Abstract
The transcription factor GLI3 is a member of the Hedgehog (Hh/HH) signaling pathway that can exist as a full length (Gli3-FL/GLI3-FL) or repressor (Gli3-R/GLI3-R) form. In response to HH activation, GLI3-FL regulates HH genes by targeting the GLI1 promoter. In the absence of HH signaling, GLI3 is phosphorylated leading to its partial degradation and the generation of GLI3-R which represses HH functions. GLI3 is also involved in tissue development, immune cell development and cancer. The absence of Gli3 in mice impaired brain and lung development and GLI3 mutations in humans are the cause of Greig cephalopolysyndactyly (GCPS) and Pallister Hall syndromes (PHS). In the immune system GLI3 regulates B, T and NK-cells and may be involved in LPS-TLR4 signaling. In addition, GLI3 was found to be upregulated in multiple cancers and was found to positively regulate cancerous behavior such as anchorage-independent growth, angiogenesis, proliferation and migration with the exception in acute myeloid leukemia (AML) and medulloblastoma where GLI plays an anti-cancerous role. Finally, GLI3 is a target of microRNA. Here, we will review the biological significance of GLI3 and discuss gaps in our understanding of this molecule.
Video Abstract
Keywords: GLI, GLI3, Hedgehog signaling, Development, Cancer, Genetic disease
Background
The GLI gene was first identified in humans as a highly expressed gene in human glioma [1]. Using cDNA probes for the zinc finger region of the GLI gene, Ruppert et al (1988), identified two additional GLI family members, GLI2 and GLI3 [2]. Further characterization of human GLI3 revealed it to be a 190 kDA protein located on chromosome 7p13 and binds to consensus sequences similar to those of GLI1 [3]. The most updated data on the National Center for Biotechnology Information (NCBI) and new publications, mapped human GLI3 to chromosome 7p14.1 (Gene ID:2737, 4]. GLI3 was identified as a gene in which mutations in GLI3 cause GCPS, a disease leading to craniofacial and limb maldevelopment. In a study by Vortkamp et al (1991), 2 translocations in GLI3 were identified, which interrupt GLI3 expression and cause GCPS [5]. Point mutations in the human GLI3 locus in GCPS patients were identified as a main cause of GCPS disease manifestation [6]. In 1996, GLI3 was described as a protein that is regulated in response to the sonic hedgehog (SHH) signaling pathway where it was described to compete in binding with GLI1 [7]. In the same study, GLI3 was characterized as a negative regulator of SHH signaling [7]. In the following year, GLI3 was recognized as the cause of PHS, a disease characterized by developmental malformations including polydactyly (extra digits) [8]. Follow-up studies described Gli3 as both an activator and repressor, similar to the Gli2 family member, in response to Shh signaling [9]. Since then, research on mouse and human Gli3/GLI3 mostly focused on its role in brain and limb development with certain exceptions of Gli3/GLI3’s role in angiogenesis, colorectal and liver cancer, TRAIL-dependent apoptosis and its role in regulating the IL-6/JAK2 pathway [10–14].
Regulation and structure
Hedgehog ligands and their function
The Hh signaling pathway plays a role in embryonic development and homeostasis of stem cells in normal tissues [15]. Dysregulations of Hh signaling cause genetic defects such as holoprosencephaly and polydactyly and are tightly linked to cancer development and progression [16, 17]. In addition, a role for Hh signaling in hematopoiesis and in the immune system has been described [18–20]. The Hh signaling pathway is activated by 3 ligands: Sonic Hedgehog (Shh/SHH), Indian Hedgehog (Ihh/IHH) or Desert Hedgehog (Dhh/DHH) (mouse/human respectively) [21]. These ligands are roughly 45 kDA with a N-terminal biologically active domain and an autocatalytic C-terminus, which is cleaved to generate the final Hh ligand form [22, 23]. After cleavage, a cholesterol moiety is added to the C-terminus and palmitate is linked to the N-terminus [24]. This allows exogenous Hh ligands to travel far distances to activate Hh signaling in various cells/tissues in the body [25]. Shh is the ligand with the highest expression and therefore is the major inducer of most Hh-related functions such as brain, limb and spinal cord development [26, 27]. Ihh was linked to chondrogenesis and negatively regulates chondrocyte differentiation [28]. Dhh null male mice are infertile while there was no visible effect in female mice suggesting a role for Dhh in spermatogenesis [29]. Additionally, Dhh was shown to play a role in peripheral nerve ensheathment [30]. Shh is mostly expressed in epithelia while Dhh is expressed in Schwann and Sertolli precursors and Ihh is expressed in the cartilage and in the gut [31]. All Hh ligands bind to the same receptor Ptch1 and initiate Hh-related signaling. However, Shh has been shown to be the most potent inducer of this pathway [32].
Classic (canonical) Hh signaling
Many components known to be involved in vertebrate Hh signaling were initially identified in Drosophila. The main components of Hh signaling in Drosophila are 1) Patched (Ptc) a 12-transmembrane protein which binds Hh ligand; 2) Smoothened (Smo), a receptor that is repressed by Ptc and released to activate the pathway once Hh ligand binds Ptc; and 3) Cubitus interruptus (Ci) which is the Drosophila analog of Gli proteins in vertebrates [33]. In the absence of Hh signaling Ci, Costal-2 (cos-2) and Fused (Fu) form the Hedgehog Signaling complex (HSC). This leads to proteasomal degradation of Ci from a 155 to 75 kDA form through phosphorylation of Protein Kinase A (Pka/PKA), Glycogen synthase kinase-3 beta (Gsk3β/GSK3β) and Casein Kinase 1 (Ck-1/CK-1) [34]. The degraded form of Ci subsequently acts as a repressor of Hh signaling. Once Hh signaling is activated by ligand binding to Ptc, and inhibits its repressor function, Smo is activated which leads to stabilization of Ci and expression of Hh related genes. The signaling cascade is well conserved between invertebrates and vertebrates, therefore the information gained from Drosophila is useful in predicting the signaling events in mice and humans.
In mammals
In mammals, Hh signaling occurs at the primary cilia of cells [35]. The intraflagellar transport (IFT) proteins: Intraflagellar transport protein 172 homolog (Ift172), Kinesin-like protein KIF3A (Kif3a), dynein cytoplasmic heavy chain 2 (Dnchc2) and Polaris, are extremely important for the formation and function of cilia, and are essential for the regulation of Hh activation or inactivation [36–38]. In addition, the kinesin-like protein Kif7, which is involved in cilia tip assembly, was shown to be essential for proper activation of this pathway and is therefore involved in positively and negatively regulating Hh-dependent functions [39, 40]. Several co-receptors for Ptch1 have been identified and were shown to be indispensable for proper Hh function. These include growth-arrest-specific 1 (Gas1), CAM-related/downregulated by oncogenes (Cdo), and brother of Cdo (Boc) which positively regulate Hh-related signaling events [41]. In addition, the Hh interaction protein (Hhip) binds and sequesters all three Hh ligands and therefore acts as a negative regulator of the Hh pathway [42].
In the absence of binding of one of the Hh ligands (Shh, Dhh or Ihh) to Ptch1, Smo is localized intracellularly [43]. The mechanism by which Ptch1 inhibits Smo is not completely understood. It was hypothesized to occur by either binding of a Smo activator or delivery of a Smo small molecule inhibitor [44]. Another hypothesis suggested that in the absence of Hh ligand, Ptch1 translocates to the base of the primary cilia and inhibits localization of Smo to the cilia [45, 46]. However, further experiments are needed to elucidate the mechanism by which Ptch1 inhibits Smo. Without SMO being activated, suppressor of fused (Sufu/SUFU) sequesters GLI transcription factors in the cytoplasm [47]. These GLI proteins are essential effectors of the HH pathway. There are 3 members of the GLI family of transcription factors: GLI1, GLI2 and GLI3. In the absence of Hh activation by ligand binding to Ptch1, Sufu is phosphorylated and recruits G-protein coupled receptor 161 (Gpr161/GPR161), PKA, GSK3β, and beta-tranducin repeat containing E3 ubiquitin protein ligase (βTrCP) complex to proteolytically cleave Gli2 and Gli3 into smaller proteins with a repressor function [48, 49]. Ck-1 negatively regulates Hh signaling in Drosophila [50]. Since many Hh signaling molecules in Drosophila are evolutionary conserved in mammals, it is probable that Ck-1 negatively regulates mammalian HH signaling as well. In fact, mutations of conserved Ck-1 sequences from Drosophila showed less phosphorylation of mammalian GLI3 and less GLI3 processing into its repressor form [51]. However, based on our current understanding, it remain unclear what role CK-1 plays in vertebrate HH signaling since there are also reports of positive regulation of HH signaling by CK-1α and CK-1ε [52–54]. Interestingly, in Drosophila, inhibition of Ck-1α or Ck-1ε alone did not significantly affect Ci processing [55]. However, when both Ck-1 subtypes were inhibited, a significant decrease of Ci repressor was observed. Therefore, it is probable that CK-1α and CK-1ε alone positively regulate HH signaling, while when forming an alpha-epsilon complex, they negatively regulate this pathway by phosphorylating GLI3. This however, is highly speculative, and more research is needed to validate the role of CK-1 in HH signaling. GLI1 is not cleaved in this process but completely degraded [56]. While Gli2 is not very stable in its repressor form and completely degrades after cleavage, the Gli3 repressor form is more stable and translocates to the nucleus where it inhibits Hh-related responses [57]. As such, in the context of Hh/HH signaling, GLI3 is known to be the most potent inhibitor of HH signaling within the GLI family.
Upon ligand binding to Ptch1, Smo translocates to the primary cilia, a process mediated by β-arrestin [58]. This leads to the association of Smo with Ellis-van Creveld syndrome protein/Ellis-van Creveld syndrome protein 2 (Evc/Evc2) complex (a complex that regulates chondrocyte proliferation and differentiation of osteoblasts) in which Sufu-Gli2/3 complex moves to the tip of the cilia where Sufu dissociates from Gli2/3 [57, 59, 60]. Sufu is then degraded and Gli2/3 full-length accumulate at the ciliary tip and can translocate into the nucleus to activate a Hh-related gene expression program [49, 61]. Gli1, Gli2-FL, and Gli3-FL have been shown to induce Hh related gene functions. Because GLI2-FL and GLI3-FL induce GLI1 expression, GLI1 therefore acts as an amplifier of the HH signal [62]. However, Gli2 is considered the most potent activator of Hh signaling [9, 63]. All members of the GLI protein family (GLI1, GLI2 and GLI3) contain five zinc finger domains which are very similar to those of Ci [64]. Although all GLI members share biochemical domains, GLI1 does not contain a N-terminal repressor domain which is present in both GLI2 and GLI3 (Fig. 1). Therefore, biochemically, GLI1 can only act as transcriptional activator while GLI2 and GLI3 can potentially act as transcriptional activators or repressors.
Fig. 1.
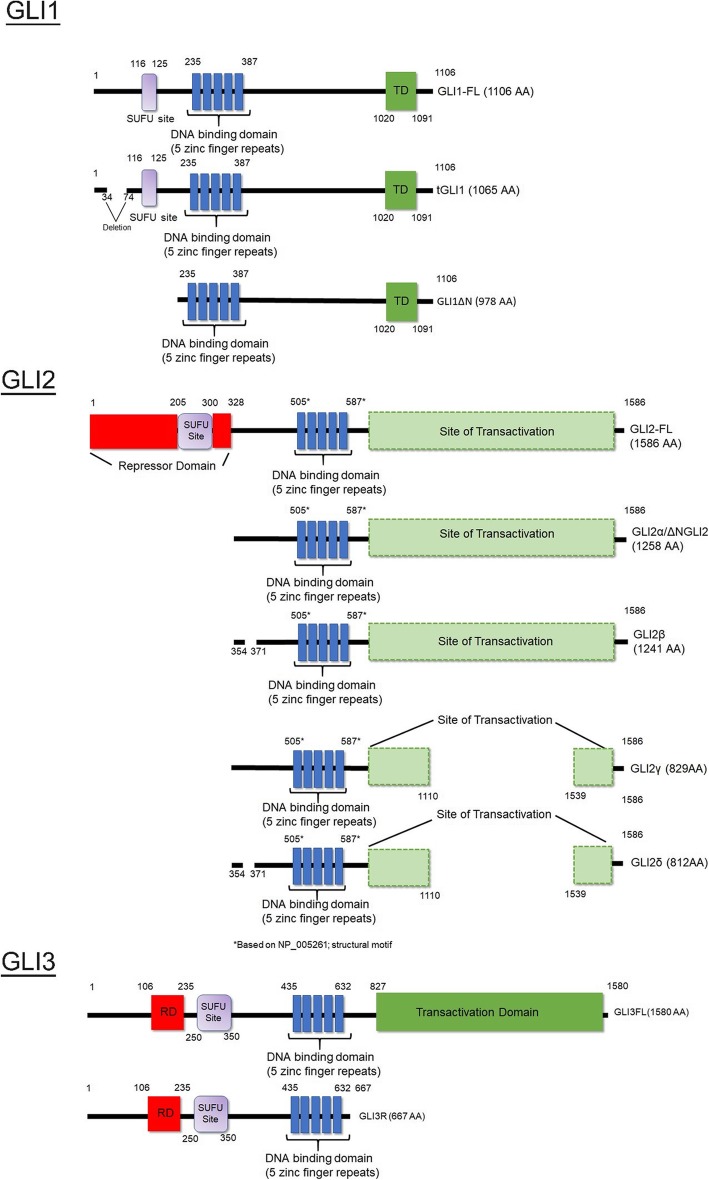
Biochemical domains of GLI family members. Schematic diagram of the biochemical domains of human GLI1, GLI2 and GLI3 showing the relative repressor (RD), DNA binding and transactivation (TD) domains. HH-related transcriptional activator GLI1 is the smallest protein of the GLI family with 1106 AA (GLI2: 1586 AA; GLI3: 1580 AA). In comparison to GLI2 and GLI3, GLI1 lacks a transcriptional repressor domain. GLI1 and GLI2 isoforms have been described. There is no direct evidence to describe the exact position of the transactivation domain and zinc finger region of human GLI2
In humans, different isoforms of GLI1 and GLI2 have been reported (Fig. 1). To date, there are no reports of different GLI3 isoforms in humans. GLI1 has three isoforms: isoform 1 is 1106 amino acids (AA) long and is considered the full length GLI1 protein (NP_005260.1). GLI1 isoform 2 sequence lacks exons 2 and 3, which leads to a truncated GLI1ΔN form (978 AA) (NP_001153517.1). This isoform was reported in healthy and malignant tissue and showed no binding to SUFU [65]. The third isoform tGLI1 is a cancer-associated form of GLI1 [66], and is not detected in normal cells but is found in glioblastoma multiforme (GBM) and other cancers. It lacks exon 3 and part of exon 4 (1065 AA), but retains most of its function; however, it is associated with increased motility and invasiveness [66]. To date, four GLI2 isoforms have been described, in addition to the full length GLI2 (1586 AA) [67, 68]. These isoforms include GLI2α (1258 AA), GLI2β (1241 AA), GLI2γ (829 AA) and GLI2δ (812 AA). GLI2α is also known as ΔNGLI2 and shows significantly higher activity in vitro than GLI2 full length [69]. GLI2β mRNA was identified to be highly expressed in basal cell carcinoma and both GLI2α and GLI2β were shown to regulate the expression of the human T cell lymphoma virus 1 gene [67, 70]. To date, no functions for GLI2γ and GLI2δ have been reported.
