Abstract
Mosquito-borne flaviviruses include several important agents of human disease and have provided striking examples of emerging infections. In this study we present the design and validation of a single tube RT-PCR assay using a pair of consensus primers for the detection of mosquito-borne flaviviruses. Sequencing of the amplicons permits the species identification. The assay was validated using RNA from the yellow fever virus vaccine strain and from representative strains of dengue viruses 1, 2, 3 and 4, West Nile virus, Kunjin virus (a clade of West Nile virus), and St. Louis encephalitis virus.
Keywords: Mosquito-borne flaviviruses, West Nile virus, RT-PCR
1. Introduction
Within the family Flaviviridae, the genus Flavivirus comprises at least 73 species, 40 of which have been associated with human disease (Burke and Monath, 2001). Nucleotide sequence analysis has shown that the genus can be divided in three clusters (Kuno et al., 1998, Lindenbach and Rice, 2001), namely the mosquito-borne viruses, the tick-borne viruses and a third cluster for which no vectors have been identified. In turn the mosquito-borne cluster encompasses nine clades, including the clades of the yellow fever virus (YFV), and of dengue fever viruses (the four serotypes of which also form a serocomplex). The Japanese encephalitis virus (JEV) serocomplex include viruses from three clades; the species in that complex include most notably JEV itself, West Nile virus (WNV), Kunjin encephalitis virus, Murray Valley encephalitis virus (MVE), St. Louis encephalitis virus (SLE) and Usutu virus (Burke and Monath, 2001, Kuno et al., 1998, Lindenbach and Rice, 2001).
The epidemiology of these viruses is complex and depends on the ecology of their vectors in relations with human habitats. Consequently, many human-dependant factors, such as increase in population/density of population, massive population displacement, unplanned urbanization, deforestation, establishment of urban mosquito populations, and deterioration of public health surveillance and infrastructure, can all influence the spread and activity of these viruses, which have frequently provided striking examples of emerging infections in both the developing world and industrialized nations (Gubler, 2004, Mackenzie et al., 2004, Tomori, 2004).
Yellow fever virus, the prototype of the genus, originated from Africa and spread to South America. At least 200,000 cases, causing 30,000 deaths, occur every year worldwide; 90% of cases occur in Africa. These numbers are widely acknowledged to be underestimates because of the insensitive surveillance in endemic areas (Gubler, 2004, Mackenzie et al., 2004, Tomori, 2004).
Dengue fever viruses have undergone a dramatic increase in geographic spread and frequency of infections over the past 30 years. Currently, the incidence is estimated at 50–100 million cases per year, causing 20,000 deaths. In all tropical areas, more than one serotype circulate and re-introduction of these viruses in the continental USA cannot be ruled out (Gubler, 2004, Mackenzie et al., 2004).
Japanese encephalitis virus (JEV) is encountered throughout Southeast Asia and is the most important cause of viral encephalitis in these countries. The current estimate of 30,000–50,000 cases/year is likely an underestimate and JEV activity is on the increase (Kabilan et al., 2004, Mackenzie et al., 2004, Promed-mail, 2005a, Promed-mail, 2005b).
West Nile virus (WNV) underwent a dramatic expansion of its ecological niche over the past few years, first noticed with an encephalitis outbreak in New York City in 1999 and followed by spread throughout the USA and Canada, as well as Mexico and the Carribean. In 2002 and 2003 the virus was responsible for two of the largest arboviral epidemics ever observed and has now become endemic in large areas of the Western Hemisphere (Davis et al., 2005, Drebot and Artsob, 2005, Hayes, 2005, Mackenzie et al., 2004). Kunjin virus, recovered from cases of encephalitis in Western Australia, has been reclassified as a clade of West Nile virus (Burke and Monath, 2001, Mackenzie et al., 2004).
St. Louis encephalitis virus has been recognized for several decades as a cause of viral encephalitis in the USA (Burke and Monath, 2001). Recently a large outbreak in Cordoba, Argentina was documented (Promed-mail, 2005a, Promed-mail, 2005b).
In the late summer of 2001, a sudden increase in mortality in several species of birds in Central Europe was demonstrated to be caused by the emergence of the Usutu virus, a flavivirus which had previously remained confined to Africa. However, the virus has not so far been linked to human disease (Weissenbock et al., 2002).
