Abstract
Connexins form gap junction channels that link neighboring cells into an intercellular communication network. Many cells that express multiple connexins produce heteromeric channels containing at least two connexins, which provides a means to fine tune gap junctional communication. Formation of channels by multiple connexins is controlled at two levels: by inherent structural compatibilities that enable connexins to hetero-oligomerize and by cellular mechanisms that restrict the formation of heteromers by otherwise compatible connexins. Here, I discuss roles for secretory compartments beyond the endoplasmic reticulum in connexin oligomerization and evidence that suggests that membrane microdomains help regulate connexin trafficking and assembly.
Introduction
Gap junctions interconnect cells by forming a direct link to enable the diffusion of small aqueous molecules and ions from one cell to its nearest neighbor [1]. This enables the flow of specific intercellular signals and metabolic cooperation between communicating cells in a tissue. The importance of such communication is shown by the increasing number of human diseases that are directly attributable to mutants and deficiencies of the gap junction protein, connexin [2]. Mutations range from those that subtly change gap junctional permeability [3] to mutations that cause severe changes in connexin trafficking and that completely inhibit gap junctional coupling 4, 5, 6, 7.
Connexins are a multigene family of transmembrane proteins [8] (Figure 1 ). Each connexin forms a channel with unique permeability characteristics, which is reflected in the types of metabolite and signaling molecule that flow through it [9]. A complete channel is formed from two hexameric hemichannels, one in each cell, that meet at the cell surface (Figure 2 ). Gap junction channels are organized into higher order semicrystalline arrays, known as plaques. In addition, connexin hemichannels function as bona fide plasma membrane channels that enable the diffusion of ATP and other aqueous molecules from the cytosol to the extracellular environment [10].
Figure 1.
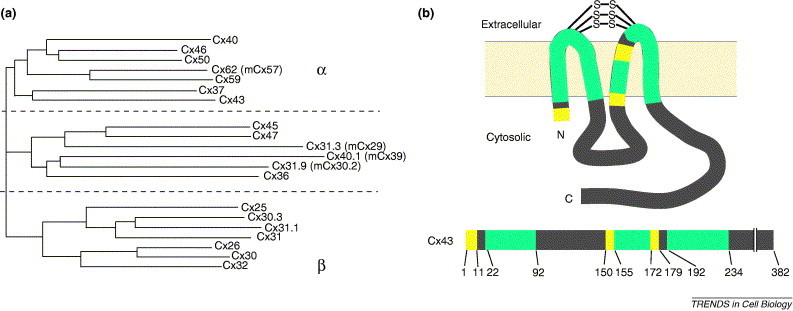
Connexin homology and structure. (a) Phylogram of 20 human connexins calculated using ClustalW and omitting C-terminal domains [85]. By protein homology, connexins form two major subgroups, α and β, with an additional group of connexins with intermediate homology [8]. Different connexins are denoted by Cx plus a number corresponding to the predicted molecular mass based on the amino acid sequence. Mouse connexin names that differ from their human orthologs are shown in parentheses*. (b) Line diagram corresponding to an individual connexin. The two extracellular loop domains are interconnected by three disulfide bridges. Green denotes regions with high amino acid sequence homology within the entire connexin protein family, yellow denotes regions in which amino acids are homologous within a subset of connexins and gray denotes divergent regions of connexins that vary in amino acid sequence and size. Numbers correspond to positions of Cx43 amino acids that define different classes of connexin homology domains.
Figure 2.
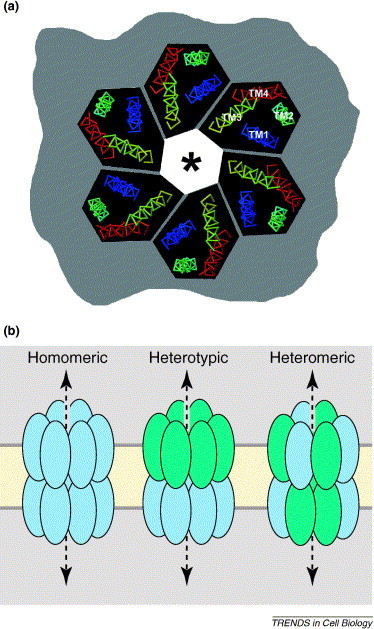
Gap junction channels. (a)En face view of a connexin hemichannel, showing only the transmembrane helical domains as viewed from the outside of the cell looking into the permeability pore (*). The image was generated with RasMol using coordinates from [12]. In this model, transmembrane (TM) domains correspond to TM1 (blue), TM2 (aqua), TM3 (yellow) and TM4 (red). (b) Types of gap junction channel. Ovals represent individual connexins and the dashed line indicates the path of the aqueous portion of the channel. Shown from left to right are a homomeric gap junction channel composed of a single type of connexin, a heterotypic channel composed of two different connexins expressed by two adjacent cells and a heteromeric channel composed of two different connexins co-expressed in the same cell.
Connexins span the membrane bilayer four times with the N and C termini oriented towards the cytoplasm. The four connexin transmembrane domains are largely α helical [11]. Fleishman et al. [12] used a best-fit algorithm to assign the transmembrane domains of connexin 32 (Cx32) mathematically to the corresponding α helices in the structural model (Figure 2). On the basis of their model, the best fit was obtained by assigning the third transmembrane domain as the predominant pore-lining helix [12], although this is controversial [13].
Different types of cell express different connexins and cells frequently express two or more connexins 14, 15, 16. Because an individual gap junction channel is composed of 12 connexins, cells that express multiple connexins can produce mixed channels, provided that the connexins are able to hetero-oligomerize. The formation of gap junction channels by two or more connexins enables cells to produce channels that have unique permeability and gating characteristics that could not be obtained using a single connexin [17]. However, the rules that govern connexin oligomerization and compatibility are complex.