Non-classical (non-canonical) HH signaling
PTCH1 and SMO are major regulators of the canonical HH signaling pathway and therefore of GLI transcription factors. However, several studies report that GLIs (especially GLI1 and GLI2) can be regulated by the cross talk with other signaling pathways in various types of cancers including melanoma, gastric cancer, colon cancer, multiple myeloma, medulloblastoma, pancreatic cancer, glioblastoma, and osteosarcoma [71–78].
The EGFR-RAS-RAF-MEK1 pathway has been frequently investigated in regulating GLI1 and GLI2 expression levels. Constitutively active MEK1 and oncogenic KRASV12 induce GLI1 protein expression and activity [71, 79]. In addition to upregulating positive regulators of HH signaling, GLI2 was shown to be stabilized upon activation of EGF-ERK1/2 signaling [80]. Utilization of the A3 adenosine receptor agonist, resulted in a decrease in breast cancer stem cell survival which correlated with a decrease in activated ERK1/2 and GLI1 [81, 82]. Therefore, EGFR signaling through MAPK can regulate GLI1 and GLI2 at the level of expression and activity.
The PI3K pathway has been closely linked to HH-related functions. Several studies suggest a role of PI3K-AKT axis in the regulation of GLI1/2 transcriptional activators [83–85]. PI3K-AKT activity induced an increase in the expression of GLI1 and GLI2 in renal cell carcinoma and chronic lymphocytic leukemia (CLL), leading to increased cancer cell growth and progression [83, 84]. The regulation of GLI by PI3K-AKT axis was shown to be induced by CCL5-CCR3 signaling in the tumor microenvironment (TME) as well [85]. In addition, the interaction between PI3K and HH has been reported to be important for tamoxifen resistant breast cancer and combined targeting of both GLI1/2 (using the GLI antagonist GANT61) and PI3K/mTOR (using PI103) synergistically increased apoptosis and reduced tumor growth [86, 87].
In addition to ERK1/2 and PI3K, TGF-β has been shown to be involved in regulating GLI1/2 expression without the involvement of SMO signaling [88–92]. In the last few years several studies have reported a role for the TGF-β-GLI1/2 axis, partially Smad3 dependent, in fibroblast activation, melanoma, stroke (ischemia/reperfusion injury), hepatocellular carcinoma, and gastric cancer [88–92]. In these studies, both GLI1 and GLI2 regulate tumor growth, survival and epithelial-to-mesenchymal transition (EMT) upon stimulation by TGF-β1/2 in a SMO independent manner. Interestingly, TGF-β2, an inducer of nephrons (a unit required for functional kidneys) lies downstream of GLI3-R [93]. This suggests that a negative feedback loop exists between TGF-β-dependent HH activation in which GLI1 and GLI2 act as positive regulators while GLI3 acts as a negative regulator. However, more experimental evidence is needed to validate this hypothesis. Additionally, in a pan cancer analysis, a positive correlation between TGF-β-related gene expression and GLI1 and GLI2 expression was observed [94]. Notably, GLI1/2 expression was more of a prognostic factor for TGF-β and EMT-related genes than for HH-related genes, which might suggest that GLI1/2 regulate tumor formation by regulating TGF-β-related gene expression rather than HH-induced gene expression.
Protein kinase C (PKC), a known stimulator of cell proliferation, was also reported to regulate GLI expression and function [95, 96]. Although PKC was shown to phosphorylate and activate SMO at the beginning of the HH signaling cascade, it was also reported to be able to compensate for SMO inhibition and induce GLI activation [97, 98]. Interestingly, PKC-mediated activation of GLI proteins occurs independently of primary cilia and MEK-1 in NIH3T3 cells [99]. PKC also positively regulates GLI1-HDAC1 association [100] and targeting HDAC1 with the HDAC inhibitor Vorinostat, only resulted in a decrease in cell proliferation at high inhibitor doses. In combination with Vorionstat, PKC inhibition enhanced the therapeutic effect and increased the therapeutic window in basal cell carcinoma [101].
The intracellular membrane trafficking protein Rab23, the serine/threonine kinase involved in cell proliferation and survival CK2α, the inhibitor of transcription SLTM, the proliferative proteins Dual Specificity Tyrosine Phosphorylation Regulation Kinases 1 (DYRK1) and DYRK2, the metabolic proteins MAP 3K10 and AMPK all regulate GLI1 expression and function as well [102–106]. These regulators are, in some way, involved in cancer growth and progression, which shows the importance of studying the crosstalk between HH signaling and other pathways, particularly in GLI-directed therapeutic approaches.
Most studies investigating non-canonical HH signaling primarily focus on GLI1 and GLI2 and oftentimes did not consider GLI3 in their experimental approach. This is understandable since GLI1 and GLI2 are positive regulators of HH ligand-induced gene expression and can therefore be considered major drivers of HH-related genes [107]. However, since various studies describe GLI activation by other signaling pathways which are activated by different ligands (other than SHH, IHH or DHH), the regulation of GLIs might be different in comparison to their regulation in the canonical HH pathway. For example, PKC has been shown to activate GLIs in a cilia-independent manner and additionally, pan cancer analysis revealed that GLIs account for TGF-β-induced gene expression rather than genes induced by classical HH pathway [94, 99]. In addition, there are multiple examples that are discussed in this review of the role of GLI3 in development, in the immune system and in cancer where GLI3 often acts as a positive regulator of those pathways, although GLI3 did not show potent HH-gene induction in the canonical HH pathway. It might be that activation, regulation and function of GLIs varies depending on which pathway triggers them, which raises the question of whether or not we can apply the principles of knowledge of the canonical pathway to the non-canonical pathways. Could it be that GLI3-FL acts as a more potent gene activator than GLI2-FL depending on which non-canonical pathway is active? Are post translational changes involved in increasing GLI3-FL function or are GLI3 mRNA levels induced? Is GLI3-FL processing into its repressor form always dependent on PKA, GSK3β or βTrCP or could these factors be substituted, differentially regulated or inhibited in different pathways of non-canonical HH signaling? This report aims to give insights into different cellular functions of GLI3 which are oftentimes in crosstalk with other important signaling pathways.
Regulation of GLI3
Gene locus and domains of mouse and human Gli3/GLI3
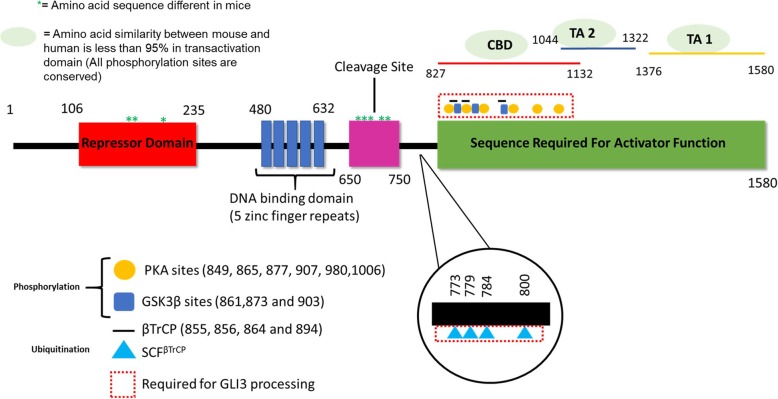
Several publications have reported that human GLI3 is located on chromosome 7p13 [5, 8, 23]. However as mentioned earlier, recent updates on NCBI have mapped human GLI3 to chromosome 7p14.1 (Gene ID: 2737) [4, 108, 109]. The human GLI3 gene is 276261 bp in length (NC_000007.14) and the mRNA sequence is composed of 15 exons that are 8405 bp in length (16 Dec, 2019) (NM_000168). However, the consensus coding sequence (CCDS) is 4743 nucleotides (nt) (nt 282-5024) with a transcription start site located in exon 2 (CCDS5465.1). Due to an error in sequencing, older literature characterized GLI3-FL to be 1596 AA in size which was later corrected to 1580 AA [110]. This is also listed in the protein database entry NP_000159.3 which shows GLI3 protein to be 1580 AA with a molecular weight of 190 kDa (GLI3-FL)(NP_000159.3). Human GLI3 contains a N-terminal repressor domain which is mapped to AA 106-235 (exons 3-6, mRNA nt 649-986) [111]. GLI3 also has a 5 zinc finger DNA-binding domain (AA 480-632 that span exons 10-13; nt 1719-2177), and a C-terminal transcriptional activation domain that is dependent on CREB-binding protein (CBP) binding (AA 827-1132 in exon 15; nt 2760-3677) (Fig. 2) in addition to transactivation domain 2 (TA2)(AA 1044-1322 in exon 15; mRNA nt 3411-4247) and transactivation domain 1 (TA1)(AA 1376-1580 in exon 15; mRNA nt 4407-5024) [62, 112]. The proteasomal cleavage site spans exons 13 and 14 (AA 650-750; nt 2229-2531) which leads to the truncated GLI3-R (677 AA with a molecular weight of 83 kDa) [113–115]. In addition to other references listed, mapping of DNA binding domain, cleavage site and activator domain was performed with the help of the publication by Krauß et al (2009) [116]. Although it can be extrapolated that the stated AA range corresponding to each domain is accurate, studies other than that describing the repressor domain [111], that precisely define GLI3’s biochemical domains, are lacking.
Fig. 2.
Biochemical domains of human GLI3 with posttranslational modifications. GLI3 can be phosphorylated by PKA and GSK3β which leads to binding by βTrCP and ubiquitination by SCFTrCP. Indicated phosphorylation and ubiquitination sites are shown and are crucial for GLI3 processing into its repressor form (red box). βTrCP binds GLI3 at the C- and N-terminus and at its core domain (activation domain; green). Only binding to GLI3 core domain is important for GLI3 processing into GLI3-R. Processing site to generate Gli3-R and zinc finger domain are completely conserved between mouse and human. Differences in amino acid sequences between mice and human are represented by asterisk and oval structure
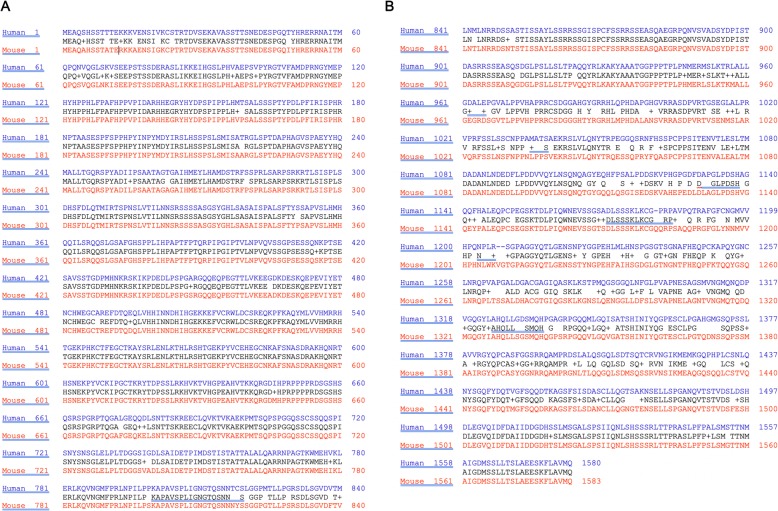
In mice, Gli3 is located on chromosome 13 with 266304 bp in length (NC_000079) (last update 19-OCT-2010). The mRNA sequence is 8427 nt long and comprises 15 exons (NM_008130) (last update: 30-Dec-2019). Between exons 2 and 15 (nt 431-5182) lies the consensus coding sequence which is 4752 nt long and encodes a 1583 AA protein (mouse GLI3-FL/mGLI3-FL) (CCDS36603.1)(NP_032156.2). Using Align Sequences Protein BLAST, we aligned amino acid sequences of mouse Gli3 and human GLI3 (Fig. 3). This revealed the biochemical domains of mouse Gli3-FL are 86% conserved in human Gli3-FL. All domains show high similarity with the exception of the transactivation domain (Similarity of human to mouse: GLI3-R domain: 98%, DNA binding domain: 100%, Cleavage site: 95%, CBP binding domain: 79%, TA2 domain: 68%, TA1 domain: 85%, Total transactivation site: 76%). Interestingly all phosphorylation sites from PKA, GSK3β and βTrCP are 100% conserved between mouse and human GLI3-FL (Fig. 2). This is not surprising since multiple studies show similar function of mouse Gli3-R in comparison to human GLI3-R regarding inhibition of HH signaling [113, 117, 118]. Amino acid sequences that are important for biochemical domains are usually conserved between species. Since the human transactivation domain shows relatively low similarity with the mouse transactivation domain of Gli3-FL, this raises the question whether or not this domain is sufficiently defined in humans yet. Hopefully, future studies will further characterize transactivation site in human GLI3 and possibly identify novel interaction partners at this protein locus.
Fig. 3.
Alignment of human and mouse GLI3/Gli3 protein sequences. Alignment of human (NP_000159.3) and mouse (NP_032156.2) GLI3/Gli3 shows 86 % similarity in amino acid sequence
Processing of GLI3-FL to GLI3-R
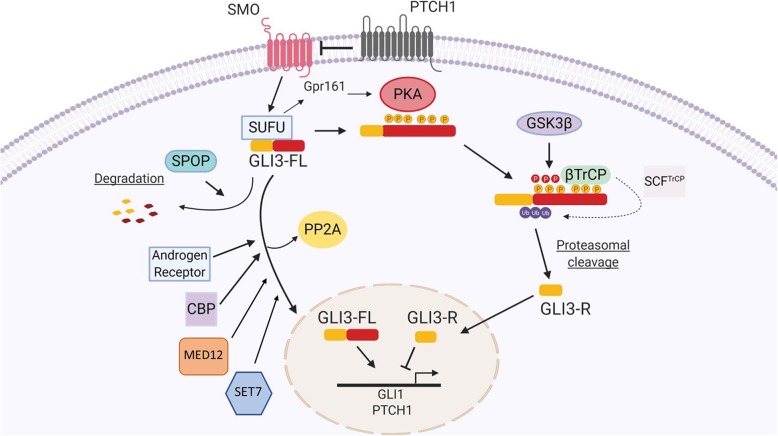
Gli2 and Gli3 show 95% amino acid similarity and both can potentially have HH activator and repressor functions [119]. However, Gli2 proteasomal cleavage is not very efficient and the cleavage product that is produced rapidly degrades while Gli3-R is very stable [120]. Therefore, GLI2 is considered an activator in response to HH signaling while GLI3 is considered a repressor in that context. In the absence of Hedgehog signaling, GLI3-FL is sequestered in the cytoplasm by Sufu (Fig. 2 and Fig. 4) [121]. Sufu recruits Gpr161 which activates Pka and allows it to phosphorylate Gli3 [122]. PKA phosphorylates human GLI3 at serine residues 849, 865, 877, 907, 980 and 1006 (PKA sites: RRXS) (Fig. 2 and Fig. 4) [122]. This leads to additional phosphorylation of GLI3 by GSK3β (at AA 861, 873 and 903)(GSK3β: SXXXpS) [123]. Subsequently, GLI3 binds βTrCP at AA 855, 856, 864 and 894, which recruits Skp1-Cul1-F-box protein-βTrCP complex (SCFTrCP) [124] (Fig. 2 and Fig. 4). This results in ubiquitination of GLI3 at lysine residues 773, 778, 784 and 800 by SCFTrCP resulting in GLI3 processing into its repressor form (GLI3-R) (Fig. 2 and Fig. 4) [124]. Binding of βTrCP was also reported to occur at the N-terminus and at the C-terminus, however this binding is PKA-independent (binding sequence: AA 1-395 and 1100-1595) [124]. GLI3-R translocates to the nucleus where it suppresses HH related functions [125].
