Abstract
The OC43 strain of human coronavirus (HCoV-OC43) is an ubiquitous respiratory tract pathogen possessing neurotropic capacities. Coronavirus structural envelope (E) protein possesses specific motifs involved in protein-protein interaction or in homo-oligomeric ion channel formation, which are known to play various roles including in virion morphology/assembly and in cell response to infection and/or virulence. Making use of recombinant viruses either devoid of the E protein or harboring mutations either in putative transmembrane domain or PDZ-binding motif, we demonstrated that a fully functional HCoV-OC43 E protein is first needed for optimal production of recombinant infectious viruses. Furthermore, HCoV-OC43 infection of human epithelial and neuronal cell lines, of mixed murine primary cultures from the central nervous system and of mouse central nervous system showed that the E protein is critical for efficient and optimal virus replication and propagation, and thereby for neurovirulence.
Keywords: HCoV-OC43, Coronavirus, E protein, Transmembrane domain, PDZ binding motif, Virus production, Pathogenesis
Highlights
-
•
Coronavirus structural envelope (E) protein specific motifs involved in protein-protein interaction or in homo-oligomeric ion channel formation are needed for optimal production of recombinant infectious virus.
-
•
Fully functional E protein of HCoV-OC43 is crucial for viral propagation in the CNS and neurovirulence.
-
•
Fully functional E protein of HCoV-OC43 is crucial for efficient viral propagation in the central nervous system and thereby for neurovirulence.
1. Introduction
Coronaviruses are widespread RNA viruses of the Nidovirales order, Coronaviridae family, most often associated with human and veterinary respiratory infections (de Groot et al., 2012). Of the six human-infecting coronavirus strains, four (HCoV-229E, HCoV-NL63, HCoV-HKU1 and HCoV-OC43) are currently co-circulating and elicit respiratory illnesses (Vabret et al., 2009). Coronaviruses also represent a significant public health concern due to the recent zoonotically emerged, highly pathogenic species, SARS corovonavirus (SARS-CoV) (Drosten et al., 2003, Ksiazek et al., 2003) in 2002–2003 and, since 2012, Middle-East respiratory syndrome coronavirus (MERS-CoV) (Zaki et al., 2012), localized to the Arabian Peninsula, but with sporadic travel-related outbreaks worldwide. In addition to their respiratory tropism, human coronaviruses have been detected concurrently with severe and acute neurological symptoms (Arabi et al., 2015, Morfopoulou et al., 2016, Yeh et al., 2004) and shown to naturally infect the central nervous system (CNS) (Arbour et al., 2000, Gu et al., 2005, Xu et al., 2005) with neurons demonstrated as the main target of infection in HCoV-OC43 (Bonavia et al., 1997, Favreau et al., 2012, Jacomy et al., 2006, Jacomy and Talbot, 2003) and SARS-CoV (Gu et al., 2005, Xu et al., 2005).
Coronaviruses represent the largest known enveloped RNA (single-stranded positive sense) viruses with a genome of approximately 30 kb (de Groot et al., 2012). The viral envelope is composed of four or five proteins, the spike (S), membrane (M), envelope (E) and hemagglutinin-esterase protein (HE), the latter in some β coronaviruses genus, such as HCoV-OC43. Coronavirus E proteins are 74–109 amino acids in length, 84 amino acids for HCoV-OC43, and share only a small amount of sequence identity between coronavirus species. However, its secondary structure, composed of a short N-terminal domain followed by a single hydrophobic transmembrane domain (TMD) and hydrophilic cytoplasmic tail, remains overall conserved and is suggested to be more important than sequence for function (Kuo et al., 2007, Torres et al., 2005). The importance of the presence of the E protein in the viral envelope is emphasized by the fact that there are only about twenty E molecules incorporated within the virion structure (Godet et al., 1992, Liu and Inglis, 1991, Yu et al., 1994) and deletion of the protein can either completely prevent the production of detectable infectious virions (Almazán et al., 2013, Curtis et al., 2002, Ortego et al., 2007, Ortego et al., 2002) or significantly reduce infectious virus titers (DeDiego et al., 2008, DeDiego et al., 2007, Kuo et al., 2007, Kuo and Masters, 2003).
The majority of the coronavirus E protein in the infected cell is localized within the secretory pathway between the membranes of the endoplasmic reticulum (ER), Golgi and intermediate compartment between them (ERGIC) (Cohen et al., 2011, Nieto-Torres et al., 2011, Venkatagopalan et al., 2015). It is in this intracellular region that additional functions mediated by various domains of the coronavirus E proteins are proposed to occur. Homo-pentameric oligomerization of the E protein TMD in membranes to form ion channels, called viroporins, has been predicted for several coronaviruses (Torres et al., 2005) and extensively studied for species such as SARS-CoV (Nieto-Torres et al., 2014, Pervushin et al., 2009) or avian infectious bronchitis virus (IBV) (Ruch and Machamer, 2012, Westerbeck and Machamer, 2015). Another domain found at the extreme C-terminal end of the E protein, a PDZ-domain binding motif (PBM), has also been predicted for several coronavirus species (Jimenez-Guardeño et al., 2014). This protein-protein interaction motif capable of interrupting normal cellular functions, has been demonstrated in other viruses to play important roles in replication, dissemination in the host and pathogenesis (Javier and Rice, 2011). The multiple properties of coronavirus E proteins have not yet been fully investigated or explained, and can at times differ between coronavirus species. The multifunctionality of the E protein could be explained by the presence of two distinct pools (monomeric versus homo-oligomeric states) present in the infected cell (Westerbeck and Machamer, 2015). Furthermore, the different motifs found within the protein (Jimenez-Guardeño et al., 2015) could mediate different specific functions. The coronavirus E protein was also recently recognized as an important virulence factor for the SARS-CoV (Jimenez-Guardeño et al., 2014), where deletion of the whole or part of the protein led to an attenuated pathology in mouse lungs (DeDiego et al., 2008, DeDiego et al., 2007), attenuation which was later linked to the E protein TMD and PBM (Jimenez-Guardeño et al., 2014, Nieto-Torres et al., 2014).
HCoV-OC43 represents a circulating strain of human coronavirus causing respiratory illness, which is naturally capable of invading the CNS where neurons are preferentially targeted for infection. In this study, we demonstrate that the fully functional HCoV-OC43 E protein (harboring specific TMD and PBM) is critical in infectious virus production and dissemination in epithelial and neuronal cell cultures and in the murine CNS and that it is a determinant of neurovirulence, a first demonstration for this coronavirus species.
2. Results
2.1. Deletion of HCoV-OC43 E protein abrogates infectious virion production, introduces a strong selection pressure for reversion in progeny production
In order to evaluate the importance of the HCoV-OC43 E protein in infectious virion production, a stop codon was introduced at the beginning of the E gene of our cDNA infectious clone, pBAC-OC43FL (St-Jean et al., 2006), preventing corresponding full-length E protein production in the resultant recombinant virus ( Fig. 1A). Transfection of BHK-21 cells with the pBAC-OC43FL led to the detection of reference HCoV-OC43 recombinant infectious virus (rOC/ATCC) whereas transfection with the pBAC-OC43-E-Stop mutant did not lead to any detectable infectious virus (rOC/E-Stop) (Fig. 1B).
Fig. 1.
The HCoV-OC43 E protein is critical for infectious particle production. (A) Representation of the full-length HCoV-OC43 genome found within the pBAC-OC43FL infectious clone (top) with a schematic representation of the HCoV-OC43 E gene to be modified at various amino acid positions indicated at their relative positions within the protein (bottom). TM, transmembrane; PBM, PDZ-binding motif. (B) Evaluation of infectious virus production corresponding to pBAC-E-Stop transfection of BHK-21 cells compared to pBAC-OC43FL. (C) Insertion of wild-type E gene into pcDNA3.1(+) expression vector, pcDNA(OC-E), yielded corresponding HCoV-OC43 E protein expression compared to empty vector. (D) Visualization of transfection efficiency on BHK-21 cells of various conditions by immunofluorescence detection of HCoV-OC43 S protein (green), nucleus staining with DAPI (blue). Subpanels: (a) mock, (b) pBAC-OC43FL (rOC/ATCC), (c) pBAC-E-Stop (rOC/E-Stop + 2 μg pcDNA(OC-E)), (d) pBAC-E-Stop (rOC/E-Stop + 2 μg pcDNA(empty)). (E) Transient co-transfection of pBAC-E-Stop and 1or 2 μg pcDNA(OC-E) in BHK-21 cells rescued detectable infectious virus in a dose-dependent manner. (* P<0.05) (F) Evaluation of infectious recombinant virus production after transient co-transfection of pBAC-E-Stop and pcDNA(OC-E) in BHK-21 cells (BHK 0). The supernatants (P0) were amplified three times by inoculation of HRT-18 cells (HRT 1–3). Infectious viral titer differences observed between experiments, revealed, by sequencing (G), the appearance of reversions at the position in the E gene where a stop codon was introduced are indicated by bold and underline. LOD, limit of detection. ┼ (cross) indicates appearance of reversion(s) in the HCoV-OC43 E gene in viral stocks as detected by sequencing.
To confirm that the inability to detect infectious viral particles was due to the lack of E protein expression, we wished to verify whether viral production could be rescued with wild-type E protein. Transfection of a plasmid containing the reference HCoV-OC43 E gene, pcDNA(OC-E), in BHK-21 cells clearly showed via Western blot assay (WB) that the E protein was produced compared to an empty plasmid condition (Fig. 1C). Subsequently, a transient co-transfection was conducted in the same cells with pBAC-E-Stop and pcDNA(OC-E) and, by making use of a monoclonal antibody against the S protein of HCoV-OC43, we confirmed that the co-transfection did not affect transfection efficiency, and that the viral S protein was produced at equivalent levels in cells transfected with pBAC-OC43FL alone or pBAC-E-Stop with pcDNA(OC-E) or empty plasmid (Fig. 1D). Following the co-transfection, infectious particles production was rescued to detectable levels in a dose-dependent manner (Fig. 1E). Viral RNA was harvested and cDNA sequenced to confirm that the infectious particles detected after transfection corresponded to rOC/ATCC and rOC/E-Stop mutant (data not shown).
As we were able to rescue infectious particles production through transient complementation, we wondered whether this resultant virus, still lacking the E gene, could be amplified further in subsequent passages. To this end, we amplified the viral stocks of all transfected plasmids three times without trans-complementation on HRT-18 epithelial cells, each time normalizing to the lowest detectable viral titer to infect cells at an identical MOI for all recombinant viruses (Fig. 1F). Throughout the amplification process we were consistently unable to detect infectious viral particles issuing from viral stocks of rOC/E-Stop. Amplifications of initially complemented viral stocks of rOC/E-Stop led to detectable titers which decreased over the course of each subsequent amplification compared to rOC/ATCC. Sequencing of viral RNA confirmed that the E gene in the viral rOC/E-Stop stocks contained the introduced stop codon at each amplification step (data not shown). These results demonstrate that production of progeny infectious HCoV-OC43 virions is still possible in the absence E protein, however the efficiency of the process is dramatically diminished.
