is crucial for circumventing the nsp1-mediated translational shutoff.
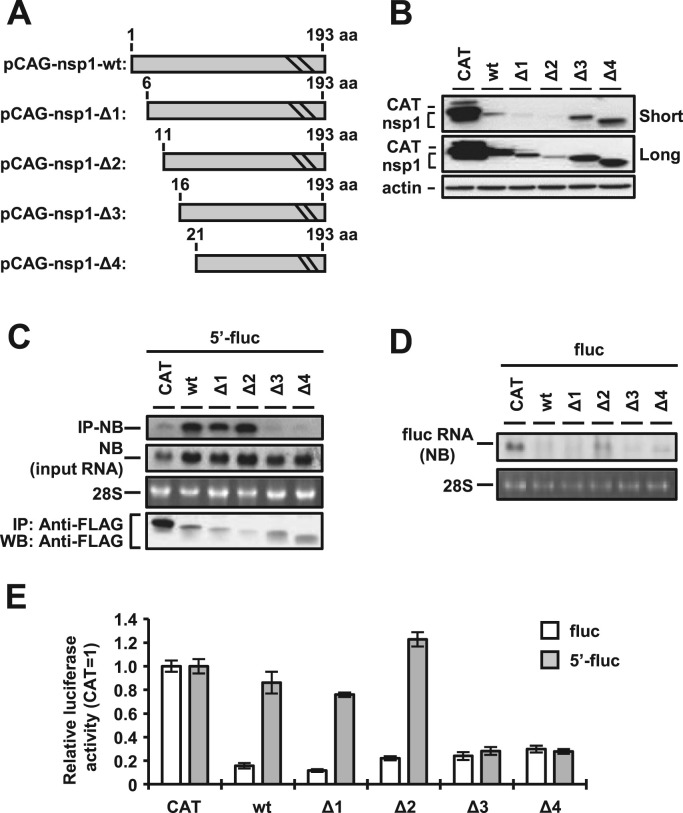
3.3. Interaction of the 5′ UTR of MERS-CoV with amino acid residues 11–15 in nsp1 participates in circumvention of the nsp1-mediated shutoff
To determine the binding region in MERS-CoV nsp1 responsible for the interaction with viral 5′ UTR, a series of N-terminal deletion nsp1 mutants based on pCAG-nsp1 were generated as shown in Fig. 3A. The name and deleted amino acid regions were as follows: pCAG-nsp1-Δ1, amino acid positions (aa) 1–5; pCAG-nsp1-Δ2, aa 1–10; pCAG-nsp1-Δ3, aa 1–15; pCAG-nsp1-Δ4, aa 1–20. First, we determined the effect of deletion on the protein expression levels. Although the expression levels of nsp1-Δ1 and nsp1-Δ2 were low, those of nsp1-Δ3 and nsp1-Δ4 were comparable to that of the nsp1-wildtype (wt) (Fig. 3B). Next, cells transfected with pcD-5′-fluc together with each of the deletion mutants were immunoprecipitated with anti-FLAG antibody at 24 h posttransfection, and total RNAs extracted from the precipitates were subjected to Northern blot analysis. Although the total amounts of the luciferase RNAs carrying the 5′ UTR in cells transfected with the nsp1 deletion mutants were comparable to those with in cells transfected with the nsp1-wt (Fig. 3C, second panel), the luciferase RNAs were efficiently co-precipitated with nsp1-wt, nsp1-Δ1 or nsp1-Δ2 but not with nsp1-Δ3 or nsp1-Δ4 (Fig. 3C, top panel), suggesting that the low levels of co-precipitation of the luciferase RNA carrying the 5′ UTR in cells co-transfected with nsp1-Δ3 or nsp1-Δ4 were not due to the low levels of intracellular expression of the RNA. In addition, the amounts of RNAs without the 5′ UTR of MERS-CoV were low in cells co-transfected with all nsp1 mutants or the nsp1-wt, suggesting that all of the nsp1 mutants retain the ability to degrade RNA (Fig. 3D). These data indicate that the N-terminal region (aa 11–15) of nsp1 participates in the interaction with the 5′ UTR of MERS-CoV.
Fig. 3.
N-terminal region of nsp1 binds to the 5′ UTR of MERS-CoV. (A) Schematic diagrams of deletion mutants derived from the pCAG-nsp1 plasmid. The numbers indicate amino acid positions. (B) Lysates of 293T cells transfected with the indicated deletion nsp1 plasmid were subjected to Western blot analysis using anti-FLAG and anti-actin antibodies. Short and Long represent short exposure and long exposure, respectively. (C) Lysates of 293T cells transfected with pcD-5′-fluc together with the indicated deletion nsp1 plasmids were immunoprecipitated with anti-FLAG antibody at 24 h posttransfection. RNAs extracted from the precipitates were subjected to Northern blot analysis using a riboprobe for the luciferase gene (top panel). The second panel represents the amount of intracellular reporter RNAs in the lysate. 28S rRNA was stained with ethidium bromide (third panel). Immunoprecipitated CAT or nsp1 proteins were detected by Western blot analysis using anti-FLAG antibody (bottom panel). (D) 293T cells were transfected with pcD-fluc together with the indicated deletion nsp1 plasmids. At 24 h posttransfection, total RNAs were extracted, and Northern blot analysis was performed by using a riboprobe for the luciferase gene (top panel). 28S rRNA was stained with ethidium bromide (bottom panel). (E) Luciferase activities in 293T cells transfected with either pcD-fluc or pcD-5′-fluc together with the indicated deletion nsp1 plasmids were determined at 24 h posttransfection after standardization with those in cells expressing CAT. The values represent the means ±SD from three independent experiments.
