Abstract
Background
Infections by both human oncoviruses, human Papillomaviruses (HPV) and Epstein–Barr virus (EBV) are very common in the adult human population and are associated with various malignancies. While HPV is generally transmitted sexually or via skin-to-skin contact, EBV is frequently transmitted by oral secretions, blood transfusions and organ transplants. This study aims to determine the prevalence and circulating genotypes of HPV and EBV in healthy blood donors in Qatar.
Methods
We explored the co-prevalence of high-risk HPVs and EBV in 378 males and only 7 females blood donors of different nationalities (mainly from Qatar, Egypt, Syria, Jordan, Pakistan, and India) residing in Qatar, using polymerase chain reaction (PCR). DNA was extracted from the buffy coat and genotyping was performed using PCR and nested-PCR targeting E6 and E7 as well as LMP-1 of HPV and EBV, respectively.
Results
We found that from the total number of 385 cases of healthy blood donors studied, 54.8% and 61% of the samples are HPVs and EBV positive, respectively. Additionally, our data revealed that the co-presence of both high-risk HPVs and EBV is 40.4% of the total samples. More significantly, this study pointed out for the first time that the most frequent high-risk HPV types in Qatar are 59 (54.8%), 31 (53.7%), 52 (49.1%), 51 (48.6%), 58 (47%) and 35 (45.5%), while the most commonly expressed low-risk HPV types are 53 (50.6%), 11 (45.5), 73 (41.7%) and 6 (41.3%), with all the cases showing multiple HPVs infection.
Conclusion
In this study, we demonstrated for the first time that HPV and EBV are commonly co-present in healthy blood donors in Qatar. On the other hand, it is important to highlight that these oncoviruses can also be co-present in several types of human cancers where they can cooperate in the initiation and/or progression of these cancers. Therefore, more studies regarding the co-presence of these oncoviruses and their interaction are necessary to understand their cooperative role in human diseases.
Keywords: HPV, EBV, Healthy blood donors, Qatar
Background
Human Papillomaviruses (HPVs) and Epstein–Barr virus (EBV) are human oncoviruses that can co-exist together. HPVs are small, double-stranded DNA viruses which tend to infect cutaneous and mucosal epithelial tissues of the ano-genital tract [1]. HPVs are categorized into high-risk or low-risk. Infections due to low-risk HPV subtypes (mostly, HPVs-6 and 11) are commonly self-limiting diseases resulting in the proliferation of epithelial cells that manifests as skin or genital warts or skin papilloma [2, 3]. On the other hand, high-risk HPV subtypes (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59 and 68) are associated with the development of several types of human carcinomas [2, 3]. In particular, cancers developing in the cervix, ano-genital tract as well as oropharynx region are frequently correlated with chronic high-risk HPV infection [4, 5]. While HPV infections are eradicated by the immune system, persistent chronic infections associated with high expression levels of E6/E7 (viral oncogenes) as well as accretion of cellular mutations may cause alterations in TP53 and RB expressions, which are known as tumor suppressor genes [6]. Additionally, it has been reported that high-risk HPVs (E5 and E6/E7) oncoproteins, can enhance cancer progression of human carcinomas via the initiation of the epithelial-mesenchymal transition (EMT) which is a hallmark of cancer metastasis [7–9]. On the other hand, it is important to note that HPVs transmissions may be enhanced by the presence of rough or macerated epithelial surfaces [10]. Several studies have demonstrated that HPV DNA can be present in circulating blood, including peripheral blood mononuclear cells (PBMCs), sera, plasma, and arterial cord blood [11–15].
Human herpesvirus 4, commonly known as Epstein–Barr virus is a DNA lymphotropic herpesvirus that causes infectious mononucleosis [16]. EBV comprises of a 172 kbp double-stranded DNA genome encoding for around 85 genes [17, 18]. There are two EBV genotypes, Type 1 and Type 2 [19]. Various viral oncogenes such as EBV-encoded nuclear antigens (EBNA [1–3]) and the latent membrane proteins (LMP [1, 2]) are encoded by the EBV [20]. LMP-1 is a 356-amino acid protein [21] and is involved in signal transduction and cell survival [18]. Since LMP1 regulates cell growth, protects cells from apoptosis, promotes cell motility, and angiogenesis, it is considered as the principal EBV-encoded oncogenic protein [22]. LMP-1 variants are divided into 7 core groups including B95-8, Alaskan, China 1, China 2, Med+ , Med− , and NC [18, 23, 24]. LMP1 can stimulate several signaling pathways especially those involved in the induction of EMT such as NF-κB, PI3K/Akt, and MAPK [7]. The other EBV onco-protein, LMP2A [25] regulates the invasive/migratory ability and stimulates changes in EMT-like cellular biomarkers [26] by targeting the rapamycin (mTOR) pathway [22]. On the other hand, EBNA1, a key product of EBV, is involved in the replication and maintenance of the EBV genome [27] and regulates EMT through the de-regulation of SLUG, SNAIL, TCF8/ZEB1, vimentin, occludins-1, as well as E-cadherin [9]. EBV is primarily transmitted through the oral route; however, blood transfusions and organ transplantations are also other possible transmission routes of EBV [28–30].
Several recent studies have demonstrated that the co-presence of high-risk HPVs and EBV mRNA in various human malignancies including cervical, breast and oral cancers, indicating their active role in cancer initiation and metastatic progression via the acceleration of EMT through numerous common signaling pathways [11, 31].
In light of these findings, the objective of this study is to explore the presence of HPV and EBV DNA in the blood of healthy individuals in the Middle East region, more specifically in Qatar.
Materials and methods
Sample collection and ethical approval
A total of 385 whole blood samples (139 from Qataris and 246 from other nationalities) were collected in EDTA tubes from healthy donors at the Blood Donor Unit of Hamad Medical Corporation (HMC) over a period of 1 year (September 2014–September 2015) as previously described [32]. Blood samples were handled and stored following standard safety procedures and guidelines. This study was approved by HMC-Institutional Review Board (HMC-IRB #14292/14) and Qatar University IRB (QU-IRB 518-EA/15) [32].