Fig. 4.
Regulation of GLI3-FL and GLI3-R. GLI3-FL (but not GLI3-R) is proteasomally degraded upon overexpression of SPOP, while binding by CBP, Androgen receptor, MED12, SET7 and release of PP2A stabilizes GLI3-FL and facilitates its translocation to the nucleus. GLI3-R generation is facilitated by SUFU recruitment of Gpr161, which activates PKA. PKA phosphorylates GLI3 at AA residues 849, 865, 877, 907, 980 and 1006. This phosphorylation leads to hyperphosphorylation by GSK3β (AA 841-880) and binding of βTrCP to GLI3. βTrCP then recruits SCFTrCP which ubiquitinates GLI3 leading to proteasomal cleavage and generation of GLI3-R which can translocate to the nucleus to regulate gene expression (Figure created with BioRender.com)
GLI3-FL, stabilization and degradation
It is generally accepted that GLI3-FL is the transcriptional activator form of GLI3 [62, 126, 127]. However, GLI3-FL has been reported to directly interact with the androgen receptor and stimulation of pancreatic cancer cells with the synthetic androgen “R1881” (methyltrienolone) leads to the nuclear translocation of GLI3-FL and subsequent promoter activation of specific GLI consensus sequences [128]. In this study, the androgen receptor was also shown to stabilize GLI3-FL. Despite the role of Sufu in sequestering Gli3 in the cytoplasm to suppress its transcriptional activity, the presence of Sufu has also been shown to stabilize Gli3 protein [129]. In addition, the Speckle-type POZ protein (SPOP) induces proteasomal degradation of GLI3-FL but not of GLI3-R [129], Protein phosphatase 2A (PP2A) directly binds GLI3-FL, and the degradation of PP2A leads to increased nuclear localization of GLI3-FL [127]. GLI3-FL has also been reported to interact with CBP in a phosphorylation-independent manner at AA 827 and 1132 in the C-terminal end of GLI3. This interaction is important for activation of the GLI1 promoter by GLI3-FL and was only detectable in the presence of SHH [62]. In addition, Mediator of RNA polymerase II transcription subunit 12 (MED12) physically interacts with GLI3 at AA 1090-1596 and facilitates gene transcription upon HH stimulation [130]. Finally, SET domain containing 7 (SET7) increases the stability of GLI3-FL and its ability to bind the GLI1 promoter region upon SHH treatment [131]. In summary, multiple studies have investigated the regulation of GLI3-FL in response to canonical HH signaling. Although this information is very valuable, it does not address the possibility that GLI3-FL is regulated in a non-canonical pathway similar to other GLI family members. Therefore, further studies are needed to determine if GLI3-FL regulation is similar in canonical (HH) and non-canonical signaling pathways.
Role in development, immune system and cancer
Development
The role of GLI3 in regulating development is frequently described. In addition to its importance in brain and lung development, GLI3 is a key player in the manifestation of GCPS, PHS and Tibial Hemimelia, [8, 132, 133]. These genetic diseases are known for the formation of an extra digit (postaxial polydactyly) consistent with the Gli3Δ699 mouse model (mice with mutated Gli3 that have similar phenotypes compared to PHS in humans) [134]. Several mutations of GLI3 which compromise GLI3 function, are tightly linked to these developmental disorders, emphasizing that GLI3 is essential for proper human development. In the following section, we provide a more detailed discussion to further elaborate on the role of GLI3 in development and genetic diseases.
GCPS, PHS and tibial hemimelia
Individuals suffering from GCPS develop postaxial polysyndactyly (limb duplication) of the hands, preaxial polysyndactyly of the feet, and macroencephaly (enlarged head). This genetic disorder is caused by a deletion of a region located on chromosome 7p14. Vortkamp et al (1992) identified GLI3 as the protein responsible for this disorder and the related phenotype [135]. That study reports GLI3 to be present on chromosome 7p13, however, as stated earlier, recent updates correctly identify the location of GLI3 on chromosome 7p14 [4, 108, 109]. SHH is responsible for limb development in the anteroposterior axis [136]. The cause for the polydactyly phenotype is a mutation within the GLI3 protein which disrupts the equilibrium between GLI3-R and GLI3 activator (GLI3-FL) forms, resulting in the lack of negative regulation of SHH signaling. In GCPS, mutations in GLI3 can occur throughout the whole protein and can vary from missense to splicing mutations, although deletions, insertions and translocations have been documented [137]. N-terminally truncated versions of GLI3 protein, which completely or partially lack the zinc finger DNA binding domain, are common in GCPS. There are also reports where families have a missense mutation in His601Arg which is located near the C-terminal end of the 5 zinc finger DNA binding domain [138]. Another reported mutation in GLI3 protein is located at AA 903-934 which are within the transactivation domain [139]. Thus, GCPS-related mutations in GLI3 occur throughout the whole protein and cannot be reduced to one mutation. In addition, the involvement of other proteins in GCPS cannot be ruled out.
PHS is clinically identified by mesoaxial (central) polydactyly and hypothalamic hamartoma (benign tumor of the hypothalamus) [140]. Mutations related to PHS are usually frameshift or nonsense mutations. These mutations lead to a truncated version of GLI3 which is 691 AA long [8]. Transgenic mice expressing truncated Gli3 (699 AA long) show PHS characteristics and this truncated Gli3 exhibits inhibitory functions similar to Gli3-R in the context of Shh signaling [134]. It can therefore be inferred that mutations in PHS lead to expression of a truncated version of Gli3 similar to Gli3-R, mimicking its function and leading to PHS (probably also due to the lack of Gli3-FL). Kang et al (1997) suggested that PHS was caused by a deletion of a guanosine at nt 2023 in exon 12 of GLI3 [8]. This exon spans the cleavage site for GLI3-FL to be processed into the GLI3-R form. Several additional mutations have been identified and can be found between AA 717-1297 [140]. This spans roughly the beginning and the end of the transactivation domain and includes ubiquitination and phosphorylation sites of GLI3 (Fig. 2).
Another clinical condition involving GLI3 is tibial hemimelia, a genetic disorder which leads to hypoplastic or absent tibia [133]. Deimling et al (2016) reported a deletion of exon 7 of GLI3 upstream of the DNA binding domain and the transactivation domain [141]. This deletion leads to a 30 kDA truncated version of GLI3 that is no longer capable of negative regulation of PTCH1 or GLI1 in response to HH signaling. These studies highlight the important role of GLI3 in limb development.
Brain development
In the late 90’s Gli3 was shown to be involved in development of the Dentate gyrus and hippocampus, 2 regions which are responsible for emotional development and memory [142]. In this study XtJ mice show a loss of Homeobox protein Emx1/2 (a transcription factor complex that regulates brain development) expression and show differences in fibroblast growth factor 8 (Fgf8) and bone morphogenetic protein 4 (Bmp4) expression compared to WT mice [142]. Gli3 also plays a role in the development of dorsal telencephalon. In a study that used the extra toes (XtJ) mouse model, an impairment in the development of dorsal di-telencephalic junction was reported [143]. This also led to a decrease in the size of the neocortex and missing parts of the hippocampus. Gli3 also controls growth and expansion of the cerebral cortex. A study using Kinesin-like protein Kif3a (Kif3a) mutant mice (where loss of Kif3a leads to the degradation of primary cilia) found a dysregulation of the expansion of cerebral cortex. The loss of primary cilia disrupts Gli3 processing which changes expression of the Shh target genes cyclin D1 and fibroblast growth factor 15 (fgf15) [144]. Petrova et al (2013) reported that the Gli3-R mediates proliferation in the dorsal and ventral subventricular zone [145]. Gli3-R mediates Shh signaling in astrocytes in the forebrain. Gli3 was also shown to be involved in regulating corpus callosum formation by interacting with Slit homolog 2 protein (Slit2) (a protein important for neural development) and therefore regulating Fgf and Wnt/β-catenin expression [146]. In mice, Gli3-R regulates the differentiation of the upper layer of cortical neurons [147]. Furthermore, using Shh-/- Gli3-/- mice embryos, Gli3 was found to play a role in the development of mature oligodendrocytes [148]. Finally, Gli3-R was shown to suppress V0 and V1 interneurons since restoration of motor neurons and V2 interneurons was detected in Shh-/- Gli3-/- embryos in comparison with single Shh-/- knockout embryos [149]. These studies define an important role for Gli3 in brain development and furthers our understanding of the developmental role of this transcription factor.
Lung development
There is evidence for a role of Gli3 in the embryonic stages of mouse lung development [150]. High levels of Gli3 expression were reported in E11.5 mouse embryos in the tip of the accessory lobe. In addition, using Gli3XtJ mice, Gli3 deficiency at E13.5 led to a reduction in size and changes in the shape of three of the five lung lobes [150]. The role of Gli3 in lung development was confirmed by another group who described the impact of Gli2 deficiency in mice lung development to be substantially increased when GLI3 was knocked out as well. They showed that Gli2-/-Gli3-/- mice embryos lacked lungs, tracheae and esophagi [151]. Therefore, in addition to its role in limb and brain development, Gli3 also regulates lung development.
Role of GLI3 in the immune system
A role for GLI3 in the immune system has been suggested in several studies [152–156]. Based on current knowledge, the regulatory role of GLI3 in the immune system spans both innate and adaptive immunity. GLI3 has been shown to regulate immune cell development and may play a role in inflammation induced by bacterial infections. A role for GLI3 in the immune system was first described in 2005, in which Gli3 regulates the immune response by modulating T cell development [152]. In this report, Gli3 regulated fetal double negative 1 (DN1) to DN2 transition and the progression of developing thymocytes from double negative (DN) to double positive (DP) state was impaired in the absence of Gli3. This regulatory effect of Gli3 occurs after pre-TCR signaling. These studies suggest that Gli3 is important for T cell development. However, whether this results in a biological effect of Gli3 loss on T cell responses to an immunological challenge remains unknown. The Cluster of Differentiation 155 (CD155; also known as poliovirus receptor) is a member of the immunoglobulin superfamily and binds to the leukocyte adhesion molecule DNAM1. Both of these molecules are expressed on the surface of NK cells [154]. Treatment of cells with the Shh ligand induced CD155 expression [155, 156]. Further analysis indicated the core promoter region of CD155 contains candidate GLI binding consensus sequences which, when mutated, results in reduced promoter activation by both Gli1 and Gli3 [156]. This suggests a role for Gli3 in regulating NK cell activation and function in the immune system via regulation of CD155 expression [156]. A role for Gli3 in fetal B cell commitment was suggested in a study where a dose-dependent reduction in CD19+, B220+ and CD19+ B220+ B cell numbers in Gli3 WT, Gli3+/- and Gli3-/- embryos was observed [157]. In addition, the total number of CD93+ B cells and the number of ckit+ CD127+, CD43+ CD19+ HSA+ BP-1+ and CD19+ HSA+ μH+ B cell subsets was significantly reduced in Gli3-/- E18.5 mouse embryos [157]. Taken together, these studies suggest Gli3 as a player in lymphocyte development. However, the biological significance of this reduced lymphocyte populations has yet to be elucidated.
In a study using RAW264.5 mouse macrophages that were challenged with lipopolysaccharide (LPS) to identify novel inducible genes, a muscle specific transcription factor Myoblast determination protein 1 (MyoD) and a major regulator of cardiac development Homeobox protein Nkx-2.5 (Nkx2.5) were reported to be upregulated. More interestingly, the expression of Gli3, but not Gli1 or Gli2, was upregulated in response to LPS suggesting a novel mechanism of regulation of Gli3 by the TLR4 signaling pathway [158]. Although this is the only report to suggest a regulatory role for Gli3 in Tlr4 signaling, it is probable that Gli3 might regulate innate inflammatory effects based on aforementioned studies. Additionally ,in a shRNA library screening during the development of myeloid cells Gli3 was suggested to play a role in cell proliferation [153].
In cancer
GLI3 regulates various biological processes that are important for cancer cell growth and progression. Several studies found that GLI3 regulates anchorage independent growth, proliferation and migration of cancer cell lines [131, 159, 160]. Studies also reported increased GLI3 expression in patient samples compared with healthy donors [160–162]. A role for GLI3 as a tumor suppressor was reported in medulloblastoma and AML [163, 164]. Although these studies are infrequent, this raises the question of how GLI3 can act as both a tumor initiator and tumor suppressor. The following section summarizes data in which GLI3 was identified as either a pro- or anti-cancerous protein and discusses what is known and what needs to be further investigated to delineate the role of GLI3 in cancer.
In glioblastoma, GLI3 was downregulated upon anti-cancer drug treatment [165]. In this study the focal adhesion kinase (FAK) autophosphorylation inhibitor ‘Y15’ effectively decreased tumor growth of the glioblastoma cell lines DBTRG and U87, especially in combination with temozolomide (a chemotherapeutic agent that induces G2/M arrest and apoptosis). In a microarray gene profiling analysis where glioblastoma cell lines DBTRG and U87 were treated with ‘Y15’ in combination with temozolomide, the expression of GLI3 was downregulated [166]. This downregulation was not visible in the presence of either drug alone suggesting that downregulation of GLI3 is a result of synergistic inhibitory effect of this drug combination [166]. It can be inferred that GLI3 downregulation is involved in the synergistic anti-tumor effect of both treatments, however the molecular mechanism resulting in this downregulation remains unclear.
In the earlier described PHS, mutations in GLI3 cause formation of polydactyly of the limbs [167]. However patients suffering from PHS also have a high recurrence of hypothalamic hamartoma, suggesting a role of GLI3 in regulating benign brain tumor formation [167–169]. In a study by Saitsu et al (2008), two somatic mutations in GLI3 in hypothalamic hamartoma patients were identified and showed reduced GLI promoter activity from the floor plate enhancer HNF3b in C3H10T1/2 cells [170]. This suppressor activity of mutated GLI3 might be related to hypothalamic hamartoma formation, however further studies are necessary to confirm the role of GLI3-R and HH signaling in this tumor development.
In addition to the involvement of GLI3 in hamartoma, GLI3 plays a role in Oral squamous cell carcinoma (OSCC) [159]. PCR array results comparing CD44high vs CD44low OSCC cell populations using SCC4 and SCC9 cells revealed a greater than 6-fold increase in GLI3 expression in the CD44high cancer stem cell subtype [159]. This upregulation of GLI3 expression was also confirmed in vivo in OSCC patient samples [159, 171]. In addition, a significant reduction in proliferation and invasion was observed in vitro and a positive correlation between GLI3 expression and tumor size was confirmed in vivo [159]. In this publication GLI3 knockdown had no effect on apoptotic cell death and there was no link between GLI3 expression and metastasis or blood lymphatic or perineural invasion. Although patients with high GLI3 expression seem to survive less, data presented in the Kaplan–Meier blot was not statistically significant. These reports show that GLI3 regulates cancer stem cells in OSCC by regulating relevant markers for EMT. Additionally, GLI3 regulates cell survival and invasion and correlates with tumor size in vivo. However, whether GLI3-FL or GLI3-R are regulating this effect, is not clear and the involvement of classical HH signaling has not been investigated. Since HH signaling is involved in regulating cancer stem cell subtypes in several cancers, it is plausible that SHH regulates this subtype partially via GLI3 in a paracrine manner [15]. However, additional studies are needed in order to reach this conclusion. Since cancer stem cells are resistant to most chemotherapy due to irregular cell division, GLI3 might be a potential target to sensitize those cells to chemotherapeutic agents. For example, since GLI3 negatively regulates S100A9 in OSCC cancer stem cells and this has been shown to induce cell death in cancer cell lines [172], targeted depletion of GLI3 in OSCC might be an effective strategy in the treatment of OSCC.
