Abstract
Pathology informatics is a relatively new field with limited structured training programs for pathologists, especially for computer programming. Here, we describe our efforts to develop and implement a training program in the department of pathology at the University of Florida to meet these additional needs of current students as well as faculty and staff. Three one-credit courses were created using a flipped classroom design. Each course was assessed with a novel survey instrument, and the impact of the program was further measured 6 months after program completion with interviews of 6 participants and thematic analysis. Course objectives were met but with room for improvement. Major factors that had a positive impact included collaborative learning and real-world practice problems. Also, it improved communication with informatics colleagues as well as job task efficiency and effectiveness. Overall, the program raised awareness of informatics professional development and career path opportunities within pathology.
Keywords: computer programming, Unix, continuing education, flipped classroom, graduate medical education, informatics, pathology, residency training
Introduction
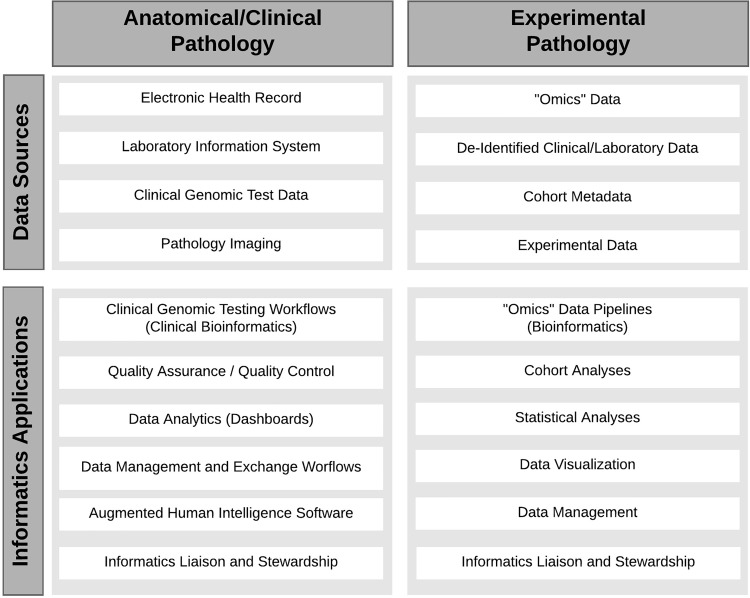
The practice of both experimental and anatomic/clinical pathology (AP/CP) has undergone a rapid digital revolution (Figure 1). In experimental pathology (EP), also known as investigative pathology, it is common to process digital/electronic data sets from large cohorts involving several thousands to hundreds of thousands of subjects. Genomic, transcriptomic, and other types of “omic” data may range from gigabytes to terabytes. Data sets in AP/CP are often highly heterogeneous as they originate from varied sources such as laboratory information systems (LISs), electronic medical record systems, clinical genomics, and pathology imaging or digital pathology.1,2
Figure 1.
Informatics within pathology. The variety of data sets used in anatomical/clinical and experimental pathology is presented with the potential informatics applications.
Informatics is now regarded as an essential skill for a community pathologist.3 Informatics-related tasks include developing enhanced laboratory reports, analyzing test ordering patterns, evaluating effectiveness of information services, laboratory quality control, and utilization management.4,5 Knowledge of informatics is critical in collecting data, storing and accessing it securely, and finally, analyzing and interpreting it.1 A high-quality analysis can fuel discovery-driven basic science research, improve AP/CP laboratory operations through development of best practices, provide insight into laboratory utilization in clinics and hospitals, and build new, intelligent pathology software workflows.
Informatics professionals typically lack adequate clinical/laboratory medicine knowledge to identify gaps or to ask the correct questions related to research and quality assurance. Although pathologists can typically identify potential gaps, without adequate informatics knowledge, often they are unable to conceptualize the solution or solve the problem. Acquiring knowledge of informatics will help the pathologist inquire about and solve problems themselves or efficiently communicate solutions to computer programmers and analysts who can then implement such solutions. Informatics training of pathologists can help bridge the current gap between informatics and pathology.
The pathology community recognizes the critical need for informatics training and created training programs such as Pathology Informatics Essentials for Residents (PIER)6,7 and Pathology Informatics Fellowships.8,9 These programs give training in broader informatics topics involving information fundamentals, information systems, workflow and processes, and governance and management.8,9 However, deficiencies have been noted in such curricula for computer programming that are critical for both pathology practice and research (AP/CP and EP divisions) in the era of “big data.”6,9 Currently at our institution, we have implementation of PIER training for residents. To meet additional informatics training needs such as custom data analysis and computational workflow development, we have developed Informatics for Pathology Practice and Research (IPPR) course series. Informatics for Pathology Practice and Research curricula is designed and targeted to meet the needs of both EP and AP/CP members including graduate students, postdoctoral fellows, residents, fellows, staff, and faculty. In this article, we describe the development and impact of IPPR involving informatics training in computer programming and Unix, which supplements the already established pathology informatics training programs such as PIER and Pathology Informatics Fellowships.
The informatics for IPPR course series is a set of 3 one-credit hands-on courses that give good background and working knowledge in 3 major aspects of informatics. The first course, IPPR 1, covered the Unix operating system (OS). The second course, IPPR 2, covered Python Programming and the last course in the series, IPPR 3, covered Advanced Data Analysis & Visualization using Python data analysis libraries—Pandas and Matplotlib. Our goal with these courses was to build foundational informatics knowledge in order to prepare the participants to later pursue advanced and specialized topics such as bioinformatics, machine learning, and clinical informatics.
