Graphical abstract
N-Substituted isatin derivatives have been prepared and their inhibition activities against SARS viral protease were evaluated.
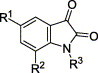
Keywords: SARS CoV protease inhibitor, Isatin
Abstract
N-Substituted isatin derivatives were prepared from the reaction of isatin and various bromides via two steps. Bioactivity assay results (in vitro tests) demonstrated that some of these compounds are potent and selective inhibitors against SARS coronavirus 3CL protease with IC50 values ranging from 0.95 to 17.50 μM. Additionally, isatin 4o exhibited more potent inhibition for SARS coronavirus protease than for other proteases including papain, chymotrypsin, and trypsin.
Severe Acute Respiratory Syndrome (SARS) is a new viral atypical pneumonia. This infectious disease originated in Southern China and Hong Kong in late 2002, and then rapidly spread to over 25 countries.1 The most common symptoms of SARS are cough, high fever (>38 °C), chills, rigor, myalgia, headache, dizziness, as well as progressive radiographic changes of the chest and lymphopenia. The disease had a quite high mortality rate (up to 15–19%) during the initial outbreak. Death may result from progressive respiratory failure owing to alveolar damage. In early 2003, a novel human coronavirus was identified as the causative agent of SARS, and the virus was named SARS coronavirus (SARS CoV).2 To date, no effective antiviral drug or vaccine has been marketed for treating SARS, although steroid and ribavirin have been used empirically in hospitals of Hong Kong,3, 4 and an HIV protease inhibitor nelfinavir could decrease the replication of SARS CoV in a preliminary in vitro examination.5
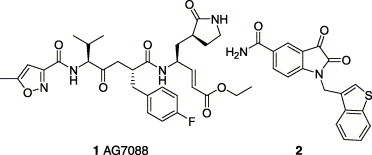
From the viewpoint of drug design, the human CoV main protease should be an attractive target due to its importance in the cleavage of the CoV polyprotein to the functional polypeptides.6 Previous studies have reported that the structure and the active site of human CoV main protease or CoV 3C-like protease (CoV 3CLpro) resemble those of picornavirus 3C protease.6, 7 One of the human rhinovirus (a subtype of picornavirus) 3C protease inhibitor, 1 (AG7088), was previously suggested as a good potential lead compound for the development of SARS CoV 3CLpro inhibitor.7 However, AG7088 failed to inhibit SARS CoV 3CLpro in vitro at this and other laboratories.8 Recently, some peptide derivatives have been reported as active inhibitors against SARS CoV 3CLpro.(a), (b) Several types of small molecules screened from various chemical libraries have been identified as potent SARS coronavirus protease inhibitors with IC50 values in low micro molar ranges.(a), (c), (d), (e), (f), (g) However, some of these small molecules may be unsuitable for structural modification because of having complex structures (e.g., sabadinine).9c Consequently, innovative scaffolds, which are easy to synthesize and chemically modify must be explored. In this report, preliminary works in the synthesis and evaluation of isatin derivatives as effective SARS CoV 3CLpro inhibitors investigated in our laboratories are described.
It is known that certain isatin (2,3-dioxindole) compounds such as 2 are potent inhibitors against rhinovirus 3C protease.10 Apparently, the isatin scaffold with derivatization may provide a good candidate for the SARS CoV 3CLpro inhibitor, because both of the proteases (human SARS CoV and rhinovirus) are cysteine protease and structurally similar at the active site.7
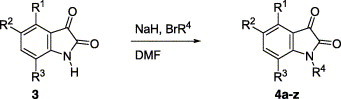
The synthesis of N-protected isatin derivatives 4 was straightforward. N-Alkylation of the corresponding isatin 3, sodium hydride, and various bromide compounds (transformed from the corresponding alcohol or methyl derivatives) in DMF provided the desired compounds 4a–z as shown in Scheme 1 .11 Moderate yields (30–60%) were achieved depending on the substituted groups in isatin.
Scheme 1.