Interestingly, when conducting independent experiments following the same experimental approach, the titers of initially complemented rOC/E-Stop sometimes increased substantially after two or three amplifications, approaching reference virus titer levels after three rounds of amplification on HRT-18 cells (Fig. 1F). Sequence analysis of the E gene of the corresponding viral stocks revealed that a reversion of sequence appeared at the position where the stop codon had been initially introduced; representing reversion to wild-type or new amino acids (Fig. 1G). Taken together, these data demonstrate that the HCoV-OC43 E protein is critical for efficient infectious virion production in epithelial cells.
2.2. Neuronal cells are susceptible to infection with HCoV-OC43 lacking E protein but progeny virus production is severely inhibited
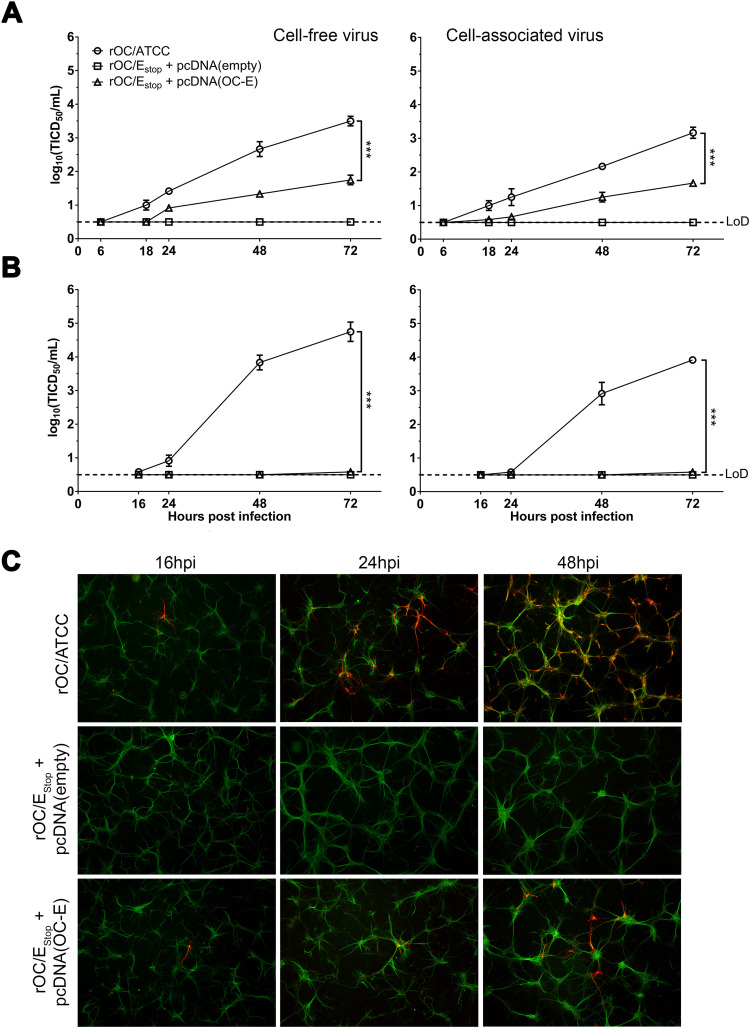
HCoV-OC43 is neuroinvasive (Arbour et al., 2000) and neurotropic, with the neuron being the main target of infection in the CNS (Jacomy et al., 2006, Jacomy and Talbot, 2003). Therefore, we sought to investigate whether the absence of the E protein would modify these neurotropic capacities by infecting a susceptible differentiated human neuronal cell line (LA-N-5) or mixed primary cultures of murine CNS cells. Initially complemented rOC/E-Stop, previously recovered from transfection on BHK-21 cells (P0), was used for infection and infectious viral titers determined over a period of 72 h post-infection (hpi). This revealed an important decrease of infectious virus production for human cells ( Fig. 2A), which was exacerbated in primary murine cells, where virus titers were under the limit of detection (Fig. 2B). However, in these primary cultures, low levels of infected cells were visualized by immunofluorescence (IFA) where we detect the viral S protein, suggesting that infection was possible even for the complemented rOC/E-Stop virus but that production of new infectious progeny and eventual propagation were severely inhibited compared to wild type virus (Fig. 2C).
Fig. 2.
HCoV-OC43 lacking E protein can infect neuronal cells but replication is severely impaired. LA-N-5 (A) and mixed primary mouse CNS cells (B) were infected with supernatant coming from BHK21 supernatant (P0) and containing virus lacking E protein, rOC/E-Stop + pcDNA(empty), or initially complemented virus, rOC/E-Stop + pcDNA(OC-E). Cell-free and cell-associated virus infectious titers were determined over 72 h post-infection. Representative of three different experiments. Statistical significance was tested at 72hpi (*** P < 0.001). (C) IFA on infected mixed primary murine CNS cells over 48 h. Green represents the microtubule associated protein 2 (MAP2) staining in neurons; red represents viral S protein.
2.3. HCoV-OC43 E protein putative transmembrane domain integrity is important for efficient infectious virion production and efficient infection of neuronal cells
The transmembrane domain (TMD) of some coronavirus E protein is known to homo-oligomerize in membranes and appears to modulate infectious virus production (Nieto-Torres et al., 2014). In order to determine the effect of HCoV-OC43 E protein TMD on virus production in cell culture, pBAC-OC43FL was modified at a key amino acid position previously identified in other coronaviruses to be critical for the stability of this specific domain (Nieto-Torres et al., 2014, Ruch and Machamer, 2012) and compared against wild-type virus during infection of cells. The large, polar glutamine at position 17 of the HCoV-OC43 E protein putative TMD was modified into a smaller, non-polar alanine (pBAC-E-TM-Q17A) in order to diminish any possible ion channel selectivity conveyed by this amino acid (Pervushin et al., 2009) at the opening of the putative ion channel.
Transfection of transmembrane mutant in BHK-21 cells yielded detectable virus titers of rOC/E-TMQ17A, which was further amplified on HRT-18 cells (P1) to significantly lower titers compared to reference virus ( Fig. 3A). Infection of human LA-N-5 cells and mixed primary cultures of mouse CNS cells showed a similar virus production kinetic with an initial delay over the first 24hpi in the cell-free fraction (Fig. 3B and C, left panels). However, in the cell-associated fractions, the amount of recovered infectious virus particles was almost identical to those of the reference virus (Fig. 3B and C, right panels), suggesting a possible defect in virus release. In order to ascertain this potential defect, human LA-N-5 cells were infected with either rOC/ATCC or rOC/E-TMQ17A, and both infectious titer and viral RNA copies (associated to total viral particles) in the cell-free fraction were quantified at 16 h post-infection (Fig. 3D). Only the infectious titer was quantified in the cell-associated fraction as (with our system of q-RT-PCR), we are not able to differentiate between viral genomic and subgenomic RNA and between viral RNA that would be inserted in virion or free in the infected cell. Similar levels of viral infectious particles were measured in the cell-associated fractions for both viruses at 16hpi indicating that the cells have been infected with the same efficiency and that the viral replication early steps are not affected by the alteration of the TMD. However infectious viral titer in the supernatant (cell-free fraction) of cells infected with rOC/E-TMQ17A was significantly reduced compared to cells infected with rOC/ATCC, although both viruses produced similar amount of total viral particles. These findings are supported by immunofluorescence assay showing a delay in propagation of rOC/E-TMQ17A over 72 hpi compared to reference virus (Fig. 3E, Figure S1A and B).
Fig. 3.
The putative HCoV-OC43 transmembrane domain plays an important role in infectious virion production, release, replication and propagation in neuronal cells. (A) Production of infectious virus after transfection of E protein transmembrane mutant in BHK-21 cells (BHK 0) and amplification on HRT-18 cells (HRT 1). (B) LA-N-5 human neuronal cells and (C) mixed primary mouse CNS cells were infected with rOC/E-TMQ17A. Cell-free and cell-associated virus fractions were recovered and tittered over 72 h. The results show a representative experiment. Statistical significance was tested at 72hpi (** P < 0.01; *** P < 0.001). (D) Infectious virus production was evaluated in the cell-associated or cell-free fraction, and total viral RNA (representative of total viral particles production) was quantified by RT-qPCR in the cell-free fraction of rOC/ATCC or rOC/E-TMQ17A infected cells following 16 h of incubation with 200 nM chloroquine to prevent re-infection. (E) The percentage of LA-N-5 cells infected by the transmembrane mutant (representative of viral propagation) was quantified by immunofluorescence and cell profiler software and compared to the reference virus over 72 h post-infection. LOD, limit of detection.
2.4. HCoV-OC43 E protein putative C-terminal protein-protein interaction motif is critical for efficient infectious virion production and dissemination
Bioinformatics analysis and modeling suggest that several coronavirus species possess a PDZ-domain binding motif (PBM) at the extreme C-terminus of their E protein that could interact with cellular and viral proteins (Jimenez-Guardeño et al., 2014). We sought to investigate whether the putative four-amino acid E protein PBM of HCoV-OC43 modulates production of infectious particles and infection of susceptible cells. To this end, we modified our cDNA infectious clone to change the two key amino acids of the putative PBM motif, at the −0 and −2 positions from the C-terminal end respectively, into inert alanines, and thereby abrogated putative motif recognition by potential interaction partner(s). Single amino acid mutants (pBAC-OC-E-PBM-D82A and pBAC-OC-E-PBM-V84 A) or double mutant (pBAC-OC-E-PBM-D82A-V84A) (Fig. 1A), were transfected in BHK-21 cells and amplified on HRT-18 cells at the same multiplicity of infection and compared to reference virus ( Fig. 4A). Amplification on HRT-18 cells demonstrated that viral titers of the double mutant were significantly decreased compared to other viruses.
Fig. 4.
A functional PDZ-domain binding motif at the C-terminal end of the HCoV-OC43 E protein is critical for efficient virus production and spread in neuronal cells. (A) Production of infectious virus after transfection of various E protein mutants with fully or partially abrogated predicted PBM after transfection in BHK-21 cells (BHK 0) and amplification on HRT-18 cells (HRT 1). The results show a representative experiment. Cell-free and cell-associated infectious virus production were determined at indicated timepoints after infection at an MOI of 0.05 on (B) LA-N-5 cells and (C) mixed primary cultures of mouse CNS cells. The results show a representative experiment. Statistical significance was tested at 72hpi (** P < 0.01; *** P < 0.001). (D) The percentage of LA-N-5 cells infected by the transmembrane mutant (representative of viral propagation) was quantified by immunofluorescence and cell profiler software and compared to the reference virus over 72 h post-infection. LOD, limit of detection.