In addition to OSCC cancer stem cells, gastric cancer stem cells, that were sorted based on CD44+ CD24+ surface expression, showed 80-fold higher GLI3 mRNA expression than the CD44lo CD24lo phenotype which positively correlated with mRNA expression of SHH and PTCH1 [161]. Since multiple studies report the involvement of SHH in gastric cancer [173, 174], it is possible that GLI3 is mediating SHH-induced effects. However additional studies are needed to confirm the specific regulatory role of GLI3 in gastric cancer development and progression. Additional evidence supporting a role for GLI3 as an oncogene was provided by Li et al (2018) who used bladder cancer tissue samples to show higher GLI3 expression in tumor samples vs benign tissue by performing microarray gene expression analysis [160]. Interestingly, GLI3 was identified as a prognostic factor for poor survival in patients suffering from bladder cancer and, consistent with aforementioned studies, GLI3 was shown to regulate cell proliferation, invasion, migration and proteins involved in EMT such as E-cadherin and N-cadherin [131, 159, 160]. miR-7-5p was identified as a negative regulator of GLI3 in bladder cancer [160]. However, no experiments were performed to identify possible downstream targets of GLI3. Additionally, given the role of GLI3 in regulating cancer stem cells in OSCC and gastric cancer, it might be useful to determine the role of GLI3 in regulating bladder cancer stem cell characteristics and survival as well.
Post translational modifications of GLI3-FL (but not GLI3-R) by SET7 has been reported to increase the stability of GLI3-FL and its ability to bind the promoter region of GLI1 upon SHH treatment [131]. SET7 lysine 436 (K436) and -K595 dependent methylation of GLI3-FL regulates cell viability and colony formation of the non-small cell lung cancer (NSCLC) cell line A549 as well. Additionally, A549 xenografts in mice, expressing GLI3-FL with mutations in SET7 methylation sites, showed less migration, invasiveness and decreased tumor volume in comparison to mice injected with A549 xenografts expressing WT GLI3. This supports the oncogenic role of GLI3 and provides information on post-translational modification-dependent tumorigenicity of GLI3 in NSCLC. Furthermore, targeting SET7 may be a viable therapeutic strategy to decrease GLI3’s oncogenic effect in NSCLC as a strategy to antagonize SHH-dependent signaling.
In colon cancer, GLI3 transcripts were significantly higher than in healthy tissue [175]. Treatment of colon carcinoma cell lines RKO and LOVO with GLI3 siRNA led to a significant decrease in cell proliferation. Furthermore, GLI3 levels inversely correlated with p53 levels within those cell lines. Additional evidence from in vitro experiments showed that siGLI3 treatment decreased p53 and MDM2 interaction and reduced ubiquitination of p53 [175]. However, it is unclear if this effect is due to GLI3-FL or GLI3-R and warrants further investigation [175]. In another study, GLI3-FL (but not GLI3-R) induced more anchor-independent growth in the human colorectal cancer cell lines HCT116, HT29, SW480 and DLD-1 which was also visible upon SHH stimulation suggesting GLI3 regulation of colony formation occurs in a paracrine manner as well [12]. In this study, the oncogenic role of GLI3 in solid tumors was further validated when GLI3-FL-overexpressing DLD-1 and HT29 mouse xenografts showed significantly increased tumor formation compared to control cells. Interestingly, GLI3 did not seem to be affected by any known cancer-related signaling molecules such as p53, WNT or MAPK signaling [12].
In pancreatic cancer cells such as PANC-1, targeting GLI3 with siRNA reduced cell viability. Additionally, siRNA mediated GLI3 knockdown appeared to sensitize PANC-1 cells to cyclopamine treatment in vitro [176]. Since cyclopmine’s reduction of cell viability is due to the initiation of pro-apoptotic cell machinery, the question arises as to what degree GLI3 is involved in regulating the apoptotic pathway in pancreatic cancer cells.
In a more recent study, GLI3-FL was suggested to be involved in the induction of castration-resistant prostate cancer (GRPC) through a mutated form of MED12 which is common in prostate cancer [177]. MED12 negatively regulates GLI3-FL activation role by directly interacting with GLI3 at AA 1090-1596 [130]. Due to a mutation in MED12, its constraining activity toward GLI3 is inactive, which leads to activation of SHH-GLI3 related gene expression in the absence of androgen [177]. Since a common therapy for prostate cancer is androgen depletion, and in the presence of mutated MED12 SHH induced GLI3 gene expression is activated when androgen is absent, new or additional therapeutic approaches might be necessary to reduce the risk of prostate cancer relapse. Furthermore, the question remains whether there is a direct interaction between GLI3 and MED12 in prostate cancer. Although this interaction between MED12 and GLI3 is probable, there are no reports of MED12 interacting with GLI3 to regulate HH related gene expression. In addition, it also remains unclear if MED12 is required for GLI3 phosphorylation. Further research is needed to elucidate the mechanism of MED12-GLI3 regulation.
It can cautiously be suggested that GLI3 may play an oncogenic role in germ cell tumors. In a study by Kuleszo et al (2017), eight germ cell tumors from children were used to perform genomic profiling and GLI3 was identified to have additional copies of its gene in the germ cell tumor group compared with healthy individuals [178]. Although no experiments were performed that directly identify GLI3 as an oncogenic protein in germ cell tumors, the higher GLI3 copy numbers in germ cell tumors is indicative of more HH activation and SHH related gene expression which is known to drive tumor growth and progression. The presence of higher copies of SMO and SHH, in addition to lower copy numbers of PTCH1 (which inhibits SMO activity) in germ cell tumors are another supporting fact of the involvement of HH signaling in germ cell cancer formation.
In addition to regulating proliferation and invasiveness in cancer, GLI3 was also reported to modulate vacuole membrane protein (VMP1), a known regulator of autophagy [179]. Lo Ré et al (2012) reported that oncogenic KRAS (KRASG12D) leads to activation of VMP1 by signaling through PI3K-AKT1-GLI3. In this pathway, GLI3 physically interacts with p300 which facilitates binding of GLI3 to the promoter region of VMP1. GLI3 mRNA expression was upregulated upon transfection with oncogenic KRASG12D and co-expression of GLI3 with constitutively active forms of PI3K and AKT1 showed higher VMP1 promoter activity. These studies show the regulation and requirement for GLI3 in autophagy; however, it is unclear which transcription factor is involved in regulating GLI3 expression downstream of AKT1. In addition, it would be interesting to determine if post translational modifications of GLI3 are required for binding and activating VMP1 promoter activity. This could be accomplished by targeting known regulators of GLI3 such as PKA or GSK3β, in addition to activating this pathway. Additionally, it is unclear if GLI3-FL or GLI3-R is responsible for the regulation of VMP1 and the biochemical domains mediating the interaction between GLI3 and p300. Depending on the binding site for the interaction between GLI3 and p300, this may determine if GLI3-FL or GLI3-R regulates the KRASG12D-PI3K-AKT1-VMP1 pathway.
In addition to the oncogenic role of GLI3 in solid tumors, GLI3 was shown to play a similar role in hematologic malignancies. Elevated GLI3 expression was reported in Hodgkin lymphoma cell lines [180]. This was validated using immunohistochemistry in primary patient biopsies where GLI3 was found in Hodgkin/Reed Sternberg cells. GLI3 was also suggested to pay a role in Diffuse large B cell lymphoma (DLBCL). This is an aggressive lymphoma that is divided into 2 subtypes based on gene expression profiling studies: activated B-cell (ABC) and germinal center B-cell (GCB) subtypes. GLI3 expression was examined in a cohort of DLBCL cell lines and was found to be elevated in cell lines belonging to the GCB DLBCL subgroup [181]. Using gene expression datasets using DLBCL patient samples, the authors also report increased GLI3 expression in the GCB subtype. Knockdown of GLI3 reduced cell proliferation suggesting that GLI3 promotes cell growth in GCB DLBCL. Further characterization of the biological role of GLI3 in these cancers will enhance our understanding of the biological significance of GLI3.
Although rare, anti-cancerous activity of GLI3 has been reported as well. GLI3 was found to be present in 94% of neuronal differentiation and glial and neuronal differentiation medulloblastoma, while it could not be detected in any of the differentiation-free medulloblastoma [163]. Additionally, patients with differentiation free medulloblastoma showed significantly less survival. This suggests a role for GLI3 as a prognostically favorable factor in medulloblastoma. The role of SHH in medulloblastoma has been reported and therapeutic success has been shown after inhibiting HH signaling [182, 183]. In theory, as a result of inhibition of HH signaling, GLI3-R should accumulate within the cell and negatively regulate HH-related gene expression and therefore cancer growth. However, whether the presence of GLI3-R form specifically correlates with a higher overall and event free survival has yet to be determined. Additionally a number of studies have reported that GLI transcription factors can be regulated in a HH independent manner [80, 85, 95, 105, 184–187]. Therefore, this raises the possibility that GLI3 may regulate gene expression in a HH-independent manner to mediate its anti-cancerous functions.
Another study reported GLI3 as a tumor suppressor protein using bone marrow from AML patients. Both GLI3-FL and GLI3-R were shown to be expressed at significantly lower levels in AML patient samples [164]. Based on their data, the absence of GLI3 in AML is due to hypermethylation of its promoter region. Global demethylation showed an increase in protein expression of both GLI3-FL and GLI3-R in vitro and ex vivo which correlated with decreased proliferation of the AML cell lines (K562 and KG1a) and primary AML blasts and with mouse survival after K562 xenograft transplantation. This increase in GLI3-FL and GLI3-R was also visible when AML cell lines were treated with the HH inhibitor PF-04449913 (a SMO inhibitor). Therefore, it might be worth investigating whether PF-04449913 might regulate demethylation of GLI3 promoter in its reported synergistic effect with decitabine in AML [164]. A cross-talk between GLI3-R, AKT1 and ERK1/2 protein has also been reported. Overexpression of GLI3-R negatively correlated with AKT1 but positively correlated with ERK1/2 [188]. AKT1 was reported to be important for GLI3-R effect on negatively influencing cell proliferation [188]. However, it was not determined if GLI3-R regulates proliferation through ERK1/2 as well. ERK1/2 is known to activate MAPK pathway however to what degree GLI-R is involved in this pathway has not been investigated. It might also be of interest to determine the role of GLI3-AKT1 axis in response to HH inhibitor treatment such as PF-04449913 or cyclopamine and if demethylation increased the activity of this pathway.
Most studies that have investigated GLI3 in cancer described the cancer supporting role of GLI3 to occur by positively regulating proliferation, survival and invasiveness. Reports suggesting GLI3’s tumor suppressive role have been rare but raise the question where the contradictory attributes of GLI3 in cancer come from. It is possible the role of GLI3 in cancer is tissue specific. In addition, the majority of studies discussed in this part of the review, describe a role for total GLI3 levels in regulating cancer cell growth and progression. However, it is unknown if it is GLI3-FL or GLI3-R that regulates these effects. This is possibly hindered by the lack of antibodies that reliably identify GLI3-FL or GLI3-R (Matissek et al, unpublished observation). Studies that focused on these two forms of GLI3 have suggested that GLI3-FL induces cancerous behavior while GLI3-R reduces cancer associated attributes [12, 131, 164, 177]. However, to date, there is only one study that solely compares the effect of GLI3-FL vs GLI3-R on cancerous characteristics [189]. In this study, GSK3 beta activity was increased which induced higher concentrations of GLI3-R and an anti-cancerous effect.
Based on previous mentioned work by Chaudhry et al (2015), hypermethylation of GLI3 promoter region leads to a loss of GLI3 and tumor suppressor function in AML [164]. In addition to epigenetic modifications that suppress GLI3 tumor function, it might also be that a disruption of GLI3-FL/GLI3-R equilibrium within the cell leads to a cancerous phenotype and tumor growth. Different expression and activation levels of GLI3-FL regulators (SPOP, androgen receptor, PP2A, CBP, SET7, MED12) or regulators of GLI3-R (PKA, GSK3β, βTrCP) in cancer cells in comparison to healthy cells might also promote cancer cell growth and survival.
GLI3 levels also positively correlated with higher expression of cancer stem cell markers and shRNA-mediated knockdown of GLI3 led to a decrease in the CD44high population (cancer stem cells) while the CD44low population was unaffected in OSCC [159]. The small cancer stem cell population within a tumor has been shown to be very potent in inducing tumor growth in OSCC [190]. Therefore, GLI3’s tumorigenic role may be due to its regulation of cancer stem cell survival. However, whether GLI3-FL or GLI3-R regulates this effect is also not fully explored. There are multiple HH inhibitors (cyclopamine, PF-04449913, GDC0449, Saridegib, Vismodegib) that showed promising results in fighting HH-driven cancers. These inhibitors target SMO to inhibit HH-related gene expression. However, since HH-independent regulation of GLI transcription factors has been reported, this might rescue the effects induced by SMO inhibition [191]. Therefore, therapies such as GANT61, which inhibit binding of GLI1 and GLI2 transcription factors to DNA, are promising since they act directly on GLI but not on SMO [192]. GLI3 expression was not affected by GANT61, therefore, targeting modulators of GLI3-R by making them more active, might decrease PTCH1-SMO-independent HH gene expression and increase the therapeutic effect of GANT61 treatment [98]. Furthermore, a thorough understanding of the signaling pathways that regulate GLI3 independent of HH will allow therapeutic targeting of GLI3 by targeting other molecular regulators of this protein.
MicroRNA (miRNA) and GLI3
As described earlier, GLI3 can interact with several different proteins, an interaction that is required for either stabilization or degradation of GLI3 protein. However, proteins are not the only molecules involved in regulating GLI3. Several lines of evidence have been reported in which GLI3 is regulated at the RNA level by microRNAs which are described below. MicroRNAs were reported to bind complementary sequences of GLI3 RNA and negatively regulate its gene expression.
In cancer, liver fibrosis and spermatogenesis
GLI3 was shown to be regulated by miR-7-5p in bladder cancer tissue. A significant upregulation of GLI3 in bladder cancer was reported by microarray gene expression profiling. Using the online software (TargetScan, PITA, miRanda) the authors found that miR7-5p was the most potent candidate for GLI3 regulation and later confirmed its regulatory role using in vitro experiments. In those studies, a physical interaction between miR-7-5p and GLI3 was identified (GLI3 interaction site: GUCUUCCA), which leads to a negative regulation of GLI3 and to a less cancerous phenotype of bladder cancer cells [160].
In other studies, miR494 and 506 were shown to regulate tGLI3 oncogenic function in pancreatic and cervical cancers [162]. In pancreatic cancer, GLI3 was regulated by miR-494 [162]. GLI3 mRNA expression was significantly upregulated in tissue from bladder cancer patients and there was an inverse correlation between GLI3 and miR-494. Furthermore, miR-494 negative regulation of GLI3 was confirmed in vitro in which inhibition of GLI3 by miR-494 led to a decrease in cell viability and cancer cell migration [162]. A similar negative regulation of GLI3 was also detected by miR-506 in cervical cancer [193]. This was reported in both primary patient biopsy samples and in cervical cancer cell lines using western blotting. GLI3 protein was increased upon miR-506 inhibition which led to reduced apoptosis and increased proliferation. In addition, mice xenografts with Casci cell stably expressing sh-miR-506 showed significantly higher tumor weights in comparison to control mice.
