Highlights
-
•
Inhibitory effects of three vivo-MOs targeting EV-71 IRES and RdRP were evaluated.
-
•
IRES targeting vivo-MOs exhibited most significant antiviral activities.
-
•
IRES targeting vivo-MOs inhibited EV-71 viral RNA translation.
-
•
Antiviral efficacy is reduced when the number of mismatches increased.
-
•
IRES targeting vivo-MOs are good antiviral candidates.
Keywords: Enterovirus 71; Hand, foot and mouth disease; Enterovirus; Morpholino oligomers; Antiviral agent
Abstract
Enterovirus 71 (EV-71) infections are generally manifested as mild hand, foot and mouth disease, but have been reported to cause severe neurological complications with high mortality rates. Treatment options remain limited due to the lack of antivirals. Octaguanidinium-conjugated morpholino oligomers (vivo-MOs) are single-stranded DNA-like antisense agents that can readily penetrate cells and reduce gene expression by steric blocking of complementary RNA sequences. In this study, inhibitory effects of three vivo-MOs that are complementary to the EV-71 internal ribosome entry site (IRES) and the RNA-dependent RNA polymerase (RdRP) were tested in RD cells. Vivo-MO-1 and vivo-MO-2 targeting the EV-71 IRES showed significant viral plaque reductions of 2.5 and 3.5 log10PFU/ml, respectively. Both vivo-MOs reduced viral RNA copies and viral capsid expression in RD cells in a dose-dependent manner. In contrast, vivo-MO-3 targeting the EV-71 RdRP exhibited less antiviral activity. Both vivo-MO-1 and 2 remained active when administered either 4 h before or within 6 h after EV-71 infection. Vivo-MO-2 exhibited antiviral activities against poliovirus (PV) and coxsackievirus A16 but vivo-MO-1 showed no antiviral activities against PV. Both the IRES-targeting vivo-MO-1 and vivo-MO-2 inhibit EV-71 RNA translation. Resistant mutants arose after serial passages in the presence of vivo-MO-1, but none were isolated against vivo-MO-2. A single T to C substitution at nucleotide position 533 was sufficient to confer resistance to vivo-MO-1. Our findings suggest that IRES-targeting vivo-MOs are good antiviral candidates for treating early EV-71 infection, and vivo-MO-2 is a more favorable candidate with broader antiviral spectrum against enteroviruses and are refractory to antiviral resistance.
1. Introduction
Enterovirus 71 (EV-71) is a single-strand, positive-sense RNA virus. EV-71 usually cause mild hand, foot and mouth disease (HFMD) characterized by fever with papulovesicular rash on the palms and soles (Ooi et al., 2010). In recent years, EV-71 infections were also associated with neurological complications with high mortalities among infants and young children < 6 years old (Solomon et al., 2010). To date, no effective antiviral agent is available for clinical use (Shang et al., 2013b, Tan et al., 2014). Thus, there is an urgent need to develop effective antiviral agents to treat EV-71 infection.
Considering the morbidity caused by EV-71, new approaches to the development of therapeutics are needed. A number of promising RNA-based therapeutics designed to inhibit EV-71 infections have shown promising results, including siRNA and shRNA (Deng et al., 2012, Sim et al., 2005, Tan et al., 2007a, Tan et al., 2007b, Wu et al., 2009). However, the limitations of RNA-based therapeutics are short half-life and it required a delivery agent which might be toxic to the host. Therefore, nucleic acid-based therapeutics should be designed to possess favorable pharmacological properties such as in vivo stability and low toxicity.
A phosphorodiamidate morpholino oligomer (PMO) is a single-stranded DNA-like compound that has the ability to bind to the mRNA and inhibit gene expression by steric blockage of complementary RNA. PMOs are highly nuclease-resistant and do not require the RNase H or other catalytic proteins for their activity (Kole et al., 2012, Summerton, 1999). PMOs have been conjugated with various cell-penetrating compounds such as cell-penetrating peptides and octaguanidinium dendrimers which are able to enhance their uptake by cells (Moulton and Jiang, 2009). Peptide conjugated-PMOs (PPMO) have been demonstrated to inhibit various viral infections, including Ebola virus (Warfield et al., 2006), West Nile virus (Deas et al., 2005), dengue virus (Kinney et al., 2005), sindbis virus (Paessler et al., 2008), coronavirus (Neuman et al., 2004), herpes simplex virus 1 (Moerdyk-Schauwecker et al., 2009), porcine reproductive and respiratory syndrome virus (Opriessnig et al., 2011, Patel et al., 2008), foot-and-mouth disease virus (Vagnozzi et al., 2007), poliovirus, rhinovirus (Stone et al., 2008), and coxsackievirus B3 (Yuan et al., 2006).
In this study, three octaguanidinium dendrimer conjugated-morpholino oligomers (vivo-MOs) targeting the EV-71 internal ribosome entry site (IRES) core sequence and the RNA-dependent RNA polymerase (RdRP) were tested for their inhibitory effects against EV-71. We demonstrated that the two vivo-MOs targeting the IRES core sequence showed significant inhibition of EV-71 infection.
2. Materials and methods
2.1. Cells and viruses
Rhabdomyosarcoma (RD, ATCC) cells were grown in Dulbecco’s modified Eagle’s medium (DMEM, Hyclone) supplemented with 10% fetal bovine serum (FBS). EV-71 strains 41 (GenBank accession number: AF316321), BrCr (GenBank accession number: AB204852) and UH1/97 (GenBank accession number: AM396587); coxsackievirus A16 (CV-A16), PV and chikungunya virus (CHIKV) strain MY/08/065 (GenBank accession number: FN295485) were propagated in RD cells.