Given the disease burden caused by these viruses and their emerging behavior, laboratory testing will become increasingly important. Yet, the laboratory diagnosis of flavivirus infection remains unsatisfactory; isolation of these viruses is time consuming, and for many of them requires a biosafety level 3 installation. Serology tests other than neutralization assays frequently display cross reactivity between species. For all these reasons, development of molecular tests for these agents, including RT-PCR, has attracted a lot of efforts and attention. In this study we present a single tube RT-PCR assay for mosquito-borne flaviviruses, using a single pair of consensus primers. We show that the assay can detect several important species of flaviviruses with high sensitivity. Species identification is provided by sequencing the amplicons; in the case of dengue viruses this also permits the serotype determination.
2. Material and methods
2.1. Viruses
Yellow fever virus vaccine strain was obtained from a vial of YF-VAX (Aventis Pasteur), reconstituted as per the manufacturer's recommendations. The preparation is certified to contain a titer of at least 2 × 105 pfu/ml.
West Nile virus strain NY, St. Louis encephalitis virus, dengue fever serotype 1, 2, 3, and 4 were grown and titrated in a biosafety level 3 facility at the National Microbiology Laboratory (Winnipeg), as described (Weingartl et al., 2003). Kunjin virus was grown in biosafety level 3 conditions, as described (Weingartl et al., 2003).
Additional samples tested consisted of mosquito pools that had been found positive for WNV using a real time RT-PCR Taqman based assay (Lanciotti et al., 2000). The samples included two pool homogenates of Culex pipiens collected in the province of Ontario, Canada in 2002, two homogenates of C. tarsalis collected in the province of Manitoba in 2003, and two homogenates of C. tarsalis from Manitoba, 2005.
For specificity studies, several viral samples were used, including clinical samples found to contain CMV and EBV DNA by PCR testing, as described (Johnson et al., 2000); a clinical isolate of influenza virus from the 2004–2005 season, typed as H3 by sequencing of the hemagglutinin gene; echovirus 11 from the laboratory collection at the Hospital for Sick Children; hepatitis C virus RNA was obtained by in vitro transcription of the infectious clone pCV-H77C (Yanagi et al., 1997) (the clone was kindly provided by Dr. J. Bukh, NIAID, National Institutes of Health) followed by DNAse I digestion, RNA purification and serial dilution; and human coronavirus OC-43 (kindly provided by Dr. P.J. Talbot, INRS, Institut Armand Frappier).
2.2. RNA extraction
RNA extraction was done using the Qiagen RNA easy kit, as per the manufacturer's recommendations, or, for the yellow fever vaccine strain, TRIzol (Life Technologies), as per the manufacturer's recommendations. The RNA was resuspended in 10 mM dithiothreitol (Promega) with 5% (v/v) RNasin (20–40 u/μl, Promega), and 10-fold serial dilutions were prepared with the same diluent.
2.3. Primers
Consensus primers were designed to target segments of the NS5 coding region conserved across several species of flaviviruses including YFV, the dengue viruses serocomplex and several members of the JEV serocomplex, (Fig. 1 ). The sequences of the primers are:
Fig. 1.

Alignments calculated from the nucleotide sequences of several flavivirus species with the primer FLAVI-1 (A) or with the reverse-complemented sequence of primer FLAVI-2 (B). Dots indicate identity with the consensus sequence on top of the alignment. West Nile virus strain NY2000 (wnny): GenBank accession #AF404756; West Nile virus strain MD2000 (wnmd): #AF404753; West Nile virus strain Eg 101 (wneg 101): #AF260968; Kunjin virus: #D00246; St. Louis encephalitis virus (sle): AF013416; Murray Valley encephalitis (mve): NC_000943; Japanese encephalitis virus (jev): AF254452; Usutu virus: #AF013412; dengue virus 1 (den1): AB074766; dengue virus type 2 (den2): NC_001474; dengue virus type 3 (den3): NC_001475; dengue virus type 4 (den4): NC_002640; yellow fever virus (yfv): #YFV54798.
FLAVI-1 (sense primer): 5′ AATGTACGCTGATGACACAGCTGGCTGGGACAC 3′ and FLAVI-2 (antisense primer): 5′ TCCAGACCTTCAGCATGTCTTCTGTTGTCATCCA 3′.
These two primers correspond to the segments [9273–9305] and [10,102–10,136], respectively, of the West Nile virus NY 2000 (GenBank accession #AF404756).