Determinants of connexin compatibility
Innate heteromeric compatibility
Figure 2 shows the different classes of channel that can be formed when cells express two or more connexins. Connexins do not ubiquitously intermix; instead, compatibility is based on their protein structure. When two connexins expressed in the same cell form a mixed channel, this is referred to as heteromeric compatibility. Examples of heteromeric channels formed by endogenously expressed connexins include Cx32–Cx26 in the liver 15, 18, Cx46–Cx50 in the lens [19] and Cx43–Cx46 in the lung [20]. Heteromeric connexin compatibility has been tested using transfected cell models, although there are many combinations that have not been examined. The heteromeric compatibility groups correspond loosely to α and β connexin subfamiles 15, 17. Connexins that do not belong in either of these subgroups have been found to form heteromers with either α connexins (e.g. Cx45–Cx43) [21] or β connexins (e.g. mCx29–Cx32) [22]. To date, no connexin has been identified that forms normal heteromeric channels with both α and β subfamily connexins.
Motifs that dictate innate heteromeric compatibility have been studied most extensively using two incompatible connexins, Cx32 and Cx43. A series of truncation and point mutants that alter connexin heteromeric specificity showed that the motifs that prevent hetero-oligomerization of Cx32 and Cx43 include pairs of amino acids in the N terminus and at the cytosolic end of the third transmembrane domain [23].
An aberrant interaction between α and β connexins can have pathologic consequences. For example, although Cx26 and Cx43 are normally incompatible [24], some Cx26 mutants that are associated with the skin disorder palmoplantar keratoderma have a dominant-negative effect on Cx43, which is probably due to formation of an aberrant heteromer [25]. However, other Cx26 mutations that are associated with non-syndromic deafness are more specifically limited to the disruption of Cx26 and other β connexins, indicating that different mutations in the same connexin can cause different human diseases.
Innate heterotypic compatibility
A gap junction channel formed by a head-to-head interaction between two different connexins is known as a heterotypic channel. Heterotypic channels have been identified in several settings in vivo, most notably in the central nervous system (CNS), in which astrocytes and oligodendrocytes are interconnected exclusively through heterotypic channels 26, 27. Although such gap junctions can contain channels with unique permeability characteristics, a more prominent role for specific heterotypic interactions is to form intercellular networks by either promoting or restricting cell interconnectivity. Disulfide bridges that interconnect the two extracellular domains are crucial for channel formation 28, 29. Two key amino acids in the second extracellular loop are the major determinants of heterotypic compatibility 15, 30, although motifs in the first extracellular loop and other parts of the protein might also have a role [31]. In contrast to innate heteromeric compatibility, innate heterotypic compatibility does not correlate well with whether a given connexin is an α or β family member 15, 32. For example, Cx32 and Cx43 cannot form heterotypic channels; however, the α connexins Cx46 and Cx50 can form heterotypic channels with either Cx32 or Cx43 [30]. In addition, some connexins, including Cx31 and Cx36, appear to only form homotypic channels 32, 33.
Regulated connexin compatibility
Although connexins have structural determinants that define their ability to form heteromeric channels, there are several examples showing that this ability is also dependant on the context of cell expression. For example, two compatible connexins, Cx43 and Cx46, are expressed by type I alveolar epithelial cells and form heteromeric channels [20]. However, these same connexins, when expressed by type II alveolar epithelial cells or osteoblastic cells, do not hetero-oligomerize (Figure 3 ) 20, 34, 35. Thus, formation of heteromeric Cx43–Cx46 channels depends on cell type and is a regulated process.
Figure 3.
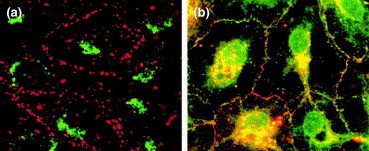
Cellular control of connexin oligomerization. Immunofluorescence images of (a) osteoblastic cells and (b) type I alveolar epithelial cells are shown, both of which express endogenous Cx43 (red) and Cx46 (green). (a) Osteoblastic cells prevent Cx43 and Cx46 from intermixing. Only Cx43 is targeted by these cells to assemble into gap junctions at the plasma membrane (red). However, Cx46 is retained by osteoblastic cells in the TGN (green perinuclear fluorescence). (b) By contrast, type I alveolar epithelial cells show co-localization of Cx43 (red) and Cx46 (green) to produce yellow fluorescence. Formation of Cx43–Cx46 heteromers was demonstrated by co-immunopurification. Adapted, with permission, from [20]. Scale bar=10 μm.
Another example of regulated connexin targeting is the myoendothelial junction between vascular endothelium and smooth muscle cells. These cells express different levels of Cx37, Cx40 and Cx43, all of which can hetero-oligomerize with one another; this has been demonstrated in transfected cell models [17]. Using a co-culture model, in which the cells maintained polarity and phenotype, Isakson and Duling [36] found that Cx37 was specifically excluded from myoendothelial junctions between endothelial cells and smooth muscle cells. Instead, Cx37 was incorporated into junctions interconnecting either two adjacent endothelial cells or two adjacent smooth muscle cells. This suggests that the vasculature regulates connexin hetero-oligomerization and uses polarized delivery of connexins to different plasma membrane domains to produce three distinct classes of cell–cell interface that influence intercellular signaling pathways. Polarized connexin targeting also has a role in regulating heteromer formation in the CNS 22, 26, 27 and oocyte–granulosa cell interfaces 37, 38. The mechanisms to control connexin hetero-oligomerization and trafficking are not well defined at present. In particular, connexin trafficking in cultured polarized cell models must be studied to define roles for differential connexin targeting to apical and basolateral secretory pathways involved in regulating the formation of heteromeric and heterotypic gap junction channels.