2.2. Vivo-MOs
All vivo-MOs were synthesized by Gene Tools LLC (USA). The 23-mer vivo-MOs were designed to be complementary to the EV-71 (strain 41) IRES stem-loop V-VI and the RdRP gene (Table 1 , Fig. 1 ). All the vivo-MOs were dissolved in phosphate buffer saline (PBS) at concentration of 0.5 mM. The cytotoxicity of the vivo-MOs were evaluated using Cell Titer 96 AQueous cell proliferation reagent (Promega) according to the manufacturer’s instructions.
Table 1.
Sequence of the 23-mer vivo-MOs and target locations in EV-71 RNA.
| Vivo-MOs | Sequence (5’–3’) | Target location in EV-71 RNA (nucleotide position) |
|---|---|---|
| 1 | CAGAGTTGCCCATTACGACACAC | IRES core (512–534) |
| 2 | GAAACACGGACACCCAAAGTAGT | IRES core (546–568) |
| 3 | AAACAATTCGAGCCAATTTCTTC | 3D Pol gene (7303–7325) |
| Control | CCTACTCCATCGTTCAGCTCTGA | – |
Fig. 1.
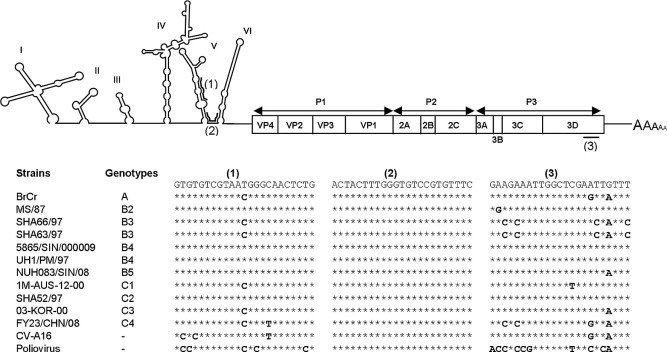
Schematic illustration of the EV-71 genomic structure. Three genomic vivo-MOs target sequences (5’ to 3’) are indicated as (1)–(3) within the proposed secondary structures of the IRES region and the RdRP gene of EV-71 RNA. The sequences of these three targeted regions were aligned across all EV-71 genotypes, CV-A16 and PV.
2.3. In vitro inhibitory effects of vivo-MOs in RD cells
RD cells were seeded at 1.5 × 104 cells or 1.5 × 105 cells within each well of a 96-well plate or 24-well plate, respectively and incubated overnight at 37 °C in 5% CO2. After overnight incubation, the growth medium was removed and replaced with EV-71 inoculum with a multiplicity of infection (MOI) of 0.1 (PFU per cell) and incubated at 37 °C for 1 h. After incubation, the inoculum was removed and replenished with maintenance medium (DMEM with 2% FBS) with or without vivo-MOs. The inhibitory effects of the vivo-MOs were evaluated by plaque assay, TaqMan real-time RT-PCR and western blot analysis 24 h post-infection (hpi) as previously described (Tan et al., 2012, Tan et al., 2013). The western blot signal was enhanced using SuperSignal® western blot enhancer (Pierce Biotechnology).
2.4. Time of addition assay
Vivo-MOs were added to RD cells at various time points relative to viral inoculation. RD cells were pre-incubated with vivo-MOs at final concentration of 5 μM for 4 h before EV-71 inoculation at a MOI of 0.1. In concurrent studies, both vivo-MOs and EV-71 were added into RD cells for 1 h followed by replacement of medium without vivo-MO. For post-infection studies, RD cells were infected with EV-71 for 1, 2, 4 and 6 h before vivo-MOs were applied. The viral titers for each experiment were quantitated 24 hpi by plaque assays.
2.5. In vitro inhibitory effects of vivo-MOs against various enteroviruses
To evaluate the efficacy of vivo-MOs against different enteroviruses including CV-A16 and PV, RD cells were pre-incubated with vivo-MOs at the final concentration of 5 μM for 4 h before viral inoculation at a MOI of 0.1 for 1 h at 37 °C. The viral titers were determined 24 hpi by plaque assay. CHIKV was used as a negative control virus in this experiment.
2.6. Construction of EV-71 infectious cDNA clones
EV-71 strain 41 infectious clone was constructed using the full-length genome PCR approach according to Yeh et al. (2011) with modifications. The primers involved in the infectious clone constructions were listed in Table S1. Full-length genome RT-PCR was performed with Superscript III reverse transcriptase (Invitrogen) and iProof High-Fidelity polymerase (Bio-Rad) using pT50/EagI-R and pSP6/EV71-F primers. EV-71 expressing enhanced green fluorescence protein (EGFP) was constructed by overlapping extension PCR strategy using Q5 High-Fidelity polymerase (NEB). The EGFP gene was fused into the EV-71 genome between the 5’ UTR and VP4 gene followed by the 2A cleavage site (-AITTL-) as previously described (Shang et al., 2013a). The full-length PCR product was then cloned into pCR-XL-TOPO (Invitrogen). In vitro transcription was performed with linearized DNA using RiboMAX-SP6 large scale RNA production system (Promega) and the RNA was transfected into RD cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
2.7. Cell free translation inhibition assay
Cell free translation assay was performed with 1 μg of in vitro transcribed RNA using 1-step human coupled IVT kit (Pierce Biotechnology) either in the presence or absence of vivo-MOs according to the manufacturer’s instructions. An aliquot of 20 μl of in vitro translated sample was subjected to SDS–PAGE and western blot analysis as described previously. The immunoblot was developed with Clarity™ Western ECL substrate (Bio-Rad) and detected by chemiluminescence.