To further confirm the effect of the N-terminal region (aa 11–15) on the evasion from the nsp1-mediated translational suppression, cells transfected with either pcD-fluc or pcD-5′-fluc together with the deletion nsp1 mutants were subjected to luciferase assay at 24 h posttransfection. The expression of the nsp1-wt and each of the mutants of nsp1, but not of CAT, suppressed the luciferase expression of pcD-fluc, suggesting that all of the nsp1 mutants retained the ability to suppress the translation of mRNA lacking the 5′ UTR of MERS-CoV (Fig. 3E). In contrast, the translational suppression of mRNA carrying 5′ UTR was abrogated by the expression of nsp1-wt, nsp1-Δ1, and nsp1-Δ2 but not by the expression of nsp1-Δ3 and nsp1-Δ4 (Fig. 3E). These results suggest that the specific interaction of the 5′ UTR of MERS-CoV with nsp1 through the N-terminal regions (aa 11–15) is required for evasion from the nsp1-mediated shutoff.
3.4. Arginine at position 13 of MERS-CoV nsp1 is critical for the recognition of viral RNA
Next, to determine the critical amino acid residue in MERS-CoV nsp1 responsible for recognition of the 5′ UTR, we constructed four alanine substitution mutants of MERS-CoV nsp1—i.e., pCAG-nsp1-G11A, pCAG-nsp1-R13A, pCAG-nsp1-G14A, and pCAG-nsp1-T15A—as shown in Fig. 4A. The expression levels of all nsp1 mutants were lower than that of CAT control and nsp1-R13A showed slightly higher amount than other nsp1 mutants and nsp1-wt (Fig. 4B). Cells transfected with each of the alanine mutants together with pcD-5′-fluc were immunoprecipitated with anti-FLAG antibody at 24 h posttransfection, and total RNAs extracted from the precipitates were subjected to Northern blot analysis. The luciferase RNAs containing the 5′ UTR of MERS-CoV were efficiently co-precipitated with nsp1-wt, nsp1-G11A, nsp1-G14A, and nsp1-T15A but not with nsp1-R13A (Fig. 4C, top panel). The total amounts of the luciferase RNAs carrying the 5′ UTR in cells transfected with the alanine substitution mutants were almost comparable to that with in cells transfected with the nsp1-wt (Fig. 4C, second panel). However, nsp1-R13A expressing cells showed a little lower amount of RNA than other nsp1 mutants or nsp1-wt expressing cells (Fig. 4C, second panel). In compared with the amount of input RNA from 293T cells co-transfected with pcD-5′-fluc and pCAG-nsp1-R13A, that of co-precipitated RNA was apparently low (Fig. 4C, top and second panel). It indicated that the amino acid residue R13 participates in the binding to the 5′ UTR of MERS-CoV (Fig. 4C, top panel). In addition, the amounts of RNA without the 5′ UTR of MERS-CoV were low in cells co-transfected with the nsp1-wt or all nsp1 mutants including nsp1-R13A, suggesting that all of the nsp1 mutants retain the ability to degrade RNA (Fig. 4D). These results suggest that R13 in MERS-CoV nsp1 plays a critical role in evasion from the nsp1-mediated RNA degradation through a specific recognition of viral RNA.
Fig. 4.
Arginine at position 13 plays a critical role in the binding with 5′ UTR. (A) The sequence from amino acids 10–16 of the wt and each alanine substitution mutant of MERS-CoV nsp1. (B) Lysates of 293T cells transfected with the indicated nsp1 plasmid were subjected to Western blot analysis using anti-FLAG (top panel) and anti-actin (bottom panel) antibodies. (C) Lysates of 293T cells transfected with pcD-5′-fluc together with the indicated nsp1 plasmids were immunoprecipitated with anti-FLAG antibody at 24 h posttransfection. RNAs extracted from the precipitates were subjected to Northern blot analysis using a riboprobe for the luciferase gene (top panel). The second panel represents the amount of intracellular reporter RNAs in the lysate. 28S rRNA was stained with ethidium bromide (third panel). Immunoprecipitated CAT or nsp1 proteins were detected by Western blot analysis using anti-FLAG antibody (bottom panel). (D) 293T cells were transfected with pcD-fluc together with the indicated nsp1 plasmids. At 24 h posttransfection, total RNAs were extracted, and Northern blot analysis was performed by using a riboprobe for the luciferase gene (top panel). 28S rRNA was stained with ethidium bromide (bottom panel). (E) Luciferase activities in 293T cells transfected with either pcD-fluc or pcD-5′-fluc together with the indicated nsp1 plasmids were determined at 24 h posttransfection after standardization with those in cells expressing CAT. The values represent the means ±SD from three independent experiments.
To further confirm the role of R13 in MERS-CoV nsp1 on the translational shutoff, the luciferase activities in cells transfected with either pCAG-CAT, pCAG-nsp1-wt, pCAG-nsp1-G11A, pCAG-nsp1-R13A, pCAG-nsp1-G14A, or pCAG-nsp1-T15A together with pcD-fluc or pcD-5′-fluc were determined at 24 h posttransfection. Luciferase activities in cells transfected with pcD-fluc exhibited a moderate reduction by the expression of nsp1-R13A compared to those expressing other mutants or the wt of nsp1, and those with pcD-5′-fluc showed no reduction by the expression of the mutants or the wt of nsp1except for nsp1-R13A (Fig. 4E). These data looked inconsistent with the translational suppression in cells expressing nsp1-Δ3 or nsp1-Δ4, as shown in Fig. 3E. However, we found the mutant which deleted 5 amino acids at C-terminus completely lost its translational shutoff activity (Supplemental Fig. 1). In compared with this mutant, nsp1-R13A mutant still retained its shutoff activity (Fig. 4E). Although the activity of translational shutoff of nsp1-R13A mutant was a little weaker than that of nsp1-wt (Fig. 4E), these data indicated that R13A mutation made nsp1 critically lost its activity to bind with 5′ UTR but retained translational shutoff activity. This contradiction might be attributable to the structure of nsp1 surrounding R13 facilitating the binding activity and/or translational suppression. In fact, the predicted secondary structure of MERS-CoV nsp1 showed that the amino acid region from 13 to 20 constitutes an alpha helix structure, as shown in Fig. 7A, suggesting that the specific interaction of MERS-CoV nsp1 with the 5′ UTR through R13 is critical for circumvention of the nsp1-mediated shutoff.