To investigate whether the ability to infect susceptible cells, replicate and disseminate is affected by the putative C-terminal PBM in the context of the CNS, LA-N-5 or mixed primary cultures of murine CNS cells were infected with single or double mutant PBM viruses and viral titers and propagation were analyzed. In LA-N-5 cells, after 18 hpi, the titers of rOC/E-PBMD82A-V84A were significantly decreased in the cell-free and cell-associated virus fraction compared to single PBM mutants or reference viruses and total infectious virus titers of the double mutant was severely altered over 72 h (Fig. 4B). This trend was exacerbated in primary mixed murine CNS cultures, which showed no detection of infectious rOC/E-PBMD82A-V84A compared to single mutant PBM and reference viruses (Fig. 4C). As no differences were observed between both single PBM mutant (rOC/E-PBMD82A and rOC/E-PBMV84A) and reference virus, we continued the characterization of the potential PBM only with the double mutant virus (rOC/E-PBMD82A-V84A). Immunofluorescence analysis indicated that cells could be infected by all viruses but we detected a significant difference in propagation for both LA-N-5 cells (Fig. 4D, Figure S2A) and primary mixed murine CNS cultures (Figure S2B), showing a significantly reduced propagation for the double PBM mutant rOC/E-PBMD82A-V84A.
2.5. Deletion of HCoV-OC43 E protein or alteration of its putative TMD and PBM alter its relative infectivity
Given that the coronavirus E protein is known to play an important role in infectious virion formation and maturation (de Groot et al., 2012), we then looked if its deletion or the alteration of its putative functional domains could also alter the total virion production. Using a RT-qPCR approach, the quantity of total viral particles in stocks of rOC/ATCC, rOC/Estop, rOC/E-TMQ17A or rOC/E-PBMD82A-V84A were evaluated and compared to the infectious titer of the corresponding viruses ( Fig. 5A). Whereas the reference virus possesses a ratio of infectious virion to total viral particles of approximately 1:100, the rOC/Estop mutant has a ratio of 1:56 000 correlating with the default in infectious virion production previously observed. Surprisingly, the quantity of total viral particles (evaluated as total RNA copy number by RT-qPCR) was close to the reference virus. The same defect was observed for rOC/E-TMQ17A but to a lesser extent with a ratio of 1:1065, but it was absent for the rOC/E-PBMD82A-V84A mutant. Indeed, even though this latter mutant produces less virions compared to the reference virus, a very high proportion of them are infectious, as the ratio of infectious over total particles is about 1:2 (Fig. 5A). These observations of modified ratio were confirmed by immunofluorescence comparing the percentage of infected cells when the quantity of virus used for the infection of LA-N-5 cells was normalized either to the number of infectious virions or to the number of total viral particles (Fig. 5B). All these findings demonstrate that the HCoV-OC43 E protein is critical for efficient replication in epithelial and neuronal cells, and that its functional domains play important and potentially distinct roles during the production of new infectious virions.
Fig. 5.
HCoV-OC43 E protein and its putative TMD and PBM modulate relative virus infectivity. (A) Infectivity assay between viruses: quantification of viral RNA in viral stocks (absolute quantity in RNA copy representative of total viral particles) and of the number of infectious particles in viral stocks. Ratio of infectious to total viral particles are indicated over each virus. (B) The percentage of LA-N-5 cells infected by the different viruses was quantified by immunofluorescence and cell profiler software at 16hpi in the presence of 200 µM Chloroquine (reinfection inhibitor). Infection were performed with amount of viruses normalized either on the quantity of viral RNA (total particles) or on the number of infectious viral particles. (** P < 0.01; *** P < 0.001).
2.6. Fully competent HCoV-OC43 E protein is not essential for neuroinvasion but is a determinant factor of neurovirulence
As HCoV-OC43 is naturally neuroinvasive and neurovirulent in mice (Brison et al., 2011, Desforges et al., 2014, Le Coupanec et al., 2015) and that the E protein is important for efficient propagation in neuronal cells (Fig. 3, Fig. 4), we wished to investigate the importance of the two functional domains of E in the process of neuroinvasion. Seven-day-old C57Bl/6 mice were infected and RT-qPCR performed at 5 days post-infection on complete brain revealed that mutant viruses were still neuroinvasive compared to the reference virus, however the copy number of viral RNA was significantly lower for both mutants ( Fig. 6A). This indicates that the TMD and PBM are not essential for HCoV-OC43 neuroinvasion. Moreover, it is interesting to note that the 4 mice (3 infected with the reference virus, and one with the TMD mutant) with a high number of viral RNA copy (>1013) were the only ones to show signs of illness at 5 dpi.
Fig. 6.
A fully functional HCoV-OC43 E protein is associated with optimal neuroinvasion and increases neurovirulence. (A) 7-day-old C57Bl/6 mice received 103 TCID50/20 µL of rOC/ATCC, rOC/E-TMQ17A or rOC/E-PBMD82A-V84A by the IN route. Neuroinvasion was detected and quantified by RT-qPCR on brain RNA at 5 days post-infection, each point represents a single mouse. (B) Twenty-two-day-old C57Bl/6 mice received 102.5 TCID50/10 µL of rOC/ATCC, rOC/E-TMQ17A or rOC/E-PBMD82A-V84A by the IC route and were observed for survival over 21 days following the injection. (C) Infected mice were weighted every 2 days for a period of 21 days following infection. The weight gain is shown as percentage of d0 set at 100%. (D) Evaluation of the clinical scores (percentage of mice at each level of the scale) of mice infected by rOC/ATCC, rOC/E-TMQ17A or rOC/E-PBMD82A-V84A based on neurological symptoms described in clinical score scale between level 0 and 4 over a period of 21 days (see Materials and methods). Representative of three different experiments. (* P < 0.05; ** P < 0.01; *** P < 0.001).
To investigate the role of the E protein in the induction of HCoV-OC43-induced neurological pathology, 22-day-old C57Bl/6 mice were intracerebrally infected with either rOC/ATCC, rOC/E-TMQ17A or rOC/E-PBMD82A-V84A, and the development of illness was monitored for 21 days after infection. During this period, only mice infected with the reference virus died, all other mice survived the infection (Fig. 6B). Moreover, mice infected with rOC/E-PBMD82A-V84A did not show any significant differences of weight gain compared to sham infected mice (Fig. 6C), nor did they show any sign of neurological disease compared to mice infected with the reference virus (Fig. 6D). However, mice infected with rOC/E-TMQ17A showed an intermediate weight gain profile between mice infected with the reference virus and the sham infected mice (Fig. 6C), suggesting that these mice were developing a disease, which was confirmed by the neurological symptoms developed by several mice, although to a lesser extent than mice infected with the reference virus (Fig. 6D). Mice infected with rOC/Estop did not show any signs of illness whether in terms of weight gain, or neurological symptoms (Figure S3A-C).
2.7. HCoV-OC43 E protein and its TMD and PBM are essential for efficient replication in the murine CNS
As HCoV-OC43 E protein and its functional domains modulate viral replication and propagation in human and murine neuronal cultures (Fig. 3, Fig. 4), we examined if the differences in neurovirulence observed (Fig. 6) were also to be associated with defective infectious virus productions and propagation in the CNS. Infection of 22-day-old C57Bl/6 mice with reference and E protein mutants revealed that the infectious titer in the brain ( Fig. 7A) and the spinal cord (Fig. 7B) was significantly reduced for the TMD mutant. This altered replication in the brain correlates with an extremely weak production in the spinal cord, where infectious virions were detected in only one mouse at 5 dpi. Production of infectious particles was under the limit of detection in mice infected with the PBM mutant, correlating with the total absence of neurovirulence (Fig. 6). However, the PBM mutant RNA was detected in the brain at 5 and 9 dpi, although at a lower level than the reference virus or the TMD mutant (Fig. 7C), indicating that this virus was capable of replicating at a low level in the brain. Similar observations were obtained following mouse infection with rOC/Estop (Fig. S3D). Viral RNA of each mutant was extracted at 5 and 9 dpi and sequenced for the E and M gene. No reversion in the targeted genes were observed (data not shown), indicating that the inserted mutations are stable during replication in the murine brain for at least 9 days. It is interesting to note than when infected with a higher viral dose (102.5 TCID50/10 µL vs 101.5 TCID50/10 µL), infectious virions of the PBM mutant could be detected in low amount in the brain of mice (Figure S4A). Our system only allows to detect infectious titer over 102.5. Thus, the apparent (but very low) increase of infectious virus production observed at 10–11 days post-infection may be explained by the fact that the PBM mutant replicates at very low level between 3 and 11 days pi and that, by doing this, this mutant could be able to avoid detection by the immune system and delay its clearance from the CNS. Viral RNA was also detected in both the brain and spinal cord (Figure S4B and C). Sequencing of the E and M genes again revealed the absence of reversion, again suggesting that the absence of detectable infectious virion production previously observed (Fig. 7) was probably due to the low infectious dose. Taken together, these data demonstrate that a fully competent E protein (with both functional TMD and PBM) is essential for HCoV-OC43 neurovirulence in association with efficient replication in the CNS, correlating with the early observations in neuronal cells (Fig. 2, Fig. 3, Fig. 4).
Fig. 7.
The E protein TM and PBM domain are essential for optimal replication in the murine brain and spinal cord. Infectious viral particles were quantified in (A) the brains and (B) the spinal cord of 22-day-old C57Bl/6 mice infected by the IC route with 101.5 TCID50/10 µL rOC/ATCC, rOC/E-TM-Q17A or rOC/E-PBM-82–84 over a period of 15 days. (C) Viral RNA was detected and quantified by RT-qPCR in the brain of infected mice at 5 and 9 days post-infection. LOD, limit of detection. Representative of three different experiments.
2.8. The putative TMD decreases HCoV-OC43 propagation in the murine brain
When infection was performed at a 101.5 TCID50/10 µL dose, only the E protein TMD mutant produced detectable virions in the brain (infectious virus is under the limit of detection for PBM mutant). However, this production was significantly lowered compared to reference virus. Therefore, based on data obtained in neuronal cultures (Fig. 2, Fig. 3, Fig. 4), we investigated if the TMD mutation also induced a defect in propagation within the brain. Brains of mice infected intracerebrally with rOC/ATCC or rOC/E-TMQ17A were harvested at 3 and 7 dpi, and viral spreading was observed by immunofluorescence. Whereas the reference virus had already infected the hippocampus at 3 days post-infection (with positive cells around the lateral ventricle and in the hypothalamus), and then continued to spread until the rest of the brain was infected at 7 dpi ( Fig. 8 left panels), the rOC/E-TMQ17A presented an important delay in spreading as only a few number of infected cells were visible at 3 dpi except for a small focus of infection around the hippocampus and the lateral ventricle. Spreading then occurred to the same brain regions compared to reference virus but to a much lower extent (Fig. 8 right panels). This suggests that although the TMD mutant seems to follow the same spreading path in the murine brain, its spreading is greatly delayed and underlines the fact that HCoV-OC43 E protein TMD is important for efficient propagation in the murine brain.
Fig. 8.
HCoV-OC43 E protein putative TM domain is required for efficient spreading in mouse CNS. Viral spreading in mice brain was examined by immunofluorescence at 3 and 7 days post-infection. The virus was detected using an antibody against the HCoV-OC43 S protein (green) and nuclei were detected using DAPI (blue). Small white arrows point to isolated infected cells. Brain regions: 1, Olfactory Bulb; 2, Lateral Ventricle; 3, Hippocampus; 4, Brainstem; 5, Hypothalamus.
3. Discussion
In this study, by modifying a full-length cDNA infectious clone of the human HCoV-OC43 virus, we demonstrate that E protein is critical for the production of infectious virions, as transient complementation with wild type E protein rescued infectious viral production and a strong selection pressure to revert to a functional E protein was observed. Moreover, mutations of specific domains revealed that a fully functional protein participate in the efficient viral spreading, associated with neuropathogenesis.