2.8. EV-71–EGFP inhibition assay
RD cells in a 96-well plate were infected with EV-71–EGFP for an hour at 37 °C. After incubation, the inoculum was removed and replaced with maintenance medium containing 2.5 μM of vivo-MOs. The EGFP expression was observed at 6 hpi using fluorescence microscopy.
2.9. Generation of vivo-MOs resistant viruses
EV-71 was passaged in RD cells with increasing concentrations of either vivo-MO-1 or vivo-MO-2. For the first passage, RD cells were infected with EV-71 at a MOI of 0.1 for 1 h at 37 °C and the inoculum was removed and replaced with maintenance medium containing 1 μM of vivo-MO-1 or vivo-MO-2. Each selection was passaged by adding 100 μl of the supernatant into new RD cells. Viruses were passaged once at each of the concentrations ranging from 1 μM to 4 μM, followed by passaging four times at 5 μM. To identify the mutation(s) which conferred resistance to vivo-MOs, viral RNA of an individual plaque population was amplified and subjected to DNA sequencing.
2.10. Reverse genetic analysis of vivo-MO resistant viruses
Point mutations were incorporated into the EV-71 infectious clone by using QuickChange Lightning site-directed mutagenesis kit (Agilent Technologies) according to the manufacturer’s instructions. Four mutants were constructed with different nucleotide substitutions at the vivo-MO-1 targeted region (Table 2 ). The degree of resistance was evaluated using inhibitory assay as described in Section 2.3.
Table 2.
Sequence of vivo-MO-1 and the in vitro transcribed infectious RNA with target sequences.
| Target | Sequences | Mismatch(es) |
|---|---|---|
| Vivo-MO-1 | 3’-CAGAGTTGCCCATTACGACACAC-3’ | – |
| MO-1-WT | 5’-GTGTGTCGTAATGGGCAACTCTG-3’ | 0 |
| MO-1-mutant-1 | 5’-GTGTGTCGTAATGGGCAACTCCG-3’ | 1 |
| MO-1-mutant-2 | 5’-GTGTGTCGTAACGGGCAACTCTG-3’ | 1 |
| MO-1-mutant-3 | 5’-GTGTGTCGTAACGGGTAACTCTG-3’ | 2 |
| MO-1-mutant-4 | 5’-GTGCGTCGTAACGGGTAACTCTG-3’ | 3 |
2.11. Statistical analysis
The data presented are the means ± standard deviations (SD) obtained from at least two independent biological replicates. Error bars represent the SD. Statistical significance was calculated using the Mann–Whitney test. A P value of <0.05 was considered statistically significant.
3. Result
3.1. Vivo-MOs complementary to EV-71 IRES stem-loop structures exhibited significant inhibitory activities
To evaluate the effects of vivo-MOs on EV-71 infectivity in RD cells, RD cells were treated with vivo-MOs an hour after infection. As shown in Fig. 2 , both vivo-MOs targeting the EV-71 IRES stem-loop region exhibited significant antiviral activity against EV-71 infection with reduction of virus-induced CPE (Fig. 2A), plaque formation (Fig. 2B), RNA (Fig. 2C) and capsid expression (Fig. 2D) in a dose-dependent manner. The vivo-MO-1 and vivo-MO-2 significantly reduced EV-71 plaque formation by up to 2.7 and 3.5 log10PFU/ml at 10 μM, respectively. Significant inhibition was observed at concentrations higher than 1 μM. The IC50 values of vivo-MO-1 and vivo-MO-2 were 1.5 μM and 1.2 μM, respectively. However, vivo-MO-3 exhibited less inhibitory effect against EV-71 infection in RD cells with a plaque reduction of only 1.2 log10PFU/ml (Fig. 2B). The vivo-MO-C which has no homologous sequence to the EV-71 genome has no inhibitory effects at all against all EV-71 strains tested. None of the vivo-MOs caused more than 20% reduction of cell viability at concentrations less than 5 μM as measured by the MTS assay (Fig. 3 ).
Fig. 2.
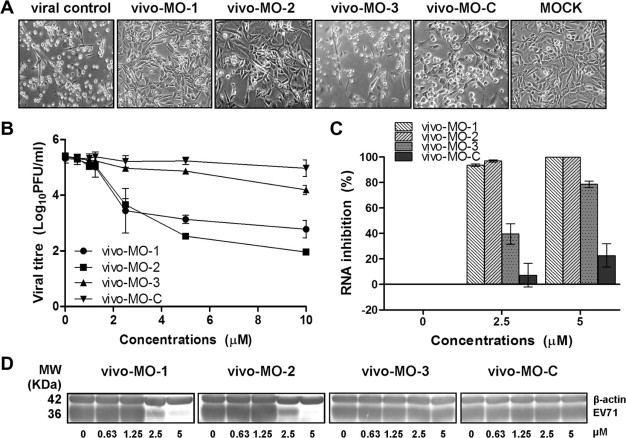
Inhibitory effect of vivo-MOs in RD cells. Various concentrations of vivo-MOs were applied to RD cells one hour post infection and (A) viral induced CPE (20 × objective) were observed 24 hpi. The total infectious particles or total viral proteins were harvested 24 hpi and evaluated by (B) plaque assay, (C) quantitative TaqMan real-time PCR, and (D) western blot analysis, respectively. EV-71 viral capsid protein was detected by mouse anti-EV-71 monoclonal antibody (Millipore) and cellular β-actin was detected using mouse anti-β-actin monoclonal antibody (Sigma). The data presented were obtained from at least two independent biological replicates.