RT-PCR with these primers would result in amplicons of similar sizes, depending on the virus species: 854 bp for dengue 1 and and dengue 2, 857 bp for dengue 3 and dengue 4, and 863 bp for YFV, WNV (including Kunjin), JEV, MVE, SLE, and Usutu.
2.4. RT-PCR
Amplification was carried out using the Qiagen One Step RT-PCR kit. Each reaction was performed in a 0.6 ml thin wall tube (Stratagene) in a total volume of 50 μl overlaid with 50 μl of mineral oil. Each reaction mix contained 10 μl of 5× Qiagen buffer, 2 μl of dNTP mix (each dNTP at 10 mM concentration), 3 μl of each primer (10 μM stock), 20 μl of molecular grade double distilled water (ddH2O) and 2 μl of the One Step enzyme mix (Qiagen). The master mix was then aliquoted in tubes, to which 10 μl of template RNA was added. The PCR thermal cycling was done on a Stratagene Robocycler 40, with an initial incubation at 50 °C for 30 min, followed by an incubation at 95 °C for 15 min, and 45 cycles consisting of denaturation at 94 °C for 1 min, annealing at 58 °C for 1 min, and elongation at 72 °C for 1 min 30 s. Extensive precautions against PCR contamination, as previously described (Johnson et al., 2000) were strictly observed. Positive and negative controls, as well as extraction controls and controls for PCR inhibition, were set up essentially as described (Johnson et al., 2000).
2.5. Electrophoresis analysis of the amplicons
A 10 μl volume of each reaction was submitted to electrophoresis on 1.8% agarose gels containing ethidium bromide. The gels were visualized on a UV transilluminator and photographed.
2.6. Sequencing
Amplicons were submitted for automated sequencing, for both strands, using the PCR primers as sequencing primers. Sequencing was performed by the DNA Sequencing Facility, The Centre for Applied Genomics, Hospital for Sick Children.
2.7. Sequences analysis
Sequence editing and analysis were done using the program Generunner for Windows ver. 3.05 (Hasting Software). Sequence alignments were calculated using ClustalX for Windows ver. 1.81 (Thompson et al., 1997), with the default parameters for gap opening and gap extension. A phylogenetic tree was established using Treecon for Windows ver. 1.3.b (Van de Peer and De Wachter, 1994). In brief, the distance was calculated without correction, with insertions and deletions taken into account. The topology was inferred using the UPGMA method. Bootstrap analysis was done using 1000 replicates.
3. Results
3.1. Primers and phylogenetic tree
Based on the sequence alignment of the NS5 coding region of several flaviviruses (Fig. 1) the consensus primers FLAVI-1 and FLAVI-2 were designed. The expected sequence of the amplicons were deduced from the genomic sequences and the primer sequences, and a phylogenetic tree was inferred (Fig. 2 ), which demonstrates that this assay permits reliable identification of the virus species by sequencing the amplicon.
Fig. 2.
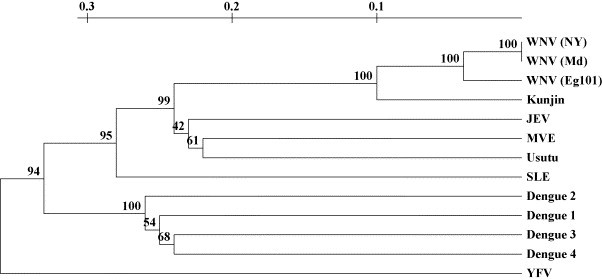
Phylogenetic tree calculated from the predicted sequences of amplicons generated by the RT-PCR assay. Bootstrap values are indicated (as percentage) above the nodes. Scale: substitution/site. WNV(NY): West Nile virus strain NY2000; WNV(Md): West Nile virus strain Md2000; WNV(Eg 101): West Nile virus strain Eg 101; JEV: Japanese encephalitis virus; MVE: Murray Valley encephalitis; SLE: St. Louis encephalitis virus; YFV: yellow fever virus.