Rationale for Choosing Unix
For IPPR 1, Unix was selected as the OS given its flexibility to customize and automate computing tasks. Unix is popular in high performance and cloud computing platforms because of easy remote accessibility and free licensing. Big data bioinformatics and health informatics software tools and workflows run on high-performance computing and cloud computing platforms (eg, Amazon Web Services, Microsoft Azure, and Google Cloud Platform) with command-line user interfaces and often have no graphical interface. Knowledge of working in a command-line Unix environment can greatly help with managing big data sets and building computational workflows and programs (eg, next-generation DNA sequencing data analysis).
Rationale for Choosing Python as a Programming Language
Python was chosen for IPPR 2 and 3, although there are many programming languages for data analytics.10,11 Some are commercial such as Microsoft Excel, SAS, Stata, SPSS, MATLAB, Mathematica; others are open source such as Google Sheets, Python, R, Octave, and Sage. Ideally, a programming language should have (1) no limits on data size, (2) ease in reproducing previously conducted analyses on new data sets, (3) adaptable to different OSs, (4) no constraints on end use, (5) presence of a community of contributors for continual addition of new functionalities, and (6) availability of code libraries for advanced analysis.10,11 R and Python are 2 popular programming languages that have these features. They are used typically for data analyses involving machine learning, large data sets, creating complex visualizations, and bioinformatics.12
As an introductory computer programming language, Python has some advantages over R. The learning curve for Python is relatively low, largely due to its clean coding syntax and indentation and good debugging/testing tools.13 Thus, it is considered a good choice for beginners, such as pathologists, who may have little to no background in computer programming. Moreover, Python, the most popular introductory programming language taught at top US universities,14 was ranked the number one programming language according to the IEEE Spectrum 2018 ranking.15 Additionally, Python comes with several thousands of readily available prebuild programs and libraries, which correspond to commonly used tasks in data science, statistics, and bioinformatics. Associated Pandas and Matplotlib libraries were used in conjunction with Python in IPPR 3. Taking advantages of these existing libraries will yield a faster software development time and shorter learning curve for newer programmers.
Anticipated Informatics Skill Set to be Obtained
After completion of this course series, the participants are expected to apply computer programming (Python) and Unix for developing custom software and computational pipelines. They can use these pipelines to process, extract, and visualize the data elements of interest by combining and manipulating data obtained from multiple sources (eg, Electronic Health Record (EHR), LIS, genomics, imaging) for pathology practice and research. Practical examples would be building custom data analytics and visualization for pathology laboratory quality indicators (eg, turnaround times) and building next-generation DNA sequencing analysis workflows for clinical genomic testing or genomics discovery.
Methods
A complementary approach (survey questions with follow-up interviews) was used to address the following research questions: (1) How does the flipped classroom impact informatics training of established pathologists and pathology students? (2) How does Unix and Python training influence pathologists’ acquisition of the skills used in conducting data science clinical/experimental research? In this section, we describe the course design and teaching style, participants, quantitative data collection and analysis, and qualitative data collection and analysis methods.
Course Design and Teaching Style
Informatics expertise of course instructor
The course instructor (Srikar Chamala, PhD) is an assistant professor with training in computer science and bioinformatics. The instructor does not have MD or direct clinical training in pathology. However, he adequately understands pathology CP/AP workflows as he spent past few years in leading pathology informatics efforts at the UF Department of Pathology, Immunology and Laboratory Medicine involving systems such as EHR, LIS, Clinical Bioinformatics (applying in Molecular Pathology Genomic test development), data analytics, and custom software development.
Course design
These courses were developed by a subject matter expert (Chamala who served as the instructor) and instructional designer (Maness); they met weekly over the period of 2 semesters. A backward design approach was followed as well as the other tenets of Understanding by Design. To create a flipped classroom environment, the university’s learning management system (Canvas) was used to house instructional materials and individual prequizzes as well as to provide flexibility about when students could attend the live class. Flipped design allowed students to review videos (lectures and tutorials), read assigned materials, and practice some sample examples before coming to the class at their own convenient time and location. During the class they worked on solving real-world practice problem sets by collaborating with other class members. Further details on how the Kim et al flipped classroom principles16 were followed are provided in Table 1. Other design elements are also described to provide design transparency for replication by other institutions and to facilitate further course design research.
Table 1.
Flipped Design Principles and IPPR Implementation.
| Flipped Design Principle | IPPR Implementation |
|---|---|
| Opportunity and incentive for students to prepare prior to class | Used a Learning Management System (LMS) for instructional materials and required preclass quizzes Flexible class times provided (offering the same class twice a week) |
| Clear guidance on in-class and out-of-class activities and connections between them | Module overview pages included student learning objectives (refer to Supplemental Tables 2-4) as well as materials and assignments for the week |
| Mechanism to assess understanding | Prequizzes and individual assignments provided participants with feedback about their learning progress Office hour availability for coaching outside of class |
| Prompt feedback | Weekly peer review Instructor/teaching assistant feedback within 1-2 weeks |
| Sufficient time to complete assignments | Assignments were released at least 1 week prior to their due date Participants worked on assignments during class |
| A learning community | Participants from all experiential backgrounds were welcomed Collaboration on assignments was encouraged |
| Purposeful, easy to access technology | Institutionally supported LMS and video conferencing were used (Canvas & Zoom) Online office hours increased instructor and teaching assistant(s) access Informatics technology (Unix and Python) is free and open source |
Abbreviation: IPPR, Informatics for Pathology Practice and Research.
Course implementation
Each one-credit course was designed as 5 one-week modules. Therefore, the 3 courses could be completed in a 15-week semester. Students accessed canvas to review videos (lectures and tutorials) and read assigned materials for each week. Prior to class, they had to take an online quiz and execute sample programming commands to enforce preparedness and check understanding, with the intent of building confidence. Weekly class time (1.5 h/wk) was devoted to solving real-world practice problems. To accommodate participant schedule needs, 2 different weekly class times were available. Students were encouraged to work together in class, while the instructor or teaching assistants (TAs) were available to coach them through solutions. Office hours provided a venue for additional assistance. By the end of the week, students were expected to submit their own solutions as well as peer review 2 solutions submitted by 2 classmates. The purpose of each course and learning objectives is shown in Supplemental Table 1, and architecture of courses by each module is presented in Supplemental Tables 2 to 4.