Fig. 7.
Alignment of the amino acid sequences of MERS-CoV and SARS-CoV nsp1 and their predicted secondary structures. (A) Nsp1 sequences of the MERS-CoV strain EMC2012 (accession no.: YP_009047229) and SARS-CoV strain Urbani (accession no.: AAP13442) were aligned using MEGA6 software (Tamura et al., 2013). Completely conserved residues are shown with an asterisk. The numbers beside the aligned sequences indicate the positions of amino acid residues. Residues indicated by a red arrow represent the functional amino acids of viral RNA recognition. Residues shown in a blue box with white characters represent the functional amino acids of RNA cleavage. Residues shown by red circles represent the conserved region in betacoronavirus nsp1. Residues shown by a green box with white characters represent the functional amino acids of translational shutoff. The secondary structures of MERS-CoV and SARS-CoV nsp1 are predicted using PSIPRED v3.3 (Buchan et al., 2013). Arrows with dashed lines represent the predicted alpha-helix structures. Black arrows represent the predicted beta-strand structures. (B) Functional amino acid residues of MERS-CoV and SARS-CoV nsp1. (a): Tanaka et al., 2012 (Tanaka et al., 2012). (b): Lokugamage et al., 2015 (Lokugamage et al., 2015). (c): Lokugamage et al., 2012 (Lokugamage et al., 2012). (d): Narayanan et al., 2008 (Narayanan et al., 2008). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
3.5. A mutant MERS-CoV carrying R13A substitution in nsp1 exhibits a growth defect
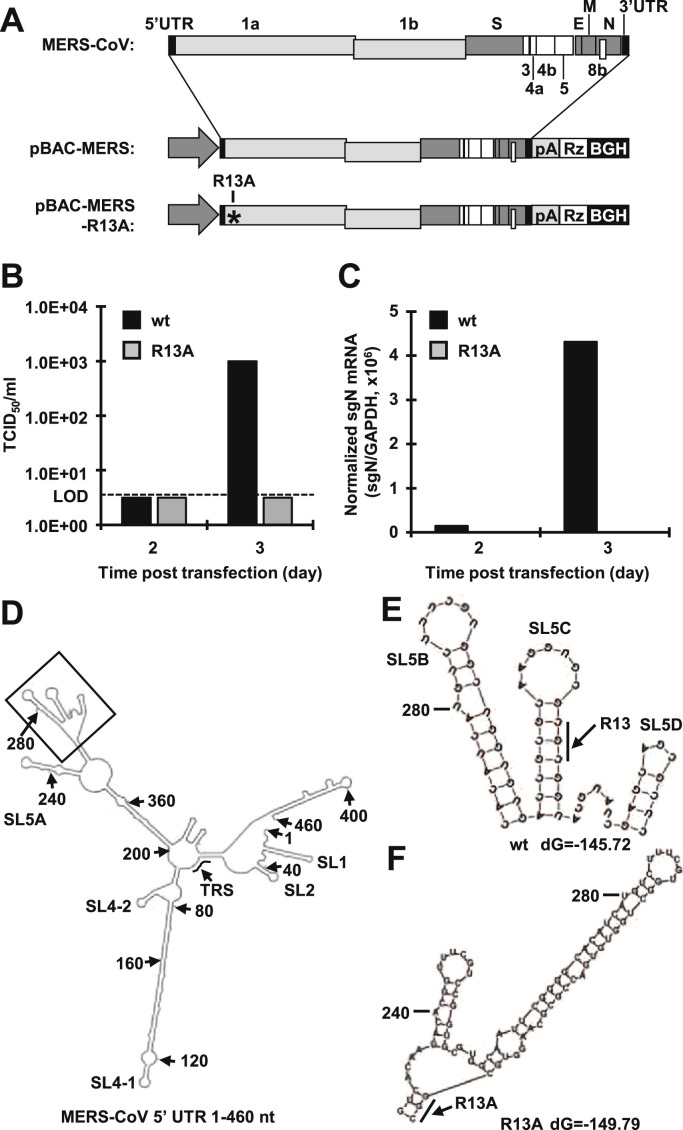
To determine the biological significance of the viral RNA recognition by MERS-CoV nsp1 in viral propagation, we employed a reverse genetics system for MERS-CoV by using a BAC system as described previously (Almazán et al., 2013). We introduced an R13A mutation into the nsp1 gene of the parental infectious cDNA (pBAC-MERS) to generate pBAC-MERS-R13A ( Fig. 5A). Culture supernatants of Huh7 cells transfected with either pBAC-MERS or pBAC-MERS-R13A were harvested at 2 and 3 days posttransfection, and infectious titers in the supernatants were determined by a TCID50 assay with Vero cells. The virus infectivity in cells transfected with pBAC-MERS reached 1.0E+03 TCID50/ml at 3 days posttransfection, while no virus production was observed in cells transfected with pBAC-MERS-R13A (Fig. 5B). Next, to determine the effect of R13A on viral RNA replication, cell lysates of Huh7 cells transfected with either pBAC-MERS or pBAC-MERS-R13A were harvested at 2 and 3 days posttransfection, and total RNAs were extracted. The level of MERS-CoV subgenomic N mRNA was determined by real-time RT-PCR. As we expected, the amounts of the subgenomic N mRNA were increased in cells transfected with pBAC-MERS, while no subgenomic N mRNA was observed in cells transfected with pBAC-MERS-R13A (Fig. 5C). These results suggest two possibilities: one is that the nsp1 protein carrying the R13A mutation is unable to bind to the 5′ UTR, and the other is that the R13A mutation disrupts cis-replication elements in the 5′ region of viral RNA. In fact, in other CoVs, the 5′ portion of the nsp1-coding region acts as a cis-acting element, and the RNA structure of this region is critical for viral replication, as described previously (Brown et al., 2007, Chen and Olsthoorn, 2010, Guan et al., 2011). Therefore, as shown in Fig. 5D, we used Mfold software to predict the MERS-CoV RNA structure from 1 to 460 nt containing the nsp1-coding region (Zuker, 2003). The predicted structure revealed that the R13A mutation in nsp1 affected the secondary structure at SL5B, C and D of MERS-CoV (Fig. 5E and F). These results suggest that the nsp1 mutation (R13A) impairs the viral replication through the disruption of SL structures in the 5′ region of MERS-CoV RNA.