Deletion of the E protein leads to varying degrees of defects for coronaviruses. Indeed, whereas murine hepatitis virus (MHV) and SARS-CoV are attenuated, showing a reduced ability to produce infectious virus without E protein, in a cell-type specific manner (DeDiego et al., 2008, DeDiego et al., 2007, Jimenez-Guardeño et al., 2015, Kuo and Masters, 2003), transmissible gastroenteritis virus (TGEV) (Ortego et al., 2002) and MERS-CoV (Almazán et al., 2013) are replication competent, but completely propagation defective, with no detectable infectious virus production when the E protein is deleted. Similarly, we were able to rescue infectious rOC/Estop production by providing wild-type E protein in trans, as the recovery of initially complemented rOC/Estop through complementation, and amplification on epithelial cells yielded detectable infectious virus (Fig. 1). As suggested for MERS-CoV (Almazán et al., 2013), the apparent low titer detected after the first passage on HRT-18 cells (especially at P1), could be due to a transfer of detached cells transfected with the initially complemented pBAC-OC43-E-Stop. Furthermore, the production of infectious particles (for initially complemented mutant lacking the E protein) was low and decreased with subsequent amplification attempts. On the other hand, these results also suggest that production of infectious virions without E protein is possible but with severely affected efficiency, underlining the requirement of a fully functional E protein. Coronavirus E protein has been suggested to allow correct virion formation in part by inhibiting M protein aggregation (Kuo et al., 2007), or by inducing scission events at the ERGIC (Fischer et al., 1998). Our data indicate that while reducing dramatically the quantity of infectious viral particles formed, the complete abrogation of HCoV-OC43 E protein did not significantly alter the quantity of total viral particles produced compared to the reference virus (Fig. 5), suggesting that the protein is important for efficient virion maturation that lead to good infectivity.
This concept is emphasized by the appearance of recombinant HCoV-OC43 E protein revertants at a very low passage number (P2 or P3) on HRT-18 cells. Indeed, the appearance of revertants with different E sequences after only a few rounds of amplification on HRT-18 cells indicates that low level of viral particles must have been produced earlier during the process as we already observed previously for HE-deleted recombinant HCoV-OC43 (Desforges et al., 2013). We observed strong selective pressure at the position where we introduced a stop codon where nucleotide changes led to reversion to reference E sequence or to another amino acid residue (tryptophan or leucine). It was previously described that SARS-CoV (Jimenez-Guardeño et al., 2015) and MHV (Kuo and Masters, 2010) E protein deletion mutants underwent compensatory mutations after a few passages in culture to utilize a partially duplicated version of the adjacent M protein to recover partial virus production. A second type of reversion was observed for SARS-CoV E protein deletion mutant after an intranasal infection of susceptible mice as the small transmembrane ion channel forming 8a was modified to incorporate a potential PBM associated with increased infectious virus production compared to E protein deletion mutant (Jimenez-Guardeño et al., 2015). Our results support the hypothesis that there is selective pressure to specifically restore the E protein functionality itself without partial duplication of the M gene (data not shown), however, it is important to note that we only introduced a stop codon at the beginning of the E gene instead of deleting part or all of the gene, as it was done for SARS-CoV and MHV (Jimenez-Guardeño et al., 2015, Kuo and Masters, 2010). A recombinant HCoV-OC43 in which the E gene would have been deleted could have used the same reversion process in other viral gene.
Production of infectious virus was reduced after infection of neuronal cell cultures with initially complemented rOC/Estop compared to reference virus. Immunofluorescence assay for viral proteins confirmed these latter results showing no defect in entry for the initially complemented rOC/Estop virus compared to reference virus at 16 hpi, while observations at 48 h indicated an important defect of viral propagation (Fig. 2). A decrease in virus spread was also reported for MHV (Kuo et al., 2007, Kuo and Masters, 2003) and SARS-CoV (DeDiego et al., 2007) ΔE mutants which formed smaller and less numerous plaques.
The coronavirus E protein is now considered as a virulence factor (reviewed extensively in (DeDiego et al., 2014)) and there have been extensive efforts to characterize the different domains of this relatively small transmembrane protein and the possibility that it acts as a viroporin, with ion channel activity. The TMD of several coronavirus E proteins (including HCoV-OC43) was predicted (Torres et al., 2005) and shown to form ion channels permeable to small cations in artificial membranes for species such as IBV, MHV, HCoV-229E (Wilson et al., 2006), MERS-CoV (Surya et al., 2015) and SARS-CoV (Wilson et al., 2004). Furthermore, it was demonstrated that the IBV E protein can exist in a penta-oligomeric state (Westerbeck and Machamer, 2015), and that for MERS-CoV (Surya et al., 2015) and SARS-CoV (Pervushin et al., 2009, Torres et al., 2006, Verdiá-Báguena et al., 2013, Verdiá-Báguena et al., 2012), it forms pentameric channels in lipid membranes. Chemical inhibition of ion channel activity (Wilson et al., 2006), destruction of the integrity of (Almazán et al., 2014, Regla-Nava et al., 2015, Ye and Hogue, 2007) or replacement of the TMD with those of other viral species (Ruch and Machamer, 2011) all led to reduced viral titers for other coronaviruses species. The E protein of coronaviruses is largely localized within the secretory pathway (Cohen et al., 2011, Nieto-Torres et al., 2011, Venkatagopalan et al., 2015) where it has recently been described for IBV to exist in two different pools; one of monomeric E proteins that disrupt the secretory pathway and a second pool in an oligomeric state, likely serving to facilitate the assembly of progeny virions (Westerbeck and Machamer, 2015). The transient delay in infectious virus release observed for the TMD mutant compared to the reference virus (Fig. 3) suggests a defect in virus release that could be the result of damaged infectious particles as it was observed for IBV, for which a TMD mutant of the E protein induces a similar defect associated with increased quantity of non-infectious viral particles released possessing a cleaved S protein near the virion surface (Ruch and Machamer, 2011). Considering the higher proportion of non-infectious virions released by cells infected with the TMD mutant compared to cells infected with the reference virus (Figs. 3D and 5A), a similar process consisting in an alteration of infectious virions in the secretory pathway may also be involved here, suggesting an important role for TMD within the cellular secretory pathway. Furthermore, mutation at homologous position T16 of IBV did not have an effect on VLP formation but rather was required for secretory pathway disruption (Ruch and Machamer, 2012, Westerbeck and Machamer, 2015). These results suggest that mutation Q17A (homologous to IBV T16A) in the HCoV-OC43 E protein putative TMD plays a role in modulating infectious virus release from infected cells.
A four-amino acid C-terminal PBM protein-protein interaction motif has been predicted for HCoV-OC43 (Jimenez-Guardeño et al., 2014). Through the replacement of the key amino acids of this motif by alanines, we demonstrated its importance in infectious virion production in the epithelial and neuronal cells tested (Fig. 4). Defective propagation without an effect on the ability to infect cells was observed with the recombinant virus with an abrogated putative PBM. Deletion of the PBM in SARS-CoV E protein led to slight decreases in viral titers in some cell types (Jimenez-Guardeño et al., 2014) while in others viral titers remained unaffected (Regla-Nava et al., 2015). Our data on HCoV-OC43 reveal significant infectious virus production defects in an epithelial cell line which was further accentuated in neurons, but only after infection by the double mutant rOC/E-PBMD82A-V84A, suggesting that HCoV-OC43 replication in cultured cells can tolerate a slight flexibility in the putative PBM sequence. Among coronavirus E proteins, only the SARS-CoV E protein has been shown to per se possess such a functional motif, which interacts with PALS1 to disturb secretory pathway membranes to alter tight junction formation (Teoh et al., 2010) and syntenin to play a role in the exacerbated inflammatory response typical of infection via p38 MAPK activation (Jimenez-Guardeño et al., 2014). For SARS-CoV E protein, the PBM is suggested to be important in two independent functions: virus stability and virulence/pathogenesis rather than virus production (Jimenez-Guardeño et al., 2015, Jimenez-Guardeño et al., 2014, Regla-Nava et al., 2015). Given that the functions of PBM is dependent on their sequence and surrounding sequence context (Ye and Zhang, 2013), further study of the HCoV-OC43 E protein neuronal interactome is warranted and could provide new insights on the precise function of the protein in infected cells. Prevention of a functional interaction between HCoV-OC43 E protein PBM and its PDZ domain-containing ligand could conceivably function in the same vein as seen in case of neurotropic encephalitic rabies virus, for which differences in disease phenotype, rapid versus attenuated spread of virus infection, was attributed to differences in PBM sequences on the C-terminal of the rabies envelope glycoprotein G leading to different cellular interaction partners to mediate either neuronal cell survival or death (Préhaud et al., 2010). Surprisingly, the PBM mutant forms less total viral particles but almost all of them are infectious. As protein-protein interaction motifs, viral PBM are involved in a variety of processes, including viral particle assembly and maturation (Javier and Rice, 2011). Abrogation of the E PBM, could prevent critical cellular and/or viral interaction necessary for the rapid and efficient formation and maturation of viral particles, making propagation highly inefficient.
The SARS-CoV E protein is critical for neuroinvasiveness in susceptible mice (DeDiego et al., 2008). HCoV-OC43 E protein seems to differ from its SARS-CoV homolog as viral RNA is present in the brain of all mice intranasally infected with either the reference virus, the TMD mutant or the PBM mutant (Fig. 6-A), indicating that the fully functional protein is not essential for neuroinvasion. On the other hand, the amount of viral RNA was significantly lower in the brain of E mutant-infected mice. Although we cannot rule out that these data represent a difference in replication and propagation once the virus is already in the brain, optimal HCoV-OC43 neuroinvasion may necessitate a fully functional E protein. The SARS-CoV E protein was recently described as an important virulence factor during infection of the lungs, being in part responsible for the immune response exacerbation (Jimenez-Guardeño et al., 2014), the lung epithelium destruction (Teoh et al., 2010), and edema accumulation in the lungs (Nieto-Torres et al., 2014). We demonstrate here that HCoV-OC43 E protein deletion as well as abrogation of the putative PBM prevented neurological symptoms following CNS infection, correlating with replication and propagation observations in epithelial and neuronal cells (Fig. 6). Disappearance of respiratory tract disease was observed for SARS-CoV in which E PBM was abrogated, preventing interaction of the viral protein with the PDZ-containing protein syntenin (Jimenez-Guardeño et al., 2014, Regla-Nava et al., 2015). The absence of neurological symptoms following infection by either rOC/Estop or rOC/E-PBMD82A-V84A could potentially be linked to glutamate excitotoxicity that we have previously observed in mice (Brison et al., 2011), possibly by interfering with PDZ-domain containing proteins found in neuronal cells (Feng and Zhang, 2009). We also demonstrate that HCoV-OC43 putative TMD plays a role in the neuropathogenesis following the CNS infection, albeit to a lesser extent than its PBM. Indeed, infected mice showed some neurological symptoms, even if their severity and frequency were lower than for the reference virus. This attenuated phenotype was also associated with reduced viral particles production and propagation in the CNS. Homologous mutation at the position 15 of SARS-CoV E protein also led to an attenuated pathology, although infectious viral particle production was not significantly affected compared to wild-type virus (Nieto-Torres et al., 2014). Taken together, these results demonstrate that HCoV-OC43 E protein is a virulence factor, with TMD and PBM being important determinants in that matter.