Fig. 3.
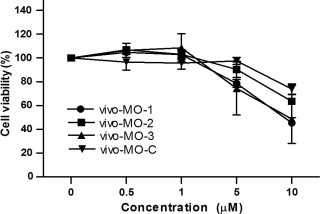
Cell viability analysis. Various concentrations of vivo-MOs were incubated with RD cells (1.5 × 104 cells) for 24 h in maintenance medium (DMEM supplemented with 2% FBS) followed by MTS assay using Cell Titer 96 AQueous One solution cell proliferation (Promega). The absorbance reading at 490 nm was obtained using a microtiter plate reader after 2 h of incubation at 37 °C. The percentage of cell viability (%) was determined by multiplying the ratio of the absorbance readings obtained from cells treated with vivo-MOs over the non-treated cells with 100%. The data presented were obtained from at least two independent biological replicates.
3.2. Vivo-MOs blocked EV-71 infections at multiple time points
To further characterize the efficacy of vivo-MOs at multiple time points relative to EV-71 infection, vivo-MOs were applied for 4 h before, or 1, 2, 4, or 6 h after EV-71 infection. Both vivo-MO-1 and vivo-MO-2 remained effective when administered before or after EV-71 infection. However, the efficacies were reduced when treatments were delayed. When vivo-MOs and EV-71 were added together into the RD cells for 1 h, the inhibitory effect was reduced, which could have resulted from the incomplete uptake of the vivo-MOs by the cells. Nonetheless, the antiviral effects were retained for both IRES-targeting vivo-MOs even when administered 6 hpi, with 92.8% plaque inhibition. Vivo-MO-3 had no observable inhibitory effects on EV-71 infection when administered 4 h before infection and 2, 4, or 6 hpi (Fig. 4 A).
Fig. 4.
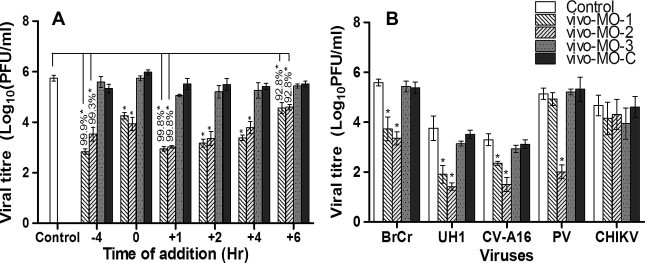
Time and specificity of vivo-MOs antiviral properties. (A) Vivo-MOs at a final concentration of 5 μM were applied to RD cells at various time points relative to EV-71 inoculation. In brief, vivo-MOs were applied for 4 h before, or 1, 2, 4, or 6 h after EV-71 infection at a MOI of 0.1. (B) RD cells were pre-treated with each of the vivo-MOs at a final concentration of 5 μM for 4 h at 37 °C before infection with other enteroviruses (EV-71 strain BrCr, UH1/97, PV and CV-A16) and CHIKV at a MOI of 0.1. The inhibitory effects of each of the vivo-MOs were evaluated by plaque assay 24 hpi and the percentages of inhibition are shown. Asterisks indicate statistically significant differences compared to the control. The data presented were obtained from at least two independent biological replicates.
3.3. Vivo-MO-2 exhibited broad-spectrum antiviral activities against various enteroviruses
Next, we investigate whether any of the vivo-MOs could inhibit different EV-71 strains as well as other picornaviruses. We evaluated each of the vivo-MOs against two other EV-71 strains (BrCr and UH1/97), PV, CV-A16 and CHIKV as the control virus. The vivo-MO-2 which targets the highly conserved region of the IRES stem-loop structure exhibited significant inhibitory activity against EV-71 strains BrCr and UH1/97, PV and CV-A16 with viral plaque reduction ranging from 1.8 to 3.1 log10PFU/ml (Fig. 4B). However, vivo-MO-1 only exhibited antiviral activities against EV-71 strains BrCr and UH1/97, CV-A16, but not against PV (Fig. 4B). EV-71 strain BrCr which has a single nucleotide mismatch in the middle of the vivo-MO-1 targeted site (Fig. 1) remained sensitive to the vivo-MO-1 treatment. The efficacy of vivo-MO-1 against CV-A16 (0.95 log10PFU/ml reduction) was significantly lower when compared to EV-71(1.86 log10PFU/ml reduction). This could be due to three nucleotide mismatches with the vivo-MO-1. PV with five nucleotide mismatches was completely resistant to the inhibitory effect of vivo-MO-1. All the vivo-MOs did not show any inhibition of CHIKV infection.
3.4. IRES-targeting vivo-MOs blocked EV-71 translation
To investigate the mechanism of action of the antiviral vivo-MOs, cell-free translation analysis was used. As depicted in Fig. 5 A, the presence of either vivo-MO-1 or vivo-MO-2 significantly blocked the IRES-dependent translation when compared to the control. Vivo-MO-3 exhibits reduced efficacies as compared to IRES-targeting vivo-MOs. In the EV-71–EGFP inhibition assay, the presence of the vivo-MO-1 or vivo-MO-2 greatly reduced EGFP expression in RD cells 6 hpi with EV-71–EGFP (Fig. 5B).