DNA extraction
DNA was extracted from PMNCs (buffy coat samples) suspended in 200 μl of PBS using Qiagen kit following the manufacturer’s instructions (Qiagen, Germany) as previously described [32]. The concentration and purity of all extracted DNA samples were measured using NanoQuant microplate reader (Tecan, Switzerland). Extracted DNA samples were then stored at − 20 °C for further testing.
HPV and EBV detection
Twenty-five nm of purified genomic DNA (Qiagen, Germany), from each sample, was analyzed for HPV and EBV by polymerase chain reaction (PCR) using specific primers as follows:
The E6/E7 and L1 region of HPV types; GP5/6, 6, 11, 16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 73 and 82 were amplified (Table 1), while, primers for GAPDH gene (Forward Primer: 5′-GAAGGC-CATGCCAGTGAGCT-3′ and Reverse Primer: 5′-CCGGGAAACTGTGGCGTGAT-3′) were used as an internal control.
Table 1.
The specific primer sets for HPV used for polymerase chain reaction (PCR) amplification
| HPV type | Forward primer (5′-3′) | Reverse primer (5′-3′) | Ta (°C) |
|---|---|---|---|
| E6/E7 region | |||
| 16 | ATGCATGGAGATACACCTACATTGCAT | GTTTCTGAGAACAGATGGGGCACAC | 61 |
| 18 | GCTTTGAGGATCCAACACGG | TGCAGCACGAATGGCACTGG | 61 |
| 31 | GGGCTCATTTGGAATCGTGTG | AACCATTGCATCCCGTCCCC | 61 |
| 33 | TGAGGATGAAGGCTTGGACC | TGACACATAAACGAACTGTG | 61 |
| 35 | CTATTGACGGTCCAGCT | TACACACAGACGTAGTGTCG | 61 |
| 45 | CCCACGCGAACCACAG | TCTAAGGTCCTCTGCCGAGC | 46.3 |
| 51 | TACGTGTTACAGAATTGAAG | AACCAGGCTTAGTTCGCCCATT | 46.3 |
| 52 | GCAGAACAAGCCACAAGCAA | TAGAGTACGAAGGTCCGTCG | 60 |
| 58 | CGAGGATGAAATAGGCTTGG | ACACAAACGAACCGTGGTGC | 40.5 |
| L1 region | |||
| GP5/GP6 | TTTGTTACTGTGGTAGATACTAC | GAAAAATAAACTGTAAATCATATT | 50.5 |
| 6 | TGTCCCATCTGCGCACCGAAGAC | CGTACACTGTTTGTGGGCGCTTC | 42.5 |
| 11 | AGTTCCGTAGATGCCAAGGGCA | TGCCTCAGGTGAGGCCCAATGC | 42.5 |
| 26 | TGGTATACAACGAGTGTCAGCTCC | GGGGCAATGATGGCCATGTCG | 40.5 |
| 39 | TGTGCAGTACCAGTGACGGATCG | ATTTTTGGCGTTGTGACTCTGTG | 40.5 |
| 53 | TTGTTCAGTGTACGGGGCTAGC | GTGACGCCATTGCAGTTATCGCCT | 43.3 |
| 56 | CTGGGCACTAGGTCAAAGCCTGCT | CAACCACGCGTAAAAGCACTCAT | 53.8 |
| 59 | AGACACCGTTACATGAGCTGCT | TCATTCTCGGAGTCGGAGTCAG | 43.3 |
| 66 | TGCGGTAGTATCCTTGGGCAGTG | TACAATAAGGGCCACACGCCAA | 46.3 |
| 68 | GTCAAAAAGACGCCCCTGCACCTA | CACACCTTAGGGTAGGGCTACAA | 48.8 |
| 73 | GGGGTGGGCAAAGGTAGGTAGC | ACAATCCAGGGGCCTCTGGTCCGA | 48.8 |
| 82 | TGTCCGTGGACACCTGCGACCA | GTAGTTAAAGGTGATGTGGCAACC | 48.8 |
On the other hand, a nested PCR of the LMP-1 gene was done as previously described [32]. In the first round of amplification, primers A1 (5′-AGTCATAGTAGCTTAGCTGAA-3′) and A2 (5′-CCATGGACAACGACACAGT -3′) amplified a fragment of 602 bp covering the C-terminus of LMP-1 gene. In the second round of amplification, primers B1 (5′-AGTCATAGTAGCTTAGCTGAA-3′) and B2 (5′-CAGTGATGAACACCACCACG-3′) amplified a 587 bp fragment. The annealing temperature for the LMP-1 gene was 53 °C.
Amplification conditions included an initial heat activation step of 15 min at 95 °C; 40 cycles of PCR amplifications at 95 °C for 30 s, annealing (Table 1) for 30 s in addition to 1 min at 72 °C; and a final extension step of 72 °C for 10 min. Analysis was performed as previously described by our group [33, 34]. Amplified products were analyzed by 1.5% agarose gel electrophoresis and visualized using iBrightCL1000 Imaging System (ThermoFisher). In each experiment, negative control (MDA-MB-453 cell line [35] and sterile water instead of DNA) and positive control (such as Hela cell line for L1 region [36] and normal oral epithelial (NOE) cell line transfected with E6/E7 of HPV type 16 for E6/E7 region [37]) were used.
Statistical analysis
To determine the relation significance between variable ratios, Chi square test was used. Results with p-value < 0.05 were considered statistically significant. The analysis was performed using Statistical Package for Social Sciences (SPSS) version 25 software (IBM SPSS).
Results
Demographic data and main findings
Of the total 385 blood samples analyzed in the present study, 378 samples (98.1%) were from males and seven samples (1.8%) from females. The majority of the samples were obtained from non-Qatari residents (66.2%), and the rest were from Qatari individuals (33.7%). The age of participants ranged between 19 and 68 years, (37.12 ± 9.3 years). Based on the study conducted previously [32], of the total 385 blood samples, 235 (61%) were positive for EBV; Table 2 summarizes the demographic data of the studied population. The present investigation revealed that 60.5% and 47.3% are positive for low-risk and high-risk HPVs, respectively. In parallel, we were able to show that EBV and HPVs are co-present in 40.4% and 50.6% for high-risk and low-risk HPVs, respectively.
Table 2.
Demographic data of the subjects positive (235) and negative (150) for EBV
| EBV positive (235) | EBV negative (150) | |
|---|---|---|
| Category | Total no. (%) | Total no. (%) |
| Nationality & gender | ||
| Qatari | 102 (43.4%) | 28 (18.66%) |
| Male | 101 | 27 |
| Female | 1 | 1 |
| Non-Qatari | 133 (56.5%) | 122 (81.33%) |
| Male | 131 | 119 |
| Female | 2 | 3 |
| Age group | ||
| 20–39 | 52 (22.12%) | 46 (30.66%) |
| 30–39 | 95 (40.4%) | 63 (42%) |
| 40–49 | 60 (25.5%) | 34 (22.66%) |
| 50–59 | 28 (11.91%) | 7 (4.66%) |
Prevalence of low-risk HPVs in healthy blood donors in Qatar
We herein explored the presence of low-risk HPVs in healthy blood donors from Qatar, in both EBV positive and negative groups. Table 3 summarizes the prevalence of low-risk HPVs in healthy blood donors, based on our PCR analysis using specific primers for LMP1 as well as E6/E7 genes of EBV and HPVs, respectively (Materials and Methods Section). It is worth noting that using GP5+/6+ primer set is a highly sensitive specific method for HPV DNA detection [38] as it targets L1 conserved regions of the viral genome; thus, allowing detection of a comprehensive variety of HPV genotypes [39, 40]. We found that GP5/GP6 was positive for more than 60% of the samples and hence, further analysis was performed to determine HPV subtypes.
Table 3.
Prevalence of low-risk HPVs in healthy blood donors from Qatar
| Samples | No. of cases | Low-risk HPV types (%) | |||
|---|---|---|---|---|---|
| 6 | 11 | 53 | 73 | ||
| EBV (+) | 235 | 41.3 | 45.5 | 50.6 | 41.7 |
| EBV (−) | 150 | 76 | 63.3 | 54 | 75.3 |
| Total | 385 | 54.8 | 52.5 | 51.9 | 54.8 |
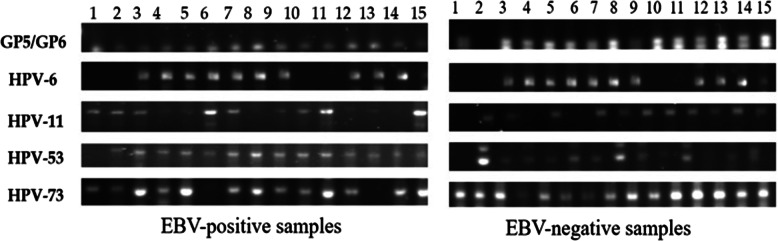
Analysis revealed that of the total 385 samples, 235 were positive and 150 were negative for EBV. On the other hand, 233 of the total samples (60.5%) were positive for low-risk HPVs, and all of the positive cases were infected with more than one type of low-risk HPV (Fig. 1). This number represents 50.6% of the 235 EBV-positive samples, and 76% of the 150 EBV-negative samples. Moreover, data revealed that the most prevalent low risk HPV types were HPV-6 and 73 (both at 54.8%), followed closely by HPV-11 (52.5%) and HPV-53 (51.9%) (Table 3).
Fig. 1.
Representative PCR reactions for low-risk HPV-subtypes in 15 different EBV-positive and EBV-negative healthy blood donors
Prevalence of high-risk HPVs in healthy blood donors in Qatar
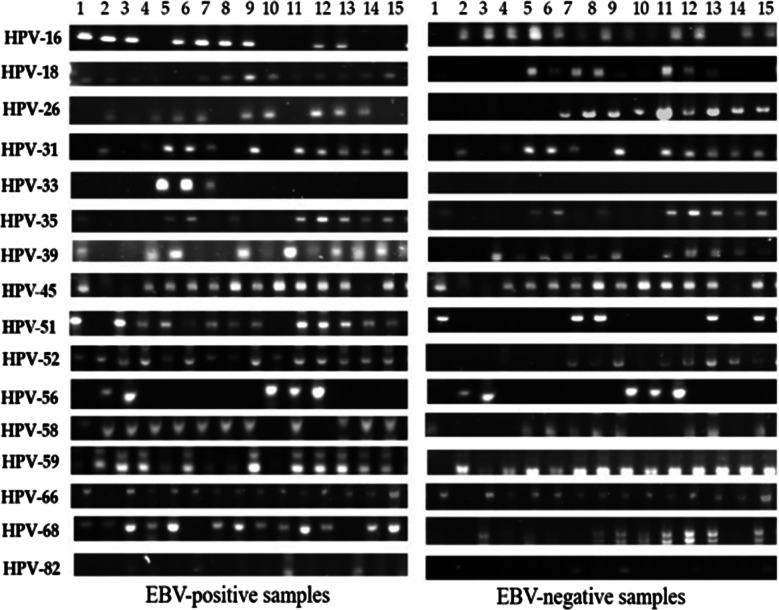
We further investigated the co-presence of high-risk HPVs and EBV in our samples by PCR using specific primers for E6/E7 as well as LMP1 genes of HPVs and EBV, respectively (Table 4). Analysis revealed that 182 of the total number of samples were positive for high-risk HPVs (47.3%), representing 40.4% of the 235 EBV-positive and 58% of the 150 EBV-negative samples. Again, all cases were infected with more than one high-risk HPV type (Fig. 2). Moreover, data revealed that the most prevalent high-risk HPV types in EBV-positive donors were HPV-59 (54.8%), HPV-31 (53.8%), HPV-52 (49.1%), HPV-51 (48.6%), HPV-58 (47%), HPV-35 (45.4%), HPV-26 (43.4%), HPV-16 (37.9%) and HPV-18 (36.1%).