The SARS CoV 3CLpro inhibition assays were conducted via fluorescence resonance energy transfer (FRET) according to the reported protocol.12a The FRET result was then confirmed through HPLC analysis of the fragment peaks cleaved by 3CLpro.13
Table 1 lists the in vitro bioactivities of compounds 4a–z against SARS CoV 3CL protease. The IC50 values (or percentage of inhibition at 20 μM if the IC50 value was exceeded 20 μM) in Table 1 were measured via FRET assay.12a The bioactivity of these compounds depended on the substituted groups in isatin scaffold and the side chain. The IC50 values demonstrated that 5-iodo such as 4o and 7-bromo such as 4k are the most potent substituted groups in isatin scaffold, and the benzothiophenemethyl side chain provides more inhibitory effect than the benzyl, heterocyclic substituted methyl, and other alkyl groups. Although thiophenecarboxylic 5-chloro-3-pyridinol ester was reported to be a potent inhibitor (IC50 = 0.5 μM) for SARS protease,9e isatins with side chain of thiophenecarboxylic amide 4u and 4z exhibited only moderate inhibition effect against SARS-CoV 3CL protease (IC50 = 12–17 μM).
Table 1.
Inhibition against SARS CoV 3CLpro activity by isatin derivatives 4
| Compound | R1 | R2 | R3 | R4 | IC50 or percentage (%) of inhibition at 20 μMa |
|---|---|---|---|---|---|
| 4a | H | C(O)NH2 | H |  |
25% inhibition at 20 μM |
| 4b | H | CN | H | 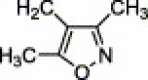 |
7.20 μM |
| 4c | H | I | H | 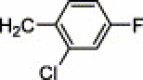 |
9.40 μM |
| 4d | H | I | H | 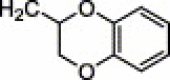 |
13.50 μM |
| 4e | H | I | H | 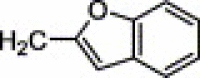 |
13% inhibition at 20 μM |
| 4f | H | OCH3 | H | 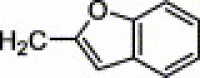 |
13% inhibition at 20 μM |
| 4g | H | NO2 | H | 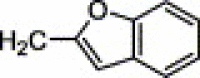 |
24% inhibition at 20 μM |
| 4h | H | H | H |  |
13.11 μM |
| 4i | H | H | NO2 |  |
2.00 μM |
| 4j | H | H | NH2 | 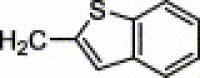 |
31% inhibition at 20 μM |
| 4k | H | H | Br |  |
0.98 μM |
| 4l | H | CH3 | NO2 |  |
39% inhibition at 20 μM |
| 4m | H | OCH3 | H |  |
27% inhibition at 20 μM |
| 4n | H | F | H |  |
4.82 μM |
| 4o | H | I | H |  |
0.95 μM |
| 4p | Cl | H | H |  |
11.20 μM |
| 4q | Cl | H | Cl | 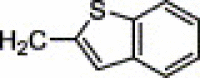 |
46% inhibition at 20 μM |
| 4r | H | I | H | 23.50 μM | |
| 4s | H | NH2 | H | 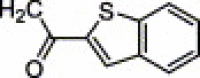 |
40% inhibition at 20 μM |
| 4t | H | CH3 | NO2 | 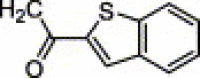 |
40% inhibition at 20 μM |
| 4u | H | I | H | 12.57 μM | |
| 4v | H | I | H | 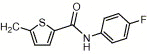 |
46% at 20 μM |
| 4w | H | I | H | 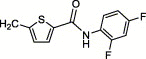 |
32% at 20 μM |
| 4x | H | I | H | 36% at 20 μM | |
| 4y | H | I | H | 47% at 20 μM | |
| 4z | H | I | H | 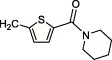 |
17.50 μM |
The IC50 values of compounds were measured by a quenched fluorescence resonance energy transfer (FRET) method:12a The mixture of SARS CoV 3CLpro (50 nM), various concentrations of the compound (from 0 to 20 μM), and a pH 7.5 buffer containing Tris–HCl (12 mM), NaCl (120 mM), EDTA (0.1 mM), DTT (1 mM), β-ME (7.5 mM) was incubated for 10 min at room temperature. Fluorogenic substrate (Dabcyl-KTSAVLQSGFRKME-Edans, 6 μM) was added. The fluorescence change resulted from the reaction was measured by continuous monitoring with Fusion-Alpha Basic Domestic System (excitation at 330 nm, emission at 515 nm). The initial velocities of the inhibited reactions were plotted against the different inhibitor concentrations to obtain the IC50.