Connexin oligomerization after the endoplasmic reticulum
Connexins do not follow the classical pathway for transmembrane protein oligomerization when forming hexamers. Similar to most channel-forming proteins [39], connexins oligomerize into a multimeric complex before delivery to the plasma membrane. However, Cx43 oligomerizes into hexamers after exit from the endoplasmic reticulum (ER) in a secretory compartment distal to the medial Golgi, which is probably the trans Golgi network (TGN) 35, 40 (Figure 4 ). This is consistent with the notion of a quality-control system in the Golgi apparatus 41, 42 that complements ER quality-control pathways [43]. However, identifying specific components of the quality-control apparatus that regulates connexin oligomerization has been difficult.
Figure 4.
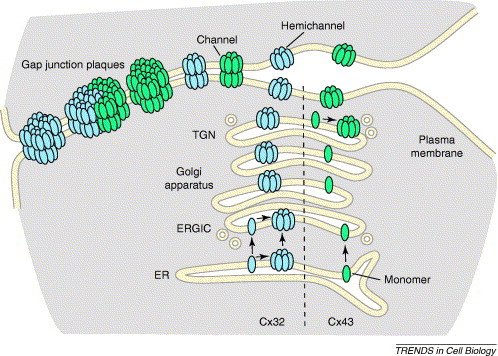
Connexin oligomerization pathways. Connexins are co-translationally inserted into the ER membrane. Depending on the connexin subtype, oligomerization can occur either in the ERGIC (Cx32, blue) or the TGN (Cx43, green). The potential for connexin oligomerization in the ER, driven by high levels of connexin expression, is also shown for Cx32. Hemichannels are subsequently transported to the plasma membrane, where they can function as channels or pair with hemichannels on adjacent cells to form complete intercellular channels. Channels at the plasma membrane further assemble into semi-crystalline arrays known as gap junction plaques, which can contain from tens to thousands of channels. Homogenous plaques are composed of either a single connexin or heteromeric connexins (not shown). Heterogeneous plaques contain regions enriched for different connexins.
This difficulty is partly due to different systems giving apparently conflicting results. For example, several connexins, including Cx26, Cx32 and Cx43, oligomerize when translated in vitro in the presence of microsomes 18, 44, 45. Although this has been cited as evidence that connexin oligomerization naturally occurs in the ER, another possibility is that microsomes lack components of the quality-control apparatus that are present in the intact ER and that inhibit connexin oligomerization. Consistent with this, addition of liver Golgi membranes to the in vitro translation microsome mix enhances Cx32 oligomerization and promotes the formation of Cx26–Cx32 heteromers [18]. Also, the ER pool of Cx32 in guinea pig liver is monomeric, although the ER pool of Cx26 does show partial oligomerization [46]. Factors provided by Golgi membranes that promote connexin oligomerization remain to be identified.
Cells do not have an unlimited capacity for quality control of protein folding and assembly. This is a particular problem when analyzing the assembly of connexins transfected into cells because overexpression could drive premature connexin oligomerization. For example, overexpression initiates the formation of intracellular plaque-like structures in the ER of transfected BHK cells suggesting aberrant connexin assembly [47]. Overexpressed newly synthesized connexins, including Cx26, Cx32 and Cx43, oligomerized in the ER, whereas endogenous Cx32 and Cx43 expressed at much lower levels did not (J.K. VanSlyke and L.S. Musil, personal communication); however, transfected HeLa cells have been used to show that Cx26 oligomerizes following exit from the ER [24]. Cx43 overexpression induces premature oligomerization [48] and it has been suggested that overexpression might underlie some of the conflicting results obtained using transfected cell models to study connexin trafficking and oligomerization 7, 49. Taken together, these results suggest that care should be taken in producing and interpreting these models. Conversely, it is plausible that there might be physiological circumstances in which high levels of connexin expression might regulate hexamer formation by inducing early connexin oligomerization in the ER, although, to date, this has not been directly demonstrated in a system examining endogenously expressed connexins.
Although unusual, transmembrane protein oligomerization in a post-ER secretory compartment is not unique to connexins. For example, targeting of coronavirus coat proteins [50] and Golgi resident enzymes [51] correlates with oligomerization in the Golgi apparatus. Remodeling and assembly of ion channels, such as ENaC and Na-K ATPases, can occur in late secretory compartments and at the plasma membrane 52, 53, 54. Another class of proteins that oligomerize in the Golgi apparatus are tetraspanins, such as CD9, CD63, CD81 and CD151 [55]. However, tetraspanins also hetero-oligomerize with several other classes of transmembrane protein, such as integrins, in the ER 56, 57 (Box 1).
Box 1. A tetraspanin–connexin connection?
Tetraspanins have been called molecular facilitators because of their ability to promote the assembly and trafficking of several classes of transmembrane proteins 86, 87. Although tetraspanins and connexins both span the bilayer four times and have similar protein orientation in the bilayer, they are structurally distinct and not homologous [88].
Intermolecular interactions involving tetraspanins are complex, because tetraspanins form homologous complexes with other tetraspanins and form heterologous complexes with other transmembrane proteins (e.g. integrins or class II major histocompatibility complex proteins). Tetraspanin–protein complexes form in the ER and the Golgi apparatus. In addition to direct interactions with other proteins, tetraspanins facilitate the formation of large-scale heterogeneous membrane lipid–protein complexes. The ability of tetraspanins to form large membrane microdomains is enhanced by palmitoylation, a post-translational modification that occurs in the Golgi apparatus.
One parallel between connexins and tetraspanins is that they partition into cholesterol- and sphingolipid-enriched membrane microdomains. Given this, and the prominent role of post-ER compartments in connexin and tetraspanin oligomerization, it is tempting to speculate that the formation of higher order complexes by these protein families might share common features. Also, given that tetraspanins function as molecular facilitators in the Golgi apparatus to regulate trafficking of other classes of junction and cell adhesion proteins, it is plausible that tetraspanins might also help regulate connexin oligomerization and/or trafficking. Further work will be required to determine whether this is the case.
