Highlights
► Atypical pestiviral Erns protein was produced in a baculovirus expression system. ► An Erns microsphere immunoassay was evaluated for detecting BVDV antibodies in 596 sera. ► The assay had a 100% sensitivity and specificity relative to ELISA.
Keywords: Atypical bovine pestivirus, BVDV, ELISA, Erns, Immunoassay, Luminex
Abstract
Atypical bovine pestiviruses are related antigenically and phylogenetically to bovine viral diarrhea viruses (BVDV-1 and BVDV-2), and may cause the same clinical manifestations in animals. Glycoprotein Erns of an atypical bovine pestivirus Th/04_KhonKaen was produced in a baculovirus expression system and was purified by affinity chromatography. The recombinant Erns protein was used as an antigen in a microsphere immunoassay for the detection of antibodies against BVDV-1 and atypical bovine pestivirus. The diagnostic performance of the new method was evaluated by testing a total of 596 serum samples, and the assay was compared with enzyme-linked immunosorbent assay (ELISA). Based on the negative/positive cut-off median fluorescence intensity (MFI) value of 2800, the microsphere immunoassay had a sensitivity of 100% and specificity of 100% compared to ELISA. The immunoassay was able to detect antibodies against both BVDV-1 and the atypical pestivirus. This novel microsphere immunoassay has the potential to be multiplexed for simultaneous detection of antibodies against different bovine pathogens in a high-throughput and economical way.
1. Introduction
The genus Pestivirus of the family Flaviviridae consists of four approved species: Bovine viral diarrhea virus 1 (BVDV-1), Bovine viral diarrhea virus 2 (BVDV-2), Classical swine fever virus (CSFV), Border disease virus (BDV); and other related viruses such as atypical bovine pestiviruses that have not been assigned to species (Pletnev et al., 2011). The pestiviral genome is a single-stranded, positive-sense RNA molecule with a standard size of approximately 12.5 kb (Colett et al., 1988, Deng and Brock, 1992). The genome consists of an open reading frame (ORF) flanked by two untranslated regions (UTRs) at the 5′ and 3′ ends. Following translation, the ORF is processed into 11 mature proteins, namely N-terminal protease (Npro), capsid protein (C), three glycoproteins (E1, E2, Erns), protein of 7 kDa (p7) and five non-structural proteins (NS2-3, NS4A, NS4B, NS5A, NS5B) (Thiel et al., 1996). Glycoprotein E2 is immunodominant in eliciting antibody response to virus infection whereas glycoprotein Erns and non-structural protein NS3 participate in immune response (Corapi et al., 1990). NS3 is conserved antigenically whereas Erns shows less antigenic diversity than E2 (Fulton et al., 2003). Phylogenetic analysis has demonstrated that the atypical bovine pestiviruses are related more closely to the recognized bovine pestivirus species BVDV-1 and BVDV-2 than to any other pestivirus species (Liu et al., 2009c, Liu et al., 2010, Decaro et al., 2011).
Bovine pestiviruses may cause subclinical infection or mild to severe diseases, depending on virus strains and host factors. An atypical pestivirus D32/00_‘HoBi’ was first detected in a batch of fetal calf serum of Brazilian origin (Schirrmeier et al., 2004) and subsequently similar viruses were detected in batches of fetal bovine serum (FBS) of different origin (Peletto et al., 2012, Liu et al., 2009b, Xia et al., 2011). Recent experiments revealed that the atypical bovine pestiviruses may cause clinical signs of the disease similar to those caused by BVDV-1 (Larska et al., 2012, Decaro et al., 2012b). Although an atypical pestivirus strain Th/04_KhonKaen was found to cause subclinical infection (Ståhl et al., 2007, Kampa et al., 2009), another atypical pestivirus was associated with an outbreak of severe respiratory disease in Italy (Decaro et al., 2011). The latter agent was detected subsequently in aborted fetuses (Decaro et al., 2012a), which is a reminiscent of a previous report (Cortez et al., 2006).
The economic losses due to BVDV infection may be high. For example, losses are estimated at 10 and 40 million $ per million calvings at the national level (Houe, 2003). In order to reduce or prevent losses caused by BVDV, national BVDV control programs have been implemented in several countries with great success, especially in Scandinavia (Lindberg et al., 2006).
