Abstract
To deliver their genetic material into host cells, enveloped viruses have surface glycoproteins that actively cause the fusion of the viral and cellular membranes. Recently determined X-ray crystal structures of the paramyxovirus fusion (F) protein in its pre-fusion and post-fusion conformations reveal the dramatic structural transformation that this protein undergoes while causing membrane fusion. Conformational changes in key regions of the F protein suggest the mechanism by which the F protein is activated and refolds.
Paramyxovirus entry
The paramyxoviruses include many important human and animal pathogens such as measles virus, mumps virus, human respiratory syncytial virus (hRSV), human parainfluenza viruses 1–4 (hPIV1–4), Nipah virus, Hendra virus, parainfluenza virus 5 (PIV5, formerly known as SV5), Newcastle disease virus (NDV) and Sendai virus. To deliver their RNA genome into host cells, these enveloped viruses have evolved a membrane fusion machine that consists of two surface glycoproteins: a receptor binding protein and a fusion (F) protein. The receptor binding protein (called the HN, H or G protein depending on its biological properties) initiates membrane fusion by binding to receptors on the surface of host cells. The atomic structures of the hemagglutinin-neuraminidase (HN) proteins of NDV 1, 2, hPIV3 [3] and PIV5 [4] have been determined in forms that are both bound and unbound to sialic acid receptor analogs. Although it remains unclear how the HN protein triggers the F protein to cause membrane fusion, the mechanism seems to involve changes in either the conformation or the oligomerization of the HN protein that transduce a signal to activate the F protein [5]. Once activated, the F protein initiates the process of membrane fusion.
The rearrangement of lipid bilayers during membrane fusion is thought to occur in a series of discrete steps that include dimpling, lipid stalk formation, hemifusion, transient pore formation and pore enlargement 6, 7. This process does not occur spontaneously; therefore, enveloped viruses have evolved surface proteins that undergo structural changes to do the work of membrane fusion. The paramyxovirus F protein is a member of the class I viral fusion proteins (Box 1). These proteins exist on the surfaces of infectious virions as trimers in metastable (high energy) conformations and become active for fusion usually only after proteolytic cleavage. For example, the paramyxovirus F protein is cleaved from an inactive precursor protein, F0, into the fusion-capable complex, F1+F2. The class I viral fusion proteins also share similar domain structures that include a hydrophobic fusion peptide, two 4–3 heptad repeat regions (HRA and HRB) and a transmembrane domain (Figure 1 a). Upon triggering, these proteins insert the fusion peptide into the host-cell membrane and refold HRA and HRB into a hairpin structure that brings the viral and cellular membranes together. X-ray crystal structures have recently been determined of the PIV5 F protein in the pre-fusion conformation [8] and the NDV and hPIV3 F proteins in the post-fusion conformation (Box 2) 9, 10. Along with the influenza virus hemagglutinin (HA) protein [11], the paramyxovirus F protein is currently the only other class I viral fusion protein for which atomic structures have been determined in both the metastable and hairpin conformations. Thus, the recently determined structures of the F protein provide fundamental insights into both paramyxovirus-mediated membrane fusion and the folding and refolding of metastable viral fusion proteins.
Box 1. Class I and class II viral fusion proteins [7].
• Class I viral fusion proteins include the paramyxovirus F protein, the influenza virus HA protein, the HIV gp160 protein, the Ebola virus GP protein and the severe acute respiratory syndrome (SARS) coronavirus S protein.
• Class I proteins cause membrane fusion by refolding into α-helical coiled-coil hairpin structures.
• Class II viral fusion proteins include the Dengue virus E protein, the West Nile virus E protein and the Semliki Forest virus E1 protein.
• The class II proteins of the flaviviruses and togaviruses cause membrane fusion by refolding their β-sheet-type domains into hairpin structures.
Figure 1.