3.2. Sensitivity and specificity
Amplicons of the expected size were readily and reproducibly obtained from RNA extracted from cultures of WNV (NY strain), SLE, Kunjin, Dengue serotypes 1, 2, 3 and 4, and YFV vaccine strain. In each case, the identity of the amplicon was confirmed by sequencing. For the stocks of viruses that had been titrated, serial dilution of RNA allowed to estimate the equivalent limit of detection in plaque forming units (pfu), as shown in Table 1 . In addition, for WNV the RNA serial dilution was used to compare the sensitivity of the assay described here with that of a commercially available quantitative RT-PCR kit (RealArt WNV LC RT-PCR kit, Artus GmbH). The sensitivity was found to be equivalent, with a measurement of approximately 16 genome copies (average of two experiments). The methods of RNA extraction used in the clinical laboratory at the Hospital for Sick Children typically extract RNA from a volume of 100–140 μl, although for WNV testing the Qiaamp Ultrasense kit (Qiagen), with a volume of sample up to 500 μl, is preferred. It can be readily seen that the sensitivity obtained for all viruses tested is similar to that reported with assays for other flaviviruses (Scaramozzino et al., 2001, Busch et al., 2005, Kao et al., 2005, Kuno, 1998).
Table 1.
Threshold of detection of the assay for different viruses
| Virus | Sensitivity threshold (pfu) |
|---|---|
| WNV (NY) | 5 |
| SLE | 0.15 |
| DEN 1 | 0.3 |
| DEN 2 | 0.3 |
| DEN 3 | 0.06 |
| DEN 4 | 2.6 |
| YFV | >0.01 |
For each virus, RNA was extracted from titrated stocks and serially diluted. For the last dilution still positive, we indicate in the table the equivalent amount of pfu.
Samples from six mosquito pools collected in different provinces in Canada over the course of 3 years were also tested. Amplicons of the expected size were obtained from all samples, and sequencing of the amplicons confirmed the viral species as WNV.
Nucleic acids extracted from preparation of cytomegalovirus, influenza H3N2 (clinical isolate, season 2004–2005), echovirus 11 or hepatitis C virus (genotype 1a) did not generate amplicons of 854–863 bp expected from flaviviruses. Similarly, nucleic acids extracted from human CSF or plasma of uninfected patients yielded negative RT-PCR.
4. Discussion
In this study the use of a single pair of consensus primers for the detection of mosquito-borne flaviviruses was validated. The targeting of segments of the NS5 coding region to detect several flaviviruses has been reported before; for example, Kuno et al. (1998) have established a precise phylogeny of the flaviviruses by sequencing large amplicons in the NS5 region from cultured viruses. The consensus primers used in this study show some similarity with two of the primers used by Kuno et al. (1998); however the version of the primers presented here and a careful optimization of the reaction allow for a sensitivity sufficient for direct detection of viruses in clinical samples and not just from cultured viruses. Scaramozzino et al. (2001) have described a highly sensitive PCR assay for the detection of several flaviviruses, but their assay used a two step RT-PCR format followed by a second heminested PCR, a procedure less convenient for clinical samples.
The analytical sensitivities are shown in Table 1; taking into account the volume of extraction, as outlined in the Section 3, the sensitivities obtained for the viruses tested compare favorably to that reported with assays for other flaviviruses (Scaramozzino et al., 2001, Busch et al., 2005, Kao et al., 2005, Kuno, 1998).
With the current sensitivity of molecular tests, laboratory diagnosis of acute yellow fever and dengue fever can be readily accomplished. Even though the viremia is short lived, it occurs during the symptomatic febrile phase and reaches high viral titers: up to 105–106 pfu/ml for YFV(Tomori, 2004), and up to 3.4 × 106 genome copies/ml and 1.49 × 107 genome copies/ml in dengue fever and dengue hemorrhagic fever, respectively (Wang et al., 2003).
The use of molecular tests for the diagnosis of other flaviviruses can be more problematic; thus for WNV, not only is the viremia of short duration but the titer is considerably lower, with a peak of no more than 1.6 × 103 pfu/ml (Hayes, 2005). A single negative RT-PCR would therefore not rule out a current infection and consequently, serological testing continues to play an important role in laboratory diagnosis in immunocompetent persons. Molecular testing would arguably be of greater usefulness in cases of suspected infection with WNV in immunocompromised patients, where the humoral response may be blunted and the viremia may be prolonged or of higher titer (Huang et al., 2002). In addition, molecular assays have been used as part of screening for blood and organ donors (CDC, 2005, Health_Canada, 2004, Kleinman et al., 2004). Finally, the use of a single polyvalent flavivirus PCR would be useful for the detection of flaviviruses in various vectors and non-human hosts, to aid in the monitoring of viral activity, including virus emergence.