Computer modeling (Fig. 1 ) showed that compound 4k and 4o were fitted into the active pocket of SARS CoV 3CLpro.14 Moreover, the isatin scaffold was docked in the S1 site, and the side chain (R4) was located in the S2 site of CoV 3CLpro. The carbonyl group of isatin and the NH groups in Cys145, Ser144, and Gly143 of protease were hydrogen bonded. On the other hand, steric effect in isatin scaffold is crucial for ensuring inhibitory potency. Therefore, the inhibitors with a bulkier side chain (4u–z) are less potent than compound 4o because of the limited space between the R4 of isatin and the His164 and Met165 of protease. Computational studies also demonstrated that the inhibitory potency depended heavily on hydrophobicity and electron affinity of the substituted groups in isatin. For example, compounds with nonpolar and more electron withdrawing groups such as bromo and nitro in R3 (4i and 4k) are significantly more potent than the compound with an amino group in R3 (4j).
Figure 1.
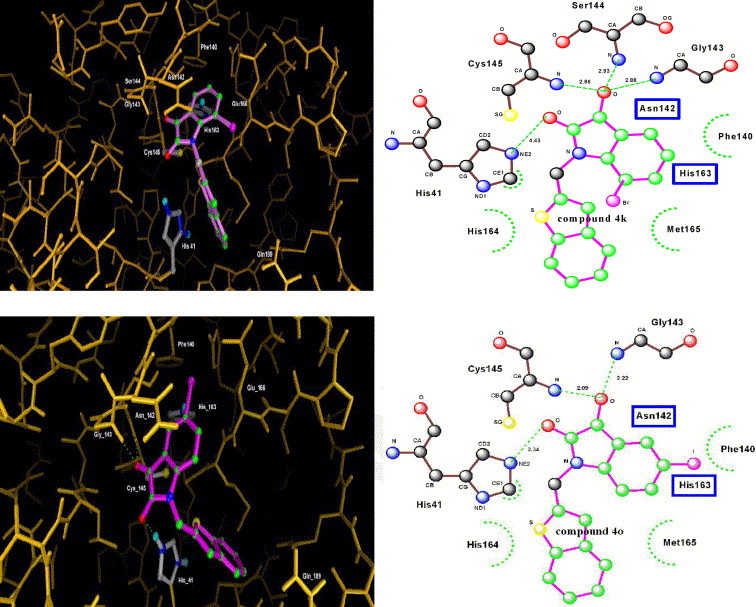
Computer modeling of compound 4k (top) and 4o (bottom) binding to SARS CoV 3CLpro (Hydrogen bonding between isatin and protease is displayed in the right figure; green: carbons in isatin, blue: nitrogen, red: oxygen.)
Table 2 shows that both isatin 4k and 4o exhibited more potent inhibition for SARS CoV 3CLpro than for other proteases such as papain (90–105 times) and trypsin (243–369 times). However, isatin 4k inhibited chymotrypsin at unexpectedly low IC50 value compared with compound 4o (10.4 μM vs 1.00 mM). Consequently, 4k is less specific for SARS CoV 3CLpro than 4o. Computer docking of compound 4o has slightly better (0.2 kcal/mol) binding affinity for SARS CoV 3CLpro. The slight difference might be the reason that compound 4o is more specific than compound 4k for other proteases.
Table 2.
Selective inhibition (IC50) against various protease by isatin derivatives 4k and 4oa
| Entry | SARS CoV 3CLprob (cysteine protease) | Papainc (cysteine protease) | Chymotrypsind (serine protease) | Trypsine (serine protease) |
|---|---|---|---|---|
| 4k | 0.98 μM | 103.00 μM | 10.40 μM | 362.40 μM |
| 4o | 0.95 μM | 87.24 μM | 1.00 mM | 243.30 μM |
The IC50 values were measured by FRET as in Table 1.
Substrate (6 μM) and SARS CoV 3CLpro (50 nM).
Substrate (10 μM) and papain (13.3 nM).
Substrate (200 μM) and chymotrypsin (5 nM).
Substrate (200 μM) and trypsin (9 nM).
In conclusion, a series of N-substituted isatin derivatives were simply prepared from isatin, sodium hydride, and various bromide derivatives. The inhibition activities of these compounds against SARS CoV 3CLpro were assessed by FRET and then confirmed via HPLC analysis. The IC50 values demonstrated that these isatin derivatives inhibited SARS CoV 3CLpro in low micro molar range (0.95–17.50 μM). To our knowledge, compound 4o is one of the most potent and selective SARS CoV 3CLpro inhibitor reported to date.