In addition to their role in tumorigenesis, micro RNAs appear to regulate GLI3 in liver fibrosis. Both miR-152 and miR-378a-3p have been shown to negatively regulate GLI3 in liver fibrosis in vitro and in vivo [194, 195]. These studies used the LX-2 hepatic stellate cells where they overexpressed miR-152 or miR-378a-3p and found a reduction in GLI3 mRNA and protein expression. Using rodent models of liver fibrosis where fibrosis was chemically induced with CCI4 in mice, GLI3 expression was increased (miR378a-3p downregulated) in tissue isolated from animals suffering from liver fibrosis suggesting GLI3 positively regulates liver fibrosis.
The role of GLI3 in spermatogenesis in mice was established in 1997 [196]. However, in 2016, a role for miR-133b in regulating Sertoli cells by targeting GLI3 was discovered [197]. miR-133b was significantly upregulated in Sertoli cells from patients suffering from Sertoli-cell-only syndrome (SCOS) and using TargetScan, GLI3 was identified as a potential target for miR-133b. This was confirmed by in vitro experiments showing the regulatory role of miR-133b in which an increase in miR-133b led to reduction of GLI3 mRNA and protein expression [197].
Conclusions
The importance of HH signaling in many biological pathways such as those related to development, cancer and inflammation, has been frequently reported. The involvement of HH signaling in positive regulation of these biological functions has focused on GLI1 and GLI2 which are known activators of HH signaling. Their role as a positive regulator in non-canonical HH pathways has been frequently described as well. However, based on this review, GLI3 also shows high potential in regulating non-canonical pathways positively. In addition to early reports of the role of GLI3 in the development of the brain, lungs, sperm and in genetic diseases, the biological significance of GLI3 was also suggested in diseases such as liver fibrosis, cancer, and in the immune system. In cancer, GLI3’s behavior seems to be bipolar since it was linked to cancer promoting and inhibitory effect in cells which can be explained by GLI3-FL activator and GLI3-R inhibitory function in gene expression. In addition, recent research shows that negative regulation of GLI3 through microRNA reverses cancerous behavior which emphasizes GLI3 as a potential oncogenic target in cancer therapy. Future studies focusing on determining the role of GLI3 in the immune response and in cancer will clarify its biological significance and lay the foundation to target this molecule to reprogram immune and cancer cells.
Acknowledgements
Not applicable
Abbreviations
- ABC
Activated B cell
- AML
Acute Myeloid Leukemia
- Bmp4
Bone morphogenetic protein 4
- Boc
Brother of Cdo
- CBP
CREB-binding-protein
- CD155
Cluster of differentiation 155
- Cdo
CAM-related/downregulated by oncogenes
- CDS
Consensus coding sequence
- Ci
Cubitus interruptus
- Ck-1/CK-1
Casein kinase 1 (mouse/human)
- CK2a
Casein kinase 2a
- CLL
Chronic lymphocytic leukemia
- Cos-2
Costal-2
- Dhh/DHH
Desert Hedgehog (mouse/human)
- DLBCL
Diffuse large B cell lymphoma
- DN
Double negative
- DNAM1
CD226, Cluster of differentiation 226
- Dnchc2
Dynein cytoplasmic heavy chain 2
- DP
Double positive
- DYRK1/DYRK2
Dual specificity tyrosine-phosphorylation-regulated kinase ½
- EMT
Epithelial-to-mesenchymal transition
- Emx1/2
Homeobox protein Emx1/2
- Evc/Evc2
Ellis-van Creveld syndrome protein/Ellis-van Creveld syndrome protein 2 complex
- FAK
Focal adhesion kinase
- Fgf15
Fibroblast growth factor 15
- Fgf8
Fibroblast growth factor 8
- Fu
Fused
- Gas1
Growth-arrest-specific 1
- GCB
Germinal center B cell
- GCPS
Greig cephalopolysyndactyly
- Gli3-FL/GLI3-FL
Full length GLI3 protein (mouse/human)
- Gli3-R/GLI3-R
Repressor form of GLI3 protein (mouse/human)
- Gpr161/GPR161
G-protein coupled receptor 161 (mouse/human)
- Gsk3β/GSK3β
Glycogen synthase kinase 3 beta (mouse/human)
- Hh/HH
Hedgehog (mouse/human)
- Hhip
Hh interaction protein
- HSC
Hedgehog Signaling Complex
- IFT
Intraflagellar transport
- Ift172
Intraflagellar transport protein 172
- Ihh/IHH
Indian Hedgehog (mouse/human)
- Kif3a
Kinesin-like protein Kif3a
- KIF3A
Kinesin-like-protein KIF3A
- Kif7
Kinesin-like protein Kif7
- MyoD
Myoblast determination protein
- Nkx2.5
Homeobox protein Nkx-2
- OSCC
Oral squamous cell carcinoma
- PHS
Pallister-Hall Syndrome
- Pka/PKA
Protein kinase A (mouse/human)
- PKC
Protein kinase C
- PP2A
Protein phosphatase 2A
- Ptc
Patched (Drosophila)
- Ptch1/PTCH1
Patched 1 (mouse/human)
- Rab23
Ras-related protein Rab23
- SCFβTrCP
Skp1-Cul1-F-box protein – βTrCP complex
- SCOS
Sertoli-cell-only syndrome
- SET7
SET domain containing 7
- Shh/SHH
Sonic Hedgehog (mouse/human)
- Slit2
Slit homolog 2 protein
- Smo/SMO
Smoothened (mouse/human)
- SPOP
Speckle-type POZ protein
- Sufu/SUFU
Suppressor of Fused (mouse/human)
- TA1
Transactivation domain 1
- TA2
Transactivation domain 2
- TME
Tumor microenvironment
- VMP1
Vacuole membrane protein 1
- βTrCP
Beta-Transducing Repeat containing Protein
Authors’ contributions
All authors read and approved the final manuscript.
Funding
This research was supported by a NIH COBRE Center of Integrated Biomedical and Bioengineering Research (CIBBR, P20 GM113131), through an institutional development award (IDeA) from the National Institute of General Medical Sciences.
Availability of data and materials
Not applicable
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Stephan J. Matissek, Email: sjm1081@wildcats.unh.edu
Sherine F. Elsawa, Email: sherine.elsawa@unh.edu
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12964-020-00540-x.
References
- 1.Kinzler KW, Bigner SH, Bigner DD, Trent JM, Law ML, O’Brien SJ, et al. Identification of an amplified, highly expressed gene in a human glioma. Science. 1987;236:70–73. doi: 10.1126/science.3563490. [DOI] [PubMed] [Google Scholar]
- 2.Ruppert JM, Kinzler KW, Wong AJ, Bigner SH, Kao FT, Law ML, et al. The GLI-Kruppel family of human genes. Mol Cell Biol. 1988;8:3104LP–313113. doi: 10.1128/mcb.8.8.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruppert JM, Vogelstein B, Arheden K, Kinzler KW. GLI3 encodes a 190-kilodalton protein with multiple regions of GLI similarity. Mol Cell Biol. 1990;10:5408 LP–5405415. doi: 10.1128/mcb.10.10.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zou Q, Tian Z, Zheng J, Zhi X, Du X, Shu J, et al. A novel missense in GLI3 possibly affecting one of the zinc finger domains may lead to postaxial synpolydactyly: case report. BMC Med Genet. 2019;20:174. doi: 10.1186/s12881-019-0889-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vortkamp A, Gessler M, Grzeschik KH. GLI3 zinc-finger gene interrupted by translocations in Greig syndrome families. Nature. 1991;352:539–540. doi: 10.1038/352539a0. [DOI] [PubMed] [Google Scholar]
- 6.Wild A, Kalff-Suske M, Vortkam A, Bornholdt D, König R, Grzeschik KH. Point mutations in human GLI3 cause Greig syndrome. Hum Mol Genet. 1997;6:1979–1984. doi: 10.1093/hmg/6.11.1979. [DOI] [PubMed] [Google Scholar]
- 7.Marigo V, Johnson RL, Vortkamp A, Tabin CJ. Sonic hedgehog differentially regulates expression of GLI and GLI3 during limb development. Dev Biol. 1996;180:273–283. doi: 10.1006/dbio.1996.0300. [DOI] [PubMed] [Google Scholar]
- 8.Kang S, Graham JM, Olney AH, Biesecker LG. GLI3 frameshift mutations cause autosomal dominant Pallister-Hall syndrome. Nat Genet. 1997;15:266–268. doi: 10.1038/ng0397-266. [DOI] [PubMed] [Google Scholar]
- 9.Sasaki H, Nishizaki Y, Hui C, Nakafuku M, Kondoh H. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development. 1999;126:3915 LP–3913924. doi: 10.1242/dev.126.17.3915. [DOI] [PubMed] [Google Scholar]
- 10.Paganelli AR, Ocaña O, Prat MI, Franco PG, López SL, Morelli L, et al. The Alzheimer-related gene presenilin-1 facilitates sonic hedgehog expression in Xenopus primary neurogenesis. Mech Dev. 2001;107:119–131. doi: 10.1016/s0925-4773(01)00458-0. [DOI] [PubMed] [Google Scholar]
- 11.Renault MA, Roncalli J, Tongers J, Misener S, Thorne T, Jujo K, et al. The hedgehog transcription factor Gli3 modulates angiogenesis. Circ Res. 2009;105:818–26. [DOI] [PMC free article] [PubMed]
- 12.Iwasaki H, Nakano K, Shinkai K, Kunisawa Y, Hirahashi M, Oda Y, et al. Hedgehog Gli3 activator signal augments tumorigenicity of colorectal cancer via upregulation of adherence-related genes. Cancer Sci. 2013;104:328–336. doi: 10.1111/cas.12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang K, Che S, Pan C, Su Z, Zheng S, Yang S, et al. The SHH/Gli axis regulates CD90-mediated liver cancer stem cell function by activating the IL6/JAK2 pathway. J Cell Mol Med. 2018;22:3679–3690. doi: 10.1111/jcmm.13651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurita S, Mott JL, Almada LL, Bronk SF, Werneburg NW, Sun SY, et al. GLI3-dependent repression of DR4 mediates hedgehog antagonism of TRAIL-induced apoptosis. Oncogene. 2010;29:4848–4858. doi: 10.1038/onc.2010.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. [DOI] [PubMed]
- 16.McMahon AP, Ingham PW, Tabin CJ. Developmental roles and clinical significance of Hedgehog signaling. Curr Top Dev Biol. 2003;53:1–114. doi: 10.1016/s0070-2153(03)53002-2. [DOI] [PubMed] [Google Scholar]
- 17.Tukachinsky H, Lopez LV, Salic A. A mechanism for vertebrate Hedgehog signaling: Recruitment to cilia and dissociation of SuFu-Gli protein complexes. J Cell Biol. 2010;191:415–428. doi: 10.1083/jcb.201004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhardwaj G, Murdoch B, Wu D, Baker DP, Williams KP, Chadwick K, et al. Sonic hedgehog induces the proliferation of primitive human hematopoietic cells via BMP regulation. Nat Immunol. 2001;2:172–180. doi: 10.1038/84282. [DOI] [PubMed] [Google Scholar]
- 19.Outram SV, Varas A, Pepicelli CV, Crompton T. Hedgehog signaling regulates differentiation from double-negative to double-positive thymocyte. Immunity. 2000;13:187–197. doi: 10.1016/s1074-7613(00)00019-4. [DOI] [PubMed] [Google Scholar]
- 20.Wang XZ, Zhang HH, Qian YL, Tang LF. Sonic hedgehog (Shh) and CC chemokine ligand 2 signaling pathways in asthma [Internet] J Chinese Med Assoc. 2019;82:343–350. doi: 10.1097/JCMA.0000000000000094. [DOI] [PubMed] [Google Scholar]
- 21.Mazzoccoli G, Keshavarzian A, Vinciguerra M. Hedgehog signaling keeps liver clock in check. J Hepatol. 2019;70:1054–1056. doi: 10.1016/j.jhep.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 22.Finco I, LaPensee CR, Krill KT, Hammer GD. Hedgehog signaling and steroidogenesis. Annu Rev Physiol. 2015;77:105–129. doi: 10.1146/annurev-physiol-061214-111754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porter JA, Young KE, Beachy PA. Cholesterol modification of hedgehog signaling proteins in animal development. Science. 1996;274:255–259. doi: 10.1126/science.274.5285.255. [DOI] [PubMed] [Google Scholar]
- 24.Guerrero I, Chiang C. A conserved mechanism of Hedgehog gradient formation by lipid modifications. Trends Cell Biol. 2007;17:1–5. doi: 10.1016/j.tcb.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 25.Zeng X, Goetz JA, Suber LM, Scott WJ, Schreiner CM, Robbins DJ. A freely diffusible form of Sonic hedgehog mediates long-range signalling. Nature. 2001;411:716–720. doi: 10.1038/35079648. [DOI] [PubMed] [Google Scholar]
- 26.Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, et al. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 27.Bale AE. Hedgehog signaling and human disease. Annu Rev Genomics Hum Genet. 2002;3:47–65. doi: 10.1146/annurev.genom.3.022502.103031. [DOI] [PubMed] [Google Scholar]
- 28.Vortkamp A, Lee K, Lanske B, Segre GV, Kronenberg HM, Tabin CJ. Regulation of rate of cartilage differentiation by Indian Hedgehog and PTH-related protein. Science. 1996;273:613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- 29.Bitgood MJ, Shen L, McMahon AP. Sertoli cell signaling by Desert hedgehog regulates the male germline. Curr Biol. 1996;6:298–304. doi: 10.1016/s0960-9822(02)00480-3. [DOI] [PubMed] [Google Scholar]
- 30.Parmantier E, Lynn B, Lawson D, Turmaine M, Namini SS, Chakrabarti L, et al. Schwann cell-derived desert hedgehog controls the development of peripheral nerve sheaths. Neuron. 1999;23:713–724. doi: 10.1016/s0896-6273(01)80030-1. [DOI] [PubMed] [Google Scholar]
- 31.Bitgood MJ, McMahon AP. Hedgehog and Bmp genes are coexpressed at many diverse sites of cell-cell interaction in the mouse embryo. Dev Biol. 1995;172:126–138. doi: 10.1006/dbio.1995.0010. [DOI] [PubMed] [Google Scholar]
- 32.Pathi S, Pagan-Westphal S, Baker DP, Garber EA, Rayhorn P, Bumcrot D, et al. Comparative biological responses to human Sonic, Indian, and Desert hedgehog. Mech Dev. 2001;106:107–117. doi: 10.1016/s0925-4773(01)00427-0. [DOI] [PubMed] [Google Scholar]
- 33.Ogden SK, Ascano M, Stegman MA, Robbins DJ. Regulation of Hedgehog signaling: a complex story [Internet] Biochem Pharmacol. 2004;67:805–814. doi: 10.1016/j.bcp.2004.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang W, Zhao Y, Tong C, Wang G, Wang B, Jia J, et al. Hedgehog-regulated Costal2-kinase complexes control phosphorylation and proteolytic processing of cubitus interruptus. Dev Cell. 2005;8:267–278. doi: 10.1016/j.devcel.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 35.Bangs F, Anderson KV. Primary cilia and Mammalian Hedgehog signaling. Cold Spring Harb Perspect Biol. 2017;9:a028175. doi: 10.1101/cshperspect.a028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci U S A. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hao L, Scholey JM. Intraflagellar transport at a glance. J Cell Sci. 2009;122:889–892. doi: 10.1242/jcs.023861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carpenter BS, Barry RL, Verhey KJ, Allen BL. The heterotrimeric kinesin-2 complex interacts with and regulates GLI protein function. J Cell Sci. 2015;128:1034–1050. doi: 10.1242/jcs.162552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liem KF, He M, Ocbina PJR, Anderson KV. Mouse Kif7/Costal2 is a cilia-associated protein that regulates Sonic hedgehog signaling. Proc Natl Acad Sci U S A. 2009;106:13377–13382. doi: 10.1073/pnas.0906944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pedersen LB, Akhmanova A. Kif7 keeps cilia tips in shape. Nat Cell Biol. 2014;16:623–625. doi: 10.1038/ncb2997. [DOI] [PubMed] [Google Scholar]