The assay presented here requires sequencing of the amplicons for confirmation and identification of the viral species. Given the increasing ease and affordability of automated sequencing, this is not a great imposition on clinical laboratories proficient in molecular techniques. Primers capable of detecting several viral species permit the use for a positive control of a viral sequence unlikely to be present in the area where the laboratory is located (for example, Kunjin virus RNA is now used at the hospital for sick children), thus eliminating a possible source of laboratory contamination of the assay. The implementation of this assay in a laboratory that must monitor, say, WNV, also means that an assay for more infrequent viruses is in fact kept ready in the laboratory. For example the diagnosis of acute infection with dengue 1 in a returning traveler could be confirmed in our laboratory with this assay (data not shown).
In summary, a single tube RT-PCR using consensus primers was designed and validated. Coupled with sequencing, it could detect with great sensitivity and identify several mosquito-borne flaviviruses including WNV, Kunjin, SLE, YFV and dengue fever viruses. It is expected that this assay will be useful for clinical diagnosis and for the ecological surveillance of flaviviruses.
Acknowledgement
This work was supported by the Department of Paediatric Laboratory Medicine, Hospital for Sick Children.
References
- Burke D.S., Monath T.P. Flaviviruses. In: Knipe D.M., Howley P.M., Griffin D.E., Lamb R.A., Martin M.A., Roizman B., editors. 4th ed. vol. 1. Lippincott Williams 7 Wilkins; Philadelphia: 2001. pp. 1043–1126. (Fields’Virology). [Google Scholar]
- Busch M.P., Tobler L.H., Saldanha J., Caglioti S., Shyamala V., Linnen J.M., Gallarda J., Phelps B., Smith R.I., Drebot M., Kleinman S.H. Analytical and clinical sensitivity of West Nile virus RNA screening and supplemental assays available in 2003. Transfusion. 2005;45:492–499. doi: 10.1111/j.0041-1132.2005.04382.x. [DOI] [PubMed] [Google Scholar]
- CDC, 2005. West Nile virus.
- Davis, C.T., Ebel, G.D., Lanciotti, R.S., Brault, A.C., Guzman, H., Siirin, M., Lambert, A., Parsons, R.E., Beasley, D.W., Novak, R.J., Elizondo-Quiroga, D., Green, E.N., Young, D.S., Stark, L.M., Drebot, M.A., Artsob, H., Tesh, R.B., Kramer, L.D., Barrett, A.D., 2005. Phylogenetic analysis of North American West Nile virus isolates, 2001–2004: evidence for the emergence of a dominant genotype. Virology. [DOI] [PubMed]
- Drebot M.A., Artsob H. West Nile virus. Update for family physicians. Can. Fam. Physician. 2005;51:1094–1099. [PMC free article] [PubMed] [Google Scholar]
- Gubler D.J. The changing epidemiology of yellow fever and dengue, 1900 to 2003: full circle? Comp. Immunol. Microbiol. Infect. Dis. 2004;27:319–330. doi: 10.1016/j.cimid.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Hayes E.B. Epidemiology and transmission dynamics of West Nile virus disease. Emerg. Infect. Dis. 2005;11:1167–1173. doi: 10.3201/eid1108.050289a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Health_Canada, 2004. Health Canada Guidance Document: Measures to prevent West Nile virus transmission through cells, tissues and organs for transplantation and assisted reproduction.
