Abstract
During the 2007–2008 influenza season global strain surveillance for antiviral resistance revealed the sudden emergence of oseltamivir resistance in influenza A H1N1 isolates. Although oseltamivir resistance rates vary from region to region, 16% of isolates tested globally were found to be oseltamivir resistant by a histidine to tyrosine mutation of residue 275 of the neuraminidase gene of influenza A. In order to implement effective resistance testing locally a novel real-time reverse-transcriptase PCR (RT-PCR) assay was developed for the detection of the H275Y mutation. To evaluate this method, 40 oseltamivir resistant and 61 oseltamivir sensitive H1N1 influenza isolates were tested using Sanger sequencing, which is the reference method for detection of resistance, pyrosequencing and the novel H275Y RT-PCR assay. In comparison to Sanger sequencing, the sensitivity and specificity of the H275Y RT-PCR assay were 100% (40/40) and 100% (61/61) respectively, while the sensitivity and specificity of pyrosequencing were 100% (40/40) and 97.5% (60/61) respectively. Although all three methods were effective in detecting the H275Y mutation associated with oseltamivir resistance, the H275Y RT-PCR assay was the most rapid and could easily be incorporated into an influenza subtyping protocol.
Keywords: Influenza, H275Y, Oseltamivir, Resistance, Molecular
The beginning of the 2007–2008 influenza season marked a significant and sudden increase in resistance of influenza A H1N1 to the neuraminidase (NA) inhibitor oseltamivir through a histidine to tyrosine mutation at residue 275 of the NA gene (Sheu et al., 2008) (residue 274 in the N2 gene). By the end of the 2007–2008 influenza season the resistance rate to oseltamivir was 16% globally. Countries with little or no previous oseltamivir resistance, such as the United States, Canada and Norway reported resistance rates of 10%, 26% and 67% respectively (World Health Organization, 2008). Early data from the Southern Hemisphere for the 2008 influenza season suggested oseltamivir resistance rates among H1N1 isolates of as high as 100% (Besselaar et al., 2008). In addition to reports of resistance in seasonal H1N1 isolates, reports of oseltamivir resistance have also emerged in patients infected with H5N1 avian influenza (de Jong et al., 2005, Kimm-Breschkin et al., 2007, Le et al., 2005), bringing into question the use of this antiviral for pandemic influenza treatment and prophylaxis (Moscona, 2005, World Health Organization, 2005). This emerging oseltamivir resistance has prompted laboratorians to look at time effective means to identify H1N1 influenza with the H275Y mutation.
Currently, the gold standard for antiviral resistance screening is phenotypic methods (Meijer et al., 2007). However, genotypic methods such as Sanger dideoxy sequence analysis of the NA gene can also be performed to detect mutations such as H275Y that are associated strongly with oseltamivir resistance. While both phenotypic and sequencing methods are effective in detecting resistance, they can be labour intensive, have a long turn-around-time and be quite expensive, suggesting the need for a rapid, high-throughput approach to influenza drug resistance testing. In this study we introduce the utilization of a novel real-time reverse-transcriptase PCR (RT-PCR) assay for detection of the histidine to tyrosine mutation at residue 275. This assay is designed to produce a sigmoidal curve when an isolate is oseltamivir susceptible and no curve when an isolate is oseltamivir resistant, thus serving as a quick screening tool that can be incorporated into the influenza subtyping protocol.
Current TaqMan technology utilizes a minor groove binder (MGB) that forms a stable complex with the minor groove of single-stranded DNA, increasing stability and specificity (Afonina et al., 1996, Uchiyama et al., 2004). Due to these qualities MGB probes are characterized by a shorter length and a higher melting temperature (T m). MGB-based probes are therefore highly applicable for detecting the presence of polymorphic sequences (Uchiyama et al., 2004); however their increased stability may result in false positives. Probe cross reactivity is therefore one reason that MGB chemistry is not generally used to detect point mutations. To avoid incorrect assignment of the H275Y point mutation an effort was made to reduce binding to oseltamivir-resistant isolates which have a C→T point mutation by designing the MGB probe to detect wild-type sequence only. To limit cross reactivity of similar sequences, the MGB probe was designed so that the MGB was linked to the mutable base pair. This was accomplished by designing the probe so that the C→T mutation was 6 bp from the 3′ of the probe, which is the site of MGB linkage.
This study compares the utility of the H275Y RT-PCR assay to Sanger sequencing, which is the current reference method for H275Y detection, as well as pyrosequencing, an emerging method for the detection of the H275Y point mutation (Duwe and Schweiger, 2008). Specimens used in this study were from a population that was moderately resistant to oseltamivir, with resistance rates of 17% in Ontario isolates in 2007–2008.
One hundred and one patient isolates were used in this study. These were influenza A H1N1 positive isolates most similar genetically to the A/Brisbane/57/2007-like lineage collected during the 2007–2008 influenza season in Ontario, Canada. Upon arrival in the laboratory, neuraminidase (N1) was detected from specimens using the Quantitect Probe RT-PCR kit (Qiagen) and published primers and probes (Schweiger et al., 2000) (Table 1 ) using an MxPro3005 real-time PCR thermocycler (Stratagene). This N1 subtyping assay has been validated previously as a sensitive and specific method for the detection of N1 in clinical specimens (Schweiger et al., 2000). It has also been used extensively in external quality assurance panels and classic phenotypic assays. Mutations at residue 275 of the N1 gene were detected using Sanger dideoxy sequencing. Forty specimens with a wild-type histidine at residue 275 and 61 specimens with a tyrosine at residue 275 were chosen randomly for analysis.
Table 1.
Primers and probes used in this study.
| Primers and probes | Sequence | Reference |
|---|---|---|
| NA1-1078 Forward | ATGGTAATGGTGTTTGGATAGGAAG | Schweiger et al. (2000) |
| NA1-1352 Reverse | AATGCTGCTCCCACTAGTCCAG | Schweiger et al. (2000) |
| NA1-1138 Probe | TGATTTGGGATCCTAATGGATGGACAG | Schweiger et al. (2000) |
| H275Y Forward | GGCCGCCTCGTACAAAATT | This study |
| H275Y Reverse | CCAGTGTCTGGGTAACAGGAGC | This study |
| H275Y Probe | CACCCAATTTTCATTATGA | This study |
| AN1B-505 Forward | TTGCTTGGTCAGCAAGTGCA | Takao et al. (2002) |
| N1-2 Reverse | CCAGTCCACCCATTTGGATCC | Yuen et al. (1998) |
| NA-Forward | AGATCGAGAAGGGGAAGGTTACTA | Reisdorf et al. (2008) |
| NA-Reverse | Bio-GTCYCTGCATACACACATCACT | Reisdorf et al. (2008) |
| NA-Forward sequencing | AAATGCACCCAAT | Reisdorf et al. (2008) |
Patient specimens were grown in RMK cells and nucleic acid was extracted from viral culture isolates using the NucliSens® easyMAG™ system (bioMérieux) according to the manufacturer's instructions. RNA was extracted from 250 μl of Eagle's Minimum Essential media (EMEM) (Biowhittaker) with an elution volume of 25 μl. To control for extraction all specimens were tested for human target gapdh by using the gapdh TaqMan control reagents (Applied Biosystems).
To perform Sanger sequencing, reverse transcription and amplification of the N1 gene was performed using the OneStep RT-PCR kit (Qiagen). Three μl of RNA were combined with 5 μl 5× OneStep RT-PCR buffer, 1 μl (200 μM) of dNTPs, 1 μl of OneStep RT-PCR enzyme mix, 0.3 μl (10 U) of RNase inhibitor, 1 μl of each primers AN1B-505 Forward (Takao et al., 2002) and N1-2 Reverse (Yuen et al., 1998) at a final concentration of 0.6 μM (Table 1) and 11.7 μl of water. Reverse transcription and amplification was performed using an iCycler (BioRad) conventional PCR thermocycler at the following conditions: 1 cycle at 50 °C for 30 min and 1 cycle at 95 °C for 15 min, followed by 40 cycles of amplification at 95 °C for 45 s, 55 °C for 45 s and 72 °C for 2 min, and a final extension cycle of 72 °C for 10 min. Following visualization of the PCR products using gel electrophoresis, labelling of the PCR products was performed using the Big Dye Terminator Cycle Sequencing Kit v3.1 (Applied Biosystems). One μl of DNA was combined with 2 μl of BigDye, 3 μl of 5× BigDye buffer, 1 μl of 10 μM AN1B-505 Forward primer and 13 μl of water. An iCycler (BioRad) was utilized for labelling using 1 cycle at 96 °C for 1 min and 25 cycles of 96 °C for 10 s, 55 °C for 5 s, 60 °C for 4 min. Labelled PCR products were purified using the DyeEx column kit (Qiagen) according to the manufacturer's instructions. Nucleotide sequences were determined using the ABI3100 genetic analyzer (Applied Biosystems) and analyzed using FinchTV (Geospiza) and Chromas (Technelysium) DNA sequencing software.
For pyrosequencing, reverse transcription and amplification of the NA gene from the extracted RNA of influenza A H1N1 isolates was performed using the OneStep RT-PCR kit (Qiagen). Three μl of RNA was combined with 5 μl of OneStep RT-PCR buffer, 1 μl (200 μM) of dNTPs, 1 μl of OneStep RT-PCR enzyme mix, 0.3 μl (10 U) of RNase inhibitor, 1 μl of each primers NA-Forward and NA-Reverse (Reisdorf et al., 2008) at a final concentration of 0.6 μM (Table 1). Reverse transcription and amplification was performed using an iCycler (BioRad) conventional PCR thermocycler at the following conditions: 1 cycle of 60 °C for 1 min, 42 °C for 5 s, 42 °C for 10 min, 50 °C for 30 min and 95 °C for 15 min, followed by 40 cycles of amplification at 95 °C for 30 s, 52 °C for 30 s and 72 °C for 1 min, and one extension cycle of 72 °C for 10 min. After visualization on an agarose gel, 20 μl of PCR product was combined with 60 μl of binding mix composed of 40 μl of binding buffer (Biotage), 20 μl of water and 3 μl of streptavidin-sepharose HP beads (Amersham Biosciences). Samples were shaken using a plate shaker (Labnet) at 1400 rpm for 10 min at room temperature and then combined with 40 μl of annealing mix composed of 44 μl of annealing buffer (Biotage) and 0.2 μl of 100 μM of sequencing primer NA-Forward Sequencing (Table 1). Pyrosequencing was carried out using a Pyromark™ ID (Biotage) according to the manufacturer's instructions.
To design a novel H275Y RT-PCR assay, reference sequences were aligned using Clustal W (Larkin et al., 2007). Primers and probe were designed by eye using the consensus sequence. The assay design was evaluated using Primer Express 3 (Applied Biosystems) in order to test for T m and secondary structure. The sequences of the primers and probe for the H275Y assay are listed in Table 1. RT-PCR was carried out using 1 μl of RNA was combined with 10 μl of 2× QuantiTect Probe RT-PCR Master Mix, 0.2 μl of QuantiTect RT Mix, 1 μl of 20× H275Y primers and probes and 7.8 μl of water. Reverse transcription and RT-PCR was carried out on the Stratagene MxPro3005 using 1 cycle of 50 °C for 30 min, 1 cycle of 95 °C for 15 min, and 45 cycles of 94 °C for 15 s and 60 °C for 1 min. To ensure that H275Y probe failure was due to mutation at residue 275 and not sample degradation or inhibition, the N1 subtyping RT-PCR was run in parallel on the same plate.
To determine the limit of detection (LOD) of the H275Y and N1 control RT-PCR assay, serial ten-fold dilutions of nucleic acid from an oseltamivir-sensitive clinical influenza A/Brisbane/57/2007 H1N1 isolate were tested. The starting concentration of virus for this assay was 1500 TCID50/mL. This dilution series was tested by both methods in triplicate to determine reproducibility as well as in singles to simulate clinical testing. The LOD for the H275Y and N1 assays were determined by probit regression (95% confidence interval) using SPSS version 15.
To assess the cross-reactivity of the H275Y RT-PCR assay with non-target respiratory pathogens, a panel of nucleic acid from 62 respiratory pathogens was tested using the H275Y assay. Nucleic acid used in this panel was derived either from clinical specimens or from ATCC strains. Extraction and inhibition controls for the clinical specimens were performed upon initial specimen testing using the Seeplex RV Detection Kit (Seegene) (Drews et al., 2008, Roh et al., 2008).
The LOD of the H275Y RT-PCR assay and the N1 control assay were determined using serial ten-fold dilutions of nucleic acid from a sequence-confirmed oseltamivir-sensitive clinical influenza H1N1 Solomon Islands specimen (Fig. 1A and B). The LOD was calculated using probit regression (95% confidence interval) and found to be 0.025 TCID50/mL for the H275Y assay and 0.25 TCID50/mL for the N1 control assay. Inter-assay and intra-assay variability experiments indicated that the assays were reproducible. The intra-assay coefficient of variation (%CV) was found to be no higher than 2.81% for the H275Y assay and 1.19% for the N1 assay. The inter-assay %CV was found to be no higher than 3.18% for the H275Y assay and 3.68% for the N1 assay.
Fig. 1.
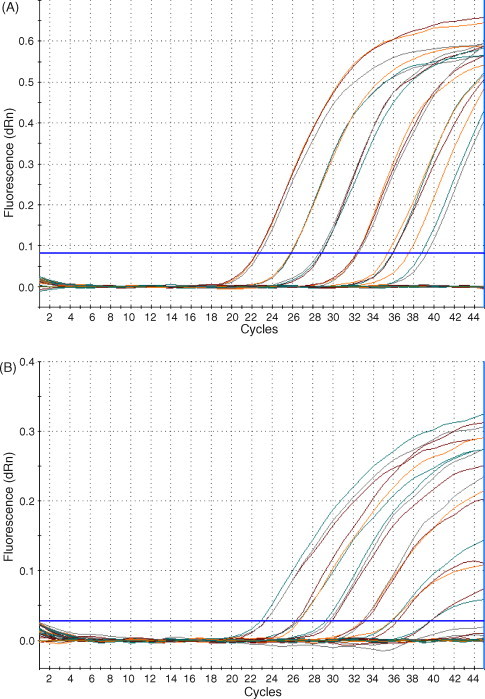
Determination of the LOD of the H275Y and N1 assays: nucleic acid extracted from an influenza A H1N1 oseltamivir-sensitive isolate was used to determine the LOD of the H275Y and N1 assays. The starting concentration of virus was 1500 TCID50/mL. Amplification curves for (A) H275Y and (B) N1 are shown.
The cross reactivity of the H275Y RT-PCR assay was assessed using a panel of nucleic acid from respiratory pathogens. Viral nucleic acid was derived from clinical specimens while bacterial nucleic acid was extracted from clinical specimens or ATCC strains. This pathogen panel included influenza A/Brisbane/10/2007 H3N2, influenza B/Florida/4/2006, parainfluenza virus 1, 2, and 3, rhinovirus, adenovirus, coronavirus, respiratory syncytial virus A and B, human metapneumovirus, Streptococcus pyogenes, Streptococcus pneumoniae, Streptococcus agalactiae, Streptococcus gordonii, Klebsiella pneumonia, Pseudomonas aeruginosa, Haemophilus parainfluenza and Haemophilus influenza A–F. Positive results (Ct values) were only obtained with oseltamivir-sensitive influenza A H1N1 nucleic acid, which was used as a positive control. Specificity of the N1 probe has been published previously (Schweiger et al., 2000) and was not repeated in this study.
The sensitivity and specificity of the H275Y RT-PCR assay were evaluated using 101 influenza A H1N1 isolates, including 40 H275Y-positive and 61 H275-positive isolates which were tested previously for the H275Y mutation using Sanger sequencing (the reference method) and pyrosequencing. All isolates were collected from Ontario patients during the 2007–2008 influenza season and were most similar genetically to A/Brisbane/57/2007-like lineage. The sensitivity and specificity of the H275Y RT-PCR assay were found to be 100% (40/40) and 100% (61/61) respectively (Table 2 ). This compared favourably to the sensitivity and specificity of pyrosequencing, which were 100% (40/40) and 97.5% (60/61) respectively. In contrast to H275-positive isolates, approximately 80% of H275Y-positive isolates produced no cycle threshold (Ct) value, while approximately 20% produced a non-sigmoidal, low fluorescence amplification curves when tested using the H275Y RT-PCR assay (Fig. 2 ). Interpretation of these curves was not problematic as the difference between amplification curves of oseltamivir-sensitive isolates and the non-sigmoidal flat curves of a subset of the resistant isolates was apparent. The TAT of the H275Y RT-PCR assay was 3 hrs after nucleic acid extraction.
Table 2.
Number of isolates used in this study and comparison of specificity and sensitivity of pyrosequencing and H275Y RT-PCR assay with Sanger sequencing.
| Assay | No. of specimens |
Percent |
||
|---|---|---|---|---|
| Resistant | Susceptible | Sensitivity | Specificity | |
| Sanger sequencing | 40 | 61 | – | – |
| H275Y RT-PCR | 40 | 61 | 100 | 100 |
| Pyrosequencing | 40 | 60 | 100 | 97.5 |
Fig. 2.
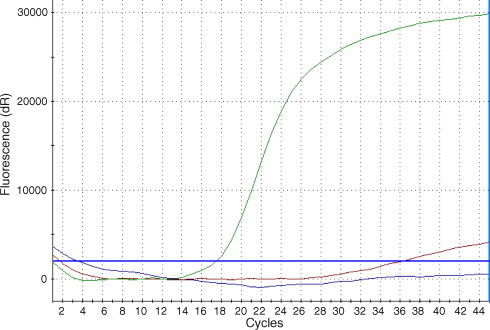
Determination of oseltamivir resistance in influenza A H1N1 isolates using the H275Y RT-PCR assay. Representative amplification curves are shown of both oseltamivir susceptible and resistant clinical isolates. Isolates which were oseltamivir susceptible produced a sigmoidal curve (green) while oseltamivir-resistant isolates produced no curve (blue). Approximately 20% of resistant isolates tested produced a low-fluorescence, non-sigmoidal curve (red).
This study was initiated during the 2007–2008 influenza season, the first season during which significant oseltamivir resistance was observed globally in influenza A H1N1 strains at residue 275 (Sheu et al., 2008, World Health Organization, 2008). As a result of this sudden increase in global resistance to the drug it was felt that an in addition to national and global surveillance programs, an oseltamivir resistance detection protocol should be implemented in Ontario clinical laboratories to assess resistance locally. This study evaluated the use of a novel H275Y RT-PCR assay for detection of H275Y-positive isolates in comparison to Sanger sequencing and pyrosequencing, and found that while all three methods may have a role in influenza diagnostic testing, the H275Y RT-PCR assay is a rapid and effective test for the detection of oseltamivir resistance through mutation at residue 275.
Sanger sequencing is the most established method for detection of point mutations and was therefore used as the reference method for this study. Although it was accurate in determining mutations at residue 275, it was quite expensive and not time-efficient to sequence every H1N1 isolate in a moderately resistant population like Ontario, where only 17% of isolates were oseltamivir resistant. In addition to time restrictions, sequence alignment analyses for Sanger-sequenced isolates must be done using a separate program that generally requires some expertise for operation.
When compared to Sanger sequencing, pyrosequencing was also quite effective in determining H275Y mutations, with a sensitivity and specificity of 100% and 97.5% respectively. Pyrosequencing was less time consuming and provided unambiguous sequence from the 5′ primer binding site, in contrast with Sanger sequencing, which does not provide sequence for the first 30-50 bp of sequence (Pourmand et al., 2006). Unlike Sanger sequencing instruments, the Biotage pyrosequencer has built-in alignment capabilities making sequence analysis less difficult. While in this study pyrosequencing was found to be highly sensitive, the success of pyrosequencing is dependent on the sequence to be analyzed since homopolymeric sequences (repetition of the same nucleotide three or more times) cannot always be discerned (Parameswaran et al., 2007).
Both Sanger sequencing and pyrosequencing are hampered by the need to PCR amplify the NA gene of every isolate before sequencing. This lengthens the turn-around-time (TAT) for oseltamivir-resistance testing and adds an extra step in the influenza testing algorithm. The H275Y RT-PCR assay was found to be as sensitive and specific as Sanger sequencing while decreasing the TAT. The TAT for the H275Y RT-PCR assay was only 3 h after nucleic acid extraction, compared to 11 h for Sanger sequencing and 5.5 h for pyrosequencing. Designing the MGB probe to detect wild type and not mutant sequence allowed for confirmatory sequencing of a minority of resistant isolates as opposed to a majority of wild type isolates, which would not have been time or cost efficient. Implementation of the RT-PCR assay during the 2007–2008 influenza season would have reduced time and cost of isolate sequencing by 83%, since only isolates which did not produce a sigmoidal curve would have to be sequenced using Sanger or pyrosequencing technologies to confirm the presence of a mutation at H275Y.
Although this assay was validated using virus from cultured cells and carried out separately from the influenza A subtyping assay, it has also been performed successfully using nucleic acid extracted directly from specimens. This assay can therefore be incorporated easily into the current subtyping protocol, thus decreasing the TAT even more.
In addition to detecting oseltamivir resistance in seasonal influenza, the H275Y RT-PCR assay can also be utilized during a pandemic influenza outbreak, since it is thought that oseltamivir resistance, which has already been reported in H5N1 avian influenzas strains (de Jong et al., 2005, Kimm-Breschkin et al., 2007, Le et al., 2005) will have a significant impact if an influenza pandemic occurs (Lipsitch et al., 2007). Currently, oseltamivir is the recommended treatment and prophylaxis for avian influenza (Schunemann et al., 2007), and has been stockpiled by most countries. The H275Y RT-PCR assay will serve as a screening tool for resistance and could be performed rapidly enough to affect treatment decisions. The short TAT and high-throughput nature of this assay will help to conserve resources during a pandemic.
The global emergence of H275Y-mediated oseltamivir resistance in influenza A H1N1 specimens has resulted in the recent development of several molecular methods for resistance detection in addition to Sanger sequencing and pyrosequencing. For example, an allelic-discrimination (SNP) assay to detect the H275Y mutation has been published by Carr et al. (2008). The novel H275Y RT-PCR assay reported here compares favourably with the Carr assay, exhibiting parallel test characteristics, no cross reactivity and a similar TAT. However, since the H275Y RT-PCR assay requires only one probe while SNP assays by nature utilize two probes, the H275 RT-PCR assay has a lower cost per test and can be multiplexed easily into influenza detection algorithms that are real-time PCR based.
A drawback of the H275Y RT-PCR assay is that oseltamivir resistance conferred by mutations in residues other than 275 will not be detected. In addition, compensatory mutations in other genes, such as the hemagglutinin gene (Abed et al., 2002) or in conserved genes would not be recognized using this method. However, since oseltamivir resistance is mediated currently by the H275Y mutation globally, with emerging resistance in many jurisdictions (World Health Organization, 2008), a rapid and cost efficient screen targeted at H275Y is the only method for high-throughput resistance testing of clinical specimens.
In conclusion, the emergence of H275Y mediated oseltamivir resistance globally in influenza A H1N1 isolates has indicated the need for a rapid molecular test for the detection of this mutation. The rapid screening approach presented in this study may allow for high-throughput testing for oseltamivir resistance in a timely fashion. The ability to incorporate this method into real-time PCR-based influenza detection testing algorithms and the potential of this assay for use in an influenza pandemic make this test a valuable tool for use in the clinical microbiology laboratory.
References
- Abed Y., Bourgault A.M., Fenton R.J., Morley P.J., Gower D., Owens I.J., Tisdale M., Boivin G. Characterization of 2 influenza A(H3N2) clinical isolates with reduced susceptibility to neuraminidase inhibitors due to mutations in the hemagglutinin gene. J Infect. Dis. 2002;186(8):1074–1080. doi: 10.1086/344237. [DOI] [PubMed] [Google Scholar]
- Afonina I., Kutyavin I., Lukhtanov E., Meyer R.B., Gamper H. Sequence-specific arrest of primer extension on single-stranded DNA by an oligonucleotide-minor groove binder conjugate. Proc. Natl. Acad. Sci. U.S.A. 1996;93(8):3199–3204. doi: 10.1073/pnas.93.8.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besselaar T.G., Naidoo D., Buys A., Gregory V., McAnerney J., Manamela J.M., Blumberg L., Schoub B.D. Widespread oseltamivir resistance in influenza A viruses (H1N1), South Africa. Emerg. Infect. Dis. 2008;14(11):1809–1810. doi: 10.3201/eid1411.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr M.J., Sayre N., Duffy M., Connell J., Hall W.W. Rapid molecular detection of the H275Y oseltamivir resistance gene mutation in circulating influenza A (H1N1) viruses. J Virol. Methods. 2008;153(2):257–262. doi: 10.1016/j.jviromet.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong M.D., Thanh T.T., Khanh T.H., Hien V.M., Smith G.J.D., Chau N.V., Cam B.V., Qui P.T., Ha D.Q., Guan Y., Peiris J.S.M., Hien T.T., Farrar J. Oseltamivir resistance during treatment of influenza A (H5N1) infection. N. Engl. J. Med. 2005;353(25):2667–2672. doi: 10.1056/NEJMoa054512. [DOI] [PubMed] [Google Scholar]
- Drews S.J., Blair J., Lombos E., DeLima C., Burton L., Mazzulli T., Low D.E. Use of the Seeplex RV Detection kit for surveillance of respiratory viral outbreaks in Toronto, Ontario, Canada. Ann. Clin. Lab Sci. 2008;38(4):376–379. [PubMed] [Google Scholar]
- Duwe S., Schweiger B. A new and rapid genotypic assay for the detection of neuraminidase inhibitor resistant influenza A viruses of subtype H1N1, H3N2, and H5N1. J Virol. Methods. 2008;153(2):134–141. doi: 10.1016/j.jviromet.2008.07.017. [DOI] [PubMed] [Google Scholar]
- Kimm-Breschkin J.L., Selleck P.W., Usman T.B., Johnson M.A. Reduced sensitivity of influenza A (H5N1) to oseltamivir. Emerg. Infect. Dis. 2007;13(9):1354–1357. doi: 10.3201/eid1309.07-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Le Q.M., Kiso M., Someya K., Sakai Y.T., Nguyen T.H., Nguyen K.H.L., Pham N.D., Ngyen H.H., Yamada S., Muramoto Y., Horimoto T., Takada A., Goto H., Suzuki T., Suzuki Y., Kawaoka Y. Avian flu: isolation of drug-resistant H5N1 virus. Nature. 2005;437(7062):1108–11108. doi: 10.1038/4371108a. [DOI] [PubMed] [Google Scholar]
- Lipsitch M., Cohen T., Murray M., Levin B.R. Antiviral resistance and the control of pandemic influenza. PLoS Med. 2007;4(1):e15. doi: 10.1371/journal.pmed.0040015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer A., Lackenby A., Hay A., Zambon M. Influenza antiviral susceptibility monitoring activities in relation to national antiviral stockpiles in Europe during the winter 2006/2007 season. Euro. Surveill. 2007;12(4):E3–E4. [PubMed] [Google Scholar]
- Moscona A. Oseltamivir resistance—disabling our influenza defenses. N. Engl. J. Med. 2005;353(25):2633–2636. doi: 10.1056/NEJMp058291. [DOI] [PubMed] [Google Scholar]
- Parameswaran P., Jalili R., Tao L., Shokralla S., Gharizadeh B., Ronaghi M., Fire A.Z. A pyrosequencing-tailored nucleotide barcode design unveils opportunities for large-scale sample multiplexing. Nucl. Acids Res. 2007:760. doi: 10.1093/nar/gkm760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourmand N., Diamond L., Garten R., Erickson J.P., Kumm J., Donis R.O., Davis R.W. Rapid and highly informative diagnostic assay for H5N1 influenza viruses. PLoS ONE. 2006;1(1):e95. doi: 10.1371/journal.pone.0000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisdorf, E., Marshall, S., Van, T., Whyte, T., Gubareva, L., Deyde, V., Shult, P., Warshauer, D., 2008. Identification of mutations in the influenza A genome associated with adamantane and neuraminidase inhibitor resistance directly from clinical specimens. 2008 25th Clinical Virology Symposium, Daytona Beach, FL, USA.
- Roh K.H., Kim J., Nam M.H., Yoon S., Lee C.K., Lee K., Yoo Y., Kim M.J., Cho Y. Comparison of the Seeplex reverse transcription PCR assay with the R-mix viral culture and immunofluorescence techniques for detection of eight respiratory viruses. Ann. Clin. Lab Sci. 2008;38(1):41–46. [PubMed] [Google Scholar]
- Schunemann H.J., Hill S.R., Kakad M., Bellamy R., Uyeki T.M., Hayden F.G., Yazdanpanah Y., Beigel J., Chotpitayasunondh T., Del M.C., Farrar J., Tran T.H., Ozbay B., Sugaya N., Fukuda K., Shindo N., Stockman L., Vist G.E., Croisier A., Nagjdaliyev A., Roth C., Thomson G., Zucker H., Oxman A.D. WHO Rapid Advice Guidelines for pharmacological management of sporadic human infection with avian influenza A (H5N1) virus. Lancet Infect. Dis. 2007;7(1):21–31. doi: 10.1016/S1473-3099(06)70684-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger B., Zadow I., Heckler R., Timm H., Pauli G. Application of a fluorogenic PCR assay for typing and subtyping of influenza viruses in respiratory samples. J Clin. Microbiol. 2000;38(4):1552–1558. doi: 10.1128/jcm.38.4.1552-1558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu T.G., Deyde V.M., Okomo-Adhiambo M., Garten R., Xu X., Bright R., Butler E., Wallis T.R., Klimov A.I., Gubareva L.V. Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide in 2004–2008. Antimicrob. Agents Chemother. 2008 doi: 10.1128/AAC.00555-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao S., Shimazu Y., Fukuda S., Kuwayama M., Miyazaki K. Neuraminidase subtyping of human influenza a viruses by RT-PCR and its application to clinical isolates. Jpn. J Infect. Dis. 2002;55(6):204–205. [PubMed] [Google Scholar]
- Uchiyama M., Maesawa C., Yashima-Abo A., Tarusawa M., Satoh M., Satoh T., Ishida Y., Ito S., Murai K., Enomoto S., Utsugisawa T., Masuda T. Short consensus probes with 3[prime]-minor groove binder of the immunoglobulin heavy-chain gene for real-time quantitative PCR in B-cell non-Hodgkin lymphomas. Lab Invest. 2004;84(7):932–936. doi: 10.1038/labinvest.3700092. [DOI] [PubMed] [Google Scholar]
- World Health Organization, 2005. WHO intercountry consultation. Influenza A/H5N1 in humans in Asia.
- World Health Organization, 2008. Influenza A(H1N1) virus resistance to oseltamivir – 2008 influenza season, southern hemisphere.
- Yuen K.Y., Chan P.K., Peiris M., Tsang D.N., Que T.L., Shortridge K.F., Cheung P.T., To W.K., Ho E.T., Sung R., Cheng A.F. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet. 1998;351(9101):467–471. doi: 10.1016/s0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]