Fig. 5.
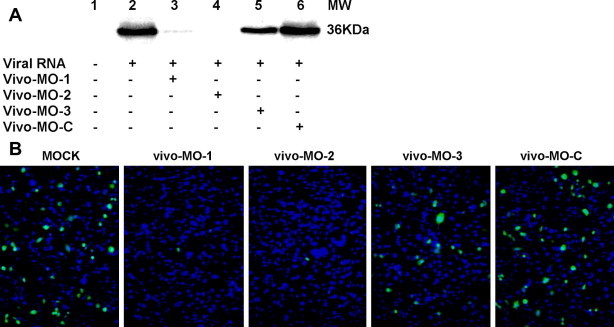
Translation inhibition assay. (A) In vitro translation was performed with 1 μg of RNA using 1-step human coupled IVT kit (Pierce Biotechnology) either in the presence of vivo-MOs at a final concentration of 10 μM. Aliquots of 20 μl of translated product was subjected to SDS–PAGE and western blot analysis. EV-71 viral protein was detected by anti-EV-71 monoclonal antibody through chemiluminescence. (B) EV-71–EGFP inhibition assay. RD cells were first infected with EV-71–EGFP for one hour, followed by addition of maintenance medium containing 2.5 μM of vivo-MOs. The EGFP expression was evaluated 6 hpi using a fluorescence microscope. Nuclei were stained with DAPI. EGFP expression and nuclei are shown in green and blue, respectively. The data presented were obtained from at least two independent biological replicates. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5. Tolerance of vivo-MO-1 to mismatches within the target RNA
We explored whether EV-71 could become resistant to vivo-MO treatments. EV-71 was serially passaged in the presence of increasing concentrations of either vivo-MO-1 or vivo-MO-2. Interestingly, only EV-71 mutants resistant to vivo-MO-1 were isolated after eight passages. We failed to isolate EV-71 mutants that were resistant to vivo-MO-2.
To investigate the determinant(s) of resistance, viral RNA was isolated from the resistant population and was sequenced. A single point mutation from T to C at position 533 was sufficient to confer resistance to vivo-MO-1 (Fig. 6 A). To characterize the loss of inhibitory activity by vivo-MO-1, we constructed EV-71 mutants carrying different mismatches at the vivo-MO-1 target site (Table 2), and the inhibitory effects of vivo-MO-1 against each of the mutants were evaluated. The mismatched RNA target sequences were designed to reflect the most likely natural variations that would arise in the EV-71 sequence. As shown in Fig. 6B, the EV-71 MO-1-mutant-1, which carried a single point mutation at position 533 (T to C substitution), required higher vivo-MO-1 concentrations to achieve a similar inhibitory effect when compared to the wild type. The EV-71 MO-1-mutant-2 with a single point mutation in the middle of the targeted sequence (a T to C substitution at position 523) remained sensitive to vivo-MO-1, but vivo-MO-1 had reduced inhibitory efficacy when compared with the wild type. Increasing the number of mutations on the targeted sequence significantly reduced the inhibitory efficacy of vivo-MO-1. The viral plaque inhibition at 2 μM of vivo-MO-1 against EV-71 MO-1-WT was 98.9% ± 0.1, and reduced to 89.6% ± 3.4 for the MO-1-mutant-1 that carried a point mutation at the 3’ end of the target sequence. The viral plaque inhibition by vivo-MO-1 reduced to 78.0% when the number of nucleotide mismatches increased to three.
Fig. 6.
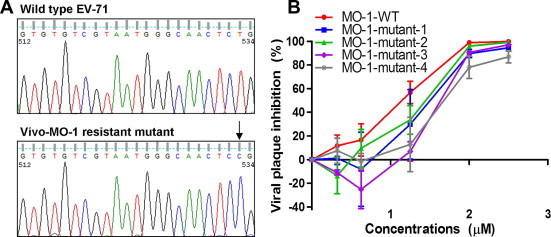
DNA sequence analysis of vivo-MO-1. (A) EV-71 was serially passaged in the presence of vivo-MO-1 from 1 μM to 5 μM. EV-71 populations that showed resistance to vivo-MO-1 were plaque purified, and subjected to DNA sequencing. Analysis of the vivo-MO-1 resistant mutant showed a T to C substitution at position 533. (B) The effect of sequence mismatches between vivo-MO-1 and target RNA on inhibition assay. Several in vitro transcribed RNA, each having a different number of nucleotide mismatches in the target region as described in Section 2 were analyzed. Viral titers were quantitated 24 hpi using standard plaque assay. The data presented were obtained from at least two independent biological replicates.
4. Discussion
To date, there is no FDA approved vaccine available to prevent infection and treatment options remain limited due to a lack of effective antivirals (Chong et al., 2012, Shang et al., 2013b, Tan and Chan, 2013). Highly negatively-charged compounds like suramin or its analog, NF449 that disrupt the virus-host receptor interactions are ineffective when EV-71 mutants acquire Glu98Gln and Lys244Arg substitutions in the VP1 capsid protein (Arita et al., 2008, Tan et al., 2013). A single point mutation, Val192Met in the VP1, is sufficient to confer resistance to the capsid binder BPROZ-194 (Shih et al., 2004). Therefore, there is an urgent need to develop effective antiviral agents to treat patients with severe EV-71 infection.