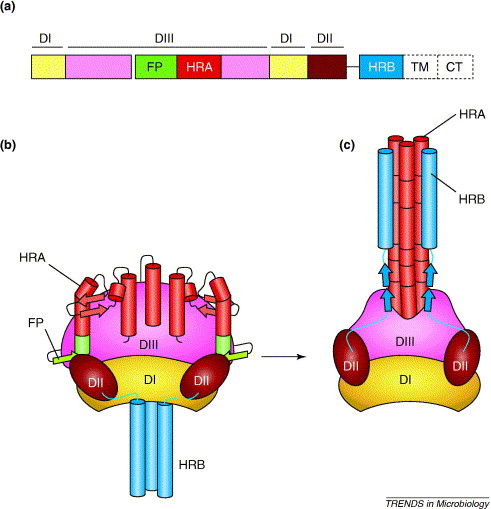
Schematic representation of the paramyxovirus F protein. (a) Domain structure of the F protein. The locations of domain I (yellow), domain II (brown) and domain III (pink) are shown as solid lines above the diagram. The fusion peptide (FP, green), heptad repeat A (red), heptad repeat B (blue), transmembrane domain (TM, dotted line) and cytoplasmic tail (CT, dotted line) are also shown. (b) Model of the pre-fusion structure of the PIV5 F protein. The colors of the domains and regions are the same as in (a). HRA (red) is in a spring-loaded conformation and HRB (blue) forms the coiled-coil stalk. (c) Model of the post-fusion structure of the hPIV3 F protein. In the transition between the pre-fusion and post-fusion structures, domains I and II keep the same fold but reorient within the molecule. Domain III undergoes a dramatic refolding process, which leads to the formation of the coiled-coil hairpin structure that causes membrane fusion. In panels (b) and (c), α-helices are depicted as cylinders and β-strands as colored arrows.
Box 2. Isolation of the pre-fusion F protein conformation.
After high-resolution structures were solved for the HR cores of the PIV5 and hRSV F proteins in the post-fusion hairpin conformation 12, 13, structural biologists sought to isolate the entire ectodomain of the F protein in its metastable pre-fusion conformation. Chen and coworkers [9] constructed a secreted form of the NDV F protein that has a stop codon inserted upstream of its transmembrane anchor and a mutated cleavage site to prevent processing of the F0 precursor by host-cell proteases. However, the NDV F protein that ultimately formed crystals was cleaved 22 amino acid residues downstream of the natural cleavage site and the resulting F protein structure was in the post-fusion conformation [9]. Yin et al. [10] subsequently generated an analogous construct of secreted hPIV3 F0 protein that also lacked its transmembrane anchor. Despite the F protein in these crystals remaining uncleaved, the hPIV3 F0 structure was also in the post-fusion conformation. This unexpected result shows that F protein cleavage is not required for its structural transition to the post-fusion form and suggests that the transmembrane anchor has an important role in stabilizing the metastable pre-fusion conformation [10]. To circumvent this problem, Yin and coworkers [8] generated a construct of the PIV5 F0 protein in which its transmembrane domain was replaced with a GCN4 trimeric coiled-coil domain [25]. In the resulting PIV5 F0 protein structure, which is presumed to be in the pre-fusion conformation [8], the GCN4 sequence fused to the C terminus of HRB helps to stabilize the triple-stranded coiled-coil conformation of HRB. It is possible, although unlikely, that the addition of the foreign sequence alters the native structure of the F protein. However, amino acid residues in the PIV5 F protein that have important roles in regulating activation of the metastable form of the full-length F protein (including residues 443, 447 and 449 at the nucleation site of HRB) also make important interactions that seem to stabilize what we consider to be the PIV5 F protein pre-fusion structure. If the HRB domains of other class I viral fusion proteins (such as the HIV gp41 protein and the SARS coronavirus S protein) form analogous coiled-coil structures in the pre-fusion conformation, secreted forms of these proteins could, potentially, also be stabilized by replacement of the viral transmembrane domain with a GCN4 trimeric coiled-coil domain.