In summary, the current study demonstrates the critical importance of a fully functional HCoV-OC43 E protein in infectious virus production and efficient spread in both epithelial and neuronal cells. Modifications to key amino acids in putative functionally important domains modulated infectious virion production and delayed virus spread in human and murine neuronal cells and within mouse CNS. This points towards the presence of a true TMD, which has a role in the secretory pathway, as seen for other coronaviruses and that the HCoV-OC43 E protein putative C-terminal PBM plays a significant role in infectious virion production as well as efficient virus spread. In addition, this study establishes a clear link between both putative functional domains of HCoV-OC43 E protein and CNS pathogenesis. Functional viroporins and viral PBM are associated with viral pathogenesis for a growing number of viruses and their studies to better understand virus-host interaction represent an emerging field (Javier and Rice, 2011, Nieva et al., 2012, Scott and Griffin, 2015). Considering that the HCoV-OC43 E protein seems to possess both functions and that they seem to be important for the induction of disease, future studies regarding their functionality and underlying mechanisms resulting in HCoV-OC43 neuropathogenesis previously described (Brison et al., 2011, Jacomy et al., 2006, Le Coupanec et al., 2015) are warranted and necessary as they will help to identify virus-host interfaces which could represent therapeutic target.
4. Materials and methods
4.1. Ethics statement
All animal experiments were approved by the Institutional Animal Care and Use Ethics Committee (IACUC) of the Institut National de la Recherche Scientifique (INRS) and conform to the Canadian Council on Animal Care (CCAC). Animal care and used protocols numbers 1304-02 and 1604-02 were issued by the IACUC of INRS for the animal experiments described herein.
4.2. Cell lines and mixed primary murine CNS cells
The BHK-21 cell line (ATCC-CCL10) was cultured in minimal essential medium alpha (MEM-α; Life Technologies) supplemented with 10% (vol/vol) fetal bovine serum (FBS; PAA GE Healthcare) and used for transfection. The HRT-18 cell line (a gift from the late David Brian, University of Tennessee) was cultured in the same medium and used for virus infections/amplifications. The LA-N-5 cell line (a kind gift of Stephan Ladisch, George Washington University School of Medicine) was cultured in RPMI medium supplemented with 15% (vol/vol) fetal bovine serum (FBS), 10 mM HEPES, 1 mM sodium pyruvate, and 100 μM non-essential amino acids (Gibco - Invitrogen). The LA-N-5 cells were differentiated into human neurons as previously described (Hill and Robertson, 1998). Briefly, cells were seeded in 24-well plates pre-coated with 0.1% gelatin (1.25 × 103 cells/well) in RPMI medium supplemented with 10% (vol/vol) FBS, 10 mM HEPES, 1 mM sodium pyruvate, and 100 μM non-essential amino acids. The next day and every 2 days for 6 days, the medium was replaced with the same medium supplemented with 10% (vol/vol) FBS and 10 μM all-trans retinoic acid (Sigma-Aldrich).
Mixed primary cultures of mouse CNS cells were prepared as previously described (Le Coupanec et al., 2015). Briefly, embryos at 15 days of gestation were removed from pregnant anesthetized CD1 mice and their cortex and hippocampus were harvested and placed in Hanks balanced salt solution (HBSS) medium, without Ca2+ and Mg2+, supplemented with 1 mM sodium pyruvate and 10 mM HEPES buffer. Tissues were gently pipetted up and down with a Pasteur pipette to dissociate the cells. After a decantation step of 3 min at room temperature, supernatants were transferred into a 50-ml tube with 36 ml of Neurobasal Medium (Invitrogen) supplemented with 0.5 mM GlutaMAX-I (Life Technologies), 10 mM HEPES buffer, B27 supplement (Life Technologies), gentamycin and 10% (vol/vol) of horse serum (Life Technologies). Cells were then seeded at 1 × 105 cells/cm2 and grown on 50 µM poly-d-lysine-treated 12-well plates containing glass coverslips (for immunofluorescence) or not (for evaluation of infectious virus production) in the same medium, which was replaced by fresh Neurobasal Medium without horse medium the next day. The medium was changed every 2 days after and the cultures were ready for infection after 7 days in culture.
4.3. Site-directed mutagenesis
Using our full-length, cDNA infectious clone pBAC-OC43FL (St-Jean et al., 2006) the recombinant HCoV-OC43 virus (rOC/ATCC) was generated. In parallel, a series of recombinant mutant viruses were produced by site-directed mutagenesis using the QuikChange Multi Site-Directed Mutagenesis Kit (Stratagene) and a variety of primers ( Table 1, Section A) to introduce nucleotide substitutions in the E gene. These substitutions in the cDNA clone introduced were: nucleotide mutations at positions 24 and 25, corresponding to a change of amino acid position 9 into a stop codon (pBAC-OC43-E-Stop, plasmid; rOC/Estop, recombinant virus) or mutations at nucleotide positions 49 and 50 corresponding to amino acid 17 (pBAC-OC43-E-TM-Q17A; rOC/E-TMQ17A), nucleotide position 245 and 246; amino acid 82 (pBAC-OC43-E-PBM-D82A; rOC/E-PBMD82A) or nucleotide position 251; amino acid 84 (pBAC-OC43-E-PBM-V84A; rOC/E-PBMV84A). A double mutant cDNA clone pBAC-OC43-E-PBM-D82A-V84A (rOC/E-PBMD82A-V84A) was also produced using the pBAC-OC43-E-PBM-D82A as a DNA template for a second-round of mutagenesis reaction with OC-E-PBM-D82A-V84A primer to introduce a second mutation at nucleotide position 251; amino acid 84. Prior to transfection of BHK-21 cells, all samples were sequenced to make sure that only the introduced mutations were present and that no other mutations appeared.
Table 1.
Primers used to introduce nucleotide substitutions in the E gene of pBAC-OC43FL (St-Jean et al., 2006) for recombinant HCoV-OC43 virus production with amino acid modifications within the E protein (Section A). Primers used to verify sequences of the full HCoV-OC43 E and M gene of recombinant viruses (Section B). Bold and underlined sections represent newly introduced nucleotide substitutions. Underlined section in mutant E-PBM 82–84 represents previously introduced nucleotide substitutions.
|
A – Primers for site-directed mutagenesis | ||
|---|---|---|
| Recombinant Virus | Primer Name | Primer Sequence |
| rOC/Estop | OC-E-stop | 5’-CT GAT GCT TAT CTT TGA GAC ACT GTG TGG-3’ |
| rOC/E-TMQ17A | OC-E-TM-Q17A | 5’-GTG TGG TAT GTG GGG GCA ATA ATT TTT ATA GTT GCC-3’ |
| rOC/E-PBMD82A | OC-E-PBM-D82A | 5’-CA GTC CTT GAT GTG GCC GAC GTT TAG GTA ATC-3’ |
| rOC/E-PBMV84A | OC-E-PBM-V84A | 3’-GT TTG GAT TAC CTA AGC GTC ATC CAC ATC AAG-5’ |
| rOC/E-PBMD82A-V84A | OC-E-PBM-D82V-V84A | 5’-CTT GAT GTG GCC GAC GCT TAG GTA ATC CAA AC-3’ |
| B – Primer for sequence verification | ||
| Primer Name | Primer Sequence | |
| OC-ns5-116-E-For | 5’-GTA GAG TTC CTA GTC ATG CTT G-3’ | |
| OC-E-222-For | 5’-GAT GTA AAA CCA CCA GTC CT-3’ | |
| OC-M-127-E-Rev | 5’-ACA TAC TGC GAC TTG TAT AGC C-3’ | |
| OC-M-172-For | 5’-ATT TTG TGG CTT ATG TGG CCC-3’ | |
| OC-M-241-E-Rev | 5’-CTA TAG AAA GGC CAA GAT ACA C-3’ | |
| OC-M-439-For | 5’-GTC ACA ATA ATA CGC GGC CA-3’ | |
| OC-M-514-Rev | 5’-CCT TAG CAA CAG TCA TAT AAG C-3’ | |
| OC-N-60-Rev | 5‘-CAT TAC CAG AAC GAT TTC C-3’ | |
| GAPDH-For | 5’-CGG AGT CAA CGG ATT TGG TCG TAT-3’ | |
| GAPDH-Rev | 5’-AGC CTT CTC CAT GGT GGT GAA GAC-3’ | |
4.4. Plasmid for transient co-transfection rescue
In order to insert the HCoV-OC43 E gene into the pcDNA3.1(+) expression vector (pcDNA; Invitrogen) and allow for E protein expression upon transient co-transfection with pBAC-OC43 infectious clones in BHK-21 cells, restriction enzymes NheI and BamHI were added to the 5’ (primer: 5’-GCTAGC ATG TTT ATG GCT GAT GCT TA-3’) and 3’ (primer: 5’-GGATCC CTA AAC GTC ATC CAC AT-5’) ends of the E gene respectively. The E gene with added restriction enzyme sites was PCR-amplified from cDNA originating from a HCoV-OC43 reference strain (ATCC) infection on HRT-18 cells using Accuprime Pfx Supermix (Life Technologies) with 1 cycle at 95 °C for 5 min, followed by 35 cycles at 94 °C for 15 s, 48 °C for 30 s and 68 °C for 1 min and 1 cycle at 68 °C for 4 min and then introduced into the pcDNA plasmid.
4.5. Recombinant virus production, transient co-transfection and virus amplification
The BHK-21 cells were cultured in MEM-α supplemented with 10% (vol/vol) FBS and used for transfection of pBAC-OC43 cDNA infectious clones with Lipofectamine 3000 Reagent (Life Technologies) according to the manufacturer's instructions. Briefly, for production of recombinant viruses, BHK-21 cells were seeded in 6-well cell culture plates at 6×105 cells/well. The next day, when cells were 70–90% confluent, the medium was replaced and cells were transfected with 7.5 µL Lipofectamine 3000 Transfection Reagent, 10 µL P3000 Reagent, 5 μg of pBAC-OC43FL or other modified pBAC DNA, and 2 μg of pcDNA(OC-E) or empty pcDNA plasmid per well. For semi-quantitative determination of transfection efficiency by immunofluorescence assay (IFA), BHK-21 cells were seeded at 5 × 104 cells/well onto glass coverslips in 24-well plates and transfected with 1.5 µL Lipofectamine 3000 Transfection Reagent, 6 µL P3000 Reagent, 1.5 μg of pBAC-OC43FL or other modified pBAC DNA, and 500 ng of pcDNA(OC-E) or empty pcDNA plasmid per well. The plates were incubated at 37 °C for 8 h and then medium replaced with MEM-α supplemented with 10% (vol/vol) FBS and 0.01% (vol/vol) gentamycin and incubated for 3 days.