Quantitative Data Collection and Analysis
The 40-item questionnaire was developed to gather information about student experiences and course design preferences. Questions were designed to obtain data related to participant demographics, preparedness for class, experience with online versus blended courses, online meeting preferences, and self-assessment of computer skills. The questionnaire used in the second and third courses included additional questions related to the completion of the other courses. Response choices for closed-ended items included Likert-type scale, rank, and multiple choice and open-ended questions.
Students were recruited to complete the questionnaire 1 week prior to the end of each course via personalized invitation e-mails from the instructional designer (who was unaffiliated with the grading process). The e-mails contained a link to the questionnaire, hosted by Qualtrics, and outlined the confidential survey process, risks, the institutional review board (study #201602565) approval, and researcher contact information. To incentivize, students were offered an extra credit opportunity of 20 points (2% of the final grade) for submitting a screenshot of the survey completion message. To maximize the completion rate, reminder e-mails were sent, 2 to 4 days apart over the next week to those who had not yet finished the questionnaire.
Descriptive statistics (ie, frequencies, central tendencies, and variability) were computed using SPSS. Demographic variables included pathology specialization (AP/CP or experimental), prior computing experience, and prior flipped hybrid experience were used to assess potential group differences. Learning objectives achievement was determined by analyzing the retrospective pretest to posttest change.
Qualitative Data Collection and Analysis
Following analysis of the free-text survey data, and 6 months after the third course ended, the instructional designer developed a semistructured interview guide (see Supplemental Appendix 1) and recruited participants by e-mail. The purpose of the interview was to describe whether the participants had applied information they learned in the courses.
The interviews were recorded and transcribed by an external agency that was unaffiliated with data collection. Pseudonyms are used to protect their anonymity. The free-text comments and interviews were analyzed using the constant comparative method.17 A thematic analysis approach was used to code data and identify patterns in responses (Table 2).
Table 2.
Interview Themes With Conceptual Definitions and Examples.
| Theme | Conceptual Definition | Example Quotes |
|---|---|---|
| Approach to Learning | Attitude toward learning informatics (eg, connecting material to personal experience, anticipation of lifelong learning, motivation/lack of, valuing hands-on experience, appreciation for learning community). | 1. “There were a couple times when I went into the homework assignment and was attempting to do it as it was outlined and found new packages that I wasn’t aware of, or something that was just really neat, and I made a cool figure out of that on the side. It was nice to have a structured experience but on my own I also went a little beyond that.” (MD/PhD student) 2. “those were really helpful, to look at how somebody else approached the same question as me. I used those a lot…[and would] refer back to them.” (AP/CP Faculty) |
| Skill Development Limitations | Constraints to learning, such as insufficient time for applying discrete informatics skills to many varied and complex real-world problems. | 3. “class, I thought, went really fast. So we would learn something and apply it right away and then move onto something else. I would probably need to do each of the exercises 5 times before I’d be able to do them myself and we just went really fast.” (AP/CP Faculty) 4. “when you get to the end of it I don’t think there’s a definite feeling of achievement or completeness for everybody…but, honestly, it’s excellent practice. It’s just really good. And of course, like anything, you’ve gotta keep practicing it.” (Resident) |
| Practical Career Impacts | Application of acquired informatics skills to improve job task efficiency and effectiveness or, pursuit of new career path. | 5. “With the same level of access to the data, it allowed me to much further drill down and analyze data, analyze turnaround times, and find trends and gaps than before. Before I was struggling with Excel and this gave me another tool.” (AP/CP Faculty) 6. “before getting into programming and I was basically a cell biologist working in the hoods in the lab and I’ve definitely switched to being more of a computer now…People are generating bigger and bigger data sets and they’re harder and harder to handle. So that was definitely a big motivation for me.” (Postdoctoral fellow) |
Abbreviation: AP/CP, anatomic/clinical pathology.
Results
Course Participants and Survey Respondents
Participants
Twenty-two individuals enrolled (Table 3) in one or more of the courses, including 12 graduate students, 3 AP/CP and 2 EP faculty members, 2 postdoctoral fellows, 1 medical resident, 1 MD-PhD student, and 1 EP staff member. Most participants (n = 14) were from the Department of Pathology, Immunology, and Laboratory Medicine (the advertisement was only sent to this department), 3 from the Department of Medicine, and 1 from Neurosurgery. Additionally, 4 participants did not provide department information. Overall, there were 17 experimental and 5 AP/CP participants.
Table 3.
Comparison of Registrants to Respondents.
| Course | Title | Duration* | Registrants | Respondents |
|---|---|---|---|---|
| IPPR 1 | Unix Operating System | First 5 weeks | 21 | 20 |
| IPPR 2 | Python Programming | Second 5 weeks | 22 | 19 |
| IPPR 3 | Advanced Data Analysis & Visualization | Third 5 weeks | 22 | 15 |
Abbreviation: IPPR, Informatics for Pathology Practice and Research.
* All 3 courses are held during single semester that runs for 15 weeks.
Survey response rate
A comparison of the registrants to respondents is presented in Table 3. Of the 22 individuals who enrolled in one or more of the IPPR courses, 21 completed at least 1 of the 3 course survey instruments. Additionally, the 14 people who took all 3 courses completed all 3 survey instruments.