Diagnosis of bovine viral diarrhea (BVD) is based on both direct and indirect detection methods. In indirect detection methods, enzyme-linked immunosorbent assay (ELISA) is used widely for monitoring herd seroprevalence. As an alternative, a microsphere-based immunoassay has been developed (Xia et al., 2010). This assay is based on the Luminex xMAP technology and it detects NS3 antibodies with high sensitivity and specificity (Xia et al., 2010). Anderson et al. (2011) reported a Luminex multiplex serology assay for the detection of antibodies to four different bovine viruses, namely, bovine herpes virus 1 (BHV-1), parainfluenza 3 virus (PI3V), bovine respiratory syncytial virus (BRSV) and BVDV. In that multiplex assay the use of the recombinant BVDV E2 protein as a potent antigen led to an increase in signal strength.
ELISAs have not been reported for specific detection of the emerging atypical bovine pestiviruses. It has been observed that commercial ELISAs, based on either crude preparation or recombinant antigens of BVDV-1/BVDV-2 are able to detect antibodies against the atypical bovine pestiviruses or their antigens to some extent (Kampa et al., 2007). The glycoprotein Erns of BVDV has also been used as an antigen for serological detection of BVDV (Grego et al., 2007, Kuhne et al., 2005) but the cross reactivity against the atypical bovine pestiviruses is still unclear.
The objectives of this study were to produce the Erns protein of an atypical bovine pestivirus using a baculovirus expression system and to evaluate the potential use of this recombinant Erns protein as an antigen in a microsphere immunoassay for the detection of antibodies raised against BVDV-1 and against atypical bovine pestiviruses.
2. Materials and methods
2.1. RNA extraction, cDNA synthesis, PCR amplification and cloning
Total RNA was extracted from an isolate of atypical bovine pestivirus Th/04_KhonKaen (Liu et al., 2009a) using TRIzol Reagent (Invitrogen, Carlsbad, CA). cDNA synthesis was primed randomly by 1 μM hexamer in a 20-μl volume using 200 units of Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA). The reaction was performed at 50 °C for 60 min and the enzyme was inactivated at 85 °C for 5 min. The RNA strand of the cDNA was removed by 2 units of Escherichia coli RNase H (Invitrogen, Carlsbad, CA) at 37 °C for 20 min. Two microliters of cDNA were used for amplification of the Erns gene region in a 25-μl reaction volume, including 12.5 μl of water, 2.5 μl of 10× buffer, 2 μl of 2.5 mM dNTP mix, 0.5 μl of dimethyl sulfoxide (Sigma, MO, USA), 2.5 μl of 10 μM each primer and 1.25 U PfuUltra DNA polymerase (Agilent Technologies Inc., Santa Clara, CA). The nucleotide sequences of primers were 5′-GAGAACATAACACAATGGAATTTGAAAG-3′ (forward) and 5′-TGCATGAGCCCCGAACC-3′ (reverse). The thermo profile was 35 cycles of 94 °C for 40 s, 49 °C for 40 s, 72 °C for 40 s. The amplified PCR product (681 bp) was separated on a 1.5% agarose gel. The excised fragment was purified using PCR cleanup system kit (Promega, Co., Madison, WI) and cloned into pFastBac/HBM-TOPO vector (Invitrogen, Carlsbad, CA), and the positive plasmids designed as pFastBacHBM-Erns were verified by DNA sequencing.
2.2. Expression of Erns protein in a baculovirus/insect cell system
The plasmid pFastBacHBM-Erns was transformed to MAX Efficiency DH10Bac competent E. coli (Invitrogen, Carlsbad, CA) to generate recombinant bacmid, which was further verified based on the phenotypic characteristics and analytical PCR according to manufacturer's instructions. One microgram of bacmid DNA and 6 μl of Cellfectin II reagent (Invitrogen, Carlsbad, CA) were used for transfection of 2.4 × 106 Spodoptera frugiperda (Sf9) cells. The cells were incubated at 27 °C for 72 h. Sf9 cells were grown in serum free medium Sf-900 II SFM (Invitrogen, Carlsbad, CA). The recombinant viruses were harvested and the virus titer was determined by BacPAK Baculovirus Rapid Titer kit (Clontech Laboratories, Inc., Mountain View, CA) as per instructions of the manufacturer. To optimize the protein expression, a multiplicity of infection (MOI) value of 0.5 and 1 was used to infect cells at a density of 2 × 106/ml. After 24 h post infection, 0.5% FBS was added and the cells were collected at 48, 72 and 96 h post infection and centrifuged at 800 × g for 5 min. Pellets were washed with 1 ml phosphate buffered saline (PBS) and stored at −20 °C if not used immediately. To analyze proteins, 75 μl of lysis buffer (200 mM Tris, 20% glycerol, 5 mM ethylenediaminetetraacetic acid (EDTA), 4% sodium dodecyl sulfate (SDS), 50 mM dithiothreitol (DTT), 6 units of benzonase) was added to the cell pellet and incubated at 4 °C for 30 min. The pellet and supernatant were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot using anti-His monoclonal antibody (Sigma, St. Louis, MI).
2.3. Purification of the recombinant Erns protein
One milliliter of cell lysis buffer (25 mM Hepes, pH 7.4, 100 mM NaCl, 1% Triton X-100, 1× protease inhibitor) was added to 1.2 × 107 cells and incubated on ice for 45 min. The cells were sheared by brief sonication and soluble proteins were recovered in the supernatant following ultra-centrifugation at 40,000 × g for 30 min at 4 °C. The His-Trap Chelating HP column (GE Healthcare, Uppsala, Sweden) was equilibrated with 10 column volumes of buffer A (50 mM Hepes, pH 7.4, 500 mM NaCl, 10 mM Imidazole). After loading the sample, the column was washed with 10 volumes of buffer B (50 mM Hepes, pH 7.4, 400 mM NaCl, 30 mM Imidazole). The protein was eluted from the column with 100, 300 and 500 mM of imidazole, respectively. The elution fractions were concentrated with a 10-kDa molecular weight cut-off Amicon Ultra-4 Centrifugal Filter Devices (Millipore Ireland B.V., County Cork, Ireland).
2.4. Microsphere immunoassay
Coupling of the recombinant Erns protein to carboxylated microspheres (Luminex Cooperation, Austin, TX) was mediated by 1-ethyl-3-3-dimethylaminopropyl carbodiimide (EDC) (Pierce, Rockford, IL), as described previously (Xia et al., 2010). The optimal amount of the protein, dilution of the serum, concentration of secondary antibody and conjugate, and blocking solutions were determined experimentally. The beads were resuspended in 200 μl of PBS–TBN (PBS, 0.1% bovine serum albumin (BSA), 0.02% Tween-20, 0.05% Azide, pH 7.4) and stored at 4 °C in darkness. Coupling reaction was confirmed by testing a mix of two Erns monoclonal antibodies (MAbs) (WS363 and WS433, AHVLA, UK) on a Luminex 200 analyzer, as described previously (Xia et al., 2010).
The immunoassay was performed in 96-well plates. The microspheres coupled with the recombinant Erns protein were resuspended in PBS with 1% BSA and 50 μl of microspheres were transferred to each well of the plates. Fifty microliter of sera and 50 μl of PBS with 1% BSA were added to the respective wells and incubated at 37 °C for 30 min on a plate shaker. The microspheres were washed twice with PBS containing 1% BSA using a HydroFlex™ microplate washer (Tecan Group Ltd., Männedorf, Switzerland). Thereafter, 50 μl of 2 μg/ml biotinylated anti-bovine IgG (Jackson ImmunoResearch, West Grove, USA) in PBS with 1% BSA were added to each well and incubated for another 30 min. Following washing as before, 50 μl of 10 μg/ml streptavidin-R-phycoerythrin conjugate (ProZyme, Inc., Hayward, CA) in PBS with 1% BSA were added to each well and incubated at 37 °C for 30 min on the plate shaker. After washing twice, the microspheres were resuspended in 100 μl of PBS with 1% BSA and analyzed by the Luminex 200 analyzer. Median fluorescence intensity (MFI) was calculated based on the measurement of 100 beads per sample. Receiver operating characteristic (ROC) analysis of the microsphere immunoassay and ELISA were performed using Medcalc software (MedCalc Software, Mariakerke, Belgium) to determine a cut-off value of the microsphere immunoassay.
2.5. Optimization of microsphere immunoassay
Several parameters of the immunoassay were optimized, including dilution of the serum, concentration of anti-bovine IgG/streptavidin, number of coupled beads and different blocking solutions.
2.6. Bovine serum samples
A total of 596 serum samples were evaluated in this study. These included 24 batches of FBS from ten different commercial suppliers (Xia et al., 2011), 306 bovine sera that were collected recently in Sweden, 246 bovine sera that had been described previously (Xia et al., 2010), and 20 samples that were collected from an experimental infection with a BVDV-1 strain Hon916, the atypical bovine pestivirus strain Th/04_KhonKaen, both viruses, or mock infection with cell culture medium (Larska et al., 2012). The FBS (n = 24) and recent collection of bovine sera (n = 306) were tested for the presence of antibodies against BVDV by a commercial ELISA kit (Svanova Biotech AB, Uppsala, Sweden) according to the manufacturer's instruction. The kit had a sensitivity of 100% and a specificity of 98.2% compared with virus neutralization test using BVDV-1 strains (Svanova Biotech AB, Uppsala, Sweden).
3. Results
3.1. Expression and purification of the recombinant Erns protein
The expression of the recombinant Erns protein in Sf9 cells was driven by a strong polyhedrin promoter of Autographa californica multiple nuclear polyhedrosis virus (AcMNPV). The expression conditions at 72 h post transfection and at an MOI of 1 were found optimal and the recombinant Erns protein (around 40 kDa) was detected by Western blot using anti-His MAb (Fig. 1A). By affinity chromatography, the Erns protein was eluted from the Ni2+ column with 300 mM imidazole (Fig. 1B). To verify the antigenicity of the recombinant Erns protein, a BVDV positive serum was used as primary antibody. As shown in Fig. 1C, the recombinant Erns protein reacted well with the BVDV positive serum, indicating that the recombinant protein maintained its antigenicity.
Fig. 1.
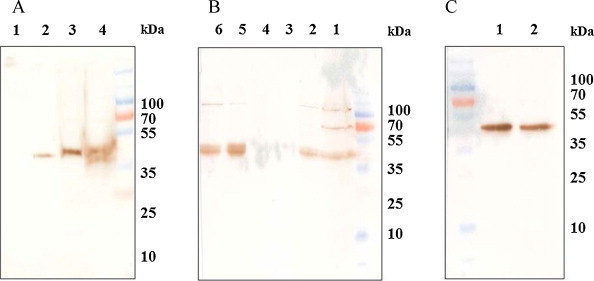
Western blot analysis of the Erns protein using anti-His MAb (A and B) or positive serum (C) as primary antibody. (A) Expression of the Erns protein. Lane 1: Sf9 cell lysate, lane 2: 48 h cell lysate, lane 3: 72 h cell lysate, lane 4: 96 h cell lysate. (B) Purification of the Erns protein, lane 1: sonication supernatant, lane 2: ultacentrifuge supernatant, lane 3: flowthrough, lane 4: wash fraction, lane 5: eluted fraction (300 mM imidazole), lane 6: concentrated protein (10 μg). (C) BVDV serum was used as primary antibody to verify antigenicity of the Erns protein, lane 1: eluted fraction (300 mM imidazole), lane 2: concentrated protein (10 μg).
3.2. Optimization of the microsphere immunoassay
As a first step, the amount of the recombinant Erns protein was determined for coupling microspheres. To test coupling efficiency, a mixture of two MAbs (WS363 and WS433) and 4 μg/ml of anti-mouse IgG conjugated with R-phycoerythrin were used in the immunoassay. Fifty micrograms of the recombinant Erns protein were found optimal for coupling 1.2 million microspheres, which gave an MFI value of around 11,000, whereas further increasing Erns protein to 80 μg had lower MFI values (Fig. 2A). A 5-fold dilution of sera was found optimal (Fig. 2B).
Fig. 2.
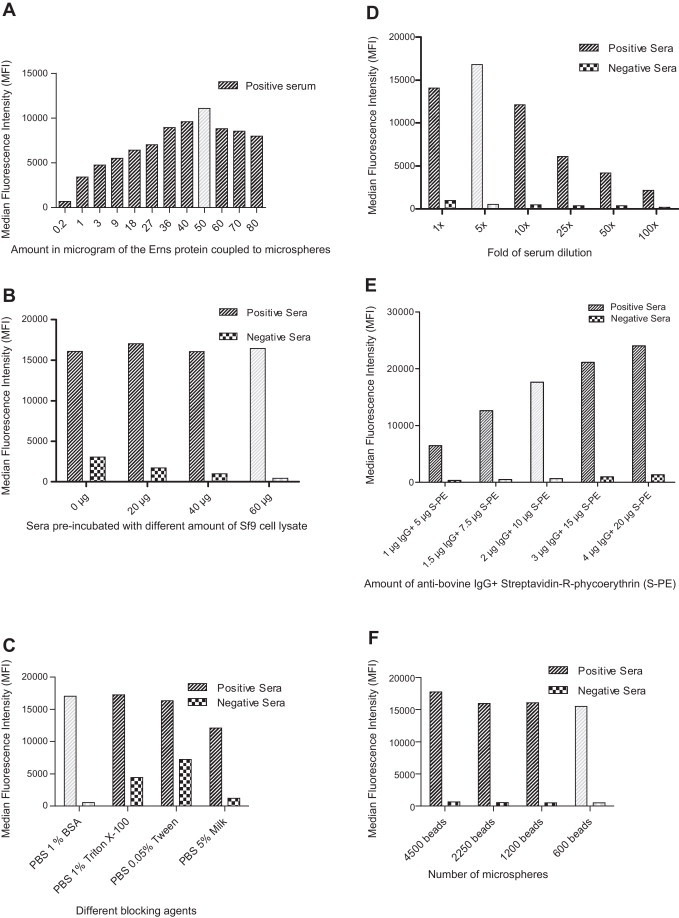
Optimization of the microsphere immunoassay. (A) Selection of coupled recombinant Erns to microsphere. (B) Blocking of Sf9 cell protein binding antibodies in 5-fold diluted bovine sera, the MFI value are average values of four positive and negative sera. (C–F) Selection of blocking agents, optimal serum dilution, amount of detection antibody and microspheres according to mean MFI value of four positive and four negative sera. Light-shaded columns indicate the optimal conditions for the immunoassay.
To reduce unspecific binding to the recombinant protein, four different blocking solutions, PBS with 1% BSA, PBS with 1% Triton X-100, PBS with 0.05% Tween-20, and PBS with 5% milk were tested in four pairs of positive and negative samples. Two solutions (PBS with 1% BSA and PBS with 5% milk) had the lowest MFI value for the negative serum while the MFI value for the positive sample remained high (Fig. 2C). Pre-incubation of sera with Sf9 cell lysate (27 mg/ml) also helped to reduce the unspecific binding effectively. Incubation of 60 μg Sf9 cell lysate with 50 μl 5-fold diluted serum at room temperature for 30 min was most successful in reducing unspecific binding (Fig. 2D). The optimal amount of secondary antibody was 2 μg/ml of biotinylated anti-bovine IgG when reacted with 10 μg/ml of streptavidin-R-phycoerythrin conjugate (Fig. 2E). Finally, 600 microspheres were found sufficient for antibody detection (Fig. 2F).
3.3. Comparison of microsphere immunoassay and ELISA
Once optimized, the microsphere immunoassay was evaluated by testing a total of 596 serum samples, which were tested by ELISA either in previous studies (Xia et al., 2010, Larska et al., 2012) or in this study. Based on the non-parametric ROC analysis, the cut-off MFI value of the microsphere immunoassay was set to 2800. Under this cut-off value, 369 samples were found negative and 227 samples were positive. The positive samples included 212 BVDV-1 clinical samples, a total of 15 samples originated from the experimental infection with atypical bovine pestivirus (n = 5), BVDV-1 (n = 5), and both BVDV-1 and atypical bovine pestivirus (n = 5). The MFI values for negative samples were between 115 and 1822 with a mean value of 357, while the values for the positive samples ranged from 3812 to 26,453 with a mean value of 13,727. The immunoassay results matched the ELISA results, showing 100% sensitivity and 100% specificity relative to ELISA. The confidence interval (CI) for sensitivity was 98–100%, and the CI for specificity was 99–100%.
4. Discussion
This study described expression and purification of the Erns glycoprotein of an atypical bovine pestivirus and subsequent evaluation of the recombinant protein as the antigen in a microsphere immunoassay for the detection of antibodies against BVDV-1 and atypical bovine pestivirus. While it is evident that atypical bovine pestiviruses cause respiratory disease and abortion, there is a need for a serological assay for the specific detection of antibodies against this atypical pestivirus. ELISA that is based on E2 and NS3 are shown to cross react with antibodies against atypical pestiviruses (Larska et al., 2012, Bauermann et al., 2012), whereas there is no report of detection of antibodies against atypical pestivirus by an Erns ELISA. Therefore, the Erns glycoprotein was selected as the antigen in our microsphere immunoassay. The test was evaluated for the detection of specific antibodies against BVDV-1. Also, possibility of detection of specific antibodies against atypical bovine pestivirus was investigated. According to obtained results, this test has the potential to be implemented in diagnosis of the disease caused by atypical bovine pestiviruses.
Production of recombinant viral protein as the antigen is an important step in developing this microsphere immunoassay. To maintain proper post-translational modifications of the Erns protein and, likely to maintain its antigenicity, a baculovirus/insect cell system was exploited in this study, resulting in high MFI values for the positive samples. In the Luminex assay described by Anderson et al. (2011), an MFI value close to 30,000 was obtained for the 1/450 dilution of a positive sample when recombinant E2 protein was used as the antigen, whereas it was only around 2000 for the same diluted sample when crude viral lysate was used as the antigen. As pointed out by Feng et al. (2004), using a recombinant protein as an antigen in a multiplex Luminex assay has improved significantly sensitivities (94% and 100%, respectively) and specificities (100% and 95%, respectively) than using bacterial membrane extracts (78% sensitivity and 65% specificity).
Various blocking agents were tested to reduce unspecific binding. It was found that PBS containing 1% BSA or 5% milk was able to reduce the unspecific binding effectively. However, blocking with PBS containing 5% milk in the immunoassay created certain technical problems: it clogged the needle in the Luminex analyzer such that the probe had to be cleaned four or five times during analysis. The microspheres were also likely to be aggregated and covered by the milk, and were lost during washing steps. Therefore, it took the analyzer longer time to count 100 beads per sample and the test could fail completely. The PBS containing 1% BSA worked well for some samples but high MFI values were also observed in several negative samples. As the microspheres itself gave neglect MFI values, it is likely that even in the purified protein there were still some impurities that caused the unspecific binding to bovine antibodies. To reduce the unspecific binding, sera were incubated with Sf9 cell lysate before being tested in the immunoassay, which brought down MFI values for the negative samples whereas the MFI values for the positive samples were not affected. Cell lysate has been used to reduce unspecific binding in a Luminex serological assay (Waterboer et al., 2006).
The diagnostic performance of the microsphere immunoassay was compared to that of commercial BVDV antibody ELISA by testing a large panel of 596 serum samples. Both results matched 100%. Two serum samples, which had been tested negative by ELISA but positive by a blocking microsphere immunoassay targeting NS3 antibodies (Xia et al., 2010), remained negative in this Erns immunoassay. It seemed likely that the two serum samples were either truly negative or very weak positive that was only detected by the blocking immunoassay. While 15 serum samples from all three infected groups were tested positive, a slight difference in the MFI values was observed. The highest MFI value (average) was 21,956 for the sera against atypical pestivirus, where the viral Erns protein was the antigen; the lowest MFI value (average) was 17,818 for the BVDV-1 sera that were heterologous to the antigen; and 20,471 for the sera of the mixed infection. Interestingly, when the Erns protein was replaced by that of BVDV-1 strain CP7, the average MFI values of these three groups changed, with the highest (22,915) for the homologous sera (BVDV-1) and the lowest (17,425) for the heterologous sera (atypical pestivirus) and middle value (20,715) for the mixed infection. More studies are needed in order to investigate whether this correlation is consistent or it is just a coincidence. Furthermore, the immunoassay could be expanded readily for the simultaneous detection of antibodies against multiple bovine viruses in the Bovine Respiratory and Enteric Disease (BRD and BED, respectively) complexes, such as bovine coronavirus, bovine respiratory syncytial virus, and bovine parainfluenza-3 virus.
5. Conclusion
The recombinant Erns protein produced in the baculovirus/insect cell system maintained its antigenicity. The Erns microsphere immunoassay was able to detect antibodies against bovine viral diarrhea viruses with performance similar to commercial ELISAs. Thus, the microsphere immunoassay, using the recombinant Erns protein of an atypical bovine pestivirus, offers a powerful novel alternative to the current diagnostic methods for bovine viral diarrhea and related diseases. This microsphere immunoassay platform opens new possibilities for the rapid and complex diagnosis of respiratory and enteric diseases, with great multiplexing potential and flexibility.
Acknowledgments
The authors wish to thank Ms. Pia Fällgren, National Veterinary Institute (SVA), Uppsala, Sweden for providing the serum samples. The work was supported by the Award of Excellence (Excellensbidrag) provided to SB by the Swedish University of Agricultural Sciences (SLU), and by the Royal Swedish Academy of Agriculture and Forestry (KSLA) to LL.
References
- Anderson S., Wakeley P., Wibberley G., Webster K., Sawyer J. Development and evaluation of a Luminex multiplex serology assay to detect antibodies to bovine herpes virus 1, parainfluenza 3 virus, bovine viral diarrhoea virus, and bovine respiratory syncytial virus, with comparison to existing ELISA detection methods. Journal of Immunological Methods. 2011;366:79–88. doi: 10.1016/j.jim.2011.01.010. [DOI] [PubMed] [Google Scholar]
- Bauermann F.V., Flores E.F., Ridpath J.F. Antigenic relationships between Bovine viral diarrhea virus 1 and 2 and HoBi virus: possible impacts on diagnosis and control. Journal of Veterinary Diagnostic Investigation. 2012;24:253–261. doi: 10.1177/1040638711435144. [DOI] [PubMed] [Google Scholar]
- Colett M.S., Larson R., Gold C., Strick D., Anderson D.K., Purchio A.F. Molecular cloning and nucleotide sequence of the pestivirus bovine viral diarrhoea virus. Virology. 1988;165:191–199. doi: 10.1016/0042-6822(88)90672-1. [DOI] [PubMed] [Google Scholar]
- Corapi W.V., Donis R.O., Dubovi E.J. Characterization of a panel of monoclonal antibodies and their use in the study of the antigenic diversity of bovine viral diarrhoea virus. American Journal of Veterinary Research. 1990;51:1388–1394. [PubMed] [Google Scholar]
- Cortez A., Heinemann M.B., De Castro M.G., Soares R.M., Pinto A.M., Alfieri A.A., Flore S.E.F., Cerqueira L.R., Richtzenhain L.J. Genetic characterization of Brazilian bovine viral diarrhea virus isolates by partial nucleotide sequencing of the 5′-UTR region. Pesquisa Veterinaria Brasileira. 2006;26:211–216. [Google Scholar]
- Decaro N., Lucente M.S., Mari V., Cirone F., Cordioli P., Camero M., Sciarretta R., Losurdo M., Lorusso E., Buonavoglia C. Atypical pestivirus and severe respiratory disease in calves, Europe. Emerging Infectious Diseases. 2011;17:1549–1552. doi: 10.3201/eid1708.101447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Lucente M.S., Mari V., Sciarretta R., Pinto P., Buonavoglia D., Martella V., Buonavoglia C. Hobi-like pestivirus in aborted bovine fetuses. Journal of Clinical Microbiology. 2012;50:509–512. doi: 10.1128/JCM.05887-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Mari V., Lucente M.S., Sciarretta R., Moreno A., Armenise C., Losurdo M., Camero M., Lorusso E., Cordioli P., Buonavoglia C. Experimental infection of cattle, sheep and pigs with ‘Hobi’-like pestivirus. Veterinary Microbiology. 2012;155:165–171. doi: 10.1016/j.vetmic.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng R., Brock K.V. Molecular cloning and nucleotide sequence of a pestivirus genome, noncytopathic bovine viral diarrhea virus strain SD-1. Virology. 1992;191:867–869. doi: 10.1016/0042-6822(92)90262-n. [DOI] [PubMed] [Google Scholar]
- Feng S., Kendall L.V., Hodzic E., Wong S., Lorenzana E., Freet K., Ku K.S., Luciw P.A., Barthold S.W., Khan I.H. Recombinant Helicobacter bilis protein P167 for mouse serodiagnosis in a multiplex microbead assay. Clinical and Diagnostic Laboratory Immunology. 2004;11:1094–1099. doi: 10.1128/CDLI.11.6.1094-1099.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton R.W., Step D.L., Ridpath J.F., Saliki J.T., Confer A.W., Johnson B.J., Briggs R.E., Hawley R.V., Burge L.J., Payton M.E. Response of calves persistently infected with noncytopathic bovine viral diarrhea virus (BVDV) subtype 1b after vaccination with heterologous BVDV strains in modified live virus vaccines and Mannheimia haemolytica bacterin-toxoid. Vaccine. 2003;21:2980–2985. doi: 10.1016/S0264-410X(03)00118-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grego E., Uslenghi F., Strasser M., Luzzago C., Frigerio M., Peletto S., Rosati S. Development and application of an enzyme-linked immunosorbent assay for detection of bovine viral diarrhea antibody based on Erns glycoprotein expressed in a baculovirus system. Journal of Veterinary Diagnostic Investigation. 2007;19:21–27. doi: 10.1177/104063870701900104. [DOI] [PubMed] [Google Scholar]
- Houe H. Economic impact of BVDV infection in dairies. Biologicals. 2003;31:137–143. doi: 10.1016/s1045-1056(03)00030-7. [DOI] [PubMed] [Google Scholar]
- Kampa J., Alenius S., Emanuelson U., Chanlun A., Aiumlamai S. Bovine herpesvirus type 1 (BHV-1) and bovine viral diarrhoea virus (BVDV) infections in dairy herds: self clearance and the detection of seroconversions against a new atypical pestivirus. Veterinary Journal. 2009;182:223–230. doi: 10.1016/j.tvjl.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Kampa J., Ståhl K., Renström L.H., Alenius S. Evaluation of a commercial Erns-capture ELISA for detection of BVDV in routine diagnostic cattle serum samples. Acta Veterinaria Scandinavica. 2007;49:7. doi: 10.1186/1751-0147-49-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhne S., Schroeder C., Holmquist G., Wolf G., Horner S., Brem G., Ballagi A. Detection of bovine viral diarrhoea virus infected cattle – testing tissue samples derived from ear tagging using an Erns capture ELISA. Journal of Veterinary Medicine Series B: Infectious Diseases and Veterinary Public Health. 2005;52:272–277. doi: 10.1111/j.1439-0450.2005.00861.x. [DOI] [PubMed] [Google Scholar]
- Larska M., Polak M.P., Riitho V., Strong R., Belák S., Alenius S., Uttenthal Å., Liu L. Kinetics of single and dual infection of calves with an Asian atypical bovine pestivirus and a highly virulent strain of bovine viral diarrhoea virus 1. Comparative Immunology, Microbiology and Infectious Diseases. 2012;35:381–390. doi: 10.1016/j.cimid.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Lindberg A., Brownlie J., Gunn G.J., Houe H., Moennig V., Saatkamp H.W., Sandvik T., Valle P.S. The control of bovine viral diarrhoea virus in Europe: today and in the future. Revue Scientifique et Technique. 2006;25:961–979. [PubMed] [Google Scholar]
- Liu L., Kampa J., Belák S., Baule C. Virus recovery and full-length sequence analysis of atypical bovine pestivirus Th/04_KhonKaen. Veterinary Microbiology. 2009;138:62–68. doi: 10.1016/j.vetmic.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Liu L., Xia H., Baule C., Belák S. Maximum likelihood and Bayesian analyses of a combined nucleotide sequence dataset for genetic characterization of a novel pestivirus, SVA/cont-08. Archives of Virology. 2009;154:1111–1116. doi: 10.1007/s00705-009-0419-4. [DOI] [PubMed] [Google Scholar]
- Liu L., Xia H., Baule C., Belák S., Wahlberg N. Effects of methodology and analysis strategy on robustness of pestivirus phylogeny. Virus Research. 2010;147:47–52. doi: 10.1016/j.virusres.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Liu L., Xia H., Wahlberg N., Belák S., Baule C. Phylogeny, classification and evolutionary insights into pestiviruses. Virology. 2009;385:351–357. doi: 10.1016/j.virol.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Peletto S., Zuccon F., Pitti M., Gobbi E., Marco L.D., Caramelli M., Masoero L., Acutis P.L. Detection and phylogenetic analysis of an atypical pestivirus, strain IZSPLV_To. Research in Veterinary Science. 2012;92:147–150. doi: 10.1016/j.rvsc.2010.10.015. [DOI] [PubMed] [Google Scholar]
- Pletnev A., Gould E., Heinz F.X., Meyers G., Thiel H.J., Bukh J., Stiasny K., Collett M.S., Becher P., Simmonds P., Rice C.M., Monath T.P. Flaviviridae. In: King A.M., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus Taxonomy. Oxford; Elsevier: 2011. pp. 1003–1020. [Google Scholar]
- Schirrmeier H., Strebelow G., Depner K., Hoffmann B., Beer M. Genetic and antigenic characterization of an atypical pestivirus isolate, a putative member of a novel pestivirus species. Journal of General Virology. 2004;85:3647–3652. doi: 10.1099/vir.0.80238-0. [DOI] [PubMed] [Google Scholar]
- Ståhl K., Kampa J., Alenius S., Persson Wadman A., Baule C., Aiumlamai S., Belák S. Natural infection of cattle with an atypical ‘HoBi’-like pestivirus—implications for BVD control and for the safety of biological products. Veterinary Research. 2007;38:517–523. doi: 10.1051/vetres:2007012. [DOI] [PubMed] [Google Scholar]
- Thiel H.J., Plagemann P.G.W., Moenning V. Pestiviruses. In: Fields B.N., Knipe D.M., Howley P.M., editors. Field Virology. 3rd ed. Lippincott-Raven; Philadelphia: 1996. pp. 1059–1073. [Google Scholar]
- Waterboer T., Sehr P., Pawlita M. Suppression of non-specific binding in serological Luminex assays. Journal of Immunological Methods. 2006;309:200–204. doi: 10.1016/j.jim.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Xia H., Liu L., Nordengrahn A., Kiss I., Merza M., Eriksson R., Blomberg J., Belák S. A microsphere-based immunoassay for rapid and sensitive detection of bovine viral diarrhoea virus antibodies. Journal of Virological Methods. 2010;168:18–21. doi: 10.1016/j.jviromet.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Vijayaraghavan B., Belák S., Liu L. Detection and identification of the atypical bovine pestiviruses in commercial foetal bovine serum batches. PLoS One. 2011;6:e28553. doi: 10.1371/journal.pone.0028553. [DOI] [PMC free article] [PubMed] [Google Scholar]