F protein structures
The PIV5 F0 protein in its pre-fusion conformation has a mushroom-like shape formed by a large globular head attached to a rod-like stalk (Figure 1b) [8]. The stalk is formed exclusively by HRB, which has a triple-stranded coiled-coil conformation. The base of the head is formed by two domains (DI and DII), which consist of residues that are predominantly located between HRA and HRB in the primary sequence. These domains are unique to the paramyxovirus F protein because the other class I viral fusion proteins have only short linker regions between HRA and HRB. In both the pre-fusion and post-fusion structures, DI forms a highly twisted β-barrel-like assembly and DII forms an immunoglobulin-like β-sandwich domain. Overall, DI and DII form a molecular scaffold that seems to help stabilize and then refold HRB and domain III (DIII).
The top of the head of the pre-fusion conformation of the F protein is formed by DIII, which consists of HRA, the fusion peptide and residues that are nearby in the primary sequence (Figure 1a). HRA is in a spring-loaded conformation at the top of the head, aimed towards the target membrane, and the fusion peptide is located on the side of the head, held in place by interactions with other residues in DIII and residues from DII. Such a configuration is reminiscent of the pre-fusion structure of the influenza virus HA protein [11]. However, HRA forms a helix–loop–helix structure in the HA protein and an 11-segment structure consisting of α-helices, β-strands and turns in the F protein (Figure 1b).
The structures of the F proteins of NDV [9] and hPIV3 [10] in the post-fusion conformation reveal that DIII and HRB undergo dramatic rearrangements during membrane fusion. HRA refolds into a triple-stranded coiled coil that propels the fusion peptide ∼115 Å from the side of the molecule into the target membrane. The HRB segments must also separate and then swing around the base of the head formed by DI and DII to bind to the HRA coiled coil and form the fusogenic hairpin structure 12, 13 (Figure 1c).
Changes in F protein conformation during membrane fusion
The F protein is thought to adopt five distinct conformations during membrane fusion (Figure 2 ). The recently determined F protein structures represent the initial and final conformations of the F protein and suggest the mechanism by which it refolds into distinct intermediates. After cleavage of the F0 precursor, the metastable conformation of the F protein is triggered to refold by binding of the HN protein to its receptor. The triggering of the F protein seems to involve a conformational change in HRB [14], and HRB has been shown to physically interact with the HN protein [15]. In the pre-fusion structure of the PIV5 F protein, residues at the nucleation site of the HRB coiled coil (residues 443, 447 and 449) interact with residues in the base of the F protein head. The importance of these HRB residues in the formation of an early F protein intermediate is confirmed by previous biochemical experiments, which show that mutations to residues 443, 447 and 449 alter the energy required to trigger the PIV5 F protein 16, 17. The next step in F protein refolding is the formation of a pre-hairpin intermediate in which HRA forms a triple-stranded coiled coil and the fusion peptide is inserted into the target membrane 14, 17 (Figure 2d). Mutations to HRA residues 132 and 161 and fusion peptide residues 105 and 109 increase the fusogenic activity of the PIV5 F protein 18, 19, 20. The positioning of these residues at key structural points in the pre-fusion conformation of the F protein is consistent with mutations to these residues that destabilize the spring-loaded conformation and cause engagement of the F protein refolding process even in the absence of the HN protein trigger. The last conformational change in the F protein during membrane fusion involves binding of the HRB segments into the grooves of the HRA coiled coil to generate a hairpin structure (Figure 2e). Because the fusion peptide is contiguous with HRA and the transmembrane domain is adjacent to HRB, the formation of the hairpin brings together the fusion peptide and transmembrane domain and, as a result, actively causes the fusion of the cellular and viral membranes [14].
Figure 2.