The use of antisense mechanisms to inhibit pathogen replication has been under investigation and has shown promising results. Fomivirsen, a 21-mer phosphorothioate oligonucleotide antisense agent used for intravitreal treatment of cytomegalovirus retinitis, is the only antisense agent that has been approved by the FDA to date (de Smet et al., 1999). There are advantages of vivo-MOs over RNA-based antisense oligomers as they are, nuclease-resistant and readily penetrate the cells (Kole et al., 2012, Moulton and Jiang, 2009). These have an advantage over peptide-based cell penetrating molecules, which are susceptible to protease degradation that can impact the effectiveness of PPMOs (Youngblood et al., 2007).
Amongst the three vivo-MOs being examined, the two vivo-MOs targeting the EV-71 IRES stem-loop region exhibited significant antiviral activity. The vivo-MOs blocked EV-71 viral RNA translation in a cell-free inhibition assay, as well as in a cell-based EGFP reporter inhibition assay. Previous studies have also reported that only PMOs targeting the positive strand IRES regions or the AUG start codon sites were effective when compared to PMOs targeting other regions of the viral RNA (Stone et al., 2008, Vagnozzi et al., 2007, Yuan et al., 2006). The PMO may bind with the complementary viral RNA and disrupt the integrity of the IRES stem-loop secondary or tertiary structures, hence arresting IRES-dependent translation (Kole et al., 2012, Warren et al., 2012). The PMOs are likely to block 40S ribosomal subunit binding the viral RNA in the vicinity of the stem-loop V in the picornaviral type I IRES (Belsham, 2009). Furthermore, it has been suggested that the IRES stem-loop V-VI of EV-71 interacts with IRES-specific trans-acting factors such as FUSE-binding protein and heterogeneous nuclear ribonucleoprotein Al (Lin et al., 2009a, Lin et al., 2009b, Shih et al., 2011). Unlike synthetic double-stranded siRNA which involve cleavage of mRNA, PMOs are translation-suppressing oligonucleotides which do not lead to RNA degradation (Kole et al., 2012, Warren et al., 2012). This could explain why siRNA targeting the similar viral sequences as vivo-MO-3 was effective, but not acting as a translation suppression oligonucleotide which down-regulates gene expression via steric blockage. IRES-targeting vivo-MO-2 exhibited broad-spectrum activities against multiple enteroviruses, but with different antiviral efficacies. This could have resulted from the IRES stem-loop structure differences between enteroviruses which affect the accessibility of the vivo-MOs to the targeted sites (Dias and Stein, 2002).
In this study, an EV-71 mutant that was resistant to vivo-MO-1 carried a single substitution of T to C at position 533. We failed to isolate any EV-71 resistant to vivo-MO-2 even after eight passages in vitro, implying that the vivo-MO-2 targeted region is intolerant to mutation in nature. Our mutagenesis studies further confirm that the position of mutation and the number of mutations affect the antiviral efficacies in RD cells. A single mismatch present in the middle of the targeted sequence (T to C substitution at position 523) was more tolerable than a mutation appearing at the end of the targeted site (T to C substitution at position 533). These findings correlated well with previous findings that PV, FMDV and WNV that showed resistance to PPMO also carry a single point mutation at the end of the PPMO-targeted sequence (Stone et al., 2008, Vagnozzi et al., 2007, Zhang et al., 2008). A similar observation has been made for influenza virus as the degree of inhibition was associated with the number of mismatches (Ge et al., 2006).
5. Conclusion
In summary, we have established two critical locations that are involved in viral translation initiation in the EV-71 genome as targets for vivo-MO antiviral intervention. The vivo-MOs worked well with low micromolar concentrations to inhibit EV-71 infection and showed little cytotoxicity in RD cells. The potent inhibition of several enteroviruses by vivo-MO-2 raises the possibility that it could be developed as a broad-spectrum antiviral agent. Furthermore, the degree of tolerance of mismatches by vivo-MOs suggests a favorable characteristic for their use as a potential antiviral agent.
Acknowledgements
This work was supported by a Ministry of Education E-Science Fund (02-02-16-SF0022) and Sunway University Research Grant (INT-FST-BIOS-0312-02) awarded to CLP, and a University Malaya High Impact Research grant (UM.C/625/1/HIR/MOHE/MED/41), postgraduate research Grant (PV013-2012A) and Exploratory Research Grant Scheme (ER017-2013A) awarded to YFC.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.antiviral.2014.04.004.