- 41.Allen BL, Tenzen T, McMahon AP. The Hedgehog-binding proteins Gas1 and Cdo cooperate to positively regulate Shh signaling during mouse development. Genes Dev. 2007;21:1244–1257. doi: 10.1101/gad.1543607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bishop B, Aricescu AR, Harlos K, O’Callaghan CA, Jones EY, Siebold C. Structural insights into hedgehog ligand sequestration by the human hedgehog-interacting protein HHIP. Nat Struct Mol Biol. 2009;16:698–703. doi: 10.1038/nsmb.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Zhou Z, Walsh CT, McMahon AP. Selective translocation of intracellular Smoothened to the primary cilium in response to Hedgehog pathway modulation. Proc Natl Acad Sci U S A. 2009;106:2623–2628. doi: 10.1073/pnas.0812110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tukachinsky H, Petrov K, Watanabe M, Salic A. Mechanism of inhibition of the tumor suppressor Patched by Sonic Hedgehog. Proc Natl Acad Sci U S A. 2016;113:E5866–E5875. doi: 10.1073/pnas.1606719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goetz SC, Anderson KV. The primary cilium: A signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pietrobono S, Gagliardi S, Stecca B. Non-canonical hedgehog signaling pathway in cancer: Activation of GLI transcription factors beyond smoothened. Front Genet. 2019;10:556. doi: 10.3389/fgene.2019.00556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dunaeva M, Michelson P, Kogerman P, Toftgard R. Characterization of the physical interaction of Gli proteins with SUFU proteins. J Biol Chem. 2003;278:5116–5122. doi: 10.1074/jbc.M209492200. [DOI] [PubMed] [Google Scholar]
- 48.Li J, Wang C, Wu C, Cao T, Xu G, Meng Q, et al. PKA-mediated Gli2 and Gli3 phosphorylation is inhibited by Hedgehog signaling in cilia and reduced in Talpid3 mutant. Dev Biol. 2017;429:147–157. doi: 10.1016/j.ydbio.2017.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Y, Yue S, Xie L, Pu XH, Jin T, Cheng SY. Dual phosphorylation of suppressor of fused (Sufu) by PKA and GSK3β regulates its stability and localization in the primary cilium. J Biol Chem. 2011;286:13502–13511. doi: 10.1074/jbc.M110.217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Price MA, Kalderon D. Proteolysis of the Hedgehog signaling effector Cubitus interruptus requires phosphorylation by Glycogen Synthase Kinase 3 and Casein Kinase 1. Cell. 2002;108:823–835. doi: 10.1016/s0092-8674(02)00664-5. [DOI] [PubMed] [Google Scholar]
- 51.Wang B, Li Y. Evidence for the direct involvement of βTrCP in Gli3 protein processing. Proc Natl Acad Sci U S A. 2006;103:33–38. doi: 10.1073/pnas.0509927103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi Q, Li S, Li S, Jiang A, Chen Y, Jiang J. Hedgehog-induced phosphorylation by CK1 sustains the activity of Ci/Gli activator. Proc Natl Acad Sci U S A. 2014;111:E5651–E5660. doi: 10.1073/pnas.1416652111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Y, Sasai N, Ma G, Yue T, Jia J, Briscoe J, et al. Sonic Hedgehog dependent Phosphorylation by CK1α and GRK2 is required for ciliary accumulation and activation of smoothened. PLoS Biol. 2011;9:1–16. [DOI] [PMC free article] [PubMed]
- 54.Varghese RT, Young S, Pham L, Liang Y, Pridham KJ, Guo S, et al. Casein kinase 1 epsilon regulates glioblastoma cell survival. Sci Rep. 2018;8:1–13. doi: 10.1038/s41598-018-31864-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jia J, Zhang L, Zhang Q, Tong C, Wang B, Hou F, et al. Phosphorylation by double-time/CKIε and CKIα targets cubitus interruptus for Slimb/β-TRCP-mediated proteolytic processing. Dev Cell. 2005;9:819–830. doi: 10.1016/j.devcel.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 56.Schrader EK, Harstad KG, Holmgren RA, Matouschek A. A Three-part signal governs differential processing of Gli1 and Gli3 proteins by the proteasome. J Biol Chem. 2011;286:39051–39058. doi: 10.1074/jbc.M111.274993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeng H, Jia J, Liu A. Coordinated translocation of mammalian gli proteins and suppressor of fused to the primary cilium. PLoS One. 2010;5:e15900. doi: 10.1371/journal.pone.0015900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kovacs JJ, Whalen EJ, Liu R, Xiao K, Kim J, Chen M, et al. β-arrestin-mediated localization of smoothened to the primary cilium. Science. 2008;320:1777–1781. doi: 10.1126/science.1157983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang C, Chen W, Chen Y, Jiang J. Smoothened transduces Hedgehog signal by forming a complex with Evc/Evc2. Cell Res. 2012;22:1593–1604. doi: 10.1038/cr.2012.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Caparrós-Martín JA, Valencia M, Reytor E, Pacheco M, Fernandez M, Perez-Aytes A, et al. The ciliary EVC/EVC2 complex interacts with smo and controls hedgehog pathway activity in chondrocytes by regulating Sufu/Gli3 dissociation and Gli3 trafficking in primary cilia. Hum Mol Genet. 2013;22:124–139. doi: 10.1093/hmg/dds409. [DOI] [PubMed] [Google Scholar]
- 61.Wen X, Lai CK, Evangelista M, Hongo J-A, de Sauvage FJ, Scales SJ. Kinetics of Hedgehog-dependent full-length Gli3 accumulation in primary cilia and subsequent degradation. Mol Cell Biol. 2010;30:1910–1922. doi: 10.1128/MCB.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dai P, Akimaru H, Tanaka Y, Maekawa T, Nakafuku M, Ishii S. Sonic hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J Biol Chem. 1999;274:8143–8152. doi: 10.1074/jbc.274.12.8143. [DOI] [PubMed] [Google Scholar]
- 63.Niewiadomski P, Kong JH, Ahrends R, Ma Y, Humke EW, Khan S, et al. Gli protein activity is controlled by multisite phosphorylation in vertebrate hedgehog signaling. Cell Rep. 2014;6:168–181. doi: 10.1016/j.celrep.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kinzler KW, Ruppert JM, Bigner SH, Vogelstein B. The GLI gene is a member of the Kruppel family of zinc finger proteins. Nature. 1988;332:371–374. doi: 10.1038/332371a0. [DOI] [PubMed] [Google Scholar]
- 65.Shimokawa T, Tostar U, Lauth M, Palaniswamy R, Kasper M, Toftgård R, et al. Novel human glioma-associated oncogene 1 (GLI1) splice variants reveal distinct mechanisms in the terminal transduction of the hedgehog signal. J Biol Chem. 2008;283:14345–14354. doi: 10.1074/jbc.M800299200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lo HW, Zhu H, Cao X, Aldrich A, Ali-Osman F. A novel splice variant of GLI1 that promotes glioblastoma cell migration and invasion. Cancer Res. 2009;69:6790–6798. doi: 10.1158/0008-5472.CAN-09-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tanimura A, Dan S, Yoshida M. Cloning of novel isoforms of the human Gli2 oncogene and their activities to enhance tax-dependent transcription of the human T-cell leukemia virus type 1 genome. J Virol. 1998;72:3958–3964. doi: 10.1128/jvi.72.5.3958-3964.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Babu D, Fanelli A, Mellone S, Muniswamy R, Wasniewska M, Prodam F, et al. Novel GLI2 mutations identified in patients with combined pituitary hormone deficiency (CPHD): evidence for a pathogenic effect by functional characterization. Clin Endocrinol. 2019;90:449–456. doi: 10.1111/cen.13914. [DOI] [PubMed] [Google Scholar]
- 69.Roessler E, Ermilov AN, Grange DK, Wang A, Grachtchouk M, Dlugosz AA, et al. A previously unidentified amino-terminal domain regulates transcriptional activity of wild-type and disease-associated human GLI2. Hum Mol Genet. 2005;14:2181–2188. doi: 10.1093/hmg/ddi222. [DOI] [PubMed] [Google Scholar]
- 70.Tojo M, Kiyosawa H, Iwatsuki K, Nakamura K, Kaneko F. Expression of the GLI2 oncogene and its isoforms in human basal cell carcinoma. Br J Dermatol. 2003;148:892–897. doi: 10.1046/j.1365-2133.2003.05284.x. [DOI] [PubMed] [Google Scholar]
- 71.Stecca B, Mas C, Clement V, Zbinden M, Correa R, Piguet V, et al. Melanomas require HEDGEHOG-GLI signaling regulated by interactions between GLI1 and the RAS-MEK/AKT pathways. Proc Natl Acad Sci U S A. 2007;104:5895–5900. doi: 10.1073/pnas.0700776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seto M, Ohta M, Asaoka Y, Ikenoue T, Tada M, Miyabayashi K, et al. Regulation of the hedgehog signaling by the mitogen-activated protein kinase cascade in gastric cancer. Mol Carcinog. 2009;48:703–712. doi: 10.1002/mc.20516. [DOI] [PubMed] [Google Scholar]
- 73.Mazumdar T, DeVecchio J, Agyeman A, Shi T, Houghton JA. The GLI genes as the molecular switch in disrupting Hedgehog signaling in colon cancer. Oncotarget. 2011;2:638–645. doi: 10.18632/oncotarget.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu Z, Li T, Reinhold MI, Naski MC. MEK1-RSK2 contributes to hedgehog signaling by stabilizing GLI2 transcription factor and inhibiting ubiquitination. Oncogene. 2014;33:65–73. doi: 10.1038/onc.2012.544. [DOI] [PubMed] [Google Scholar]
- 75.Lu J, Liu L, Zheng M, Li X, Wu A, Wu Q, et al. MEKK2 and MEKK3 suppress Hedgehog pathway-dependent medulloblastoma by inhibiting GLI1 function. Oncogene. 2018;37:3864–3878. doi: 10.1038/s41388-018-0249-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Deng W, Vanderbilt DB, Lin CC, Martin KH, Brundage KM, Ruppert JM. SOX9 inhibits β-TrCP-mediated protein degradation to promote nuclear GLI1 expression and cancer stem cell properties. J Cell Sci. 2015;128:1123–1138. doi: 10.1242/jcs.162164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Malatesta M, Steinhauer C, Mohammad F, Pandey DP, Squatrito M, Helin K. Histone acetyltransferase PCAF is required for hedgehog-Gli-dependent transcription and cancer cell proliferation. Cancer Res. 2013;73:6323–6333. doi: 10.1158/0008-5472.CAN-12-4660. [DOI] [PubMed] [Google Scholar]
- 78.Yoon JW, Gallant M, Lamm MLG, Iannaccone S, Vieux KF, Proytcheva M, et al. Noncanonical regulation of the hedgehog mediator GLI1 by c-MYC in Burkitt lymphoma. Mol Cancer Res. 2013;11:604–615. doi: 10.1158/1541-7786.MCR-12-0441. [DOI] [PubMed] [Google Scholar]
- 79.Ji Z, Mei FC, Xie J, Cheng X. Oncogenic KRAS activates hedgehog signaling pathway in pancreatic cancer cells. J Biol Chem. 2007;282:14048–14055. doi: 10.1074/jbc.M611089200. [DOI] [PubMed] [Google Scholar]
- 80.Kasper M, Schnidar H, Neill GW, Hanneder M, Klingler S, Blaas L, et al. Selective modulation of Hedgehog/GLI target gene expression by epidermal growth factor signaling in human keratinocytes. Mol Cell Biol. 2006;26:6283–6298. doi: 10.1128/MCB.02317-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jafari SM, Panjehpour M, Aghaei M, Joshaghani HR, Enderami SE. A3 adenosine receptor agonist inhibited survival of breast cancer stem cells via GLI-1 and ERK1/2 pathway. J Cell Biochem. 2017;118:2909–2920. doi: 10.1002/jcb.25945. [DOI] [PubMed] [Google Scholar]
- 82.Jafari SM, Joshaghani HR, Panjehpour M, Aghaei M, Zargar Balajam N. Apoptosis and cell cycle regulatory effects of adenosine by modulation of GLI-1 and ERK1/2 pathways in CD44 + and CD24 − breast cancer stem cells. Cell Prolif. 2017;50:e12345. doi: 10.1111/cpr.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou J, Zhu G, Huang J, Li L, Du Y, Gao Y, et al. Non-canonical GLI1/2 activation by PI3K/AKT signaling in renal cell carcinoma: A novel potential therapeutic target. Cancer Lett. 2016;370:313–323. doi: 10.1016/j.canlet.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 84.Kern D, Regl G, Hofbauer SW, Altenhofer P, Achatz G, Dlugosz A, et al. Hedgehog/GLI and PI3K signaling in the initiation and maintenance of chronic lymphocytic leukemia. Oncogene. 2015;34:5341–5351. doi: 10.1038/onc.2014.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Elsawa SF, Almada LL, Ziesmer SC, Novak AJ, Witzig TE, Ansell SM, et al. GLI2 transcription factor mediates cytokine cross-talk in the tumor microenvironment. J Biol Chem. 2011;286:21524–21534. doi: 10.1074/jbc.M111.234146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ramaswamy B, Lu Y, Teng KY, Nuovo G, Li X, Shapiro CL, et al. Hedgehog signaling is a novel therapeutic target in tamoxifen-resistant breast cancer aberrantly activated by PI3K/AKT pathway. Cancer Res. 2012;72:5048–5059. doi: 10.1158/0008-5472.CAN-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Graab U, Hahn H, Fulda S. Identification of a novel synthetic lethality of combined inhibition of hedgehog and PI3K signaling in rhabdomyosarcoma. Oncotarget. 2015;6:8722–8735. doi: 10.18632/oncotarget.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liang R, Šumová B, Cordazzo C, Mallano T, Zhang Y, Wohlfahrt T, et al. The transcription factor GLI2 as a downstream mediator of transforming growth factor-β-induced fibroblast activation in SSc. Ann Rheum Dis. 2017;76:756–764. doi: 10.1136/annrheumdis-2016-209698. [DOI] [PubMed] [Google Scholar]
- 89.Pierrat MJ, Marsaud V, Mauviel A, Javelaud D. Transcriptional repression of the tyrosinase-related protein 2 gene by transforming growth factor-β and the Kruppel-like transcription factor GLI2. J Dermatol Sci. 2019;94:321–329. doi: 10.1016/j.jdermsci.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 90.Peng L, Yang C, Yin J, Ge M, Wang S, Zhang G, et al. TGF-β2 induces Gli1 in a Smad3-dependent manner against cerebral ischemia/reperfusion injury after isoflurane post-conditioning in rats. Front Neurosci. 2019;13:7688–95 Frontiers Media S.A. [DOI] [PMC free article] [PubMed]