Qualitative interview respondents and themes
Eight people responded to a request to be interviewed. A quarter of the course registrants participated in interviews—2 faculty, 1 resident, 1 postdoctoral student, 1 MD-PhD student, and 1 graduate student (n = 6). The first of 3 overarching themes identified from the interviews (all of which are described further in Table 2) was Approach to Learning which included, among other things, students’ motivation or lack of, how they connected material to personal experience, and their value of authentic assessments and the learning community. Second was their discussion of Skill Development Limitations such as time constraints for complex problem-solving. The final theme was Practical Career Impacts where participants discussed how they applied skills to improve job task efficiency and effectiveness or their pursuit of a new career path. The qualitative findings are presented in detail throughout the results, in conjunction with quantitative data, highlighting both similarities and differences in respondents’ points of view.
Prior experience
Programming
Half of the participants had no prior experience with Unix (n = 11) or Python (n = 12) before the course series. Four people did not provide a response. The rest chose “a little” or “a moderate amount” to describe their experience except for one student in IPPR 1 who chose “a great deal” of Unix experience.
Online learning environment
Half of the participants had not taken a blended/hybrid course (n = 11) or a fully online course (n = 11). Therefore, the online learning environment was new to many. Others had varying experience levels of 1 to 5 or more previous courses with some components online.
Course Design and Implementation
Twice-a-week offering
Offering the same class twice a week was appreciated by the participants as it helped to accommodate their schedule needs (Table 4, quote 1). The course participants range from graduate students to full-time working professions with very different daily work schedule demands and priorities.
Table 4.
Course Design and Implementation Participant Sample Quotes.
| Course Design and Implementation (sample quotes) | |
|---|---|
| 1 | Twice-a-week offering. “It was really [tough] making time to go attend the course and work on the stuff with the group…Having 2 days of the week to do it was good, but it’s still tough.”(AP/CP faculty) |
| 2 | Flipped course design. “there’s something different about when you’re sitting next to someone, like, ‘Hey, look at my screen and let’s figure this problem out.’” (AP/CP faculty) |
| 3 | Flipped course design. “there were frustrating parts about it because it felt like you were on your own learning it sometimes…The big pro of it is that it fit my lifestyle where I’ve got a job and everything else to worry about.” (Postdoctoral fellow) |
Abbreviation: AP/CP, anatomic/clinical pathology.
Flipped course design
Participants remarked that the flipped design was advantageous in balancing scheduling demands. They also valued their time learning together with others in the class which helped mitigate their frustration in learning or solving challenging concepts and exercises (Table 4, quotes 2 and 3).
Weekly effort
Given the academic accreditation standards for a 3-credit course, students are expected to spend 9 hours each week to successfully complete each course. The median time spent was closer to 8 hours. Additionally, students were evenly distributed across the range of 3 to 12 hours each week, except for 2 students who reported they spent around 18 hours a week on IPPR 2. Although we hoped to gain more information on these outliers, these participants did not respond to our interview request.
Skill Development
Course satisfaction
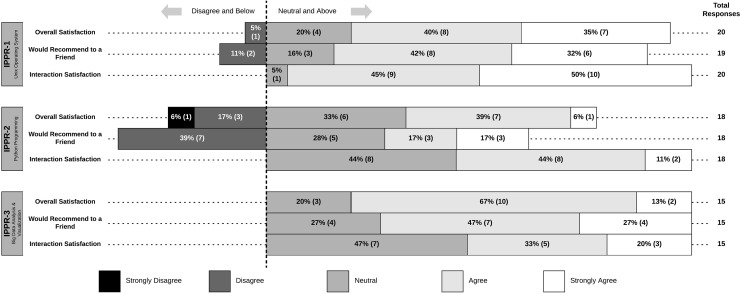
Impressively, there were no reports of dissatisfaction with the level of interaction participants had with their classmates, instructor, and TAs (Figure 2). Additionally, most respondents were satisfied overall and would recommend the experience to a friend (Figure 2). Disagreement was mostly related to IPPR 2 (Figure 2) so this finding was explored in the interviews. Students reported that it was not until the third course (IPPR 3) that they recognized and appreciated the potential power of applying Python skills that they have learned in IPPR 2 (Table 5, quote 1). Since the quantitative survey data were collected at the end of each course, students may have rated IPPR 2 differently at the end of the series. The fact that 100% of respondents for IPPR 3 chose neutral or above in overall satisfaction and would recommend it to a friend suggests that they recognized the utility of IPPR 2 skill sets since this is the foundation of IPPR 3.
Figure 2.
Course satisfaction metrics. Respondents’ agreement levels for each course with the 3 course satisfaction elements (overall, recommendation, and interaction).
Table 5.
Sample Quotes for Skill Development.