Fig. 5.
R13A mutation caused the loss of viral replication capacity. (A) Schematic diagrams of the genome structure of MERS-CoV (top). Black boxes represent 5′ and 3′ UTR. Light gray boxes represent ORF 1a and 1b. Gray and white boxes represent ORFs encoding structural and accessary proteins, respectively. The middle and bottom diagrams represent the schematic diagrams of BAC DNA of MERS-CoV. pA represents the polyA tail. Rz represents the HDV ribozyme. BGH represents the BGH termination and polyadenylation sequence. An asterisk represents the position of the mutated amino acid. (B, C) Huh7 cells were transfected with either pBAC-MERS or pBAC-MERS-R13A, and the virus titers of culture supernatants (B) and levels of subgenomic N mRNA in transfected cells (C) were determined at 2 and 3 days posttransfection. The dashed line represents the limit of detection (LOD). (D, E) Secondary RNA structures from 1 to 460 nt at the 5′ UTR of MERS-CoV were predicted using Mfold software (Zuker, 2003). The nucleotide position of each 40 nt is indicated by an arrow with the position number. TRS represents the transcription regulatory sequence. (E) The partial RNA structure of the wt MERS-CoV represents the black box in panel D. A bold line represents the codon of arginine at position 13. (F) Partial RNA structure of the R13A mutant of MERS-CoV. The bold line represents the codon of alanine at position 13.
3.6. A secondary structure in the 5′ region of MERS-CoV is crucial for efficient viral propagation through the interaction with nsp1
To examine the possibility that the absence of R13A mutant-virus production is due to disruption of the SL structure in the 5′ region of viral RNA, we generated an additional mutant possessing substitutions of A9 and R13 to G and A, respectively ( Fig. 6A), in order to retain the secondary structure of SLs of MERS-CoV. The deduced secondary structures showed that SL5B, 5C, and 5D were retained in the 5′ region of the viral RNA possessing the substitutions of A9G and R13A, as shown in Fig. 6B. First, we investigated the biological properties of nsp1-A9G/R13A mutant by immunoprecipitation (Fig. 6C) and luciferase assay (Fig. 6D). The results showed that nsp1-A9G/R13A mutant possessed low activity of binding with 5′ UTR and induced translational shutoff against both reporter plasmids with or without 5′ UTR (Fig. 6C and D). These results indicated that nsp1-A9G/R13A mutant possessed similar properties with nsp1-R13A mutant (Fig. 4C and E). To determine the effect of A9G and R13A mutations in nsp1 on the viral propagation, supernatants of Huh7 cells transfected with either pBAC-MERS or pBAC-MERS-A9G/R13A were harvested at 72 h posttransfection and passaged once on Vero cells. Preservation of the mutations in the recovered viruses was confirmed by direct sequencing (data not shown). The recovered wt and A9G/R13A mutant MERS-CoV were inoculated into Vero cells at a multiplicity of infection (MOI) of 0.001, and infectious titers in the culture supernatants and intracellular viral RNA were determined. Although the propagation efficiency was lower compared to that of the wt MERS-CoV, the mutant virus carrying two substitutions of A9G/R13A recovered the production of infectious particles in the supernatants and replication in cells (Fig. 6E and F). We also observed similar results using Huh7 cells (Fig. 6G and H). In addition, the A9G/R13A mutant MERS-CoV showed smaller plaque size than wt virus (Fig. 6I). These results suggest that 1) the secondary structure in the 5′ region of MERS-CoV is crucial for an efficient viral propagation and 2) viral RNA recognition by nsp1 is also important for efficient viral propagation.
Fig. 6.
Rigorous viral RNA recognition by nsp1 in viral replication. (A) The predicted SL5C in MERS-CoV structures of the wt (left) and A9G/R13A mutant (right). Introduced nucleotide mutations are shown in a gray box with white letters. (B) Partial RNA structures of the wt (left) and A9G/R13A mutant (right) of MERS-CoV from the region in the black square in Fig. 5D. Free energies (dG) are shown below. (C) Lysates of 293T cells transfected with either pCAG-CAT, pCAG-nsp1 or pCAG-nsp1-A9G/R13A together pcD-5′-fLuc were immunoprecipitated with anti-FLAG antibody at 24 h posttransfection. RNAs extracted from the precipitates were subjected to Northern blot analysis using a riboprobe for the luciferase gene. (D) Luciferase activities in 293T cells transfected with either pcD-fluc or pcD-5′-fluc together with the indicated expression plasmids were determined at 24 h posttransfection after standardization with those in cells expressing CAT. The values represent the means ±SD from three independent experiments. (E, F) Vero cells were infected with each recombinant MERS-CoV at an MOI of 0.001, and the virus titers of culture supernatants (E) and levels of subgenomic N mRNA in infected cells (F) were determined at 12, 24 and 48 h postinfection. Because the values of mock samples were under LOD, the data of mock were not shown. (G, H) Huh7 cells were infected with each recombinant MERS-CoV at an MOI of 0.001, and the virus titers of culture supernatants (G) and levels of subgenomic N mRNA in infected cells (H) were determined at 6, 12 and 24 h postinfection. Because the values of mock samples were under LOD, the data of mock were not shown. (I) Plaque morphology of the wt and nsp1-A9G/R13A mutant MERS-CoVs following agarose overlay plaque assay using Vero cell. Infected cells were fixed with phosphate-buffered formaldehyde at 3 days postinfection and stained using crystal violet.