Appendix A. Supplementary data
References
- Arita M., Wakita T., Shimizu H. Characterization of pharmacologically active compounds that inhibit poliovirus and enterovirus 71 infectivity. J. Gen. Virol. 2008;89:2518–2530. doi: 10.1099/vir.0.2008/002915-0. [DOI] [PubMed] [Google Scholar]
- Belsham G.J. Divergent picornavirus IRES elements. Virus Res. 2009;139:183–192. doi: 10.1016/j.virusres.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Chong P., Hsieh S.Y., Liu C.C., Chou A.H., Chang J.Y., Wu S.C., Liu S.J., Chow Y.H., Su I.J., Klein M. Production of EV71 vaccine candidates. Hum. Vaccine Immunother. 2012;8:1775–1783. doi: 10.4161/hv.21739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smet M.D., Meenken C.J., van den Horn G.J. Fomivirsen – a phosphorothioate oligonucleotide for the treatment of CMV retinitis. Ocul. Immunol. Inflamm. 1999;7:189–198. doi: 10.1076/ocii.7.3.189.4007. [DOI] [PubMed] [Google Scholar]
- Deas T.S., Binduga-Gajewska I., Tilgner M., Ren P., Stein D.A., Moulton H.M., Iversen P.L., Kauffman E.B., Kramer L.D., Shi P.Y. Inhibition of flavivirus infections by antisense oligomers specifically suppressing viral translation and RNA replication. J. Virol. 2005;79:4599–4609. doi: 10.1128/JVI.79.8.4599-4609.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J.X., Nie X.J., Lei Y.F., Ma C.F., Xu D.L., Li B., Xu Z.K., Zhang G.C. The highly conserved 5’ untranslated region as an effective target towards the inhibition of enterovirus 71 replication by unmodified and appropriate 2’-modified siRNAs. J. Biomed. Sci. 2012;19:73. doi: 10.1186/1423-0127-19-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias N., Stein C.A. Antisense oligonucleotides: basic concepts and mechanisms. Mol. Cancer Ther. 2002;1:347–355. [PubMed] [Google Scholar]
- Ge Q., Pastey M., Kobasa D., Puthavathana P., Lupfer C., Bestwick R.K., Iversen P.L., Chen J., Stein D.A. Inhibition of multiple subtypes of influenza A virus in cell cultures with morpholino oligomers. Antimicrob. Agents Chemother. 2006;50:3724–3733. doi: 10.1128/AAC.00644-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney R.M., Huang C.Y., Rose B.C., Kroeker A.D., Dreher T.W., Iversen P.L., Stein D.A. Inhibition of dengue virus serotypes 1 to 4 in vero cell cultures with morpholino oligomers. J. Virol. 2005;79:5116–5128. doi: 10.1128/JVI.79.8.5116-5128.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole R., Krainer A.R., Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 2012;11:125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.Y., Li M.L., Shih S.R. Far upstream element binding protein 2 interacts with enterovirus 71 internal ribosomal entry site and negatively regulates viral translation. Nucleic Acids Res. 2009;37:47–59. doi: 10.1093/nar/gkn901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.Y., Shih S.R., Pan M., Li C., Lue C.F., Stollar V., Li M.L. HnRNP A1 interacts with the 5’ untranslated regions of enterovirus 71 and Sindbis virus RNA and is required for viral replication. J. Virol. 2009;83:6106–6114. doi: 10.1128/JVI.02476-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerdyk-Schauwecker M., Stein D.A., Eide K., Blouch R.E., Bildfell R., Iversen P., Jin L. Inhibition of herpes simplex virus 1 ocular infection with morpholino oligomers targeting ICP0 and ICP27. Antiviral Res. 2009;84:131–141. doi: 10.1016/j.antiviral.2009.07.020. [DOI] [PubMed] [Google Scholar]
- Moulton J.D., Jiang S. Gene knockdowns in adult animals: PPMOs and vivo-morpholinos. Molecules. 2009;14:1304–1323. doi: 10.3390/molecules14031304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Stein D.A., Kroeker A.D., Paulino A.D., Moulton H.M., Iversen P.L., Buchmeier M.J. Antisense morpholino-oligomers directed against the 5’ end of the genome inhibit coronavirus proliferation and growth. J. Virol. 2004;78:5891–5899. doi: 10.1128/JVI.78.11.5891-5899.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi M.H., Wong S.C., Lewthwaite P., Cardosa M.J., Solomon T. Clinical features, diagnosis, and management of enterovirus 71. Lancet Neurol. 2010;9:1097–1105. doi: 10.1016/S1474-4422(10)70209-X. [DOI] [PubMed] [Google Scholar]
- Opriessnig T., Patel D., Wang R., Halbur P.G., Meng X.J., Stein D.A., Zhang Y.J. Inhibition of porcine reproductive and respiratory syndrome virus infection in piglets by a peptide-conjugated morpholino oligomer. Antiviral Res. 2011;91:36–42. doi: 10.1016/j.antiviral.2011.04.012. [DOI] [PubMed] [Google Scholar]
- Paessler S., Rijnbrand R., Stein D.A., Ni H., Yun N.E., Dziuba N., Borisevich V., Seregin A., Ma Y., Blouch R., Iversen P.L., Zacks M.A. Inhibition of alphavirus infection in cell culture and in mice with antisense morpholino oligomers. Virology. 2008;376:357–370. doi: 10.1016/j.virol.2008.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D., Opriessnig T., Stein D.A., Halbur P.G., Meng X.J., Iversen P.L., Zhang Y.J. Peptide-conjugated morpholino oligomers inhibit porcine reproductive and respiratory syndrome virus replication. Antiviral Res. 2008;77:95–107. doi: 10.1016/j.antiviral.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang B., Deng C., Ye H., Xu W., Yuan Z., Shi P.Y., Zhang B. Development and characterization of a stable eGFP enterovirus 71 for antiviral screening. Antiviral Res. 2013;97:198–205. doi: 10.1016/j.antiviral.2012.12.010. [DOI] [PubMed] [Google Scholar]