- 91.Sun SL, Wang XY. TGF-β1 promotes proliferation and invasion of hepatocellular carcinoma cell line HepG2 by activating GLI-1 signaling. Eur Rev Med Pharmacol Sci. 2018;22:7688–7695. doi: 10.26355/eurrev_201811_16389. [DOI] [PubMed] [Google Scholar]
- 92.Liang M, Liu XC, Liu T, Li WJ, Xiang JG, Xiao D, et al. GLI-1 facilitates the EMT induced by TGF-β1 in gastric cancer. Eur Rev Med Pharmacol Sci. 2018;22:6809–6815. doi: 10.26355/eurrev_201810_16148. [DOI] [PubMed] [Google Scholar]
- 93.Rowan CJ, Li W, Martirosyan H, Erwood S, Hu D, Kim YK, et al. Hedgehog-GLI signaling in Foxd1-positive stromal cells promotes murine nephrogenesis via TGFβ signaling. Development. 2018;145:1–13. [DOI] [PubMed]
- 94.de Reynies A, Javelaud D, Elarouci N, Marsaud V, Gilbert C, Mauviel A. Large-scale pan-cancer analysis reveals broad prognostic association between TGF-β ligands, not Hedgehog, and GLI1/2 expression in tumors. bioRxiv Cancer Biol. [cited 2019 Dec 15]; Available from: http://biorxiv.org/cgi/content/short/728949v1?rss=1. [DOI] [PMC free article] [PubMed]
- 95.Atwood SX, Li M, Lee A, Tang JY, Oro AE. GLI activation by atypical protein kinase C ι/λ regulates the growth of basal cell carcinomas. Nature. 2013;494:484–488. doi: 10.1038/nature11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Riobo NA, Haines GM, Emerson CP. Protein kinase C-δ and mitogen-activated protein/extracellular signal-regulated kinase-1 control GLI activation in hedgehog signaling. Cancer Res. 2006;66:839–845. doi: 10.1158/0008-5472.CAN-05-2539. [DOI] [PubMed] [Google Scholar]
- 97.Jiang K, Liu Y, Fan J, Epperly G, Gao T, Jiang J, et al. Hedgehog-regulated atypical PKC promotes phosphorylation and activation of Smoothened and Cubitus interruptus in Drosophila. Proc Natl Acad Sci U S A. 2014;111:E4842–E4850. doi: 10.1073/pnas.1417147111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wickström M, Dyberg C, Shimokawa T, Milosevic J, Baryawno N, Fuskevåg OM, et al. Targeting the hedgehog signal transduction pathway at the level of GLI inhibits neuroblastoma cell growth in vitro and in vivo. Int J Cancer. 2013;132:1516–1524. doi: 10.1002/ijc.27820. [DOI] [PubMed] [Google Scholar]
- 99.Lauth M, Bergström Å, Toftgård R. Phorbol esters inhibit the Hedgehog signalling pathway downstream of suppressor of fused, but upstream of Gli. Oncogene. 2007;26:5163–5168. doi: 10.1038/sj.onc.1210321. [DOI] [PubMed] [Google Scholar]
- 100.Canettieri G, Di Marcotullio L, Greco A, Coni S, Antonucci L, Infante P, et al. Histone deacetylase and Cullin3-REN KCTD11 ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat Cell Biol. 2010;12:132–142. doi: 10.1038/ncb2013. [DOI] [PubMed] [Google Scholar]
- 101.Mirza AN, Fry MA, Urman NM, Atwood SX, Roffey J, Ott GR, et al. Combined inhibition of atypical PKC and histone deacetylase 1 is cooperative in basal cell carcinoma treatment. JCI Insight. 2017;2:1–12 American Society for Clinical Investigation. [DOI] [PMC free article] [PubMed]
- 102.Eggenschwiler JT, Bulgakov OV, Qin J, Li T, Anderson KV. Mouse Rab23 regulates Hedgehog signaling from Smoothened to Gli proteins. Dev Biol. 2006;290:1–12. doi: 10.1016/j.ydbio.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 103.Zhang S, Wang Y, Mao JH, Hsieh D, Kim IJ, Hu LM, et al. Inhibition of CK2α down-regulates Hedgehog/Gli signaling leading to a reduction of a stem-like side population in human lung cancer cells. PLoS One. 2012;7:e38996. doi: 10.1371/journal.pone.0038996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang Z, Zhan X, Kim B, Wu J. A proteomic approach identifies SAFB-like transcription modulator (SLTM) as a bidirectional regulator of GLI family zinc finger transcription factors. J Biol Chem. 2019;294:5549–5561. doi: 10.1074/jbc.RA118.007018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Singh R, Dhanyamraju PK, Lauth M. DYRK1B blocks canonical and promotes non-canonical Hedgehog signaling through activation of the mTOR/AKT pathway. Oncotarget. 2017;8:833–845. doi: 10.18632/oncotarget.13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Varjosalo M, Björklund M, Cheng F, Syvänen H, Kivioja T, Kilpinen S, et al. Application of active and kinase-deficient kinome collection for identification of kinases regulating hedgehog signaling. Cell. 2008;133:537–548. doi: 10.1016/j.cell.2008.02.047. [DOI] [PubMed] [Google Scholar]
- 107.Pandolfi S, Stecca B. Cooperative integration between HEDGEHOG-GLI signalling and other oncogenic pathways: Implications for cancer therapy. Expert Rev Mol Med. 2015;17:e5. doi: 10.1017/erm.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Abdullah YM, Azeem Z, Bilal M, Liaqat K, Hussain S, et al. Variants in GLI3 Cause Greig Cephalopolysyndactyly Syndrome. Genet Test Mol Biomarkers. 2019;23:744–750. doi: 10.1089/gtmb.2019.0071. [DOI] [PubMed] [Google Scholar]
- 109.Tang Y, Ge J, Li T. Greig cephalopolysyndactyly syndrome caused by novel GLI3 mutation: a family report. Cancer Cell Res. 2019;22:597–601. [Google Scholar]
- 110.Biesecker LG. Greig cephalopolysyndactyly syndrome. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, et al., editors. Seattle (WA); 1993. [PubMed]
- 111.Tsanev R, Tiigimägi P, Michelson P, Metsis M, Østerlund T, Kogerman P. Identification of the gene transcription repressor domain of Gli3. FEBS Lett. 2009;583:224–228. doi: 10.1016/j.febslet.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ito S, Kitazawa R, Haraguchi R, Kondo T, Ouchi A, Ueda Y, et al. Novel GLI3 variant causing overlapped Greig cephalopolysyndactyly syndrome (GCPS) and Pallister-Hall syndrome (PHS) phenotype with agenesis of gallbladder and pancreas. Diagn Pathol. 2018;13:1–4 [cited 2019 Dec 19] Available from: http://genetics.bwh.harvard.edu/pph2. [DOI] [PMC free article] [PubMed]
- 113.Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- 114.Pan Y, Wang B. A novel protein-processing domain in Gli2 and Gli3 differentially blocks complete protein degradation by the proteasome. J Biol Chem. 2007;282:10846–10852. doi: 10.1074/jbc.M608599200. [DOI] [PubMed] [Google Scholar]
- 115.Meyer NP, Roelink H. The amino-terminal region of Gli3 antagonizes the Shh response and acts in dorsoventral fate specification in the developing spinal cord. Dev Biol. 2003;257:343–355. doi: 10.1016/s0012-1606(03)00065-4. [DOI] [PubMed] [Google Scholar]
- 116.Krauß S, So J, Hambrock M, Köhler A, Kunath M, Scharff C, et al. Point mutations in GLI3 lead to misregulation of its subcellular localization. PLoS One. 2009;4:1–13 [cited 2019 May 28] Available from: www.plosone.org. [DOI] [PMC free article] [PubMed]
- 117.Cao T, Wang C, Yang M, Wu C, Wang B. Mouse limbs expressing only the Gli3 repressor resemble those of Sonic hedgehog mutants. Dev Biol. 2013;379:221–228. doi: 10.1016/j.ydbio.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Huang Y, Roelink H, McKnight GS. Protein kinase A deficiency causes axially localized neural tube defects in mice. J Biol Chem. 2002;277:19889–19896. doi: 10.1074/jbc.M111412200. [DOI] [PubMed] [Google Scholar]
- 119.Hui CC, Slusarski D, Platt KA, Holmgren R, Joyner AL. Expression of three mouse homologs of the drosophila segment polarity gene cubitus interruptus, Gli, Gli-2, and Gli-3, in ectoderm-and mesoderm-derived tissues suggests multiple roles during postimplantation development. Dev Biol. 1994;162:402–413. doi: 10.1006/dbio.1994.1097. [DOI] [PubMed] [Google Scholar]
- 120.Pan Y, Bai CB, Joyner AL, Wang B. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol Cell Biol. 2006;26:3365–3377. doi: 10.1128/MCB.26.9.3365-3377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kogerman P, Grimm T, Kogerman L, Krause D, Undén AB, Sandstedt B, et al. Mammalian Suppressor-of-Fused modulates nuclear-cytoplasmic shuttling of GLI-1. Nat Cell Biol. 1999;1:312–319. doi: 10.1038/13031. [DOI] [PubMed] [Google Scholar]
- 122.Mukhopadhyay S, Wen X, Ratti N, Loktev A, Rangell L, Scales SJ, et al. The ciliary G-protein-coupled receptor Gpr161 negatively regulates the sonic hedgehog pathway via cAMP signaling. Cell. 2013;152:210–223. doi: 10.1016/j.cell.2012.12.026. [DOI] [PubMed] [Google Scholar]
- 123.Kise Y, Morinaka A, Teglund S, Miki H. Sufu recruits GSK3β for efficient processing of Gli3. Biochem Biophys Res Commun. 2009;387:569–574. doi: 10.1016/j.bbrc.2009.07.087. [DOI] [PubMed] [Google Scholar]
- 124.Tempe D, Casas M, Karaz S, Blanchet-Tournier M-F, Concordet J-P. Multisite protein kinase A and glycogen synthase kinase 3 phosphorylation leads to Gli3 ubiquitination by SCF TrCP. Mol Cell Biol. 2006;26:4316–4326. doi: 10.1128/MCB.02183-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hatsell SJ, Cowin P. Gli3-mediated repression of Hedgehog targets is required for normal mammary development. Development. 2006;133:3661 LP–3663670. doi: 10.1242/dev.02542. [DOI] [PubMed] [Google Scholar]
- 126.Wiegering A, Petzsch P, Köhrer K, Rüther U, Gerhardt C. GLI3 repressor but not GLI3 activator is essential for mouse eye patterning and morphogenesis. Dev Biol. 2019;450:141–154. doi: 10.1016/j.ydbio.2019.02.018. [DOI] [PubMed] [Google Scholar]
- 127.Krauß S, Foerster J, Schneider R, Schweiger S. Protein phosphatase 2A and rapamycin regulate the nuclear localization and activity of the transcription factor GLI3. Cancer Res. 2008;68:4658–4665. doi: 10.1158/0008-5472.CAN-07-6174. [DOI] [PubMed] [Google Scholar]
- 128.Li N, Truong S, Nouri M, Moore J, Al Nakouzi N, Lubik AA, et al. Non-canonical activation of hedgehog in prostate cancer cells mediated by the interaction of transcriptionally active androgen receptor proteins with Gli3. Oncogene. 2018;37:2313–2325. doi: 10.1038/s41388-017-0098-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang C, Pan Y, Wang B. Suppressor of fused and Spop regulate the stability, processing and function of Gli2 and Gli3 full-length activators but not their repressors. Development. 2010;137:2001–2009. doi: 10.1242/dev.052126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhou H, Kim S, Ishii S, Boyer TG. Mediator modulates Gli3-dependent sonic hedgehog signaling. Mol Cell Biol. 2006;26:8667–8682. doi: 10.1128/MCB.00443-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fu L, Wu H, Cheng SY, Gao D, Zhang L, Zhao Y. Set7 mediated Gli3 methylation plays a positive role in the activation of sonic hedgehog pathway in mammals. Elife. 2016;5:e15690. doi: 10.7554/eLife.15690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Duncan PA, Klein RM, Wilmot PL, Shapiro LR. Greig cephalopolysyndactyly syndrome. Am J Dis Child. 1979;133:818–821. doi: 10.1001/archpedi.1979.02130080058010. [DOI] [PubMed] [Google Scholar]
- 133.Weber M. New classification and score for tibial hemimelia. J Child Orthop. 2008;2:169–75 [cited 2019 Jun 5] Available from: https://online.boneandjoint.org.uk/doi/pdf/10.1007/s11832-008-0081-5. [DOI] [PMC free article] [PubMed]
- 134.Bose J. Pallister-Hall syndrome phenotype in mice mutant for Gli3. Hum Mol Genet. 2002;11:1129–1135. doi: 10.1093/hmg/11.9.1129. [DOI] [PubMed] [Google Scholar]
- 135.Vortkamp A, Franz T, Gessler M, Grzeschik KH. Deletion of GLI3 supports the homology of the human Greig cephalopolysyndactyly syndrome (GCPS) and the mouse mutant extra toes (Xt) Mamm Genome. 1992;3:461–463. doi: 10.1007/BF00356157. [DOI] [PubMed] [Google Scholar]
- 136.Al-Qattan MM, Kozin SH. Update on embryology of the upper limb. J Hand Surg [Am] 2013;38:1835–1844. doi: 10.1016/j.jhsa.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 137.Johnston JJ, Olivos-Glander I, Turner J, Aleck K, Bird LM, Mehta L, et al. Clinical and molecular delineation of the Greig cephalopolysyndactyly contiguous gene deletion syndrome and its distinction from acrocallosal syndrome. Am J Med Genet. 2003;123A:236–242. doi: 10.1002/ajmg.a.20318. [DOI] [PubMed] [Google Scholar]
- 138.Volodarsky M, Langer Y, Birk OS. A novel GLI3 mutation affecting the zinc finger domain leads to preaxial-postaxial polydactyly-syndactyly complex. BMC Med Genet. 2014;15:1–4 [cited 2019 Jun 1] Available from: http://www.biomedcentral.com/1471-2350/15/110. [DOI] [PMC free article] [PubMed]
- 139.Elson E, Perveen R, Donnai D, Wall S, Black GCM. De novo GLI3 mutation in acrocallosal syndrome: Broadening the phenotypic spectrum of GLI3 defects and overlap with murine models. J Med Genet. 2002;39:804–806. doi: 10.1136/jmg.39.11.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Johnston JJ, Sapp JC, Turner JT, Amor D, Aftimos S, Aleck KA, et al. Molecular analysis expands the spectrum of phenotypes associated with GLI3 mutations. Hum Mutat. 2010;31:1142–1154. doi: 10.1002/humu.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Deimling S, Sotiropoulos C, Lau K, Chaudhry S, Sturgeon K, Kelley S, et al. Tibial hemimelia associated with GLI3 truncation. J Hum Genet. 2016;61:443–446. doi: 10.1038/jhg.2015.161. [DOI] [PubMed] [Google Scholar]
- 142.Theil T. Gli3 and dorsal forebrain development. 1999. [Google Scholar]
- 143.Kuschel S, Rüther U, Theil T. A disrupted balance between Bmp/Wnt and Fgf signaling underlies the ventralization of the Gli3 mutant telencephalon. Dev Biol. 2003;260:484–495. doi: 10.1016/s0012-1606(03)00252-5. [DOI] [PubMed] [Google Scholar]