Table 4.
Prevalence of high-risk HPVs in samples from healthy blood donors in Qatar
| Samples | No. of cases | High-risk HPV types (%) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16 | 18 | 26 | 31 | 33 | 35 | 39 | 45 | 51 | 52 | 56 | 58 | 59 | 66 | 68 | 82 | ||
| EBV (+) | 235 | 35.7 | 34.5 | 32.8 | 39.6 | 4.6 | 35.3 | 32.8 | 30.6 | 36.2 | 37.9 | 27.7 | 37.4 | 40.4 | 29.4 | 25.5 | 20.8 |
| EBV (−) | 150 | 41.3 | 38.6 | 60 | 76 | 0 | 61.3 | 62 | 56.6 | 68 | 66.7 | 22.6 | 62 | 77.3 | 39.3 | 71.3 | 11.3 |
| Total | 385 | 37.9 | 36.1 | 43.4 | 53.8 | 2.8 | 45.4 | 44.2 | 40.7 | 48.6 | 49.1 | 25.7 | 47 | 54.8 | 33.3 | 43.3 | 17.1 |
Fig. 2.
Representative PCR reactions for high-risk HPV-subtypes in 15 different EBV-positive and EBV-negative samples of healthy blood donors
Demographic association of HPV-Subtypes and EBV in Healthy Donors in Qatar
A demographic analysis of the samples from healthy donors by classifying them into Qataris and Non-Qataris showed that, in EBV-negative samples, HPV type 35 is the only highly prevalent subtype in both populations (65.4% and 84%, respectively, p = 0.029). However, in EBV-positive samples, as shown in Table 5, the most prevalent HPV-subtypes include HPV types 52, 56, 66 and 68 (p = 0.02, p = 0.03, p = 0.03, p = 0.02, respectively).
Table 5.
Association of HPV subtypes in 235 EBV-positive samples from Qatari vs non-Qatari healthy blood donors
| QATARIS v/s NON-QATARIS | ||||
|---|---|---|---|---|
| Samples | HPV-subtypes (%) | |||
| 52 | 56 | 66 | 68 | |
| Qataris (102) | 47 | 37.2 | 39.2 | 19.6 |
| Non-Qataris (133) | 30.8 | 24.8 | 26.3 | 32.3 |
| p-value | 0.022* | 0.034* | 0.0355* | 0.029* |
Discussion
Viral infections are the most common etiologic causes of infection-related cancer agents (~ 15%) [41, 42]. Although these infections predominantly affect developing countries, their frequency cannot be ignored in developed countries [43, 44]. In 38% of virus-related cancers, there is co-presence of HPVs along with EBV [42]. However, these cancers develop after a relatively long latent period (15–40 years) [45]. Viral infections within cells are not limited and their combined oncogenic effects can lead to the onset and/or progression of cancer disease [42, 46].
In the present study, we investigated the co-presence of both low- and high-risk HPVs and EBV in the blood of healthy individuals from the middle east region, more specifically in Qatar. Several previous studies have already shown the occurrence of HPVs in subjects with unidentified HPV-related disease or cancer; viral DNA/RNA being identified in several tissues including sperm cells [47, 48], placental tissue [49, 50] as well as peripheral blood cells [51, 52]; indicating that HPV can disseminate through the bloodstream [13, 47].
Additionally, based on few studies, HPV DNA was shown to circulate in the bloodstream during ephemeral asymptomatic infections [53–55]; however, it was indicated that detection of HPV DNA in blood samples is a useful and potential biomarker of severe HPV-related diseases including cancer. Furthermore, studies have detected HPVs in blood samples of both males and females with non-cancer related HPV urogenital infections. While an earlier study in asymptomatic women with urogenital infections reported HPV positivity in peripheral blood cells [52], another study on male patients’ blood showed HPV DNA detection in semen infection [47]. In the present study, we revealed that the presence of both low-risk and high-risk HPVs is ~ 60.5% and 47.3% in healthy blood donors in a diverse population group from Qatar, respectively. Similar to our study, a study among healthy Australian male donors conducted by Chen et al. revealed the prevalence of 8.3% for HPV infection [51]. Metagenomics analysis using whole-genome shotgun sequencing in 748 samples collected from a cohort of 103 healthy North American human subjects was conducted by the NIH Human Microbiome Project. From each subject, four organs (skin, vagina, mouth, and gut) were used for sample collection. This study in healthy humans, who did not display classical HPV-associated diseases, revealed the overall HPV prevalence of 68.9%; the highest being present in the skin (61.3%), followed by the vagina (41.5%), mouth (30%), and gut (17.3%). The co-existence of multiple HPV types was found in 48.1% of HPV-positive samples [56]. This high prevalence of HPV and coexistence of HPV subtypes in the blood of healthy subjects could be attributed to its presence at multiple body sites, as reported previously [56].
Our study has some limitations: Given that it included a very low number of female participants due to socio-cultural factors and religious/fatalistic barriers, in addition to low levels of hemoglobin in women which is reported as a cause of donor loss [57–60], it is essential to perform other studies of a larger number of cases that may ensure a higher female participation in Qatar in order to clarify gender variations in HPV and EBV distribution in this population. Such studies are essential for the region in order to allow for the selection of the right HPV and EBV vaccines as a necessary preventive tool against several serious diseases associated with these oncoviruses, including cancer. In addition, we cannot anticipate the real clinical impact of the observed frequencies of HPV and EBV in the population of Qatar. However, our group is currently exploring the co-presence of HPVs and EBV in different types of cancers in Qatar including breast, cervical as well as colorectal; analysis in breast and cervical cancers can aid in providing clear data about role of high-risk HPVs and EBV in female cases.