| Skill Development (sample quotes) | |
|---|---|
| 1 | Course Satisfaction—Significance of Python skills (IPPR 2) unclear until IPPR 3. “being that you have to learn to walk before you run, [IPPR 2] might have been slow, it might have been…basic I guess. You know where it’s just like ‘Oh well this is neat but its not as powerful as I thought.’ And then when you introduce Pandas [IPPR 3] in the third portion its power is magnified exponentially.” (Resident) |
| 2 | Motivation. “There’s a need for data scientists, a big need and I think that that’s, I recognize that and that’s why I invested my efforts heavily into learning programming.” (Postdoctoral fellow) |
| 3 | Motivation. “People are generating bigger and bigger data sets and they’re harder and harder to handle. So that was definitely a big motivation for me.” (Postdoctoral fellow) |
| 4 | Motivation. “I just really fell into it [final project], loved it, and made it the best I possibly could with error proofing and parsing input from the user…It was a lot of fun.” (Resident) |
| 5 | Motivation. “There were a couple times when I went into the homework assignment and was attempting to do it as it was outlined and found new packages that I wasn’t aware of, or something that was just really neat, and I made a cool figure out of that on the side. It was nice to have a structured experience but on my own I also went a little beyond that.” (MD/PhD student) |
| 6 | Real-world course material. “Python [Jupiter] notebook is, is how people get a lot of research done. A lot of buildings scripts done. This is what’s used in the industry. And so to be familiar with it and to understand like what’s the process. You might not realize that you’re learning that but you are. And I think that’s what’s kind of what was important.” (Resident) |
| 7 | Peer review. “That’s probably the best way to get people to become better at coding and to understand what’s written on the screen; is to just peer review, and then to do it yourself also.” (Graduate student) |
| 8 | Peer review. “you definitely get exposure to a lot of different ways to code. I definitely learned a lot of code just by looking at other people’s good and bad examples and it was very, very helpful quite honestly.” (Resident) |
| 9 | Peer review. “those [peer reviews] were really helpful, to look at how somebody else approached the same question as me. I used those a lot [and would] refer back to them.” (AP/CP Faculty) |
| 10 | Instructor/TA. “the professor’s attentive, so he’s helpful if you have questions. He tries to answer them quickly.” (Graduate student) |
| 11 | Instructor/TA. “He’s so supportive and encouraging and the TAs were so wonderful and they never made me feel bad at all for not understanding it. They were actually really encouraging and said things like that they really admire me for going out of my comfort zone, and those little comments really go a long way when you’re frustrated because you have no idea what’s going on. I appreciate those things.” (AP/CP Faculty) |
| 12 | Overview videos. “I think a few minutes at the beginning of each course with the recorded, sort of recap of the lecture in a short summary would be nice. That way if you’re not able to attend, you can watch that recording or if you are able to attend you can be there for that. That would be helpful.” (MD-PhD student) |
| 13 | Integrate Unix and Python. “one of the things that I guess I would change is to try to incorporate the first part [IPPR 1 Unix O/S]…to integrate that more with the rest of the portions [IPPR 2 and 3] I think could be beneficial and helpful for learning some tricks.” (Resident) |
| 14 | Additional exercises on laboratory management. “For me, for instance, if [the homework] was lab turnaround times and volumes or prices of things, people in the lab that would be taking the course would know that already, understand that data. Seeing it manipulated in the system in Pandas, that would give people ideas for how to apply it and I think that would be pretty cool.” (AP/CP Faculty) |
Abbreviations: AP/CP, anatomic/clinical pathology; IPPR, Informatics for Pathology Practice and Research; TA, teaching assistant.
Achievement of the course objectives
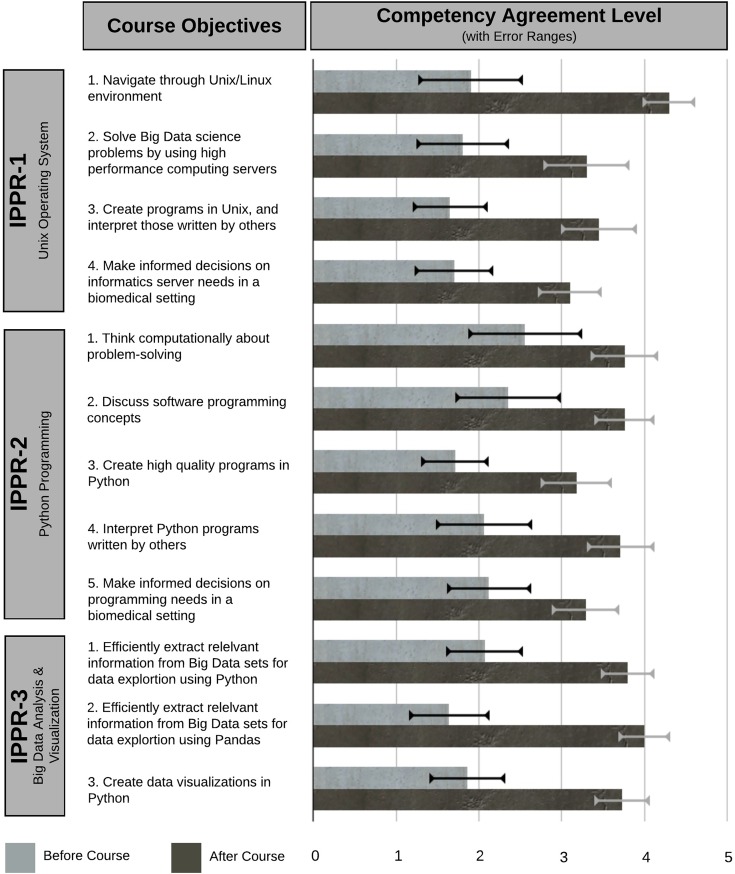
Compared to the beginning of the course, participants reported increased levels of agreement at the end that they could competently complete every course objective (Figure 3). However, as they didn’t strongly agree overall that they achieved the learning objectives (scores <5, Figure 3), we used the interviews to further gain insight. Participants mostly had a positive approach to learning where hands-on and problem-solving experience was valued and were able to connect material to their personal experience. Achievement of course objectives was influenced by the participants’ lack of time to apply their newly acquired discrete skills in an integrated manner to solving diverse, relevant real-world problems. Thus, below we describe contributing factors to successful achievement of course objectives as well as limitations to content mastery.
Figure 3.
Achievement of course objectives. Respondents’ agreement level that they achieved competency with the course objectives (retrospective preassessment and postcourse).
Successful learning factors
Motivation
The majority of the students interviewed were motivated to apply this informatics knowledge to their line of work (Table 5, quotes 2 and 3). There were many examples of participants going above and beyond their assignments to challenge themselves to improve proficiency and frequently made connections from the assignments to their work (Table 5, quotes 4 and 5).