Acknowledgments
The financial support from the Ministry of Economy Affairs of ROC and the NMR analytical support provided by Ms. M. H. Lee are greatly appreciated.
References and notes
- 1.Lee N., Hui D., Wu A., Chan P., Cameron P., Joynt G.M., Ahuja A., Yung M.Y., Leung C.B., To K.F., Lui S.F., Szeto C.C., Chung S., Sung J.J.Y. N. Engl. J. Med. 2003;348:1986. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 2.Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.-R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A.M., Berger A., Burguière A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.-C., Müller S., Rickerts V., Stürmer M., Vieth S., Klenk H.-D., Osterhaus A.D.M.E., Schmitz H., Doerr H.W. N. Engl. J. Med. 2003;348:1967. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 3.So L.K.-Y., Lau A.C.W., Yam L.Y.C., Cheung T.M.T., Poon E., Yung R.W.H., Yuen K.Y. Lancet. 2003;361:1615. doi: 10.1016/S0140-6736(03)13265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hon K.L.E., Leung C.W., Cheng W.T.F., Chan P.K.S., Chu W.C.W., Kwan Y.W., Li A.M., Fong N.C., Ng P.C., Chiu M.C., Li C.K., Tam J.S., Fok T.F. Lancet. 2003;361:1701. doi: 10.1016/S0140-6736(03)13364-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamamoto N., Yang R., Yoshinaka Y., Amari S., Nakano T., Cinatl J., Rabenau H., Doerr H.W., Hunsmann G., Otaka A., Tamamura H., Fujii N., Yamamoto N. Biochem. Biophys. Res. Commun. 2004;318:719. doi: 10.1016/j.bbrc.2004.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Science. 2003;300:1763. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 7.Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L.M., Guan Y., Rozanov M., Spaan W.J.M., Gorbalenya A.E. J. Mol. Biol. 2003;331:991. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jenwitheesuk E., Samudrala R. Bioorg. Med. Chem. Lett. 2003;13:3989. doi: 10.1016/j.bmcl.2003.08.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Wu C.-Y., Jan J.-T., Ma S.-H., Kuo C.-J., Juan H.-F., Cheng Y.-S.E., Hsu H.-H., Huang H.-C., Wu D., Brik A., Liang F.-S., Liu R.-S., Fang J.-M., Chen S.-T., Liang P.-H., Wong C.-H. PNAS. 2004;101:10012. doi: 10.1073/pnas.0403596101. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Jain R.P., Pettersson H.I., Zhang J., Aull K.D., Fortin P.D., Huitema C., Eltis L.D., Parrish J.C., James M.N.G., Wishart D.S., Vederas J.C. J. Med. Chem. 2004;47:6113. doi: 10.1021/jm0494873. [DOI] [PubMed] [Google Scholar]; (c) Toney J.H., Navas-Martin S., Weiss S.R., Koeller A. J. Med. Chem. 2004;47:1079. doi: 10.1021/jm034137m. [DOI] [PubMed] [Google Scholar]; (d) Bacha U., Barrila J., Velazquez-Campoy A., Leavitt S.A., Freire E. Biochemistry. 2004;43:4906. doi: 10.1021/bi0361766. [DOI] [PubMed] [Google Scholar]; (e) Blanchard J.E., Elowe N.H., Fortin P.D., Huitema C., Cechetto J.D., Eltis L.D., Brown E.D. Chem. Biol. 2004;43:1445. doi: 10.1016/j.chembiol.2004.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Kao R.Y., Tsui W.H.W., Lee T.S.W., Tanner J.A., Watt R.M., Huang J.-D., Hu L., Chen G., Chen Z., Zhang L., He T., Chan K.-H., Tse H., To A.P.C., Ng L.W.Y., Wong B.C.W., Tsoi H.-W., Yang D., Ho D.D., Yuen K.-Y. Chem. Biol. 2004;43:1293. doi: 10.1016/j.chembiol.2004.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Hsu J.T.-A., Kuo C.-J., Hsieh H.-P., Wang Y.-C., Huang K.-K., Lin C.P.-C., Huang P.-F., Chen X., Liang P.-H. FEBS Lett. 2004;574:116. doi: 10.1016/j.febslet.2004.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Webber S.E., Tikhe J., Worland S.T., Fuhrman S.A., Hendrickson T.F., Matthews D.A., Love R.A., Patick A.K., Meador J.W., Ferre R.A., Brown E.L., DeLisle D.M., Ford C.E., Binford S.L. J. Med. Chem. 1996;39:5072. doi: 10.1021/jm960603e. [DOI] [PubMed] [Google Scholar]