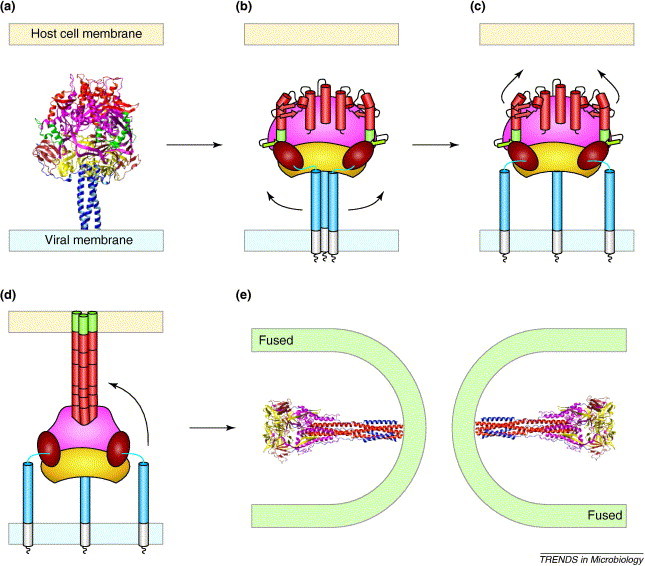
Model of the paramyxovirus-mediated membrane fusion. The F protein is thought to adopt five conformations during membrane fusion: (a) an inactive F0 precursor protein; (b) a metastable, native conformation of the cleaved F1+F2 protein; (c) an early fusion intermediate in which the HRB strands open up; (d) a pre-hairpin intermediate in which HRA forms a triple-stranded coiled coil and the fusion peptide inserts into the target membrane; and (e) the fusogenic hairpin structure that actively brings together the viral and cellular membranes. Ribbon diagrams are included of the pre-fusion PIV5 F0 protein structure [part (a), modified from [8]] and the post-fusion hPIV3 F protein structure [part (e), modified from [10]]. Conformations of the F protein that have not yet been determined at atomic resolution (b–d) are represented schematically as in Figure 1.
Concluding remarks
High-resolution structures of the paramyxovirus F protein in its pre-fusion and post-fusion conformations now provide a structural basis for the analysis of the membrane fusion mechanism of the paramyxoviruses. In addition to explaining the phenotypes of previously characterized F protein mutants, these structures will enable future identification of residues that were previously unrecognized as important in regulating the progression of discrete refolding intermediates. A key unresolved issue in paramyxovirus fusion is the nature of the interaction between the HN protein and the F protein during fusion activation. Structures of both the HN protein and the F protein will certainly assist in the generation and testing of novel hypotheses on the nature of this protein–protein interaction. The pre-fusion F protein structure will undoubtedly be used as a target in the structure-based design of fusion inhibitors. Peptides and small molecules that are designed to bind to the pre-hairpin intermediate of the F protein are potent inhibitors of virus replication 21, 22, 23. It is possible that compounds that bind to the longer-lived native structure might have enhanced therapeutic properties. The successful use of the HIV fusion inhibitor enfuvirtide (T-20) in drug combination therapies is proof of the concept that fusion inhibitors can be clinically effective [24]. Although the metastable structure of the paramyxovirus F protein differs substantially from that of the influenza virus HA protein, key structural similarities such as the spring-loaded configuration of HRA and the positioning of the fusion peptide on the side of the molecule suggest that other class I viral fusion proteins such as HIV gp41 could also use similar strategies to stabilize and refold their pre-fusion conformations. Ultimately, further insights into how viruses invade host cells will depend on obtaining more structural information and performing additional biochemical experiments on the membrane fusion mechanisms of enveloped viruses.
Acknowledgements
We thank the Cancer Center Support Grant (CA 21765), the American Lebanese Syrian Associated Charities (ALSAC) and the Children's Infection Defense Center (CIDC) for supporting the work of our laboratory. We thank Betsy Williford in Biomedical Communications for help preparing the figures.