Di-lysine-tagged connexins as probes to define sites of oligomerization
Most studies to characterize early events in connexin oligomerization require membrane trafficking inhibitors, such as brefeldin A. Although these are useful tools for identifying early events in protein assembly, they can also alter the composition of the secretory pathway [58] and can induce stress responses that are due to the accumulation of unfolded proteins in the ER [59]. Given these potential pitfalls, connexin chimeras containing a di-lysine-based C-terminal ER retention–retrieval sequence, HKKSL, were used to study early events in connexin oligomerization 60, 61. Consistent with Cx43 oligomerization in the Golgi apparatus or TGN, ER-localized Cx43–HKKSL is monomeric. In contrast to Cx43, ER-localized Cx32–HKKSL oligomerizes. Importantly, this difference was not due to protein overexpression [61].
Given that Cx32–HKKSL that was localized to the ER was oligomerized, does Cx32 normally oligomerize in the ER? This is possible; however, although the HKKSL motif is a strong ER retention signal, it also acts as a retrieval signal for proteins that have escaped from the ER [62]. Access and retrieval of proteins containing di-lysine motifs from the ER–Golgi intermediate compartment (ERGIC) and, to a lesser extent, the cis Golgi stack form a retrograde recycling pathway, which contributes to quality control in the secretory pathway 63, 64. By contrast, ER retrieval of di-lysine-containing proteins from late Golgi compartments and the TGN is relatively rare [65]. Consistent with this, a small fraction of Cx43–HKKSL escapes from the ER retrieval pathway and forms functional gap junction channels [60]. Thus, an alternative explanation to Cx32–HKKSL oligomerization in the ER is that Cx32–HKKSL oligomerizes in a compartment such as the ERGIC, where they are efficiently retrieved by the di-lysine tag. Given this and the arguments presented here that support connexin oligomerization after exit from the ER, I propose a model that favors the ERGIC as a major intracellular compartment in which Cx32 oligomerizes (Figure 4).
Identifying the precise subcompartments involved in connexin oligomerization requires further testing and is likely to be difficult, given the complex interrelationships and architecture of the ER, ERGIC, Golgi stacks and TGN [66]. Nonetheless, the notion that different connexins can oligomerize in different intracellular compartments provides a potential mechanism for regulating whether connexins form heteromeric hemichannels, as prior oligomerization into homomers could preclude subsequent formation of heteromers. This type of control is more important for inherently compatible connexins, such as Cx37 and Cx43 or Cx43 and Cx46, as opposed to inherently incompatible connexins, such as Cx32 and Cx43. Identification of the sites of oligomerization for other connexins is required to confirm if this is the case.
Structural determinants that regulate connexin oligomerization
The third transmembrane domain and second extracellular loop are necessary and sufficient to prevent premature Cx43 oligomerization [61]. Charged amino acid residues at both membrane-interface regions of the third transmembrane domain are crucial elements of this Cx43 motif, whereas Cx32 and other β connexins have bulky hydrophobic groups at comparable positions. Intriguingly, one of the key amino acids required to regulate the site of Cx43 oligomerization, Arg153, is also crucial for the innate heteromeric incompatibility of Cx43 and Cx32 described earlier [23]. This provides a potential link that connects the relative stability of Cx43 as a monomer and the innate compatibility of Cx43 to hetero-oligomerize with other connexins.
Importantly, mutations in amino acid residues that are localized to membrane interfaces interfere with connexin trafficking and cause connexins to accumulate in the ER and Golgi apparatus 4, 6, 67. Disease-related connexin mutations can interfere with oligomerization. For example, a mutant form of Cx50 that is associated with human cataract, P88S, forms cytoplasmic accumulations in the ER with partial plaque-like characteristics [68]. In addition, two mutant Cx32 proteins that are associated with Charcot-Marie-Tooth disease and do not traffic to the plasma membrane remain monomeric and are subsequently processed by the ER-associated degradation (ERAD) quality-control pathway [67].
Connexin partitioning into membrane microdomains
Increasing evidence suggests that cholesterol- and sphingolipid-enriched membrane microdomains, or lipid ‘rafts’ [69], might have roles in connexin trafficking and assembly. Consistent with a potential role for microdomains in regulating connexin assembly, treatment of cells with extracellular cholesterol enhances gap junction formation and intercellular communication [70]. Several connexins partition into biochemically isolated microdomains 71, 72, 73, 74, 75 and contain binding motifs for a microdomain-associated protein, caveolin-1 [73]. Whether connexins associate with microdomains enriched for caveolin-1 depends on the preparation technique. For example, hemichannels containing Cx32 are found in Triton X-100-insoluble microdomain fractions, whereas homomeric Cx26 hemichannels are not [73]. However, both Cx32- and Cx26-containing hemichannels are found in Brij-58-insoluble microdomain fractions [73]. Whether this reflects the in vivo partitioning of Cx26, Cx32 and Cx26–Cx32 heteromers into different classes of membrane microdomain remains to be determined. It also demonstrates the complexity of using different fractionation techniques to study the formation of membrane microdomains.
As caveolin-1 has been co-localized to gap junction plaques [72], it has been suggested that the plaques themselves are located in microdomains. However, accumulating evidence suggests that connexins are present in membrane microdomains before incorporation into gap junctions 71, 73. For example, connexin-containing microdomain fractions are enriched in glycosphingolipids, whereas the glycosphingolipid content of gap junction plaques is low [73]. Also, in studies of ZO-1, a connexin-binding scaffold protein 76, 77, Laing et al. [71] showed that cells expressing a dominant-negative ZO-1 construct had an increase in microdomain-associated Cx43, which localized to the region of the Golgi apparatus. By contrast, overexpressing wild-type functional ZO-1 enhanced Cx43 transport to the plasma membrane and enhanced gap junction plaque formation. Although this suggests that functional ZO-1 might enhance the transition of Cx43 from microdomains to gap junction plaques, ZO-1 binding is not an absolute requirement for their formation, because connexins that do not bind to ZO-1 can form plaques 73, 77, 78. In fact, ZO-1 might also serve to limit plaque size [77]. In general, studies that focus strictly on binary connexin–ZO-1 interactions should be interpreted with caution. For example, the effect of mutant or wild-type ZO-1 on connexins might be indirect because these proteins might interfere with or promote the binding of other proteins to the connexin C terminus.