On the other hand, our present study showed for the first time the co-presence of high and low-risk HPVs with EBV in 40.4% and 50.6% of the examined samples, respectively. More specifically, in EBV-negative samples, approximately more than half of the Qataris were positive for low-risk skin HPV-types HPV-6, 11, 53 and 73 (Table 3) that are frequently expressed in asymptomatic infections of normal, healthy skin [61, 62]. The rest were high-risk HPV types that are linked with cancer development. Moreover, the most significantly expressed high-risk HPV subtypes in Qatari healthy donors are in line with previous studies on HPV prevalence in the Middle East [63–65], thus they included HPV-52, 56, 66 and 68 (p = 0.02, p = 0.03, p = 0.03, p = 0.02, respectively, Table 4). Although the percentage of Qatari healthy donors in this study is small, this HPV DNA prevalence in healthy males’ merits further investigation.
Conclusions
Our study provides evidence that HPV and EBV can be detected and quantified in blood samples of healthy blood donors without any obvious HPV infection, suggesting potential viral persistence at different anatomical sites. This random distribution of HPV subtypes along with EBV, suggests potential facilitative or competitive interactions among these onco-viruses, which has been recently reported by several studies [31, 34, 46, 66]. Since these oncoviruses can be found in various cancers; our group has previously demonstrated that E6/E7 of high-risk HPVs can convert non-invasive cancer cells to invasive phenotypes [22]. Additionally, HPVs genotyping can help select the most relevant HPV vaccine to prevent HPV-associated cancers. However, further studies are required to assess the role of HPV DNA detection in the bloodstream of people with asymptomatic infection to elucidate the underlying mechanisms of the co-presence of HPVs and EBV in healthy donors; especially since vaccines for these onco-viruses are available or are currently under clinical trial [67–69]. This is a vital step as vaccinating individuals can aid in lowering the risk of viral transmission in the common population and ultimately prevent the onset of HPVs- and EBV-related diseases, including cancer. The co-prevalence of HPVs and EBV among healthy blood donors should draw the attention of clinicians, researchers and healthcare workers in Qatar, which could aid in promoting safety practices in health care centers, especially in blood banks and organ transplant centers.
In conclusion, our study revealed that both low-risk and high-risk HPVs are frequently co-present with EBV among healthy blood donors in the heterogeneous population of Qatar. Further observational and clinical studies are required to assess the full relevance of the obtained data.
Acknowledgements
The authors would like to thank Mrs. A. Kassab for her critical reading of the manuscript.
Abbreviations
- DNA
Deoxyribonucleic acid
- EBV
Epstein–Barr virus
- EBNA
EBV-encoded nuclear antigens
- HPV
Human Papilloma virus
- LMP
Latent membrane protein
- PCR
Polymerase chain reaction
Authors’ contributions
Conceptualization: AEAM, MA; Methodology: IG, MKS; data curation: IG, MKS, AS, AJ; writing: IG; writing—review and editing: AS, AJ, SV, HAT, AEAM; Supervision: AEAM, SV, GKN; funding acquisition: AEAM. All authors read and approved the final manuscript.
Funding
This work is supported by Qatar University grants# GCC-2017-002 QU/KU and QUCG-CMED-2018\2019-3.
Availability of data and materials
All data generated or analyzed during this study are included in this published article [and its additional files].
Ethics approval and consent to participate
This study was approved by the Hamad Medical Corporation IRB (HMC-IRB #14292/14) and Qatar University IRB (QU-IRB 518-EA/15).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
1/29/2022
A Correction to this paper has been published: 10.1186/s12935-022-02459-4
Contributor Information
Semir Vranic, Email: semir.vranic@gmail.com.
Ala-Eddin Al Moustafa, Email: aalmoustafa@qu.edu.qa.
References
- 1.Stanley MA. Genital human papillomavirus infections: current and prospective therapies. J Gen Virol. 2012;93(4):681–691. doi: 10.1099/vir.0.039677-0. [DOI] [PubMed] [Google Scholar]
- 2.de Villiers E-M, Fauquet C, Broker TR, Bernard H-U. zur Hausen H. Classification of papillomaviruses. Virology. 2004;324(1):17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 3.Bernard H-U. The clinical importance of the nomenclature, evolution and taxonomy of human papillomaviruses. J Clin Virol. 2005;32:1–6. doi: 10.1016/j.jcv.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 4.Bosch F, Lorincz A, Munoz N, Meijer CJ, Shah KV. The causal relation between human papillomavirus and cervical cancer. J Clin Pathol. 2002;55:244–265. doi: 10.1136/jcp.55.4.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zur Hausen H. Papillomaviruses in Human Cancers. Proc Assoc Am Physician. 1999;111(6):581–587. doi: 10.1046/j.1525-1381.1999.99723.x. [DOI] [PubMed] [Google Scholar]
- 6.Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10:550. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 7.Chen X, Bode AM, Dong Z, Cao Y. The epithelial–mesenchymal transition (EMT) is regulated by oncoviruses in cancer. FASEB J. 2016;30(9):3001–3010. doi: 10.1096/fj.201600388R. [DOI] [PubMed] [Google Scholar]