The cells from 6-well culture plates were harvested either to recover total RNA or total protein while the supernatant (P0) was recovered by aspiration after centrifugation at 500 × g for 7 min and then clarified at 1000 × g for 10 min. The supernatant (P0) served to inoculate HRT-18 cells in order to amplify the viral stocks. The supernatant from this first-round amplification (P1) served for a second round of viral amplification on HRT-18 cells from which supernatant was recovered (P2) and in some cases, was repeated again for a third round of amplification (P3). The production of infectious viral particles corresponding to the different pBAC-OC43 cDNA clones was titrated by an immunoperoxidase assay (IPA) prior to each amplification step in order that titers could be normalized to the lowest detectable titer and replication rates be compared.
4.6. Infection of human cell lines and of primary mouse CNS cultures
The HRT-18 and LA-N-5 cells as well as mixed primary cultures of mouse CNS cells were infected at a MOI equivalent to the lowest detectable titer of the series of recombinant virus stock used during each experiment or mock-infected and then incubated at 33 °C (HRT-18) or 37 °C (LA-N-5 cell line and primary CNS cultures), for 2 h (for virus adsorption), and incubated at 33 °C with fresh MEM-α supplemented with 1% (vol/vol) FBS (for HRT-18 cells), at 37 °C with fresh RPMI medium supplemented with 2.5% (vol/vol) FBS (for LA-N-5 cells) or at 37 °C with fresh Neurobasal Medium with B27-GlutaMAX-I (for primary murine CNS cell cultures) for different periods of time before fixing cells for immunofluorescence detection or harvesting the cell-associated and/or cell-free medium fractions for infectious virus titer determination by IPA. For relative infectivity and release assay, LA-N-5 cells were infected at a MOI equivalent to the lowest titer of the compared recombinant viruses and incubated at 37 °C for 16 h with fresh RPMI medium supplemented with 2.5% (vol/vol) FBS and 200 nM chloroquine (N4-(7-Chloro-4-quinolinyl)-N1,N1-dimethyl-1,4-pentanediamine diphosphate salt, Sigma, CAS number 50–63-5) in order to prevent re-infection.
4.7. Mice, survival curves, body weight variations and evaluation of clinical scores
Infection of 22-day-old female or 7-day-old male and female C57Bl/6 mice (Charles River) were performed as previously described (Le Coupanec et al., 2015). Briefly, mice were inoculated respectively by the IC route with 101.5 or the intranasal route with 103 of 50% tissue culture infective doses (TCID50) recombinant virus. Groups of 10 mice infected by each recombinant virus were observed on a daily basis over a period of 21 dpi, and survival and weight variations were evaluated. Clinical scores were evaluated using a scale with 5 distinctive levels 0–4); where 0 was equivalent to the asymptomatic mouse; 1 for mice with early hunched backs; 2 for mice presenting slight social isolation, weight loss, and abnormal gait; 3 for mice presenting total social isolation, ruffled fur, hunched backs, weight loss and almost no movement; and number 4 was attributed to mice that were in moribund state or dead.
4.8. Titration of infectious virus using an immunoperoxidase assay (IPA)
The IPA was performed on HRT-18 cells, as previously described (Lambert et al., 2008). Briefly, the primary antibody used was mAb 4.3E4 (hybridoma supernatant; ½ dilution) directed against the S protein of HCoV-OC43. The secondary antibody was horseradish peroxidase-conjugated goat anti-mouse immunoglobulin (KPL; 1/500). Immune complexes were detected by incubation with 0.025% (w/v) 3,3’-diaminobenzidine tetrahydrochloride (Bio-Rad) and 0.01% (vol/vol) hydrogen peroxide in PBS and infectious virus titers were calculated by the Karber method, as previously described (Lambert et al., 2008).
4.9. Immunofluorescence for semi-quantification of transfection efficiency or virus propagation
BHK-21 and LAN-5 cells as well as mixed primary cultures of mouse CNS were fixed onto glass coverslips with 4% (wt/vol) paraformaldahyde for 30 min at room temperature and permeabilized for 5 min with 100% methanol at −20 °C. For LA-N-5 and BHK-21 cells, to detect HCoV-OC43 spike (S) protein, one-hour incubations of primary 4.3.E.4 (hybridoma supernatant; ½ dilution;) followed by secondary antibody AlexaFluor 488 donkey anti-mouse IgG (H + L) (1/1000; Life Technologies-Molecular probes) were conducted with three PBS washes between steps. For primary mouse CNS cultures, after blocking with a PBS-BSA 2% (wt/vol) solution for one hour at room temperature, primary antibody polyclonal rabbit anti-S protein (dilution 1/1000) and mouse monoclonal antibody against the neuron-specific MAP2 protein (1/1000; BD Pharmagen, catalog no. 556320) were diluted in PBS + 0.1% Triton X-100 and incubated on cells for one hour at room temperature followed by three PBS washes. Cells were then incubated one hour at room temperature with anti-rabbit Alexa Fluor 568- and anti-mouse Alexa Fluor 488-conjugated secondary antibodies (1/1000; Life Technologies-Molecular probes) in PBS. For all cell types, nucleus detection was accomplished by a 5-min incubation with 4',6-diamidino-2-phenylindole (DAPI; 1 μg/ml; Life Technologies). Triplicate samples were mounted on glass slides with Immuno-Mount medium (Fisher Scientific). Immunofluorescent staining was observed under a Nikon Eclipse E800 microscope with a QImaging Retiga-EXi Fast 1394 digital camera using Procapture system software. Percentage of infected cells was quantified from immunofluorescence pictures with the CellProfiler software (Carpenter et al., 2006).
For immunofluorescence on brain section, perfusion with 4% (wt/vol) paraformaldehyde (PFA) was performed on infected C57Bl/6 mice for each virus, at 3 and 7 dpi. Murine brains were carefully harvested and conserved in 30% (wt/vol) sucrose at 4 °C for 72 h. Prior to section, harvested brains were embedded in Tissue-Tek OCT compound (Sakura Finetek, WWR) at −20 °C. Sagittal brain sections were prepared at a thickness of 60 µm with a microtome cryostat HM 525; Microm. Serial sections were collected and Prior to staining, sections were incubated with a solution of two droplets of H2O2 in PBS for 10 min at RT and washed with PBS. Sections were then permeabilized with a solution of 0,1% Triton in PBS for 2 h at RT, and blocked with a solution of PBS containing 1 droplet of horse normal serum according to the manufacturer's protocol (ABC kit Vectastain, Vector Laboratories) for 1 h at RT. For detection of viral antigens, sections were incubated overnight at 4 °C in a 1/500 dilution of polyclonal rabbit anti-S protein of bovine coronavirus (BCoV). After three washes with PBS, sections were incubated in the dark for 1 h at room temperature with the secondary fluorescent antibodies Alexa Fluor 488 anti-mouse (1/500; Life Technologies). After three PBS washes, sections were incubated for 5 min at room temperature with 4’,6-diamidino-2 phenylindole (DAPI; 1 μg/ml; Life Technologies), washed once with PBS and water and then mounted with Immuno-Mount mounting medium (Fisher Scientific). Immunofluorescent staining was observed under a Zeiss LSM780 confocal microscope.
4.10. RNA extraction, cDNA synthesis and gene amplification
After transfection of BHK-21 cells or infection of HRT-18 cells, cells were scraped from wells or plates, centrifuged at 500 × g for 7 min at 4 °C, medium was removed and cell pellet resuspended with 1.5 ml ice-cold PBS and centrifuged at 1000 × g for 10 min at 4 °C. PBS was aspirated and the dry pellet stored at −80 °C until use. Total RNA was extracted using the RNeasy Mini Kit (QIAGEN) with QIAshredder spin columns (QIAGEN) to lyse cells according to manufacturer's instructions. RNA quality was verified using the Agilent 2100 Bioanalyzer using the Agilent RNA 6000 Nano Assay protocol according to manufacturer's instructions and concentration measured using a ND1000 spectrophotometer (Nanodrop). To produce cDNA, 5 μg of total extracted RNA was reverse transcribed using the SuperScript III First-Strand Synthesis Supermix Kit using oligo(dT) primer (Invitrogen) according to manufacturer's instructions.
PCR was conducted using Accuprime Pfx Supermix (Life Technologies) with one cycle at 95 °C for 1 min, followed by 40 cycles at 95 °C for 35 s, 50 °C for 45 s and 68 °C for 2 min, followed by one cycle at 68 °C for 7 min using several sets of primers to amplify the E gene (forward primer, OC-ns5-116-E-For, OC-E-222-For, OC-M-172-For or OC-M-439-For; reverse primers, OC-M127-E-Rev, OC-M-514-Rev, OC-M241-E-Rev or OC-N-60-Rev) and GAPDH gene as control (forward primer, GAPDH-For; reverse primer, GADPH-Rev). A complete list and description of all primers is presented in Table 1, section B.
4.11. RNA extraction for infectivity assay and quantification of viral RNA in mouse tissue
Real time RT-PCR for the absolute quantification of viral RNA in viral stocks and during infection of murine CNS, was modified from Vijgen and collaborators (Vijgen et al., 2005) using the Taqman technology and the use of cRNA standards for the generation of a standard curve and to evaluate the copy number of viral RNA in samples with the MEGAshortscript kit (Ambion/Life Technologies) (Fronhoffs et al., 2002, Vijgen et al., 2005). Briefly, total RNA was extracted with the Qiazol reagent (Qiagen) for HRT-18 cell culture supernatant and mouse tissue to evaluate the amount of viral RNA in virus stock and in mouse tissue respectively. cRNA standards were constructed exactly as described elsewhere made as previously described (Vijgen et al., 2005). RNA concentrations were evaluated in all samples and quantified using a ND1000 spectrophotometer (Nanodrop). Real-time quantitative RT-PCR was performed with the TaqMan-RNA-to-CT 1-Step kit (Applied Biosystems/Life Technologies) in a 20 µL reaction mixture with 10 µL of 2× TaqMan RT-PCR Mix (containing ROX as a passive reference dye), 900 nM of forward and reverse primers, and 200 nM of FAM BHQ1-TP probe. Four µL of RNA for supernatant samples and cRNA standards (serial dilutions), were used for the reaction. Amplification and detection were performed in a StepOnePlus Realtime PCR system apparatus and analysis were performed with the StepOne software version 2.3 (Applied Biosystems).
4.12. Protein extraction and Western blot analysis
To confirm E protein production after transfection of pcDNA(OC-E) in BHK-21 cells, proteins from whole cell lysates were extracted. Harvested cells were pipetted up and down into RIPA buffer (150 mM NaCl, 50 mM Tris, pH 7.4, 1% (v/v) NP-40, 0.25% (wt/vol) sodium deoxycholate, 1 mM EDTA) supplemented with protease cocktail inhibitor (Sigma). Lysates were incubated on ice for 20 min and centrifuged at 17,000 × g for 10 min at 4 °C. Supernatants were harvested, aliquoted and stored at −80 °C until further analyzed.