Real-world course material
Usage of data sets, assignments, computational infrastructure, and programming platforms based on real-world applications and setting were appreciated by students (eg, Table 5, quote 6). They enjoyed using the same tools as industry, such as a high-performance computing server and a web-based interactive computational environment called Jupyter Notebook for Python programming (from Anaconda Navigator).
Peer-reviewing coding assignments
Even though students were given individual programming and coding assignments, students were encouraged to discuss the problems in groups (of their choice) during class and collaborate outside of class with the caveat that they were only exchanging the programming concepts and not copying the code from other students in the group. Comparing their work to their peers helped students evaluate programming solution efficiency. This was greatly appreciated by students (Table 5, quotes 7-9) and the instructor perceived the classroom environment as a very happy and productive atmosphere.
Instructor and TA
Another community aspect of the course mentioned by participants as being valuable for learning was the encouragement, attentiveness, and prompt response that they received from the instructor and the TA (Table 5, quotes 10 and 11).
Suggestions for improvements
Overview videos or material
Some survey participants suggested having an overview video for the classes. One student suggested that each module begin with a summary video of key points from the previous class (Table 5, quote 12). They felt reviews would benefit all students, especially those who were unable to attend. This would also help address one of the participant’s concern that, at the beginning, they “could not even understand how to read the textbook with computer code examples.”
Need for integration of Unix and Python courses
Although the Unix OS course (IPPR 1) was well received by the majority of participants, some of the interviewees noted that this course seemed disconnected from the other 2 python-based courses (IPPR 2 and IPPR 3). They advised having a greater integration of Unix OS and Python skills, or revisiting Unix applications at the end of the series (Table 5, quote 13).
Additional exercises on laboratory management
Although assignment exercises were based on practical applications using synthetic/publicly available genomic and medical data sets, one of the AP/CP faculty suggested having assignments include more on common laboratory management issues (Table 5, quote 14).
Application of Learning
Usability of acquired skills
Impact on current work
The interviewees provided specific accounts of how they were able to apply what they learned upon completing the course series. Their descriptions about their research or clinical work mostly included anecdotes of improved data analysis. Utility included processing, manipulation, and visualization of large data sets ranging from research projects to identifying pathology laboratory operational issues (Table 6, quotes 1-3). For a postdoctoral fellow, learning programming means “you start thinking a certain way,” so when he went to apply the concepts to his data “it wasn’t too hard [for him] to do Google searches and look back at [his] notes and catch on pretty quickly.” Exemplifying improved skills, he was able to use Python skills to make better data visualizations. An AP/CP faculty member used these new skills to find workflow solutions and track quality improvement for their required reporting for continued certification (Table 6, quote 3). However, one graduate student interviewed did not make the same level of connection to laboratory work. For him, the assignments “taught you little tidbits of things, of how to code or how to understand the code” which were “definitely helpful for the future, just understanding the basics” but lacked cumulative application for creating your own “program to analyze raw data.”
Table 6.
Sample Quotes for Postcourse Application of Learning.
| Application of Learning (sample quotes) | |
|---|---|
| 1 | Impact on current work. “People are generating data sets everywhere and [he has a role in] trying to centralize that with the deidentified patient information so that all of the data can be tied together and analyzed as a whole.” (Postdoctoral fellow) |
| 2 | Impact on current work. “using a little bit of [SAP] business objects to pull out a giant file and then using Python to program my script and hack through it to get these statistical analyses going.” (Resident) |
| 3 | Impact on current work. “this [course] ultimately has led to a project that I’m working on right now to revamp and automate the way that we look [understand/visualize] at that data because before the data was poorly accessible and…the analysis of the data was not [programmed] correct[ly]. This allowed me to fine-tune that analysis and now…it generates a report for the supervisors and other people in the lab so that they can digest it more easily.” (AP/CP faculty) |
| 4 | Improved communication with information technology colleagues. “To be able to talk to programmers that know it very well and to understand what they are talking back about so that you can facilitate the discussion between your physician peers and the programming team.” (Resident) |
| 5 | Improved communication with information technology colleagues “[T]he whole lab generates a lot of data, and that’s all stored in our lab information system, but access to it and understanding it is not trivial, and not everybody that’s good at the IT/IS side of it is necessarily good at the analysis because they don’t necessarily know what we’re doing with all the data, all the tasks, all the labs, all the studies that we’re doing. [The course series] gave me some tools to help bridge that gap, and I really found it to be incredibly valuable.” (AP/CP faculty) |
| 6 | Broader marketability and work flexibility “before getting into programming and I was basically a cell biologist working in the hoods in the lab and I’ve definitely switched to being more of a computer now…People are generating bigger and bigger data sets and they’re harder and harder to handle. So that was definitely a big motivation for me.” (Postdoctoral fellow) |
Abbreviation: AP/CP, anatomic/clinical pathology.
Improved communication with information technology colleagues
In addition to the direct use of programming skills for analyzing their own data after completion of IPPR, participants expressed that they now had a foundation of knowledge to better communicate with informatics colleagues (Table 6, quotes 4 and 5) and “understand informatics literature,” including “comprehending already analyzed data from other labs or other institutions.” These professional relationships occurred because the information technology team is “providing supplementary support for writing code” or are the only ones who “have direct access to [protected] information.”
Career path impacts
Formal informatics career pursuit
Two students discussed how the course impacted their pursuit of a Clinical Informatics Fellowship or an Informatics Medical subspecialty board certification. Although one faculty member had already decided to take the board exam, they were surprised by how much the course helped them prepare. An AP/CP faculty member recounted, “there was a whole chapter that I probably wouldn’t have had any idea what I was looking at if I hadn’t taken the course.” In addition, a resident explained how she had an interest in informatics but it was a postresidency career path that she could consider “someday.” Her experience in the course, however, convinced her that “this is where you need to be and this is what needs to be in your future,” so she is now actively pursuing both career avenues.