- 8.Al Moustafa A-E. E5 and E6/E7 of high-risk HPVs cooperate to enhance cancer progression through EMT initiation. Cell Adh Migr. 2015;9(5):392–393. doi: 10.1080/19336918.2015.1042197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaur N, Gandhi J, Robertson ES, Verma SC, Kaul R. Epstein-Barr virus latent antigens EBNA3C and EBNA1 modulate epithelial to mesenchymal transition of cancer cells associated with tumor metastasis. Tumour Biol. 2015;36(4):3051–3060. doi: 10.1007/s13277-014-2941-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oriel JD. Natural history of genital warts. Br J Vener Dis. 1971;47(1):1–13. doi: 10.1136/sti.47.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tseng C-J, Pao CC, Lin J-D, Soong Y-K, Hong J-H, Hsueh S. Detection of human Papillomavirus Types 16 and 18 mRNA in peripheral blood of advanced cervical cancer patients and its association with prognosis. J Clin Oncol. 1999;17(5):1391. doi: 10.1200/JCO.1999.17.5.1391. [DOI] [PubMed] [Google Scholar]
- 12.Tsai H-J, Peng Y-W, Lin L-Y, Chou M-C, Lee H, Chiou H-L. An association between human papillomavirus 16/18 deoxyribonucleic acid in peripheral blood with p16 protein expression in neoplastic cervical lesions. Cancer Detect Prev. 2005;29(6):537–543. doi: 10.1016/j.cdp.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Bodaghi S, Wood LV, Roby G, Ryder C, Steinberg SM, Zheng Z-M. Could human papillomaviruses be spread through blood? J Clin Microbiol. 2005;43(11):5428–5434. doi: 10.1128/JCM.43.11.5428-5434.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong SM, Pai SI, Rha S-H, Hildesheim A, Kurman RJ, Schwartz PE, et al. Detection and quantitation of human Papillomavirus DNA in the plasma of patients with cervical carcinoma. Cancer Epidemiol Biomark Prev. 2002;11(1):3–6. [PubMed] [Google Scholar]
- 15.Rombaldi RL, Serafini EP, Mandelli J, Zimmermann E, Losquiavo KP. Transplacental transmission of Human Papillomavirus. Virol J. 2008;5:106. doi: 10.1186/1743-422X-5-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmer R, Quinton W, Gray J. Addendum. The Lancet. 1964;283(7335):702. [PubMed] [Google Scholar]
- 17.```.Santpere G, Darre F, Blanco S, Alcami A, Villoslada P, Mar Albà M, et al. Genome-wide analysis of wild-type Epstein-Barr virus genomes derived from healthy individuals of the 1,000 Genomes Project. Genome Biol Evol. 2014;6(4):846–860. doi: 10.1093/gbe/evu054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, Benbrahim-Tallaa L, Guha N, Freeman C, Galichet L, Cogliano V. WHO International Agency for Research on Cancer Monograph Working Group. A review of human carcinogens–Part B: biological agents. Lancet Oncol. 2009;10(4):321–322. doi: 10.1016/s1470-2045(09)70096-8. [DOI] [PubMed] [Google Scholar]
- 19.Sample J, Young L, Martin B, Chatman T, Kieff E, Rickinson A, et al. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J Virol. 1990;64(9):4084–4092. doi: 10.1128/jvi.64.9.4084-4092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guidry JT, Scott RS. The interaction between human papillomavirus and other viruses. Virus Res. 2017;231:139–147. doi: 10.1016/j.virusres.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Chang YS. Epstein-Barr virus latent membrane protein 1: structure and functions. J Biomed Sci. 2003;10(5):490–504. doi: 10.1007/BF02256110. [DOI] [PubMed] [Google Scholar]
- 22.Cyprian FS, Al-Farsi HF, Vranic S, Akhtar S, Al Moustafa A-E. Epstein-Barr Virus and Human Papillomaviruses interactions and their roles in the initiation of epithelial-mesenchymal transition and cancer progression. Front Oncol. 2018;8:111. doi: 10.3389/fonc.2018.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tzellos S, Farrell PJ. Epstein-barr virus sequence variation-biology and disease. Pathogens. 2012;1(2):156–174. doi: 10.3390/pathogens1020156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yakovleva L, Senyuta NB, Goncharova EV, Scherback LN, Smirnova RV, Pavlish OA, Gurtsevitch VE. Epstein-Barr Virus LMP1 oncogene variants in cell lines of different origin. Mol Biol. 2015;49(5):800–810. doi: 10.7868/S0026898415050213. [DOI] [PubMed] [Google Scholar]
- 25.Kong Q-L, Hu L-J, Cao J-Y, Huang Y-J, Xu L-H, Liang Y, et al. Epstein-Barr virus-encoded LMP2A induces an epithelial-mesenchymal transition and increases the number of side population stem-like cancer cells in nasopharyngeal carcinoma. PLoS Pathog. 2010;6(6):e1000940. doi: 10.1371/journal.ppat.1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin Z, Wan X, Jiang R, Deng L, Gao Y, Tang J, et al. Epstein-Barr virus-encoded latent membrane protein 2A promotes the epithelial-mesenchymal transition in nasopharyngeal carcinoma via metastatic tumor antigen 1 and mechanistic target of rapamycin signaling induction. J Virol. 2014;88(20):11872–11885. doi: 10.1128/JVI.01867-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mui UN, Haley CT, Tyring SK. Viral oncology: molecular biology and pathogenesis. J Clin Med. 2017;6(12):111. doi: 10.3390/jcm6120111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerber P, Walsh J, Rosenblum E, Purcell R. Association of EB-Virus infection with the post-perfusion syndrome. Lancet. 1969;293(7595):593–596. doi: 10.1016/s0140-6736(69)91532-3. [DOI] [PubMed] [Google Scholar]