Protein concentrations were determined using a bicinchoninic acid (BCA) protein assay kit (Novagen) according to the manufacturer's instructions. Ten μg of protein was loaded on a Tris-Glycine 4–15% gradient gel, transferred to PVDF membrane with a semi-dry trans-blot apparatus (Bio-Rad). Membranes were blocked overnight at 4 °C with TBS buffer containing 1% (vol/vol) Tween (TBS-T) and 5% (wt/vol) non-fat milk. The following day all steps were conducted at room temperature, with, or solutions diluted in, TBS-T and milk. A primary rabbit polyclonal antibody was used to detect either the HCoV-OC43 E protein (1/5000) or GAPDH (1/10 000) for 1 h, followed by three 10 min washes. Anti-rabbit IgG horseradish peroxidase linked whole antibody (from donkey) (GE Healthcare) was the secondary antibody used, followed by three 10 min washes. Detection was performed using 1/1 solution of Clarity Western ECL Substrate (Bio-Rad) for one minute followed by membrane exposure on CL-X-Posure Film (Thermo Scientific).
4.13. Statistical tests
For cell experiments (percentage of infection), statistical analysis were conducted by one-way analysis of variance (ANOVA), followed by Tukey's post hoc test, or a t-test. For mice experiments, results were compared using two non-parametric statistical tests: Kruskal-Wallis and Mann-Whitney. Statistical significance was defined as p < 0.05 and is indicated with * (Student's t-test p value < 0.05), ** (Student's t-test p value < 0.01) or *** (Student's t-test p value < 0.001).
Acknowledgements
We thank Jessie Tremblay for excellent technical assistance with confocal microscopy. J.K.S. and G.D. wishes to thank Mathieu Meessen-Pinard, Mathieu Dubé, and especially Marc Desforges and Pierre J. Talbot for their advice and support throughout the project. Additionally, J.K.S. and G.D. wishes to thank M.D. and P.J.T. for guidance in the preparation of this manuscript. This study was supported by Discovery grant 42619-2009 from the National Sciences and Engineering Research Council of Canada and operating grant MT-9203 from the Institute of Infection and Immunity of the Canadian Institutes for Health Research to P.J.T. who is the holder of the Tier-1 (Senior) Canada Research Chair in Neuroimmunovirology award. J.K.S. gratefully acknowledges a masters studentship from the Fondation Universitaire Armand-Frappier de l’INRS. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgments
Funding
This study was supported by Discovery grant 42619-2009 from the National Sciences and Engineering Research Council of Canada and operating grant MT-9203 from the Institute of Infection and Immunity of the Canadian Institutes for Health Research to P.J.T. who is the holder of the Tier-1 (Senior) Canada Research Chair in Neuroimmunovirology award. J.K.S. gratefully acknowledges a masters studentship from the Fondation Universitaire Armand-Frappier de l’INRS. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Competing interests
The authors have declared that no competing interests exist.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2017.12.023.
Appendix A. Supplementary material
Sup. Figure 1.
HCoV-OC43 E protein transmembrane (TM) domain is important for its efficient propagation in human and murine neuronal cells. (A) Differentiated LA-N-5 and (B) mixed primary murine CNS cells were infected with either rOC/ATCC or rOC/E-TMQ17A at the same MOI and cells were fixed at several time points and HCoV-OC43 S protein (red) was revealed by immunofluorescence staining to follow viral propagation. Green represents the microtubule associated protein 2 (MAP2) staining in neurons of the mixed primary murine CNS cells. Nucleus were stained with DAPI (blue).
Sup. Figure 2.
HCoV-OC43 E protein PBM motif greatly improves its propagation in human and murine neuronal cells. (A) Differentiated LA-N-5 and (B) mixed primary murine CNS cells were infected with either rOC/ATCC or rOC/E-TMD82A-V84A at the same MOI and cells were fixed at several time points and HCoV-OC43 S protein (red) was revealed by immunofluorescence staining to follow viral propagation. Green represents the microtubule associated protein 2 (MAP2) staining in neurons of the mixed primary murine CNS cells. Nucleus were stained with DAPI (blue).
Sup. Figure 3.
HCoV-OC43 E protein is required for pathogenesis in the CNS. (A) Survival curves of mice in percentage over 21 days following the injection. (B) Infected mice were weighted every 2 days for a period of 21 days following infection. The weight gain is shown as percentage of j0 set at 100%. (C) Evaluation of the clinical scores (percentage of mice at each level of the scale) of mice infected by rOC/ATCC or rOC/Estop based on neurological symptoms described in clinical score scale between level 0 and 4 (see Materials and methods). (D) Viral RNA was detected and quantified by RT-qPCR in the brain of infected mice at 5 and 9 days post-infection. Representative of three different experiments. (** P<0.01; *** P<0.001).
Sup. Figure 4.
rOC/E-PBMD82A-V84Ais able to produce infectious virions in the brain and to propagate to the spinal cord after an infection at a higher dose. (A) Infectious viral particles were quantified in the brains of 22-day-old C57Bl/6 mice infected by the IC route by 102.5 TCID50/10 µL rOC/ATCC, or rOC/E-PBM-82–84 over 11 days post-infection. Viral RNA was detected and quantified by RT-qPCR in the (B) brain and (C) spinal cord of infected mice over 11 days post-infection. LOD, limit of detection.
References
- Almazán F., Dediego M.L., Sola I., Zuñiga S., Nieto-torres J.L., Marquez-jurado S., Andrés G. A vaccine candidate east respiratory syndrome coronavirus as a vaccine candidate. MBio. 2013;4:1–11. doi: 10.1128/mBio.00650-13.Editor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazán F., Sola I., Zuñiga S., Marquez-Jurado S., Morales L., Becares M., Enjuanes L. Coronavirus reverse genetic systems: infectious clones and replicons. Virus Res. 2014;189:262–270. doi: 10.1016/j.virusres.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi Y.M., Harthi A., Hussein J., Bouchama A., Johani S., Hajeer A.H., Saeed B.T., Wahbi A., Saedy A., AlDabbagh T., Okaili R., Sadat M., Balkhy H. Severe neurologic syndrome associated with Middle East respiratory syndrome corona virus (MERS-CoV) Infection. 2015;43:495–501. doi: 10.1007/s15010-015-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbour N., Day R., Newcombe J., Talbot P.J. Neuroinvasion by human respiratory coronaviruses. J. Virol. 2000;74:8913–8921. doi: 10.1128/JVI.74.19.8913-8921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonavia A., Arbour N., Yong V.W., Talbot P.J. Infection of primary cultures of human neural cells by human coronaviruses 229E and OC43. J. Virol. 1997;71:800–806. doi: 10.1128/jvi.71.1.800-806.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brison E., Jacomy H., Desforges M., Talbot P.J. Glutamate excitotoxicity is involved in the induction of paralysis in mice after infection by a human coronavirus with a single point mutation in its spike protein. J. Virol. 2011;85:12464–12473. doi: 10.1128/JVI.05576-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J., Golland P., Sabatini D.M. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J.R., Lin L.D., Machamer C.E. Identification of a Golgi complex-targeting signal in the cytoplasmic tail of the severe acute respiratory syndrome coronavirus envelope protein. J. Virol. 2011;85:5794–5803. doi: 10.1128/JVI.00060-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis K., Yount B., Baric R. Heterologous gene expression from transmissible gastroenteritis virus replicon particles. J. Virol. 2002;76:1422–1434. doi: 10.1128/JVI.76.3.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeDiego M.L., Alvarez E., Almazan F., Rejas M.T., Lamirande E., Roberts A., Shieh W.J., Zaki S.R., Subbarao K., Enjuanes L. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J. Virol. 2007;81:1701–1713. doi: 10.1128/JVI.01467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeDiego M.L., Pewe L., Alvarez E., Rejas M.T., Perlman S., Enjuanes L. Pathogenicity of severe acute respiratory coronavirus deletion mutants in hACE-2 transgenic mice. Virology. 2008;376:379–389. doi: 10.1016/j.virol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeño J.M., Regla-Nava J.A., Castaño-Rodriguez C., Fernandez-Delgado R., Usera F., Enjuanes L. Coronavirus virulence genes with main focus on SARS-CoV envelope gene. Virus Res. 2014;194:124–137. doi: 10.1016/j.virusres.2014.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desforges M., Desjardins J., Zhang C., Talbot P.J. The acetyl-esterase activity of the hemagglutinin-esterase protein of human coronavirus OC43 strongly enhances the production of infectious virus. J. Virol. 2013;87:3097–3107. doi: 10.1128/JVI.02699-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desforges M., Le Coupanec A., Stodola J.K., Meessen-Pinard M., Talbot P.J. Human coronaviruses: viral and cellular factors involved in neuroinvasiveness and neuropathogenesis. Virus Res. 2014;194:145–158. doi: 10.1016/j.virusres.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Günther S., Preiser W., Werf S., van der, Brodt H.-R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A.M., Berger A., Burguière A.-M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.-C., Müller S., Rickerts V., Stürmer M., Simon Vieth H.-D.K., Osterhaus A.D.M.E., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Favreau D.J., Meessen-Pinard M., Desforges M., Talbot P.J. Human coronavirus-induced neuronal programmed cell death is cyclophilin D dependent and potentially caspase dispensable. J. Virol. 2012;86:81–93. doi: 10.1128/JVI.06062-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W., Zhang M. Organization and dynamics of PDZ-domain-related supramodules in the postsynaptic density. Nat. Rev. Neurosci. 2009;10:87–99. doi: 10.1038/nrn2540. [DOI] [PubMed] [Google Scholar]
- Fischer F., Stegen C.F., Masters P.S., Samsonoff W.A. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 1998;72:7885–7894. doi: 10.1128/jvi.72.10.7885-7894.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fronhoffs S., Totzke G., Stier S., Wernert N., Rothe M., Bruning T., Koch B., Sachinidis A., Vetter H., Ko Y. A method for the rapid construction of cRNA standard curves in quantitative real-time reverse transcription polymerase chain reaction. Mol. Cell Probes. 2002;16:99–110. doi: 10.1006/mcpr.2002.0405. [DOI] [PubMed] [Google Scholar]
- Godet M., L’Haridon R., Vautherot J.F., Laude H. TGEV corona virus ORF4 encodes a membrane protein that is incorporated into virions. Virology. 1992;188:666–675. doi: 10.1016/0042-6822(92)90521-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Baker S.C., Baric R., Enjuanes L., Gorbalenya A.E., Holmes K.V., Perlman S., Poon L., Rottier P.J.M., Talbot P.J., Woo P.C.Y., Ziebuhr J. Coronaviridae. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus Taxonomy. Elsevier; New York, NY: 2012. pp. 806–828. [DOI] [Google Scholar]
- Gu J., Gong E., Zhang B., Zheng J., Gao Z., Zhong Y., Zou W., Zhan J., Wang S., Xie Z., Zhuang H., Wu B., Zhong H., Shao H., Fang W., Gao D., Pei F., Li X., He Z., Xu D., Shi X., Anderson V.M., Leong A.S.-Y. Multiple organ infection and the pathogenesis of SARS. J. Exp. Med. 2005;202:415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D.P., Robertson K.A. Differentiation of LA-N-5 neuroblastoma cells into cholinergic neurons: methods for differentiation, immunohistochemistry and reporter gene introduction. Brain Res. Protoc. 1998;2:183–190. doi: 10.1016/S1385-299X(97)00041-X. [DOI] [PubMed] [Google Scholar]