4. Discussion
Many viruses suppress host protein synthesis and modify host cell environments to promote virus-specific translation (Lloyd, 2006, Schneider and Mohr, 2003). The CoV nsp1 is a multifunctional protein involved in viral replication, pathogenesis, evasion from antiviral signaling, and suppression of host protein synthesis (Brockway and Denison, 2005, Kamitani et al., 2009, Kamitani et al., 2006, Lokugamage et al., 2012, Narayanan et al., 2008, Wang et al., 2010, Wathelet et al., 2007, Züst et al., 2007). In alphacoronaviruses, the nsp1s of HCoV-229E, HCoV-NL63 and TGEV induce suppression of protein synthesis in mammalian cells (Huang et al., 2011a, Wang et al., 2010, Züst et al., 2007). In betacoronaviruses, the nsp1s of MHV, SARS-CoV and several bat CoVs also have the ability to induce translational suppression (Kamitani et al., 2006, Tohya et al., 2009, Züst et al., 2007). The nsp1 of SARS-CoV is the most studied among CoVs. The nsp1 of SARS-CoV predominantly localizes in the cytoplasm of cells upon infection or expression (Kamitani et al., 2006), while the nsp1 of MERS-CoV diffuses into both the nucleus and cytoplasm (Lokugamage et al., 2015). Although the nsp1s of SARS-CoV and MERS-CoV have been suggested to suppress host protein synthesis through inhibition of the translational machinery and digestion of mRNA by endonuclease activity (Huang et al., 2011b, Lokugamage et al., 2015), the molecular mechanisms of translational shutoff and RNA degradation induced by MERS-CoV nsp1 are largely unknown. Bovine coronavirus nsp1 has been shown to bind viral RNA and to regulate viral replication (Gustin et al., 2009). A previous study revealed that SARS-CoV nsp1 specifically binds to the 5′ UTR of viral RNA to evade nsp1-mediated shutoff (Tanaka et al., 2012). In this study, we identified crucial amino acid residues in MERS-CoV nsp1 responsible for the interaction with the 5′ UTR of MERS-CoV RNA and demonstrated that the interaction of nsp1 with viral RNA facilitates an efficient replication. We also showed that the RNA sequence of the 5′-terminus of the nsp1-coding region plays a critical role in viral replication by acting as a cis-acting element.
MERS-CoV and SARS-CoV belong to lineages C and B in the genus Betacoronavirus, respectively (Ksiazek et al., 2003, Snijder et al., 2003, van Boheemen et al., 2012). Although the nsp1s of SARS-CoV and MERS-CoV consist of 180 and 193 amino acids, respectively, and share only 26.1% amino acid sequence identity, Almeida et al. showed that a consensus amino acid sequence, LRKxGxKG, is observed in the nsp1s of SARS-CoV and MHV (Almeida et al., 2007), and a similar sequence of LRKxGxGG was also conserved in MERS-CoV nsp1, as shown in Fig. 7A. In addition, both SARS-CoV and MERS-CoV possess RNA cleavage activity in nsp1 and the amino acid sequences of RK responsible for RNA cleavage are conserved in R124/K125 and R146/K147, respectively, within the conserved sequence, as shown in blue in Fig. 7A (Lokugamage et al., 2012, Lokugamage et al., 2015). Our previous report showed the positively charged R124 in the consensus sequence for RNA cleavage in SARS-CoV nsp1 is also crucial for binding to the viral 5′ UTR (Tanaka et al., 2012). However, R146A substitution in MERS-CoV nsp1 exhibited no effect on the interaction with the 5′ UTR of viral RNA (data not shown). Interestingly, the mutation of R13A in MERS-CoV nsp1, which is distant from the conserved sequence, abrogated the interaction with the 5′ UTR of viral RNA (Fig. 4C), suggesting that the binding domain in nsp1 with the 5′ UTR of viral RNA is different between MERS-CoV and SARS-CoV. In fact, SARS-CoV nsp1 has a deletion in the region corresponding to R13 in MERS-CoV nsp1, as shown in Fig. 7A. Although the partial structure information of SARS-CoV nsp1 (aa 13–128) revealed a novel complex beta-barrel fold by using NMR (Almeida et al., 2007), no structural information on either the N or C terminal regions was available due to their flexibilities (Almeida et al., 2007). The secondary structure of MERS-CoV nsp1 predicted by PSIPRED v3.3 (Buchan et al., 2013) suggested that R13 is located in an alpha-helix structure, as shown in Fig. 7A. Collectively, these data suggest that SARS-CoV and MERS-CoV are evolved to utilize R124 and R13 in nsp1, respectively, for interaction with the 5′ UTR of viral RNA to circumvent the nsp1-mediated shutoff. Further structural analysis studies will be needed to elucidate the molecular mechanisms underlying the interaction of MERS-CoV nsp1 with the 5′ UTR of viral RNA.