References
- 1.Crennell S. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat. Struct. Biol. 2000;7:1068–1074. doi: 10.1038/81002. [DOI] [PubMed] [Google Scholar]
- 2.Zaitsev V. Second sialic acid binding site in Newcastle disease virus hemagglutinin-neuraminidase: implications for fusion. J. Virol. 2004;78:3733–3741. doi: 10.1128/JVI.78.7.3733-3741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lawrence M.C. Structure of the haemagglutinin-neuraminidase from human parainfluenza virus type III. J. Mol. Biol. 2004;335:1343–1357. doi: 10.1016/j.jmb.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 4.Yuan P. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure. 2005;13:803–815. doi: 10.1016/j.str.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 5.Lamb R.A. Paramyxovirus membrane fusion: lessons from the F and HN atomic structures. Virology. 2006;344:30–37. doi: 10.1016/j.virol.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chernomordik L.V., Kozlov M.M. Protein-lipid interplay in fusion and fission of biological membranes. Annu. Rev. Biochem. 2003;72:175–207. doi: 10.1146/annurev.biochem.72.121801.161504. [DOI] [PubMed] [Google Scholar]
- 7.Earp L.J. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 2005;285:25–66. doi: 10.1007/3-540-26764-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin H.S. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure. 2001;9:255–266. doi: 10.1016/s0969-2126(01)00581-0. [DOI] [PubMed] [Google Scholar]
- 10.Yin H.S. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skehel J.J., Wiley D.C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 12.Baker K.A. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell. 1999;3:309–319. doi: 10.1016/s1097-2765(00)80458-x. [DOI] [PubMed] [Google Scholar]
- 13.Zhao X. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc. Natl. Acad. Sci. U. S. A. 2000;97:14172–14177. doi: 10.1073/pnas.260499197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russell C.J. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 2001;20:4024–4034. doi: 10.1093/emboj/20.15.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gravel K.A., Morrison T.G. Interacting domains of the HN and F proteins of Newcastle disease virus. J. Virol. 2003;77:11040–11049. doi: 10.1128/JVI.77.20.11040-11049.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paterson R.G. Fusion protein of the paramyxovirus SV5: destabilizing and stabilizing mutants of fusion activation. Virology. 2000;270:17–30. doi: 10.1006/viro.2000.0267. [DOI] [PubMed] [Google Scholar]
- 17.Russell C.J. A dual-functional paramyxovirus F protein regulatory switch segment: activation and membrane fusion. J. Cell Biol. 2003;163:363–374. doi: 10.1083/jcb.200305130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito M. An amino acid in the heptad repeat 1 domain is important for the haemagglutinin-neuraminidase-independent fusing activity of simian virus 5 fusion protein. J. Gen. Virol. 2000;81:719–727. doi: 10.1099/0022-1317-81-3-719. [DOI] [PubMed] [Google Scholar]
- 19.West D.S. Role of the simian virus 5 fusion protein N-terminal coiled-coil domain in folding and promotion of membrane fusion. J. Virol. 2005;79:1543–1551. doi: 10.1128/JVI.79.3.1543-1551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russell C.J. Conserved glycine residues in the fusion peptide of the paramyxovirus fusion protein regulate activation of the native state. J. Virol. 2004;78:13727–13742. doi: 10.1128/JVI.78.24.13727-13742.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cianci C. Orally active fusion inhibitor of respiratory syncytial virus. Antimicrob. Agents Chemother. 2004;48:413–422. doi: 10.1128/AAC.48.2.413-422.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambert D.M. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc. Natl. Acad. Sci. U. S. A. 1996;93:2186–2191. doi: 10.1073/pnas.93.5.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao Q., Compans R.W. Peptides corresponding to the heptad repeat sequence of human parainfluenza virus fusion protein are potent inhibitors of virus infection. Virology. 1996;223:103–112. doi: 10.1006/viro.1996.0459. [DOI] [PubMed] [Google Scholar]
- 24.Greenberg M. HIV fusion and its inhibition in antiretroviral therapy. Rev. Med. Virol. 2004;14:321–337. doi: 10.1002/rmv.440. [DOI] [PubMed] [Google Scholar]
- 25.Harbury P.B. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]