- Shang L., Xu M., Yin Z. Antiviral drug discovery for the treatment of enterovirus 71 infections. Antiviral Res. 2013;97:183–194. doi: 10.1016/j.antiviral.2012.12.005. [DOI] [PubMed] [Google Scholar]
- Shih S.R., Stollar V., Li M.L. Host factors in enterovirus 71 replication. J. Virol. 2011;85:9658–9666. doi: 10.1128/JVI.05063-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih S.R., Tsai M.C., Tseng S.N., Won K.F., Shia K.S., Li W.T., Chern J.H., Chen G.W., Lee C.C., Lee Y.C., Peng K.C., Chao Y.S. Mutation in enterovirus 71 capsid protein VP1 confers resistance to the inhibitory effects of pyridyl imidazolidinone. Antimicrob. Agents Chemother. 2004;48:3523–3529. doi: 10.1128/AAC.48.9.3523-3529.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim A.C., Luhur A., Tan T.M., Chow V.T., Poh C.L. RNA interference against enterovirus 71 infection. Virology. 2005;341:72–79. doi: 10.1016/j.virol.2005.06.047. [DOI] [PubMed] [Google Scholar]
- Solomon T., Lewthwaite P., Perera D., Cardosa M.J., McMinn P., Ooi M.H. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect. Dis. 2010;10:778–790. doi: 10.1016/S1473-3099(10)70194-8. [DOI] [PubMed] [Google Scholar]
- Stone J.K., Rijnbrand R., Stein D.A., Ma Y., Yang Y., Iversen P.L., Andino R. A morpholino oligomer targeting highly conserved internal ribosome entry site sequence is able to inhibit multiple species of picornavirus. Antimicrob. Agents Chemother. 2008;52:1970–1981. doi: 10.1128/AAC.00011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerton J. Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochim. Biophys. Acta. 1999;1489:141–158. doi: 10.1016/s0167-4781(99)00150-5. [DOI] [PubMed] [Google Scholar]
- Tan C.W., Chan Y.F. Enterovirus 71 receptors: promising drug targets? Expert Rev. Anti Infect. Ther. 2013;11:547–549. doi: 10.1586/eri.13.47. [DOI] [PubMed] [Google Scholar]
- Tan C.W., Chan Y.F., Sim K.M., Tan E.L., Poh C.L. Inhibition of enterovirus 71 (EV-71) infections by a novel antiviral peptide derived from EV-71 capsid protein VP1. PLoS One. 2012;7:e34589. doi: 10.1371/journal.pone.0034589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.W., Lai J.K.F., Sam I.C., Chan Y.F. Recent developments in antiviral agents against enterovirus 71 infection. J. Biomed. Sci. 2014;21:14. doi: 10.1186/1423-0127-21-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.W., Poh C.L., Sam I.C., Chan Y.F. Enterovirus 71 uses cell surface heparan sulfate glycosaminoglycan as an attachment receptor. J. Virol. 2013;87:611–620. doi: 10.1128/JVI.02226-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan E.L., Tan T.M., Chow V.T., Poh C.L. Enhanced potency and efficacy of 29-mer shRNAs in inhibition of Enterovirus 71. Antiviral Res. 2007;74:9–15. doi: 10.1016/j.antiviral.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Tan E.L., Tan T.M., Tak Kwong Chow V., Poh C.L. Inhibition of enterovirus 71 in virus-infected mice by RNA interference. Mol. Ther. 2007;15:1931–1938. doi: 10.1038/sj.mt.6300287. [DOI] [PubMed] [Google Scholar]
- Vagnozzi A., Stein D.A., Iversen P.L., Rieder E. Inhibition of foot-and-mouth disease virus infections in cell cultures with antisense morpholino oligomers. J. Virol. 2007;81:11669–11680. doi: 10.1128/JVI.00557-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfield K.L., Swenson D.L., Olinger G.G., Nichols D.K., Pratt W.D., Blouch R., Stein D.A., Aman M.J., Iversen P.L., Bavari S. Gene-specific countermeasures against Ebola virus based on antisense phosphorodiamidate morpholino oligomers. PLoS Pathog. 2006;2:e1. doi: 10.1371/journal.ppat.0020001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren T.K., Shurtleff A.C., Bavari S. Advanced morpholino oligomers: a novel approach to antiviral therapy. Antiviral Res. 2012;94:80–88. doi: 10.1016/j.antiviral.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Yang F., Zhao R., Zhao L., Guo D., Jin Q. Identification of small interfering RNAs which inhibit the replication of several enterovirus 71 strains in China. J. Virol. Methods. 2009;159:233–238. doi: 10.1016/j.jviromet.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Yeh M.T., Wang S.W., Yu C.K., Lin K.H., Lei H.Y., Su I.J., Wang J.R. A single nucleotide in stem loop II of 5’-untranslated region contributes to virulence of enterovirus 71 in mice. PLoS One. 2011;6:e27082. doi: 10.1371/journal.pone.0027082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblood D.S., Hatlevig S.A., Hassinger J.N., Iversen P.L., Moulton H.M. Stability of cell-penetrating peptide-morpholino oligomer conjugates in human serum and in cells. Bioconjug. Chem. 2007;18:50–60. doi: 10.1021/bc060138s. [DOI] [PubMed] [Google Scholar]
- Yuan J., Stein D.A., Lim T., Qiu D., Coughlin S., Liu Z., Wang Y., Blouch R., Moulton H.M., Iversen P.L., Yang D. Inhibition of coxsackievirus B3 in cell cultures and in mice by peptide-conjugated morpholino oligomers targeting the internal ribosome entry site. J. Virol. 2006;80:11510–11519. doi: 10.1128/JVI.00900-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Dong H., Stein D.A., Shi P.Y. Co-selection of West Nile virus nucleotides that confer resistance to an antisense oligomer while maintaining long-distance RNA/RNA base pairings. Virology. 2008;382:98–106. doi: 10.1016/j.virol.2008.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