Although connexins can associate with membrane microdomains, further studies are needed to relate this to their oligomeric state. However, given that membrane lipids and microdomains are continually remodeled throughout the secretory pathway [79], this could be an intriguing potential modulator of connexin oligomerization. In addition, microdomains could also have a role in polarized delivery of connexins to distinct plasma membrane domains, comparable to roles for microdomains in the polarized trafficking of other classes of transmembrane protein [80].
Conclusion and perspectives
The context of connexin expression is an important factor in determining the characteristics of intercellular communication. Cells control whether compatible connexins oligomerize into heteromeric gap junction channels. Oligomerization of different connexins in different intracellular compartments is likely to have an important role in regulating heteromer formation. This implies that cell-specific organelle remodeling might help determine which types of gap junction channel are produced. Cell culture models that preserve cell phenotype and polarity will be useful to study how cells contribute to regulating connexin oligomerization.
The formation of gap junction channels is balanced by other control points that regulate plasma membrane gap junction content, including connexin phosphorylation, internalization and turnover 16, 81, 82, 83. In addition, identifying a mechanistic basis for regulated connexin oligomerization will require more detailed study of other less well-characterized connexins. Whether the mechanisms that control Cx43 and Cx32 are representative of other α and β connexins, or connexins with intermediate homology, remains to be determined. In addition, little is known about the trafficking and assembly of a newly discovered class of gap junction-forming proteins, known as mammalian pannexins, which are distinct from connexins and instead are homologous to invertebrate gap junction proteins [84].
Although significant progress has been made in identifying structural connexin motifs and organelles involved in connexin oligomerization, little is known about chaperones and other protein cofactors that are required to regulate this process. Further understanding of the molecular basis for regulation of gap junction channel formation will require the identification of components of the connexin quality-control apparatus.
Acknowledgements
I am grateful to the reviewers for their critiques and I thank Drs Judy VanSlyke and Linda Musil for sharing their unpublished data. The author is supported by NIH grants GM61012, P01-HL019737–26 Project 3 and support from the University Research Committee of Emory University.
Footnotes
At the 2005 International Gap Junction Conference, a proposal to develop a revised nomenclature to unify connexin naming between species was approved, but has not been finalized. Changes to the connexin nomenclature will be published elsewhere and posted to the Human Genome Organization website (http://www.gene.ucl.ac.uk/nomenclature/).
References
- 1.Goodenough D.A. Connexins, connexons, and intercellular communication. Annu. Rev. Biochem. 1996;65:475–502. doi: 10.1146/annurev.bi.65.070196.002355. [DOI] [PubMed] [Google Scholar]
- 2.Kelsell D.P. Human diseases: clues to cracking the connexin code? Trends Cell Biol. 2001;11:2–6. doi: 10.1016/s0962-8924(00)01866-3. [DOI] [PubMed] [Google Scholar]
- 3.Beltramello M. Impaired permeability to Ins(1,4,5)P3 in a mutant connexin underlies recessive hereditary deafness. Nat. Cell Biol. 2005;7:63–69. doi: 10.1038/ncb1205. [DOI] [PubMed] [Google Scholar]
- 4.Yum S.W. Diverse trafficking abnormalities of connexin32 mutants causing CMTX. Neurobiol. Dis. 2002;11:43–52. doi: 10.1006/nbdi.2002.0545. [DOI] [PubMed] [Google Scholar]
- 5.Martin P.E. Analysis of gap junction assembly using mutated connexins detected in Charcot-Marie-Tooth X-linked disease. J. Neurochem. 2000;74:711–720. doi: 10.1046/j.1471-4159.2000.740711.x. [DOI] [PubMed] [Google Scholar]
- 6.Roscoe W. Oculodentodigital dysplasia-causing connexin43 mutants are non-functional and exhibit dominant effects on wild-type connexin43. J. Biol. Chem. 2005;280:11458–11466. doi: 10.1074/jbc.M409564200. [DOI] [PubMed] [Google Scholar]
- 7.Thomas T. Mechanisms of Cx43 and Cx26 transport to the plasma membrane and gap junction regeneration. J. Cell Sci. 2005;118:4451–4462. doi: 10.1242/jcs.02569. [DOI] [PubMed] [Google Scholar]
- 8.Sohl G., Willecke K. An update on connexin genes and their nomenclature in mouse and man. Cell Commun. Adhes. 2003;10:173–180. doi: 10.1080/cac.10.4-6.173.180. [DOI] [PubMed] [Google Scholar]
- 9.Goldberg G.S. Selective permeability of gap junction channels. Biochim. Biophys. Acta. 2004;1662:96–101. doi: 10.1016/j.bbamem.2003.11.022. [DOI] [PubMed] [Google Scholar]
- 10.Goodenough D.A., Paul D.L. Beyond the gap: functions of unpaired connexon channels. Nat. Rev. Mol. Cell Biol. 2003;4:285–294. doi: 10.1038/nrm1072. [DOI] [PubMed] [Google Scholar]
- 11.Unger V.M. Three-dimensional structure of a recombinant gap junction membrane channel. Science. 1999;283:1176–1180. doi: 10.1126/science.283.5405.1176. [DOI] [PubMed] [Google Scholar]