The 5′ UTR of CoV RNA forms higher-order structures and contains cis-acting sequences (Yang and Leibowitz, 2015). Many cis-acting sequences have been identified, and their functional roles in viral RNA transcription and replication were investigated by using defective interfering RNAs (Kim and Makino, 1995, Raman et al., 2003, Raman and Brian, 2005). The 5′ UTR of MERS-CoV possesses 4 stem-loops, SL1, SL2, SL4-1, and SL4-2, which are conserved among beta coronaviruses (Yang and Leibowitz, 2015). As previously shown for SARS-CoV SL1 (Tanaka et al., 2012), we found here that MERS-CoV SL1, which is located in both genomic and subgenomic viral RNAs, plays a critical role in viral replication through the interaction with nsp1. Although the structural significance of SL1 in both SARS-CoV and MERS-CoV remains to be investigated, SL1 is critical for the recognition of viral RNA by the nsp1s of both CoVs, suggesting that the interaction between SL1 and nsp1 is an ideal therapeutic target for infection with CoV.
The R13A mutant of MERS-CoV was replication defective and the secondary structure model predicted that R13A substitution (CGC to GCC) in viral RNA induced structural changes in SL5B, 5 C and 5D (Fig. 5E and F), suggesting that the secondary RNA structure of SL5B, 5C and 5D acts as a cis-acting RNA element. In fact, a mutant MHV possessing a deletion in the N-terminal region of nsp1 was replication defective in DBT-9 cells (Brockway and Denison, 2005). In addition, SL5 of CoV was shown to be crucial as a cis-acting element and packaging signal (Brown et al., 2007, Chen and Olsthoorn, 2010, Guan et al., 2011). Although the R13A mutant was defective in RNA replication, the A9G/R13A mutant, which retained the SL5 structure, successfully replicated in Huh7 and Vero cells (Fig. 6), suggesting that RNA structures in SL5B, C and/or D of MERS-CoV act as cis-acting elements and are critical for viral replication.
SARS-CoV nsp1 is localized in the cytoplasm, while MERS-CoV nsp1 is localized in both the nucleus and cytoplasm (Lokugamage et al., 2015), just as in TGEV nsp1 and other alphacoronaviruses (Narayanan et al., 2015). Although the functional relevance of the nuclear localization of nsp1 remains unclear, nsp1 in the nucleus has been suggested to participate in host gene transcription or mRNA export. In MHV, nsp1 localizes to the site of RNA replication through an interaction with nsp7 and nsp10 upon infection, while nsp1 translocates to the assembly site of infectious particles at a late stage of infection (Brockway et al., 2004). These data suggest that the nsp1 of CoVs participates not only in RNA replication but also in particle assembly processes. Further studies are needed to clarify the roles of nsp1 in the life cycle of CoVs.
In conclusion, the data in this study suggest that MERS-CoV nsp1, as well as SARS-CoV nsp1, has acquired a functional role to selectively recognize its own mRNA, and this role is critical for circumvention of the nsp1-mediated translational shutoff. This common strategy for viral RNA recognition by nsp1 should provide new insight into the roles of nsp1 in the CoV life cycle as well as a target for the development of novel antivirals broadly effective against coronavirus infection.
Acknowledgments
We thank Dr. Bart L. Haagmens (Erasmus Medical Center) for providing the MERS-CoV/EMC2012 strain through Dr. Makoto Takeda (National Institute of Infectious Diseases). We thank Ms. Kaede Yukawa for secretarial assistance, Ms. Kanako Yoshizawa for technical assistance, Dr. Shinji Makino (University of Texas Medical Branch) for helpful insights, Drs. Makoto Sugiyama and Naoto Ito (Gifu University) and Dr. Ken Maeda (Yamaguchi University) for kindly providing research reagents, and Drs. Luis Enjuanes and Marta L. DeDiego for kindly providing SARS-CoV-Rep DNA. WK was supported by a Grant-in-Aid for Scientific Research (16K08811) and by funds from the Takeda Science Foundation. This work was supported in part of by Research Program on Emerging and Re-emerging Infectious Diseases from Japan Agency for Medical Research and Development, AMED.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2017.08.026.
Appendix A. Supplementary material
Supplementary material
References
- Almazán F., Dediego M.L., Galán C., Escors D., Álvarez E., Ortego J., Sola I., Zuñiga S., Alonso S., Moreno J.L., Nogales A., Capiscol C., Enjuanes L. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J. Virol. 2006;80:10900–10906. doi: 10.1128/JVI.00385-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazán F., DeDiego M.L., Sola I., Zuñiga S., Nieto-Torres J.L., Marquez-Jurado S., Andrés G., Enjuanes L. Engineering a replication-competent, propagation-defective Middle East respiratory syndrome coronavirus as a vaccine candidate. MBio. 2013;4 doi: 10.1128/mBio.00650-13. (e00650-00613) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida M.S., Johnson M.A., Herrmann T., Geralt M., Wüthrich K. Novel beta-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:3151–3161. doi: 10.1128/JVI.01939-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham A., Chand M.A., Brown C.S., Aarons E., Tong C., Langrish C., Hoschler K., Brown K., Galiano M., Myers R., Pebody R.G., Green H.K., Boddington N.L., Gopal R., Price N., Newsholme W., Drosten C., Fouchier R.A., Zambon M. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Eur. Surveill. 2012;17:20290. [PubMed] [Google Scholar]
- Bradburne A.F., Bynoe M.L., Tyrrell D.A. Effects of a "new" human respiratory virus in volunteers. Br. Med. J. 1967;3:767–769. doi: 10.1136/bmj.3.5568.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockway S.M., Denison M.R. Mutagenesis of the murine hepatitis virus nsp1-coding region identifies residues important for protein processing, viral RNA synthesis, and viral replication. Virology. 2005;340:209–223. doi: 10.1016/j.virol.2005.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockway S.M., Lu X.T., Peters T.R., Dermody T.S., Denison M.R. Intracellular localization and protein interactions of the gene 1 protein p28 during mouse hepatitis virus replication. J. Virol. 2004;78:11551–11562. doi: 10.1128/JVI.78.21.11551-11562.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.G., Nixon K.S., Senanayake S.D., Brian D.A. An RNA stem-loop within the bovine coronavirus nsp1 coding region is a cis-acting element in defective interfering RNA replication. J. Virol. 2007;81:7716–7724. doi: 10.1128/JVI.00549-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan D.W., Minneci F., Nugent T.C., Bryson K., Jones D.T. Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Res. 2013;41:W349–W357. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz U., Müller M.A., Nitsche A., Sanewski A., Wevering N., Bauer-Balci T., Bonin F., Drosten C., Schweiger B., Wolff T., Muth D., Meyer B., Buda S., Krause G., Schaade L., Haas W. Contact investigation of a case of human novel coronavirus infection treated in a German hospital, October–November 2012. Eur. Surveill. 2013:18. [PubMed] [Google Scholar]
- Cencic R., Desforges M., Hall D.R., Kozakov D., Du Y., Min J., Dingledine R., Fu H., Vajda S., Talbot P.J., Pelletier J. Blocking eIF4E-eIF4G interaction as a strategy to impair coronavirus replication. J. Virol. 2011;85:6381–6389. doi: 10.1128/JVI.00078-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., Perlman S. Dysregulated Type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.C., Olsthoorn R.C. Group-specific structural features of the 5′-proximal sequences of coronavirus genomic RNAs. Virology. 2010;401:29–41. doi: 10.1016/j.virol.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Su C., Ke M., Jin X., Xu L., Zhang Z., Wu A., Sun Y., Yang Z., Tien P., Ahola T., Liang Y., Liu X., Guo D. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 2011;7:e1002294. doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor R.F., Roper R.L. Unique SARS-CoV proteinnsp1: bioinformatics, biochemistry and potential effects on virulence. Trends Microbiol. 2007;15:51–53. doi: 10.1016/j.tim.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling B.J., Park M., Fang V.J., Wu P., Leung G.M., Wu J.T. Preliminary epidemiological assessment of MERS-CoV outbreak in South Korea, May to June 2015. Eur. Surveill. 2015;20:7–13. doi: 10.2807/1560-7917.es2015.20.25.21163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly E., Debarnot C., Ferron F., Bouvet M., Coutard B., Imbert I., Gluais L., Papageorgiou N., Sharff A., Bricogne G., Ortiz-Lombardia M., Lescar J., Canard B. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011;7:e1002059. doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguière A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Müller S., Rickerts V., Stürmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Guan B.J., Wu H.Y., Brian D.A. An optimal cis-replication stem-loop IV in the 5′ untranslated region of the mouse coronavirus genome extends 16 nucleotides into open reading frame 1. J. Virol. 2011;85:5593–5605. doi: 10.1128/JVI.00263-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin K.M., Guan B.J., Dziduszko A., Brian D.A. Bovine coronavirus nonstructural protein 1 (p28) is an RNA binding protein that binds terminal genomic cis-replication elements. J. Virol. 2009;83:6087–6097. doi: 10.1128/JVI.00160-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Lokugamage K.G., Rozovics J.M., Narayanan K., Semler B.L., Makino S. Alphacoronavirus transmissible gastroenteritis virus nsp1 protein suppresses protein translation in mammalian cells and in cell-free HeLa cell extracts but not in rabbit reticulocyte lysate. J. Virol. 2011;85:638–643. doi: 10.1128/JVI.01806-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Lokugamage K.G., Rozovics J.M., Narayanan K., Semler B.L., Makino S. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 2011;7:e1002433. doi: 10.1371/journal.ppat.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani W., Huang C., Narayanan K., Lokugamage K.G., Makino S. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat. Struct. Mol. Biol. 2009;16:1134–1140. doi: 10.1038/nsmb.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani W., Narayanan K., Huang C., Lokugamage K., Ikegami T., Ito N., Kubo H., Makino S. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad. Sci. USA. 2006;103:12885–12890. doi: 10.1073/pnas.0603144103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.N., Makino S. Characterization of a murine coronavirus defective interfering RNA internal cis-acting replication signal. J. Virol. 1995;69:4963–4971. doi: 10.1128/jvi.69.8.4963-4971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler E., Thiel V. SARS-CoV and IFN: too little, too late. Cell Host Microbe. 2016;19:139–141. doi: 10.1016/j.chom.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J., Group S.W. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Lloyd R.E. Translational control by viral proteinases. Virus Res. 2006;119:76–88. doi: 10.1016/j.virusres.2005.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokugamage K.G., Narayanan K., Huang C., Makino S. Severe acute respiratory syndrome coronavirus protein nsp1 is a novel eukaryotic translation inhibitor that represses multiple steps of translation initiation. J. Virol. 2012;86:13598–13608. doi: 10.1128/JVI.01958-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokugamage K.G., Narayanan K., Nakagawa K., Terasaki K., Ramirez S.I., Tseng C.T., Makino S. Middle East respiratory syndrome coronavirus nsp1 inhibits host gene expression by selectively targeting mRNAs transcribed in the nucleus while sparing mRNAs of cytoplasmic origin. J. Virol. 2015;89:10970–10981. doi: 10.1128/JVI.01352-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M.A., Corman V.M., Jores J., Meyer B., Younan M., Liljander A., Bosch B.J., Lattwein E., Hilali M., Musa B.E., Bornstein S., Drosten C. MERS coronavirus neutralizing antibodies in camels, Eastern Africa, 1983–1997. Emerg. Infect. Dis. 2014;20:2093–2095. doi: 10.3201/eid2012.141026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailles A., Blanckaert K., Chaud P., van der Werf S., Lina B., Caro V., Campese C., Guéry B., Prouvost H., Lemaire X., Paty M.C., Haeghebaert S., Antoine D., Ettahar N., Noel H., Behillil S., Hendricx S., Manuguerra J.C., Enouf V., La Ruche G., Semaille C., Coignard B., Lévy-Bruhl D., Weber F., Saura C., Che D., investigation team First cases of Middle East respiratory syndrome coronavirus (MERS-CoV) infections in France, investigations and implications for the prevention of human-to-human transmission, France. Euro Surveill. 2013:18. [PubMed] [Google Scholar]