- Huang C., Slater B., Rudd R., Parchuri N., Hull R., Dupuis M., Hindenburg A. First Isolation of West Nile virus from a patient with encephalitis in the United States. Emerg. Infect. Dis. 2002;8:1367–1371. doi: 10.3201/eid0812.020532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson G., Nelson S., Petric M., Tellier R. Comprehensive PCR-based assay for detection and species identification of human herpesviruses. J. Clin. Microbiol. 2000;38:3274–3279. doi: 10.1128/jcm.38.9.3274-3279.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabilan L., Rajendran R., Arunachalam N., Ramesh S., Srinivasan S., Samuel P.P., Dash A.P. Japanese encephalitis in India: an overview. Indian J. Pediatr. 2004;71:609–615. doi: 10.1007/BF02724120. [DOI] [PubMed] [Google Scholar]
- Kao C.L., King C.C., Chao D.Y., Wu H.L., Chang G.J. Laboratory diagnosis of dengue virus infection: current and future perspectives in clinical diagnosis and public health. J. Microbiol. Immunol. Infect. 2005;38:5–16. [PubMed] [Google Scholar]
- Kleinman, S., Busch, M., Caglioti, S., Stramer, S.L., Dodd, R., Strong, D.M., Dickey, W., Salvidar, B., Gilchrist, M., Brend, S., Nakhasi, H., Epstein, J., Goodman, J., Chamberland, M., Kuehnert, M., Petersen, L., Crall, N., Marfin, A., Boo, T., Montgomery, S., 2004. Update: West Nile virus screening of blood donations and transfusion-associated transmission, United States, 2003. MMWR—Morbidity Mortality Weekly Report, vol. 53, p. 281-4. [PubMed]
- Kuno G. Universal diagnostic RT-PCR protocol for arboviruses. J. Virol. Meth. 1998;72:27–41. doi: 10.1016/s0166-0934(98)00003-2. [DOI] [PubMed] [Google Scholar]
- Kuno G., Chang G.J., Tsuchiya K.R., Karabatsos N., Cropp C.B. Phylogeny of the genus Flavivirus. J. Virol. 1998;72:73–83. doi: 10.1128/jvi.72.1.73-83.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti R.S., Kerst A.J., Nasci R.S., Godsey M.S., Mitchell C.J., Savage H.M., Komar N., Panella N.A., Allen B.C., Volpe K.E., Davis B.S., Roehrig J.T. Rapid detection of west nile virus from human clinical specimens, field-collected mosquitoes, and avian samples by a TaqMan reverse transcriptase-PCR assay. J. Clin. Microbiol. 2000;38:4066–4071. doi: 10.1128/jcm.38.11.4066-4071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach B.D., Rice C.M. Flaviviridae: the viruses and their replication. In: Knipe D.M., Howley P.M., editors. 4th ed. Vol.1. Lippincott Williams and Wilkins; Philadelphia: 2001. pp. 991–1042. (Fields’ Virology). [Google Scholar]
- Mackenzie J.S., Gubler D.J., Petersen L.R. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat. Med. 2004;10:S98–S109. doi: 10.1038/nm1144. [DOI] [PubMed] [Google Scholar]
- Promed-mail, 2005a. Flavivirus infection, fatal, Argentina (Cordoba) (03): St Louis encephalitis.September 4 2005, http://www.promedmail.org/.
- Promed-mail, 2005b. Japanese encephalitis, India (Uttar Pradesh) (06). September 9 2005, http://www.promedmail.org/.
- Scaramozzino N., Crance J.M., Jouan A., DeBriel D.A., Stoll F., Garin D. Comparison of flavivirus universal primer pairs and development of a rapid, highly sensitive heminested reverse transcription-PCR assay for detection of flaviviruses targeted to a conserved region of the NS5 gene sequences. J. Clin. Microbiol. 2001;39:1922–1927. doi: 10.1128/JCM.39.5.1922-1927.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomori O. Yellow fever: the recurring plague. Crit. Rev. Clin. Lab. Sci. 2004;41:391–427. doi: 10.1080/10408360490497474. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y., De Wachter R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- Wang W.K., Chao D.Y., Kao C.L., Wu H.C., Liu Y.C., Li C.M., Lin S.C., Ho S.T., Huang J.H., King C.C. High levels of plasma dengue viral load during defervescence in patients with dengue hemorrhagic fever: implications for pathogenesis. Virology. 2003;305:330–338. doi: 10.1006/viro.2002.1704. [DOI] [PubMed] [Google Scholar]
- Weingartl H.M., Drebot M.A., Hubalek Z., Halouzka J., Andonova M., Dibernardo A., Cottam-Birt C., Larence J., Marszal P. Comparison of assays for the detection of West Nile virus antibodies in chicken serum. Can. J. Vet. Res. 2003;67:128–132. [PMC free article] [PubMed] [Google Scholar]
- Weissenbock H., Kolodziejek J., Url A., Lussy H., Rebel-Bauder B., Nowotny N. Emergence of Usutu virus, an African mosquito-borne flavivirus of the Japanese encephalitis virus group, central Europe. Emerg. Infect. Dis. 2002;8:652–656. doi: 10.3201/eid0807.020094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagi M., Purcell R.H., Emerson S.U., Bukh J. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl. Acad. Sci. U.S.A. 1997;94:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]