- 12.Fleishman S.J. A Cα model for the transmembrane α helices of gap junction intercellular channels. Mol. Cell. 2004;15:879–888. doi: 10.1016/j.molcel.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 13.Sosinsky G.E., Nicholson B.J. Structural organization of gap junction channels. Biochim. Biophys. Acta. 2005;1711:99–125. doi: 10.1016/j.bbamem.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 14.Simon A.M., Goodenough D.A. Diverse functions of vertebrate gap junctions. Trends Cell Biol. 1998;8:477–483. doi: 10.1016/s0962-8924(98)01372-5. [DOI] [PubMed] [Google Scholar]
- 15.Harris A.L. Emerging issues of connexin channels: biophysics fills the gap. Q. Rev. Biophys. 2001;34:325–472. doi: 10.1017/s0033583501003705. [DOI] [PubMed] [Google Scholar]
- 16.Saez J.C. Plasma membrane channels formed by connexins: their regulation and functions. Physiol. Rev. 2003;83:1359–1400. doi: 10.1152/physrev.00007.2003. [DOI] [PubMed] [Google Scholar]
- 17.Cottrell G.T., Burt J.M. Functional consequences of heterogeneous gap junction channel formation and its influence in health and disease. Biochim. Biophys. Acta. 2005;1711:126–141. doi: 10.1016/j.bbamem.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 18.Ahmad S. Synthesis and assembly of connexins in vitro into homomeric and heteromeric functional gap junction hemichannels. Biochem. J. 1999;339:247–253. [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang J.X., Goodenough D.A. Heteromeric connexons in lens gap junction channels. Proc. Natl. Acad. Sci. U. S. A. 1996;93:1287–1291. doi: 10.1073/pnas.93.3.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Das Sarma J. Heteromeric connexin interactions prior to the trans Golgi network. J. Cell Sci. 2001;114:4013–4024. doi: 10.1242/jcs.114.22.4013. [DOI] [PubMed] [Google Scholar]
- 21.Koval M. Transfected connexin45 alters gap junction permeability in cells expressing endogenous connexin43. J. Cell Biol. 1995;130:987–995. doi: 10.1083/jcb.130.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altevogt B.M. Connexin29 is uniquely distributed within myelinating glial cells of the central and peripheral nervous systems. J. Neurosci. 2002;22:6458–6470. doi: 10.1523/JNEUROSCI.22-15-06458.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lagree V. Specific amino-acid residues in the N-terminus and TM3 implicated in channel function and oligomerization compatibility of connexin43. J. Cell Sci. 2003;116:3189–3201. doi: 10.1242/jcs.00604. [DOI] [PubMed] [Google Scholar]
- 24.Gemel J. Connexin43 and connexin26 form gap junctions, but not heteromeric channels in co-expressing cells. J. Cell Sci. 2004;117:2469–2480. doi: 10.1242/jcs.01084. [DOI] [PubMed] [Google Scholar]
- 25.Rouan F. trans-dominant inhibition of connexin-43 by mutant connexin-26: implications for dominant connexin disorders affecting epidermal differentiation. J. Cell Sci. 2001;114:2105–2113. doi: 10.1242/jcs.114.11.2105. [DOI] [PubMed] [Google Scholar]
- 26.Altevogt B.M., Paul D.L. Four classes of intercellular channels between glial cells in the CNS. J. Neurosci. 2004;24:4313–4323. doi: 10.1523/JNEUROSCI.3303-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagy J.I., Rash J.E. Connexins and gap junctions of astrocytes and oligodendrocytes in the CNS. Brain Res. Brain Res. Rev. 2000;32:29–44. doi: 10.1016/s0165-0173(99)00066-1. [DOI] [PubMed] [Google Scholar]
- 28.Bao X. Functional expression in Xenopus oocytes of gap-junctional hemichannels formed by a cysteine-less connexin 43. J. Biol. Chem. 2004;279:9689–9692. doi: 10.1074/jbc.M311438200. [DOI] [PubMed] [Google Scholar]
- 29.Foote C.I. The pattern of disulfide linkages in the extracellular loop regions of connexin 32 suggests a model for the docking interface of gap junctions. J. Cell Biol. 1998;140:1187–1197. doi: 10.1083/jcb.140.5.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White T.W. Functional analysis of selective interactions among rodent connexins. Mol. Biol. Cell. 1995;6:459–470. doi: 10.1091/mbc.6.4.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haubrich S. Incompatibility of connexin 40 and 43 hemichannels in gap junctions between mammalian cells is determined by intracellular domains. Mol. Biol. Cell. 1996;7:1995–2006. doi: 10.1091/mbc.7.12.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elfgang C. Specific permeability and selective formation of gap junction channels in connexin-transfected HeLa cells. J. Cell Biol. 1995;129:805–817. doi: 10.1083/jcb.129.3.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teubner B. Functional expression of the murine connexin 36 gene coding for a neuron-specific gap junctional protein. J. Membr. Biol. 2000;176:249–262. doi: 10.1007/s00232001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abraham V. Phenotypic control of gap junctional communication by cultured alveolar epithelial cells. Am. J. Physiol. 1999;276:L825–L834. doi: 10.1152/ajplung.1999.276.5.L825. [DOI] [PubMed] [Google Scholar]
- 35.Koval M. Connexin46 is retained as monomers in a trans-Golgi compartment of osteoblastic cells. J. Cell Biol. 1997;137:847–857. doi: 10.1083/jcb.137.4.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Isakson B.E., Duling B.R. Heterocellular contact at the myoendothelial junction influences gap junction organization. Circ. Res. 2005;97:44–51. doi: 10.1161/01.RES.0000173461.36221.2e. [DOI] [PubMed] [Google Scholar]