- Jacomy H., Talbot P.J. Vacuolating encephalitis in mice infected by human coronavirus OC43. Virology. 2003;315:20–33. doi: 10.1016/S0042-6822(03)00323-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacomy H., Fragoso G., Almazan G., Mushynski W.E., Talbot P.J. Human coronavirus OC43 infection induces chronic encephalitis leading to disabilities in BALB/C mice. Virology. 2006;349:335–346. doi: 10.1016/j.virol.2006.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javier R.T., Rice A.P. Emerging theme: cellular PDZ proteins as common targets of pathogenic viruses. J. Virol. 2011;85:11544–11556. doi: 10.1128/JVI.05410-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Guardeño J.M., Nieto-Torres J.L., DeDiego M.L., Regla-Nava J. a., Fernandez-Delgado R., Castaño-Rodriguez C., Enjuanes L. The PDZ-binding motif of severe acute respiratory syndrome coronavirus envelope protein is a determinant of viral pathogenesis. PLoS Pathog. 2014;10:e1004320. doi: 10.1371/journal.ppat.1004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Guardeño J.M., Regla-Nava J.A., Nieto-Torres J.L., DeDiego M.L., Castaño-Rodriguez C., Fernandez-Delgado R., Perlman S., Enjuanes L. Identification of the mechanisms causing reversion to virulence in an attenuated SARS-CoV for the design of a genetically stable vaccine. PLoS Pathog. 2015;11:e1005215. doi: 10.1371/journal.ppat.1005215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret L.J.A.T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.-E., Humphrey C.D., Wun P., the SARS Working Group M.D. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kuo L., Masters P.S. The small envelope protein E is not essential for murine coronavirus replication. J. Virol. 2003;77:4597–4608. doi: 10.1128/JVI.77.8.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Masters P.S. Evolved variants of the membrane protein can partially replace the envelope protein in murine coronavirus assembly. J. Virol. 2010;84:12872–12885. doi: 10.1128/JVI.01850-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Hurst K.R., Masters P.S. Exceptional flexibility in the sequence requirements for coronavirus small envelope protein function. J. Virol. 2007;81:2249–2262. doi: 10.1128/JVI.01577-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert F., Jacomy H., Marceau G., Talbot P.J. Titration of human coronaviruses using an immunoperoxidase assay. J. Vis. Exp. 2008 doi: 10.3791/751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Coupanec A., Desforges M., Meessen-Pinard M., Dubé M., Day R., Seidah N.G., Talbot P.J. Cleavage of a neuroinvasive human respiratory virus spike glycoprotein by proprotein convertases modulates neurovirulence and virus spread within the central nervous system. PLoS Pathog. 2015;11:e1005261. doi: 10.1371/journal.ppat.1005261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Inglis S.C. Association of the infectious bronchitis virus 3c protein with the virion envelope. Virology. 1991;185:911–917. doi: 10.1016/0042-6822(91)90572-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfopoulou S., Brown J.R., Davies E.G., Anderson G., Virasami A., Qasim W., Chong W.K., Hubank M., Plagnol V., Desforges M., Jacques T.S., Talbot P.J., Breuer J. Human coronavirus OC43 associated with fatal encephalitis. N. Engl. J. Med. 2016;375:497–498. doi: 10.1056/NEJMc1509458. [DOI] [PubMed] [Google Scholar]
- Nieto-Torres J.L., Dediego M.L., Alvarez E., Jiménez-Guardeño J.M., Regla-Nava J.A., Llorente M., Kremer L., Shuo S., Enjuanes L. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415:69–82. doi: 10.1016/j.virol.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres J.L., DeDiego M.L., Verdiá-Báguena C., Jimenez-Guardeño J.M., Regla-Nava J.A., Fernandez-Delgado R., Castaño-Rodriguez C., Alcaraz A., Torres J., Aguilella V.M., Enjuanes L. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 2014;10:e1004077. doi: 10.1371/journal.ppat.1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieva J.L., Madan V., Carrasco L. Viroporins: structure and biological functions. Nat. Rev. Microbiol. 2012;10:563–574. doi: 10.1038/nrmicro2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortego J., Escors D., Laude H., Enjuanes L. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 2002;76:11518–11529. doi: 10.1128/JVI.76.22.11518-11529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortego J., Ceriani J.E., Patiño C., Plana J., Enjuanes L. Absence of E protein arrests transmissible gastroenteritis coronavirus maturation in the secretory pathway. Virology. 2007;368:296–308. doi: 10.1016/j.virol.2007.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervushin K., Tan E., Parthasarathy K., Lin X., Jiang F.L., Yu D., Vararattanavech A., Soong T.W., Liu D.D.X., Torres J. Structure and inhibition of the SARS coronavirus envelope protein ion channel. PLoS Pathog. 2009;5:e1000511. doi: 10.1371/journal.ppat.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Préhaud C., Wolff N., Terrien E., Lafage M., Mégret F., Babault N., Cordier F., Tan G.S., Maitrepierre E., Ménager P., Chopy D., Hoos S., England P., Delepierre M., Schnell M.J., Buc H., Lafon M. Attenuation of rabies virulence: takeover by the cytoplasmic domain of its envelope protein. Sci. Signal. 2010;3:ra5. doi: 10.1126/scisignal.2000510. [DOI] [PubMed] [Google Scholar]
- Regla-Nava J.A., Nieto-Torres J.L., Jimenez-Guardeño J.M., Fernandez-Delgado R., Fett C., Castaño-Rodríguez C., Perlman S., Enjuanes L., DeDiego M.L. Severe acute respiratory syndrome coronaviruses with mutations in the E protein are attenuated and promising vaccine candidates. J. Virol. 2015;89:3870–3887. doi: 10.1128/JVI.03566-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruch T.R., Machamer C.E. The hydrophobic domain of infectious bronchitis virus E protein alters the host secretory pathway and is important for release of infectious virus. J. Virol. 2011;85:675–685. doi: 10.1128/JVI.01570-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruch T.R., Machamer C.E. A single polar residue and distinct membrane topologies impact the function of the infectious bronchitis coronavirus E protein. PLoS Pathog. 2012;8:e1002674. doi: 10.1371/journal.ppat.1002674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott C., Griffin S. Viroporins: structure, function and potential as antiviral targets. J. Gen. Virol. 2015;96:2000–2027. doi: 10.1099/vir.0.000201. [DOI] [PubMed] [Google Scholar]
- St-Jean J.R., Desforges M., Almazán F., Jacomy H., Enjuanes L., Talbot P.J. Recovery of a neurovirulent human coronavirus OC43 from an infectious cDNA clone. J. Virol. 2006;80:3670–3674. doi: 10.1128/JVI.80.7.3670-3674.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surya W., Li Y., Verdià-Bàguena C., Aguilella V.M., Torres J. MERS coronavirus envelope protein has a single transmembrane domain that forms pentameric ion channels. Virus Res. 2015;201:61–66. doi: 10.1016/j.virusres.2015.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoh K.-T., Siu Y.-L., Chan W.-L., Schlüter M.A., Liu C.-J., Peiris J.S.M., Bruzzone R., Margolis B., Nal B. The SARS coronavirus E protein interacts with PALS1 and alters tight junction formation and epithelial morphogenesis. Mol. Biol. Cell. 2010;21:3838–3852. doi: 10.1091/mbc.E10-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J., Wang J., Parthasarathy K., Liu D.X., Xiang D. The transmembrane oligomers of coronavirus protein E. Biophys. J. 2005;88:1283–1290. doi: 10.1529/biophysj.104.051730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J., Parthasarathy K., Lin X., Saravanan R., Kukol A., Liu D.X. Model of a putative pore: the pentameric alpha-helical bundle of SARS coronavirus E protein in lipid bilayers. Biophys. J. 2006;91:938–947. doi: 10.1529/biophysj.105.080119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret A., Dina J., Brison E., Brouard J., Freymuth F. Coronavirus humains (HCoV) Pathol. Biol. 2009;57:149–160. doi: 10.1016/j.patbio.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatagopalan P., Daskalova S.M., Lopez L.A., Dolezal K.A., Hogue B.G. Coronavirus envelope (E) protein remains at the site of assembly. Virology. 2015;478:75–85. doi: 10.1016/j.virol.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdiá-Báguena C., Nieto-Torres J.L., Alcaraz A., DeDiego M.L., Torres J., Aguilella V.M., Enjuanes L. Coronavirus E protein forms ion channels with functionally and structurally-involved membrane lipids. Virology. 2012;432:485–494. doi: 10.1016/j.virol.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdiá-Báguena C., Nieto-Torres J.L., Alcaraz A., Dediego M.L., Enjuanes L., Aguilella V.M. Analysis of SARS-CoV e protein ion channel activity by tuning the protein and lipid charge. Biochim. Biophys. Acta - Biomembr. 2013;1828:2026–2031. doi: 10.1016/j.bbamem.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Moe E., Maes P., Duson G., Ranst M., Van Development of one-step, real-time, quantitative reverse transcriptase pcr assays for absolute quantitation. J. Clin. Microbiol. 2005;43:5452–5456. doi: 10.1128/JCM.43.11.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerbeck J.W., Machamer C.E. A coronavirus E protein is present in two distinct pools with different effects on assembly and the secretory pathway. J. Virol. 2015 doi: 10.1128/JVI.01237-15. (JVI.01237-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson L., Mckinlay C., Gage P., Ewart G. SARS coronavirus E protein forms cation-selective ion channels. Virology. 2004;330:322–331. doi: 10.1016/j.virol.2004.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson L., Gage P., Ewart G. Hexamethylene amiloride blocks E protein ion channels and inhibits coronavirus replication. Virology. 2006;353:294–306. doi: 10.1016/j.virol.2006.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Zhong S., Liu J., Li L., Li Y., Wu X., Li Z., Deng P., Zhang J., Zhong N., Ding Y., Jiang Y. Detection of severe acute respiratory syndrome coronavirus in the brain: potential role of the chemokine mig in pathogenesis. Clin. Infect. Dis. 2005;41:1089–1096. doi: 10.1086/444461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F., Zhang M. Structures and target recognition modes of PDZ domains: recurring themes and emerging pictures. Biochem. J. 2013;455:1–14. doi: 10.1042/BJ20130783. [DOI] [PubMed] [Google Scholar]
- Ye Y., Hogue B.G. Role of the coronavirus E viroporin protein transmembrane domain in virus assembly. J. Virol. 2007;81:3597–3607. doi: 10.1128/JVI.01472-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E.A., Collins A., Cohen M.E., Duffner P.K., Faden H. Detection of coronavirus in the central nervous system of a child with acute disseminated encephalomyelitis. Pediatrics. 2004;113:e73–e76. doi: 10.1542/peds.113.1.e73. [DOI] [PubMed] [Google Scholar]
- Yu X., Bi W., Weiss S.R., Leibowitz J.L. Mouse hepatitis virus gene 5b protein is a new virion envelope protein. Virology. 1994;202:1018–1023. doi: 10.1006/viro.1994.1430. [DOI] [PubMed] [Google Scholar]
- Zaki A.M., Boheemen S., van, Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]