- 29.Hanto DW, Frizzera G, Purtilo DT, Sakamoto K, Sullivan JL, Saemundsen AK, et al. Clinical Spectrum of Lymphoproliferative Disorders in Renal Transplant Recipients and Evidence for the Role of Epstein-Barr Virus. Cancer Res. 1981;41:4253–4261. [PubMed] [Google Scholar]
- 30.Alfieri C, Tanner J, Carpentier L, Perpete C, Savoie A, Paradis K, et al. Epstein-Barr virus transmission from a blood donor to an organ transplant recipient with recovery of the same virus strain from the recipient’s blood and oropharynx. Blood. 1996;87(2):812–817. [PubMed] [Google Scholar]
- 31.Al Moustafa A-E, Al-Antary N, Aboulkassim T, Akil N, Batist G, Yasmeen A. Co-prevalence of Epstein-Barr virus and high-risk human papillomaviruses in Syrian women with breast cancer. Human Vaccin Immunother. 2016;12(7):1936–1939. doi: 10.1080/21645515.2016.1139255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smatti MK, Yassine HM, AbuOdeh R, AlMarawani A, Taleb SA, Althani AA, et al. Prevalence and molecular profiling of Epstein Barr virus (EBV) among healthy blood donors from different nationalities in Qatar. PLoS ONE. 2017;12(12):e0189033. doi: 10.1371/journal.pone.0189033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darnel AD, Wang D, Ghabreau L, Yasmeen A, Sami S, Akil N, et al. Correlation between the presence of high-risk human papillomaviruses and Id gene expression in Syrian women with cervical cancer. Clin Microbiol Infect. 2010;16(3):262–266. doi: 10.1111/j.1469-0691.2009.02774.x. [DOI] [PubMed] [Google Scholar]
- 34.Al-Thawadi H, Ghabreau L, Aboulkassim T, Yasmeen A, Vranic S, Batist G, et al. Co-incidence of Epstein-Barr virus and high-risk human Papillomaviruses in cervical cancer of Syrian women. Front Oncol. 2018;8:250. doi: 10.3389/fonc.2018.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heng B, Glenn WK, Ye Y, Tran B, Delprado W, Lutze-Mann L, et al. Human papilloma virus is associated with breast cancer. Br J Cancer. 2009;101(8):1345–1350. doi: 10.1038/sj.bjc.6605282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao C-Y, Fu B-B, Li Z-Y, Mushtaq G, Kamal MA, Li J-H, et al. Observations on the expression of human papillomavirus major capsid protein in HeLa cells. Cancer Cell Int. 2015;15:53. doi: 10.1186/s12935-015-0206-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Al Moustafa A, Foulkes WD, Benlimame N, Wong A, Yen L, Bergeron J, Batist G, Alpert L, Alaoui-Jamali MA. E6/E7 proteins of HPV type 16 and ErbB-2 cooperate to induce neoplastic transformation of primary normal oral epithelial cells. Oncogene. 2004;23:350–358. doi: 10.1038/sj.onc.1207148. [DOI] [PubMed] [Google Scholar]
- 38.Fuessel Haws AL, He Q, Rady PL, Zhang L, Grady J, Hughes TK, et al. Nested PCR with the PGMY09/11 and GP5+/6+ primer sets improves detection of HPV DNA in cervical samples. J Virol Methods. 2004;122(1):87–93. doi: 10.1016/j.jviromet.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 39.Rady PL, Chin R, Arany I, Hughes TK, Tyring SK. Direct sequencing of consensus primer generated PCR fragments of human papillomaviruses. J Virol Methods. 1993;43(3):335–350. doi: 10.1016/0166-0934(93)90151-g. [DOI] [PubMed] [Google Scholar]
- 40.Rady PL, Arany I, Hughes TK, Tyring SK. Type-specific primer-mediated direct sequencing of consensus primer-generated PCR amplicons of human papilloma viruses: a new approach for the simultaneous detection of multiple viral type infections. J Virol Methods. 1995;53(2):245–254. doi: 10.1016/0166-0934(95)00029-t. [DOI] [PubMed] [Google Scholar]
- 41.Weiss P. Tumour-inducing viruses. Br J Hosp Med. 2016;77(Sup10):565–568. doi: 10.12968/hmed.2016.77.10.565. [DOI] [PubMed] [Google Scholar]
- 42.Shi Y, Peng S-L, Yang L-F, Chen X, Tao Y-G, Cao Y. Co-infection of Epstein-Barr virus and human papillomavirus in human tumorigenesis. Chin J Cancer. 2016;35(1):16. doi: 10.1186/s40880-016-0079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Antonsson A, Wilson LF, Kendall BJ, Bain CJ, Whiteman DC, Neale RE. Cancers in Australia in 2010 attributable to infectious agents. Aust NZ J Public Health. 2015;39(5):446–451. doi: 10.1111/1753-6405.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118(12):3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 45.Zur Hausen H. Papillomaviruses in the causation of human cancers—a brief historical account. Virology. 2009;384(2):260–265. doi: 10.1016/j.virol.2008.11.046. [DOI] [PubMed] [Google Scholar]
- 46.Al Moustafa A-E, Cyprian FS, Al-Antary N, Yasmeen A. High-risk human Papillomaviruses and Epstein-Barr Virus presence and crosstalk in human oral carcinogenesis. In: Al Moustafa A-E, editor. Development of oral cancer: risk factors and prevention strategies. Cham: Springer; 2017. pp. 83–94. [Google Scholar]
- 47.Foresta C, Bertoldo A, Garolla A, Pizzol D, Mason S, Lenzi A, et al. Human papillomavirus proteins are found in peripheral blood and semen Cd20+ and Cd56+ cells during HPV-16 semen infection. BMC Infect Dis. 2013;13:593. doi: 10.1186/1471-2334-13-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rintala M, Grénman SE, Pöllänen PP, Suominen JJ, Syrjänen SM. Detection of high-risk HPV DNA in semen and its association with the quality of semen. Int J STD AIDS. 2004;15(11):740–743. doi: 10.1258/0956462042395122. [DOI] [PubMed] [Google Scholar]