- 37.Simon A.M. Female infertility in mice lacking connexin 37. Nature. 1997;385:525–529. doi: 10.1038/385525a0. [DOI] [PubMed] [Google Scholar]
- 38.Veitch G.I. Selective assembly of connexin37 into heterocellular gap junctions at the oocyte/granulosa cell interface. J. Cell Sci. 2004;117:2699–2707. doi: 10.1242/jcs.01124. [DOI] [PubMed] [Google Scholar]
- 39.Deutsch C. The birth of a channel. Neuron. 2003;40:265–276. doi: 10.1016/s0896-6273(03)00506-3. [DOI] [PubMed] [Google Scholar]
- 40.Musil L.S., Goodenough D.A. Multisubunit assembly of an integral plasma membrane channel protein, gap junction connexin43, occurs after exit from the ER. Cell. 1993;74:1065–1077. doi: 10.1016/0092-8674(93)90728-9. [DOI] [PubMed] [Google Scholar]
- 41.Arvan P. Secretory pathway quality control operating in Golgi, plasmalemmal, and endosomal systems. Traffic. 2002;3:771–780. doi: 10.1034/j.1600-0854.2002.31102.x. [DOI] [PubMed] [Google Scholar]
- 42.Trombetta E.S., Parodi A.J. Quality control and protein folding in the secretory pathway. Annu. Rev. Cell Dev. Biol. 2003;19:649–676. doi: 10.1146/annurev.cellbio.19.110701.153949. [DOI] [PubMed] [Google Scholar]
- 43.Ellgaard L., Helenius A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- 44.Zhang J.T. Membrane integration of in vitro-translated gap junctional proteins: co- and post-translational mechanisms. Mol. Biol. Cell. 1996;7:471–482. doi: 10.1091/mbc.7.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Falk M.M. Cell-free synthesis and assembly of connexins into functional gap junction membrane channels. EMBO J. 1997;16:2703–2716. doi: 10.1093/emboj/16.10.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diez J.A. Assembly of heteromeric connexons in guinea-pig liver en route to the Golgi apparatus, plasma membrane and gap junctions. Eur. J. Biochem. 1999;262:142–148. doi: 10.1046/j.1432-1327.1999.00343.x. [DOI] [PubMed] [Google Scholar]
- 47.Kumar N.M. Synthesis and assembly of human β(1) gap junctions in BHK cells by DNA transfection with the human β (1) cDNA. J. Cell Sci. 1995;108:3725–3734. doi: 10.1242/jcs.108.12.3725. [DOI] [PubMed] [Google Scholar]
- 48.Das Sarma, J. et al. Regulation of connexin43 oligomerization is saturable. Cell Commun. Adhes. (in press) [DOI] [PubMed]
- 49.Skerrett I.M. Aberrant gating, but a normal expression pattern, underlies the recessive phenotype of the deafness mutant Connexin26M34T. FASEB J. 2004;18:860–862. doi: 10.1096/fj.03-0763fje. [DOI] [PubMed] [Google Scholar]
- 50.Weisz O.A. Oligomerization of a membrane protein correlates with its retention in the Golgi complex. J. Cell Biol. 1993;122:1185–1196. doi: 10.1083/jcb.122.6.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Munro S. An investigation of the role of transmembrane domains in Golgi protein retention. EMBO J. 1995;14:4695–4704. doi: 10.1002/j.1460-2075.1995.tb00151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weisz O.A., Johnson J.P. Noncoordinate regulation of ENaC: paradigm lost? Am. J. Physiol. Renal Physiol. 2003;285:F833–F842. doi: 10.1152/ajprenal.00088.2003. [DOI] [PubMed] [Google Scholar]
- 53.Blanco G., Mercer R.W. Isozymes of the Na-K-ATPase: heterogeneity in structure, diversity in function. Am. J. Physiol. 1998;275:F633–F650. doi: 10.1152/ajprenal.1998.275.5.F633. [DOI] [PubMed] [Google Scholar]
- 54.DeTomaso A.W. The α and β subunits of the Na,K-ATPase can assemble at the plasma membrane into functional enzyme. J. Cell Biol. 1994;127:55–69. doi: 10.1083/jcb.127.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang X. Palmitoylation of tetraspanin proteins: modulation of CD151 lateral interactions, subcellular distribution, and integrin-dependent cell morphology. Mol. Biol. Cell. 2002;13:767–781. doi: 10.1091/mbc.01-05-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu C.C. Assembly of urothelial plaques: tetraspanin function in membrane protein trafficking. Mol. Biol. Cell. 2005;16:3937–3950. doi: 10.1091/mbc.E05-02-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang X. Palmitoylation supports assembly and function of integrin-tetraspanin complexes. J. Cell Biol. 2004;167:1231–1240. doi: 10.1083/jcb.200404100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Altan-Bonnet N. Molecular basis for Golgi maintenance and biogenesis. Curr. Opin. Cell Biol. 2004;16:364–372. doi: 10.1016/j.ceb.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 59.Chae H.J. BI-1 regulates an apoptosis pathway linked to endoplasmic reticulum stress. Mol. Cell. 2004;15:355–366. doi: 10.1016/j.molcel.2004.06.038. [DOI] [PubMed] [Google Scholar]
- 60.Sarma J.D. Targeted gap junction protein constructs reveal connexin-specific differences in oligomerization. J. Biol. Chem. 2002;277:20911–20918. doi: 10.1074/jbc.M111498200. [DOI] [PubMed] [Google Scholar]
- 61.Maza J. Defining a minimal motif required to prevent connexin oligomerization in the endoplasmic reticulum. J. Biol. Chem. 2005;280:21115–21121. doi: 10.1074/jbc.M412612200. [DOI] [PubMed] [Google Scholar]
- 62.Andersson H. Protein targeting to endoplasmic reticulum by dilysine signals involves direct retention in addition to retrieval. J. Biol. Chem. 1999;274:15080–15084. doi: 10.1074/jbc.274.21.15080. [DOI] [PubMed] [Google Scholar]