Broader marketability and work flexibility
A postdoctoral fellow described a more dramatic career transformation since the course (Table 6, quote 6). He recognized a big professional need for data scientists and this insight served as a primary motivator for him to work hard in IPPR to learn programming. Plus, acquiring informatics skills resulted in greater flexibility to balance work and home life since his work is no longer dependent on laboratory facilities.
No desire to change career trajectory
However, not everyone agreed that there was career path impact. For example, given the advanced stage of his career, one faculty member was not interested in pursuing a fellowship or board certification. Similarly, a graduate student who anticipated not using programming skills during his career maintained the same position after taking the course. Graduate student divulged further that his dissatisfaction was because of perceived pressure from his principal investigator, and some other students in their laboratory, into taking these courses. Due to his research interest area, he felt that he was “not going to really use most of this in the future.” However, for those who do foresee using bioinformatics in their career, he felt the series would be beneficial, adding “especially if you’ve never done bioinformatics or coding before, it’s definitely something that you should take.”
Continued Learning
There was clear acknowledgment among those interviewed that a single semester greatly limited their ability to thoroughly learn programming. As an AP/CP faculty quipped, how much can one really expect “to get out of a couple of credit hours?” He equated learning programming to a musical instrument because one wouldn’t expect to “immediately turn around and write a symphony.” Similarly, a resident remarked, “Obviously there is much more to learn, but I think this has prepared me to learn it.”
There was also an interest in learning more. A postdoc noted that “[the instructor is] the expert in some things that I would like to still learn.” Topics of interest for future courses frequently included statistical data visualization tools (mostly R but also Seaborn) as well as SQL and Business Objects. One person would especially like guidance with genetics data (bioinformatics).
Discussion
Systematic informatics training in Pathology is relatively new. In this article, we described our efforts to set up and assess such a training program at the University of Florida. The training was done through 3 courses (1 credit hour each): IPPR 1—Unix OS, IPPR 2—Python Programming, and IPPR 3—Advanced Data Analysis & Visualization. Overall assessment and impact of this training were assessed with qualitative and quantitative data collection and analysis.
We acknowledge that finding a faculty with interdisciplinary domain expertise in informatics and pathology is not common. An alternative approach for institutions that lack such interdisciplinary training expertise is to have multiple instructors coteach the classes. For example, an MD-trained faculty with pathology clinical expertise could coteach the course series with another faculty (PhD/Masters level) with informatics expertise in computer programming and UNIX. Based on our IPPR implementation experience, we suggest potential instructor(s) to have the following expertise and qualification for effectively teach this type of course series—(1) proficiency and hands-on experience in programming and Unix OS, (2) overall understanding of the pathology domain and informatics needs (eg, CP/AP workflows and EP research of pathology), (3) enthusiasm to teach even to members with zero programming/computational experience.
Informatics for Pathology Practice and Research course series is offered via university academic credits, which has following advantages: (1) courses are listed on the official transcripts, (2) the standard and rigor of the course follows standards set by university and educational accredited organizations. However, this comes with a burden of tuition costs for nongraduate students such as faculty, postdocs, staff, residents, and fellows. Tuition for faculty, postdocs, and staff enrolled in IPPR course series was paid by the University of Florida’s employee education benefits program. For our resident(s), paying tuition became a challenge and our department chair agreed to pay the tuition from departmental funds.
The assessment findings suggest that we were able to meet our course objectives, but with room for improvement. Major factors that contributed to learning included the collaborative environment and coding assignments. The courses were deemed to have impact in the subsequent months after training through development of new software workflows and improvement in communication with informatics colleagues. A significant impact of the course was in raising awareness of the importance of informatics, motivating participants for new career paths, and giving them flexibility in their pathology practice and research.
Interviewing selected participants rather than the entire sample is a limitation of this study. The overall qualitative findings cannot be generalized; we cannot determine how well they represent nonparticipants’ perspectives. Responses are contextualized by the individuals who participated and are situated at a particular point in time. The findings were also restricted by the available response time and individuals’ willingness to provide authentic responses.
Given the heterogeneity of the backgrounds of pathology participants, we suggest that implementing such a training program will require strong institutional support. In the absence of institutional support, participants with diverse demands on their time and workload pressures may have difficulty in participating in such a program. Awareness at the institutional level of the importance of such a program and communicating it to members will have a positive impact on the success of the program. Support in the form of tuition will also have strong, positive impact.
Conclusion
We have successfully developed and implemented a foundational training program in pathology informatics at the University of Florida. We expect to improve this training based on the feedback provided by students. We hope that participants of this informatics training program will choose to further pursue informatics training in bioinformatics, imaging informatics, machine learning, and clinical informatics. We plan to expand our training to databases, R-programming, and genetic analysis. In future versions of the program, we also plan to include ad hoc presentations, elective rotations, and research mentoring focused on specific informatics themes such as cloud computing, EHR/LIS tools, pathology informatics workflows, and so on. We are interested in exploring opportunities to partner with other institutions to extend the impact of this course series. This includes pursuing grant opportunities to provide these trainings in a fully online format including “train-the-trainer” sessions.