- Memish Z.A., Cotten M., Meyer B., Watson S.J., Alsahafi A.J., Al Rabeeah A.A., Corman V.M., Sieberg A., Makhdoom H.Q., Assiri A., Al Masri M., Aldabbagh S., Bosch B.J., Beer M., Muller M.A., Kellam P., Drosten C. Human infection with MERS coronavirus after exposure to infected camels, Saudi Arabia, 2013. Emerg. Infect. Dis. 2014;20:1012–1015. doi: 10.3201/eid2006.140402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Huang C., Lokugamage K., Kamitani W., Ikegami T., Tseng C.T., Makino S. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 2008;82:4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Ramirez S.I., Lokugamage K.G., Makino S. Coronavirus nonstructural protein 1: common and distinct functions in the regulation of host and viral gene expression. Virus Res. 2015;202:89–100. doi: 10.1016/j.virusres.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Chamberlain P., Bowden F., Joseph J. Atlas of coronavirus replicase structure. Virus Res. 2014;194:49–66. doi: 10.1016/j.virusres.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera R.A., Wang P., Gomaa M.R., El-Shesheny R., Kandeil A., Bagato O., Siu L.Y., Shehata M.M., Kayed A.S., Moatasim Y., Li M., Poon L.L., Guan Y., Webby R.J., Ali M.A., Peiris J.S., Kayali G. Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt. Eur. Surveill. 2013;18 doi: 10.2807/1560-7917.es2013.18.36.20574. (pii=20574) [DOI] [PubMed] [Google Scholar]
- Prentice E., McAuliffe J., Lu X., Subbarao K., Denison M.R. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 2004;78:9977–9986. doi: 10.1128/JVI.78.18.9977-9986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S., Bouma P., Williams G.D., Brian D.A. Stem-loop III in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 2003;77:6720–6730. doi: 10.1128/JVI.77.12.6720-6730.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S., Brian D.A. Stem-loop IV in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 2005;79:12434–12446. doi: 10.1128/JVI.79.19.12434-12446.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken C.B., Haagmans B.L., Müller M.A., Gutierrez C., Godeke G.J., Meyer B., Muth D., Raj V.S., Smits-De Vries L., Corman V.M., Drexler J.F., Smits S.L., El Tahir Y.E., De Sousa R., van Beek J., Nowotny N., van Maanen K., Hidalgo-Hermoso E., Bosch B.J., Rottier P., Osterhaus A., Gortázar-Schmidt C., Drosten C., Koopmans M.P. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect. Dis. 2013;13:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken C.B., Messadi L., Feyisa A., Ularamu H., Godeke G.J., Danmarwa A., Dawo F., Jemli M., Melaku S., Shamaki D., Woma Y., Wungak Y., Gebremedhin E.Z., Zutt I., Bosch B.J., Haagmans B.L., Koopmans M.P. Geographic distribution of MERS coronavirus among dromedary camels, Africa. Emerg. Infect. Dis. 2014;20:1370–1374. doi: 10.3201/eid2008.140590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R.J., Mohr I. Translation initiation and viral tricks. Trends Biochem. Sci. 2003;28:130–136. doi: 10.1016/S0968-0004(03)00029-X. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Kamitani W., DeDiego M.L., Enjuanes L., Matsuura Y. Severe acute respiratory syndrome coronavirus nsp1 facilitates efficient propagation in cells through a specific translational shutoff of host mRNA. J. Virol. 2012;86:11128–11137. doi: 10.1128/JVI.01700-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weißbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- Tohya Y., Narayanan K., Kamitani W., Huang C., Lokugamage K., Makino S. Suppression of host gene expression by nsp1 proteins of group 2 bat coronaviruses. J. Virol. 2009;83:5282–5288. doi: 10.1128/JVI.02485-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boheemen S., de Graaf M., Lauber C., Bestebroer T.M., Raj V.S., Zaki A.M., Osterhaus A.D., Haagmans B.L., Gorbalenya A.E., Snijder E.J., Fouchier R.A. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012:3. doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Berkhout B. Human coronavirus NL63, a new respiratory virus. FEMS Microbiol. Rev. 2006;30:760–773. doi: 10.1111/j.1574-6976.2006.00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C., Wertheim-van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Shi H., Rigolet P., Wu N., Zhu L., Xi X.G., Vabret A., Wang X., Wang T. Nsp1 proteins of group I and SARS coronaviruses share structural and functional similarities. Infect. Genet. Evol. 2010;10:919–924. doi: 10.1016/j.meegid.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathelet M.G., Orr M., Frieman M.B., Baric R.S. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 2007;81:11620–11633. doi: 10.1128/JVI.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S.R., Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 2005;69:635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Huang Y., Lau S.K., Yuen K.Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H., Poon R.W., Cai J.J., Luk W.K., Poon L.L., Wong S.S., Guan Y., Peiris J.S., Yuen K.Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamshchikov V., Mishin V., Cominelli F. A new strategy in design of +RNA virus infectious clones enabling their stable propagation in E. coli. Virology. 2001;281:272–280. doi: 10.1006/viro.2000.0793. [DOI] [PubMed] [Google Scholar]
- Yang D., Leibowitz J.L. The structure and functions of coronavirus genomic 3′ and 5′ ends. Virus Res. 2015;206:120–133. doi: 10.1016/j.virusres.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Curtis K.M., Baric R.S. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J. Virol. 2000;74:10600–10611. doi: 10.1128/jvi.74.22.10600-10611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züst R., Cervantes-Barragán L., Kuri T., Blakqori G., Weber F., Ludewig B., Thiel V. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog. 2007;3:e109. doi: 10.1371/journal.ppat.0030109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material