- 63.Klumperman J. The recycling pathway of protein ERGIC-53 and dynamics of the ER-Golgi intermediate compartment. J. Cell Sci. 1998;111:3411–3425. doi: 10.1242/jcs.111.22.3411. [DOI] [PubMed] [Google Scholar]
- 64.Hauri H.P. ERGIC-53 and traffic in the secretory pathway. J. Cell Sci. 2000;113:587–596. doi: 10.1242/jcs.113.4.587. [DOI] [PubMed] [Google Scholar]
- 65.Itin C. Targeting of protein ERGIC-53 to the ER/ERGIC/cis-Golgi recycling pathway. J. Cell Biol. 1995;131:57–67. doi: 10.1083/jcb.131.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mogelsvang S. Predicting function from structure: 3D structure studies of the mammalian Golgi complex. Traffic. 2004;5:338–345. doi: 10.1111/j.1398-9219.2004.00186.x. [DOI] [PubMed] [Google Scholar]
- 67.VanSlyke J.K. Intracellular transport, assembly, and degradation of wild-type and disease-linked mutant gap junction proteins. Mol. Biol. Cell. 2000;11:1933–1946. doi: 10.1091/mbc.11.6.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Berthoud V.M. Loss of function and impaired degradation of a cataract-associated mutant connexin50. Eur. J. Cell Biol. 2003;82:209–221. doi: 10.1078/0171-9335-00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Edidin M. The state of lipid rafts: from model membranes to cells. Annu. Rev. Biophys. Biomol. Struct. 2003;32:257–283. doi: 10.1146/annurev.biophys.32.110601.142439. [DOI] [PubMed] [Google Scholar]
- 70.Meyer R. Increased gap junction assembly between cultured cells upon cholesterol supplementation. J. Cell Sci. 1990;96:231–238. doi: 10.1242/jcs.96.2.231. [DOI] [PubMed] [Google Scholar]
- 71.Laing J.G. ZO-1 alters the plasma membrane localization and function of Cx43 in osteoblastic cells. J. Cell Sci. 2005;118:2167–2176. doi: 10.1242/jcs.02329. [DOI] [PubMed] [Google Scholar]
- 72.Schubert A.L. Connexin family members target to lipid raft domains and interact with caveolin-1. Biochemistry. 2002;41:5754–5764. doi: 10.1021/bi0121656. [DOI] [PubMed] [Google Scholar]
- 73.Locke D. Lipid rafts prepared by different methods contain different connexin channels, but gap junctions are not lipid rafts. Biochemistry. 2005;44:13027–13042. doi: 10.1021/bi050495a. [DOI] [PubMed] [Google Scholar]
- 74.Barth K. Distribution of caveolin-1 and connexin43 in normal and injured alveolar epithelial R3/1 cells. Histochem. Cell Biol. 2005;123:239–247. doi: 10.1007/s00418-004-0727-4. [DOI] [PubMed] [Google Scholar]
- 75.Lin D. Differential phosphorylation of connexin46 and connexin50 by H2O2 activation of protein kinase Cgamma. Mol. Vis. 2004;10:688–695. [PubMed] [Google Scholar]
- 76.Singh D. Connexin 43 interacts with zona occludens-1 and -2 proteins in a cell cycle stage-specific manner. J. Biol. Chem. 2005;280:30416–30421. doi: 10.1074/jbc.M506799200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hunter A.W. ZO-1 Alters Connexin43 Gap Junction Size and Organization by Influencing Channel Accretion. Mol. Biol. Cell. 2005;16:5686–5698. doi: 10.1091/mbc.E05-08-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Falk M.M. Connexin-specific distribution within gap junctions revealed in living cells. J. Cell Sci. 2000;113:4109–4120. doi: 10.1242/jcs.113.22.4109. [DOI] [PubMed] [Google Scholar]
- 79.van Meer G., Sprong H. Membrane lipids and vesicular traffic. Curr. Opin. Cell Biol. 2004;16:373–378. doi: 10.1016/j.ceb.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 80.Schuck S., Simons K. Polarized sorting in epithelial cells: raft clustering and the biogenesis of the apical membrane. J. Cell Sci. 2004;117:5955–5964. doi: 10.1242/jcs.01596. [DOI] [PubMed] [Google Scholar]
- 81.Segretain D., Falk M.M. Regulation of connexin biosynthesis, assembly, gap junction formation, and removal. Biochim. Biophys. Acta. 2004;1662:3–21. doi: 10.1016/j.bbamem.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 82.Laird D.W. Connexin phosphorylation as a regulatory event linked to gap junction internalization and degradation. Biochim. Biophys. Acta. 2005;1711:172–182. doi: 10.1016/j.bbamem.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 83.Solan J.L., Lampe P.D. Connexin phosphorylation as a regulatory event linked to gap junction channel assembly. Biochim. Biophys. Acta. 2005;1711:154–163. doi: 10.1016/j.bbamem.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 84.Panchin Y.V. Evolution of gap junction proteins – the pannexin alternative. J. Exp. Biol. 2005;208:1415–1419. doi: 10.1242/jeb.01547. [DOI] [PubMed] [Google Scholar]
- 85.Higgins D.G. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 86.Levy S., Shoham T. Protein-protein interactions in the tetraspanin web. Physiology (Bethesda) 2005;20:218–224. doi: 10.1152/physiol.00015.2005. [DOI] [PubMed] [Google Scholar]
- 87.Hemler M.E. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 2005;6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 88.Seigneuret M. Complete predicted three-dimensional structure of the facilitator transmembrane protein and hepatitis C virus receptor CD81: conserved and variable structural domains in the tetraspanin superfamily. Biophys. J. 2006;90:212–227. doi: 10.1529/biophysj.105.069666. [DOI] [PMC free article] [PubMed] [Google Scholar]