Supplemental Material
Supplemental Material, APC-19-0101_Supplemental_Appendix_1 for Informatics Training for Pathology Practice and Research in the Digital Era by Heather T. D. Maness, Linda S. Behar-Horenstein, Michael Clare-Salzler and Srikar Chamala in Academic Pathology
Supplemental Material, APC-19-0101_Supplemental_Table_1 for Informatics Training for Pathology Practice and Research in the Digital Era by Heather T. D. Maness, Linda S. Behar-Horenstein, Michael Clare-Salzler and Srikar Chamala in Academic Pathology
Supplemental Material, APC-19-0101_Supplemental_Table_2 for Informatics Training for Pathology Practice and Research in the Digital Era by Heather T. D. Maness, Linda S. Behar-Horenstein, Michael Clare-Salzler and Srikar Chamala in Academic Pathology
Supplemental Material, APC-19-0101_Supplemental_Table_3 for Informatics Training for Pathology Practice and Research in the Digital Era by Heather T. D. Maness, Linda S. Behar-Horenstein, Michael Clare-Salzler and Srikar Chamala in Academic Pathology
Supplemental Material, APC-19-0101_Supplemental_Table_4 for Informatics Training for Pathology Practice and Research in the Digital Era by Heather T. D. Maness, Linda S. Behar-Horenstein, Michael Clare-Salzler and Srikar Chamala in Academic Pathology
Acknowledgements
The authors gratefully acknowledge Vektra Casler, for contribution to refine some of the figures used in this manuscript.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Srikar Chamala  https://orcid.org/0000-0001-6367-7615
https://orcid.org/0000-0001-6367-7615
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Sinard JH, Powell SZ, Karcher DS. Pathology training in informatics: evolving to meet a growing need. Arch Pathol Lab Med. 2014;138:505–511. [DOI] [PubMed] [Google Scholar]
- 2. Mandelker D, Lee RE, Platt MY, et al. Pathology informatics fellowship training: focus on molecular pathology. J Pathol Inform. 2014;5:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Black-Schaffer WS, Morrow JS, Prystowsky MB, Steinberg JJ. Training pathology residents to practice 21st century medicine. Acad Pathol. 2016;3 doi: 10.1177/2374289516665393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hanley T, Sowder AM, Palmer CA, Weiss RL. Teaching laboratory management principles and practices through mentorship and graduated responsibility: the assistant medical directorship. Acad Pathol. 2016;3 doi: 10.1177/2374289516678972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baird G. The laboratory test utilization management toolbox. Biochem Med. 2014;24:223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Henricks WH, Karcher DS, Harrison JH, Jr, et al. Pathology informatics essentials for residents. Arch Pathol Lab Med. 2017;141:27. [DOI] [PubMed] [Google Scholar]
- 7. Henricks WH, Karcher DS, Harrison JH, Jr, et al. Pathology informatics essentials for residents: a flexible informatics curriculum linked to accreditation council for graduate medical education milestones (a secondary publication). Acad Pathol. 2016;1:3 doi: 10.1177/2374289516659051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Levy B. Clinical informatics: new board certification and fellowship for physician leaders. Physician Leadersh J. 2015;2:38. [PubMed] [Google Scholar]
- 9. Quinn AM, Klepeis VE, Mandelker DL, et al. The ongoing evolution of the core curriculum of a clinical fellowship in pathology informatics. J Pathol Inform. 2014;5:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Harrison J. Beyond the LIS: open source big data tools for laboratory operational analytics and visualization. 2016. https://attendee.gotowebinar.com/recording/1720942784226527745. Accessed June 27, 2018.
- 11. Kopf D. If you want to upgrade your data analysis skills, which programming language should you learn? 2017. https://qz.com/1063071/the-great-r-versus-python-for-data-science-debate. Accessed August 9, 2018.
- 12. Lee CH. How to choose between learning python or R first. 2015. https://blog.udacity.com/2015/01/python-vs-r-learn-first.html. Accessed August 9, 2018.
- 13. Willems K. Choosing R or python for data analysis? An infographic. 2015. https://www.datacamp.com/community/tutorials/r-or-python-for-data-analysis. Accessed August 9, 2018.
- 14. Guo P. Python is now the most popular introductory teaching language at top U.S. universities. BLOG@CACM. 2014. https://cacm.acm.org/blogs/blog-cacm/176450-python-is-now-the-most-popular-introductory-teaching-language-at-top-u-s-universities/fulltext. Accessed August 9, 2018.
- 15. Cass S, Bulusu P. Interactive: the top programming languages. 2018. https://spectrum.ieee.org/static/interactive-the-top-programming-languages-2018. Accessed August 9, 2018.
- 16. Kim MK, Kim SM, Khera O, Getman J. The experience of three flipped classrooms in an urban university: an exploration of design principles. Internet High Educ. 2014;22:37–50. [Google Scholar]
- 17. Harding J. Qualitative Data Analysis from Start to Finish. Thousand Oaks, CA: Sage; Publications, Inc; 2013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, APC-19-0101_Supplemental_Appendix_1 for Informatics Training for Pathology Practice and Research in the Digital Era by Heather T. D. Maness, Linda S. Behar-Horenstein, Michael Clare-Salzler and Srikar Chamala in Academic Pathology
Supplemental Material, APC-19-0101_Supplemental_Table_1 for Informatics Training for Pathology Practice and Research in the Digital Era by Heather T. D. Maness, Linda S. Behar-Horenstein, Michael Clare-Salzler and Srikar Chamala in Academic Pathology
Supplemental Material, APC-19-0101_Supplemental_Table_2 for Informatics Training for Pathology Practice and Research in the Digital Era by Heather T. D. Maness, Linda S. Behar-Horenstein, Michael Clare-Salzler and Srikar Chamala in Academic Pathology
Supplemental Material, APC-19-0101_Supplemental_Table_3 for Informatics Training for Pathology Practice and Research in the Digital Era by Heather T. D. Maness, Linda S. Behar-Horenstein, Michael Clare-Salzler and Srikar Chamala in Academic Pathology
Supplemental Material, APC-19-0101_Supplemental_Table_4 for Informatics Training for Pathology Practice and Research in the Digital Era by Heather T. D. Maness, Linda S. Behar-Horenstein, Michael Clare-Salzler and Srikar Chamala in Academic Pathology