- 49.Syrjänen S. Current concepts on human papillomavirus infections in children. APMIS. 2010;118(6–7):494–509. doi: 10.1111/j.1600-0463.2010.02620.x. [DOI] [PubMed] [Google Scholar]
- 50.Tseng C-J, Lin C-Y, Wang R-L, Chen L-J, Chang Y-L, Hsieh T-T, et al. Possible transplacental transmission of human papillomaviruses. Am J Obstet Gynecol. 1992;166:35–40. doi: 10.1016/0002-9378(92)91825-u. [DOI] [PubMed] [Google Scholar]
- 51.Chen AC-H, Keleher A, Kedda M-A, Spurdle AB, Antonsson A, McMillan NAJ. Human papillomavirus DNA detected in peripheral blood samples from healthy Australian male blood donors. J Med Virol. 2009;81(10):1792–1796. doi: 10.1002/jmv.21592. [DOI] [PubMed] [Google Scholar]
- 52.Pao CC, Lin S-S, Lin C-Y, Maa J-S, Lai C-H, Hsieh T-T. Identification of human Papillomavirus DNA sequences in peripheral blood Mmononuclear cells. Am J Clin Pathol. 1991;95(4):540–546. doi: 10.1093/ajcp/95.4.540. [DOI] [PubMed] [Google Scholar]
- 53.Chiou H-L, Wu M-F, Liaw Y-C, Cheng Y-W, Wong R-H, Chen C-Y, et al. The presence of human papillomavirus type 16/18 DNA in blood circulation may act as a risk marker of lung cancer in Taiwan. Cancer. 2003;97(6):1558–1563. doi: 10.1002/cncr.11191. [DOI] [PubMed] [Google Scholar]
- 54.Capone RB, Pai SI, Koch WM, Gillison ML, Danish HN, Westra WH, et al. Detection and quantitation of human Papillomavirus (HPV) DNA in the Sera of patients with HPV-associated head and neck squamous cell carcinoma. Clin Cancer Res. 2000;6(11):4171–4175. [PubMed] [Google Scholar]
- 55.Widschwendter A, Blassnig A, Wiedemair A, Müller-Holzner E, Müller HM, Marth C. Human papillomavirus DNA in sera of cervical cancer patients as tumor marker. Cancer Lett. 2003;202(2):231–239. doi: 10.1016/j.canlet.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 56.Ma Y, Madupu R, Karaoz U, Nossa CW, Yang L, Yooseph S, et al. Human Papillomavirus community in healthy persons, defined by metagenomics analysis of human microbiome project shotgun sequencing data sets. J Virol. 2014;88(9):4786–4797. doi: 10.1128/JVI.00093-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Al Shaer L, Sharma R, AbdulRahman M. Analysis of blood donor pre-donation deferral in Dubai: characteristics and reasons. J Blood Med. 2017;2017(8):55–60. doi: 10.2147/JBM.S135191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arun R, Subash S, Arumugam P. Analysis of blood donor deferral causes in Chenna Indiai, Int J Med Health Sci. 2012;3:61–65. [Google Scholar]
- 59.Okoroiwu HU, Asemota EA. Blood donors deferral prevalence and causes in a tertiary health care hospital, southern Nigeria. BMC Health Serv Res. 2019;19(1):510. doi: 10.1186/s12913-019-4352-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khurram S, Borhany M, Anwar N, Naseer I, Boota S, Mirza I, et al. Frequency and reasons of donor deferral prior to blood donation process: a single centre experience. Transfus Med. 2017;27(1):10–15. doi: 10.1111/tme.12368. [DOI] [PubMed] [Google Scholar]
- 61.Chen AC, McMillan NAJ, Antonsson A. Human papillomavirus type spectrum in normal skin of individuals with or without a history of frequent sun exposure. J Gen Virol. 2008;89(11):2891–2897. doi: 10.1099/vir.0.2008/003665-0. [DOI] [PubMed] [Google Scholar]
- 62.Antonsson A, Karanfilovska S, Lindqvist PG, Hansson BG. General acquisition of human Papillomavirus infections of skin occurs in early infancy. J Clin Microbiol. 2003;41(6):2509–2514. doi: 10.1128/JCM.41.6.2509-2514.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Elmi AA, Bansal D, Acharya A, Skariah S, Dargham SR, Abu-Raddad LJ, et al. Human Papillomavirus (HPV) infection: molecular epidemiology, genotyping, seroprevalence and associated risk factors among Arab Women in Qatar. PLoS ONE. 2017;12(1):e0169197. doi: 10.1371/journal.pone.0169197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Albawardi A, Quddus MR, Al Awar S, Almarzooqi S. Frequency of rare and multi viral high-risk HPV types infection in cervical high grade squamous intraepithelial lesions in a non-native dominant middle eastern country: a polymerase chain reaction-based pilot study. Diagn Pathol. 2018;13(1):42. doi: 10.1186/s13000-018-0716-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seoud M. Burden of Human Papillomavirus-Related Cervical Disease in the Extended Middle East and North Africa—A Comprehensive Literature Review. J Lower Genit Tract Dis. 2012;16(2):106–120. doi: 10.1097/LGT.0b013e31823a0108. [DOI] [PubMed] [Google Scholar]
- 66.Vranic S, Cyprian FS, Akhtar S, Al Moustafa A-E. The role of Epstein-Barr Virus in cervical cancer: a brief update. Front Oncol. 2018;8:113. doi: 10.3389/fonc.2018.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rajčáni J, Bánáti F, Szenthe K, Szathmary S. The potential of currently unavailable herpes virus vaccines. Expert Rev Vaccin. 2018;17(3):239–248. doi: 10.1080/14760584.2018.1425620. [DOI] [PubMed] [Google Scholar]
- 68.Luxembourg A, Moeller E. 9-Valent human papillomavirus vaccine: a review of the clinical development program. Expert Rev Vaccin. 2017;16(11):1119–1139. doi: 10.1080/14760584.2017.1383158. [DOI] [PubMed] [Google Scholar]
- 69.Lee L-Y, Garland SM. Human papillomavirus vaccination: the population impact. F1000Research. 2017;6:866. doi: 10.12688/f1000research.10691.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article [and its additional files].