- 11.All of the new compounds were characterized. The spectra of the active new compounds are reported here. Compound 4k: 1H NMR (CDCl3) δ 7.67 (m, 1H), 7.61 (m, 2H), 7.54 (d, J = 7.3 Hz, 1H), 7.25–7.18 (m, 3H), 6.94 (d, J = 7.5 Hz, 1H), 5.60 (s, 2H). Compound 4o: 1H NMR (CDCl3) δ 7.90 (m, 1H), 7.82 (dd, J = 8.3, 1.7 Hz, 1H), 7.75 (d, J = 7.7 Hz, 1H), 7.71 (d, J = 7.4 Hz, 1H), 7.40–7.25 (m, 3H), 6.77 (d, J = 8.3 Hz, 1H), 5.15 (s, 2H). MS (EI) m/z 419 M+
- 12.(a) Kuo C.-J., Chi Y.-H., Hsu J.T.-A., Liang P.-H. Biochem. Biophys. Res. Commun. 2004;318:862. doi: 10.1016/j.bbrc.2004.04.098. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Blanchard J.E., Elowe N.H., Huitema C., Fortin P.D., Cechetto J.D., Eltis L.D., Brown E.D. Chem. Biol. 2004;11:1445. doi: 10.1016/j.chembiol.2004.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan K., Wei P., Feng Q., Chen S., Huang C., Ma L., Lai B., Pei J., Liu Y., Chen J., Lai L. J. Biol. Chem. 2004;279:1637. doi: 10.1074/jbc.M310875200. Substrate (Dabcyl-KTSAVLQSGFRKME-Edans) and the fragments (Dabcyl-KTSAVLQ, Edans-EMKRFGS) were analyzed by HPLC: Photodiarray detector monitored at UV 300 and 500 nm; reversed phase column COSMOSIL 5C18-MS 4.5 mm × 150 mm; 0.1% trifluoroacetic acid in acetonitrile/0.1% trifluoroacetic acid in water (10/90, v/v) for 20 min followed by a linear gradient to 0.1% trifluoroacetic acid in acetonitrile/0.1% trifluoroacetic acid in water (45/55, v/v) at the flow rate of 1.0 mL/min. A similar result has been reported during our research: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Docking experiments were conducted by using Autodock 3.0.5 with a Lamarckian Genetic Algorithm (LGA).15 The crystal structure of SARS coronavirus 3CL protease complex with (coded 1uk4) was obtained from The Protein Databank (PDB: http://www.rcsb.org/pdb/). Only chain-A of the dimer was used in this simulation. The structures of isatin derivatives were built in CAChe (Fujitsu, Japan) and refined by performing an optimized geometry calculation in Mechanics using augmented MM2 parameters and stored in PDB format. MGLTOOLS (MGL, Scripps Institute) was used for structure preparation and parameter creation to meet the input requirements of Autodock. Briefly, essential hydrogen atoms were added to the model structure of 3CL protease (chain A) along with Kollman united atom charges, and solvation parameters. Molecules were added all-hydrogen atoms, assigned Gasteiger–Marsili charges,16 merge non-polar H atoms, and defined torsions. Autogrid tool in Autodock3.0.5 was applied to compute energy grids (60 × 60 × 70 in xyz directions with 0.375 Å spacing) of various compound atoms. Grip maps were centered at the active site. Solis and Wets’ local search method with LGA was applied to generate available conformations of compound structures within the active site. During docking experiments, each compound was kept flexible (except for the rings and amide bonds) and the protein was rigid
- 15.Morris G.M., Goodsell D.S., Halliday R.S., Huey R., Hart W.E., Belew R.K., Olson A.J. J. Comput. Chem. 1998;19:1639. [Google Scholar]
- 16.Gasteiger J., Marsili M. Tetrahedron. 1980;36:3219. [Google Scholar]