- 144.Wilson SL, Wilson JP, Wang C, Wang B, Mcconnell SK. Primary cilia and Gli3 activity regulate cerebral cortical size. Dev Neurobiol. 2012;72:1196–1212. doi: 10.1002/dneu.20985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Petrova R, Garcia ADR, Joyner AL. Titration of GLI3 Repressor Activity by Sonic Hedgehog Signaling Is Critical for Maintaining Multiple Adult Neural Stem Cell and Astrocyte Functions. J Neurosci. 2013;33:17490–17505. doi: 10.1523/JNEUROSCI.2042-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Amaniti EM, Hasenpusch-Theil K, Li Z, Magnani D, Kessaris N, Mason JO, et al. Gli3 is required in Emx1+ progenitors for the development of the corpus callosum. Dev Biol. 2013;376:113–124. doi: 10.1016/j.ydbio.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 147.Wang H, Ge G, Uchida Y, Luu B, Ahn S. Gli3 is required for maintenance and fate specification of cortical progenitors. J Neurosci. 2011;31:6440–6448. doi: 10.1523/JNEUROSCI.4892-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tan M, Hu X, Qi Y, Park J, Cai J, Qiu M. Gli3 mutation rescues the generation, but not the differentiation, of oligodendrocytes in Shh mutants. Brain Res. 2006;1067:158–163. doi: 10.1016/j.brainres.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 149.Litingtung Y, Chiang C. Specification of ventral neuron types is mediated by an antagonistic interaction between Shh and Gli3. Nat Neurosci. 2000;3:979–85. [DOI] [PubMed]
- 150.Grindley JC, Bellusci S, Perkins D, Hogan BLM. Evidence for the involvement of the Gli gene family in embryonic mouse lung development. Dev Biol. 1997;188:337–348. doi: 10.1006/dbio.1997.8644. [DOI] [PubMed] [Google Scholar]
- 151.Motoyama J, Liu J, Mo R, Ding Q, Post M, Hui CC. Essential function of Gli2 and Gli3 in the formation of lung, trachea and oesophagus. Nat Genet. 1998;20:54–57. doi: 10.1038/1711. [DOI] [PubMed] [Google Scholar]
- 152.Hager-Theodorides AL, Dessens JT, Outram SV, Crompton T. The transcription factor Gli3 regulates differentiation of fetal CD4 -CD8- double-negative thymocytes. Blood. 2005;106:1296–1304. doi: 10.1182/blood-2005-03-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yang T, Cui H, Wen M, Zuber J, Kogan SC, Wei G. TCEA1 regulates the proliferative potential of mouse myeloid cells. Exp Cell Res. 2018;370:551–560. doi: 10.1016/j.yexcr.2018.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lam RA, Chwee JY, Le Bert N, Sauer M, Pogge Von Strandmann E, Gasser S. Regulation of self-ligands for activating natural killer cell receptors. Ann Med. 2013;45:384–394. doi: 10.3109/07853890.2013.792495. [DOI] [PubMed] [Google Scholar]
- 155.Gao J, Zheng Q, Xin N, Wang W, Zhao C. CD155, an onco-immunologic molecule in human tumors [Internet] Cancer Sci. 2017;108:1934–1938. doi: 10.1111/cas.13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Solecki DJ, Gromeier M, Mueller S, Bernhardt G, Wimmer E. Expression of the human poliovirus receptor/CD155 gene is activated by Sonic hedgehog. J Biol Chem. 2002;277:25697–25702. doi: 10.1074/jbc.M201378200. [DOI] [PubMed] [Google Scholar]
- 157.Solanki A, Lau C-I, Saldaña JI, Ross S, Crompton T. The transcription factor Gli3 promotes B cell development in fetal liver through repression of Shh. J Exp Med. 2017;214:2041–2058. doi: 10.1084/jem.20160852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ravasi T, Wells C, Forest A, Underhill DM, Wainwright BJ, Aderem A, et al. Generation of diversity in the innate immune system: macrophage heterogeneity arises from gene-autonomous transcriptional probability of individual inducible genes. J Immunol. 2002;168:44–50. doi: 10.4049/jimmunol.168.1.44. [DOI] [PubMed] [Google Scholar]
- 159.Rodrigues MFSD, Miguita L, De Andrade NP, Heguedusch D, Rodini CO, Moyses RA, et al. GLI3 knockdown decreases stemness, cell proliferation and invasion in oral squamous cell carcinoma. Int J Oncol. 2018;53:2458–2472. doi: 10.3892/ijo.2018.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Li J, Qiu M, An Y, Huang J, Gong C. miR-7-5p acts as a tumor suppressor in bladder cancer by regulating the hedgehog pathway factor Gli3. Biochem Biophys Res Commun. 2018;503:2101–2107. doi: 10.1016/j.bbrc.2018.07.166. [DOI] [PubMed] [Google Scholar]
- 161.Zhang C, Li C, He F, Cai Y, Yang H. Identification of CD44+CD24+ gastric cancer stem cells. J Cancer Res Clin Oncol. 2011;137:1679–1686. doi: 10.1007/s00432-011-1038-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ma Y, Li G, Hu J, Liu X, Shi B. MicroRNA-494 regulates Gli3 expression and inhibits pancreatic cancer cells growth and migration. J Cell Biochem. 2018;119:5324–5331. doi: 10.1002/jcb.26658. [DOI] [PubMed] [Google Scholar]
- 163.Miyahara H, Natsumeda M, Yoshimura J, Ogura R, Okazaki K, Toyoshima Y, et al. Neuronal differentiation associated with Gli3 expression predicts favorable outcome for patients with medulloblastoma. Neuropathology. 2014;34:1–10. doi: 10.1111/neup.12052. [DOI] [PubMed] [Google Scholar]
- 164.Chaudhry P, Singh M, Triche TJ, Guzman M, Merchant AA. GLI3 repressor determines Hedgehog pathway activation and is required for response to SMO antagonist glasdegib in AML. Blood. 2017;129:3465–3475. doi: 10.1182/blood-2016-05-718585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Golubovskaya VM, Huang G, Ho B, Yemma M, Morrison CD, Lee J, et al. Pharmacologic blockade of FAK autophosphorylation decreases human glioblastoma tumor growth and synergizes with temozolomide. Mol Cancer Ther. 2013;12:162–172. doi: 10.1158/1535-7163.MCT-12-0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Huang G, Ho B, Conroy J, Liu S, Qiang H, Golubovskaya V. The microarray gene profiling analysis of glioblastoma cancer cells reveals genes affected by FAK inhibitor Y15 and combination of Y15 and temozolomide. Anti Cancer Agents Med Chem. 2014;14:9–17. doi: 10.2174/18715206113139990141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kuo JS, Casey SO, Thompson L, Truwit CL. Pallister-hall syndrome: clinical and MR features. Am J Neuroradiol. 1999;20:1839–1841. [PMC free article] [PubMed] [Google Scholar]
- 168.Graham JM, Saunders R, Fratkin J, Spiegel P, Harris M, Klein RZ. A cluster of Pallister-Hall syndrome cases, (congenital hypothalamic hamartoblastoma syndrome) Am J Med Genet Suppl. 1986;2:53–63. doi: 10.1002/ajmg.1320250609. [DOI] [PubMed] [Google Scholar]
- 169.Clarren SK, Alvord EC, Hall JG. Congenital hypothalamic hamartoblastoma, hypopituitarism, imperforate anus, and postaxial polydactyly - a new syndrome? Part II: Neuropathological considerations. Am J Med Genet. 1980;7:75–83. doi: 10.1002/ajmg.1320070111. [DOI] [PubMed] [Google Scholar]
- 170.Craig DW, Itty A, Panganiban C, Szelinger S, Kruer MC, Sekar A, et al. Identification of somatic chromosomal abnormalities in hypothalamic hamartoma tissue at the GLI3 locus. Am J Hum Genet. 2008;82:366–374. doi: 10.1016/j.ajhg.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Saleh A, Zain RB, Hussaini H, Ng F, Tanavde V, Hamid S, et al. Transcriptional profiling of oral squamous cell carcinoma using formalin-fixed paraffin-embedded samples. Oral Oncol. 2010;46:379–386. doi: 10.1016/j.oraloncology.2010.02.022. [DOI] [PubMed] [Google Scholar]
- 172.Nakatani Y, Yamazaki M, Chazin WJ, Yui S. Regulation of S100A8/A9 (Calprotectin) binding to tumor cells by zinc ion and its implication for apoptosis-inducing activity. 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Berman DM, Karhadkar SS, Maitra A, De Oca RM, Gerstenblith MR, Briggs K, et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- 174.Xie K, Abbruzzese JL. Developmental biology informs cancer: the emerging role of the hedgehog signaling pathway in upper gastrointestinal cancers. Cancer Cell. 2003;4:245–247. doi: 10.1016/s1535-6108(03)00246-0. [DOI] [PubMed] [Google Scholar]
- 175.Kang HN, Oh SC, Kim JS, Yoo YA. Abrogation of Gli3 expression suppresses the growth of colon cancer cells via activation of p53. Exp Cell Res. 2012;318:539–549. doi: 10.1016/j.yexcr.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 176.Steg A, Amm HM, Novak Z, Frost AR, Johnson MR. Gli3 mediates cell survival and sensitivity to cyclopamine in pancreatic cancer. Cancer Biol Ther. 2010;10:893–902. doi: 10.4161/cbt.10.9.13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Muthukumar S. MED12 mutations promote castration-resistant prostate cancer through hyperactivated GLI3/SHH signalling. 2019. Theses Diss [cited 2019 Dec 5]. Available from: https://athenaeum.uiw.edu/uiw_etds/361.
- 178.Kuleszo D, Koczkowska M, Lipska-Ziętkiewicz BS, Grajkowska W, Adamkiewicz-Drożyńska E, Dembowska-Bagińska B, et al. Comparative genomic analysis of intracranial germ cell tumors – the preliminary study focused on sonic hedgehog signaling pathway. Wspolczesna Onkol. 2017;21:279–284. doi: 10.5114/wo.2017.72390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lo Ré AE, Fernández-Barrena MG, Almada LL, Mills LD, Elsawa SF, Lund G, et al. Novel AKT1-GLI3-VMP1 pathway mediates KRAS oncogene-induced autophagy in cancer cells. J Biol Chem. 2012;287:25325–25334. doi: 10.1074/jbc.M112.370809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Greaves WO, Kim JE, Singh RR, Drakos E, Kunkalla K, Sánchez-Espiridión B, et al. Glioma-associated oncogene homologue 3, a hedgehog transcription factor, is highly expressed in Hodgkin and Reed-Sternberg cells of classical Hodgkin lymphoma. Hum Pathol. 2011;42:1643–1652. doi: 10.1016/j.humpath.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Han W, Ibarra G, Gupta M, Yin Y, Elsawa SF. Elevated GLI3 expression in germinal center diffuse large B cell lymphoma [Internet] Leuk Lymphoma. 2018;59:L2743–L2745. doi: 10.1080/10428194.2018.1439169. [DOI] [PubMed] [Google Scholar]
- 182.Ellison DW, Dalton J, Kocak M, Nicholson SL, Fraga C, Neale G, et al. Medulloblastoma: Clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta Neuropathol. 2011;121:381–396. doi: 10.1007/s00401-011-0800-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Romer JT, Kimura H, Magdaleno S, Sasai K, Fuller C, Baines H, et al. Suppression of the Shh pathway using a small molecule inhibitor eliminates medulloblastoma in Ptc1+/-p53-/- mice. Cancer Cell. 2004;6:229–240. doi: 10.1016/j.ccr.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 184.Javelaud D, Pierrat MJ, Mauviel A. Crosstalk between TGF-β and hedgehog signaling in cancer. FEBS Lett. 2012;14:2016–2025. doi: 10.1016/j.febslet.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 185.Jackson DA, Smith TD, Amarsaikhan N, Han W, Neil MS, Boi SK, et al. Modulation of the IL-6 receptor α underlies GLI2-mediated regulation of Ig secretion in Waldenström macroglobulinemia cells. J Immunol. 2015;195:2908–2916. doi: 10.4049/jimmunol.1402974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Han W, Jackson DA, Matissek SJ, Misurelli JA, Neil MS, Sklavanitis B, et al. Novel molecular mechanism of regulation of CD40 ligand by the transcription factor GLI2. J Immunol. 2017;198:4481–4489. doi: 10.4049/jimmunol.1601490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zheng X, Rumie Vittar NB, Gai X, Fernandez-Barrena MG, Moser CD, Hu C, et al. The transcription factor GLI1 mediates TGFβ1 driven EMT in hepatocellular carcinoma via a SNAI1-dependent mechanism. PLoS One. 2012;7:1–13. [DOI] [PMC free article] [PubMed]
- 188.Karin M. The regulation of AP-1 activity by mitogen-activated protein kinases. J Biol Chem. 1995;270:16483–16486. doi: 10.1074/jbc.270.28.16483. [DOI] [PubMed] [Google Scholar]
- 189.Trnski D, Sabol M, Gojević A, Martinić M, Ozretić P, Musani V, et al. GSK3β and Gli3 play a role in activation of Hedgehog-Gli pathway in human colon cancer - targeting GSK3β downregulates the signaling pathway and reduces cell proliferation. Biochim Biophys Acta Mol basis Dis. 1852;2015:2574–2584. doi: 10.1016/j.bbadis.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 190.Lee SH, Hong HS, Liu ZX, Kim RH, Kang MK, Park NH, et al. TNFα enhances cancer stem cell-like phenotype via Notch-Hes1 activation in oral squamous cell carcinoma cells. Biochem Biophys Res Commun. 2012;424:58–64. doi: 10.1016/j.bbrc.2012.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Xie H, Paradise BD, Ma WW, Fernandez-Zapico ME. Recent advances in the clinical targeting of Hedgehog/GLI signaling in cancer. Cells. 2019;8:394. doi: 10.3390/cells8050394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Agyeman A, Jha BK, Mazumdar T, Houghton JA. Mode and specificity of binding of the small molecule GANT61 to GLI determines inhibition of GLI-DNA binding. Oncotarget. 2014;5:4492–4503. doi: 10.18632/oncotarget.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Wen SY, Lin Y, Yu YQ, Cao SJ, Zhang R, Yang XM, et al. MiR-506 acts as a tumor suppressor by directly targeting the hedgehog pathway transcription factor Gli3 in human cervical cancer. Oncogene. 2015;34:717–725. doi: 10.1038/onc.2014.9. [DOI] [PubMed] [Google Scholar]
- 194.Li L, Zhang L, Zhao X, Cao J, Li J, Chu G. Downregulation of miR-152 contributes to the progression of liver fibrosis via targeting Gli3 in vivo and in vitro. Exp Ther Med. 2019;18:425–434. doi: 10.3892/etm.2019.7595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Hyun J, Wang S, Kim J, Rao KM, Park SY, Chung I, et al. MicroRNA-378 limits activation of hepatic stellate cells and liver fibrosis by suppressing Gli3 expression. Nat Commun. 2016;7:10993. doi: 10.1038/ncomms10993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Persengiev SP, Kondova II, Millette CF, Kilpatrick DL. Gli family members are differentially expressed during the mitotic phase of spermatogenesis. Oncogene. 1997;14:2259–2264. doi: 10.1038/sj.onc.1201068. [DOI] [PubMed] [Google Scholar]
- 197.Yao C, Sun M, Yuan Q, Niu M, Chen Z, Hou J, et al. MiRNA-133b promotes the proliferation of human Sertoli cells through targeting GLI3. Oncotarget. 2016;7:2201–2219. doi: 10.18632/oncotarget.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable