Highlights
-
•
The Pneumoviriniae fusion (F) protein mediates virus-cell membrane fusion.
-
•
The F protein refolds from a prefusion to a postfusion conformation during fusion.
-
•
Methods are described to isolate MAbs specific of the postfusion conformation.
-
•
Similar methods may be applicable to other type I viral fusion proteins.
Keywords: Fusion proteins, Pneumovirinae, Conformation specific antibodies
Abstract
Paramyxovirus entry into cells requires fusion of the viral and cell membranes mediated by one of the major virus glycoproteins, the fusion (F) glycoprotein which transits from a metastable pre-fusion conformation to a highly stable post-fusion structure during the membrane fusion process. F protein refolding involves large conformational changes of the protein trimer. One of these changes results in assembly of two heptad repeat sequences (HRA and HRB) from each protomer into a six-helix bundle (6HB) motif. To assist in distinguishing pre- and post-fusion conformations of the Pneumovirinae F proteins, and as extension of previous work (Palomo et al., 2014), a general strategy was designed to obtain polyclonal and particularly monoclonal antibodies specific of the 6HB motif of the Pneumovirinae fusion protein. The antibodies reported here should assist in the characterization of the structural changes that the F protein of human metapneumovirus or respiratory syncytial virus experiences during the process of membrane fusion.
1. Introduction
Human metapneumovirus (hMPV) and human respiratory syncytial virus (hRSV) are prototypes of the Metapneumovirus and the Pneumovirus genus of the Pneumovirinae subfamily within the Paramyxoviridae family, respectively (for a recent review, Collins and Karron, 2013). hRSV is the most important cause of acute lower respiratory tract infections (ALRI) in infants worldwide (Hall et al., 2009, Nair et al., 2010) and also a common cause of ALRI in the elderly and adults with cardiopulmonary disease or in whom immune responses are impaired or reduced (Whimbey and Ghosh, 2000, Falsey et al., 2005). Although less is known about hMPV, it is also recognized as a pathogen of clinical relevance, only second to hRSV as a cause of ALRI (Schuster and Williams, 2013). Two subtypes of hMPV (A and B) have been identified by comparison of sequences and by antigenic analysis of viral strains (van den Hoogen et al., 2004).
As in other paramyxoviruses, both hMPV and hRSV encode two major glycoproteins that are inserted in the lipid viral envelope (Collins and Melero, 2011). One is the attachment glycoprotein (G), responsible for the initial interaction of the virus with the target cell surface. The other is the fusion (F) glycoprotein that mediates fusion of the viral and cell membrane, facilitating virus entry (Walsh and Hruska, 1983).
The Pneumovirinae F glycoprotein is a homotrimer in which each subunit is synthesized as an inactive precursor that needs to be cleaved to become fusion competent (Gonzalez-Reyes et al., 2001, Zimmer et al., 2001). The hMPV precursor is slightly shorter (539 aa) than its hRSV counterpart (574 aa) due to a variance of their respective proteolytic processing pathways. While hMPV_F, as in other paramyxoviruses is cleaved at a single site placed immediately upstream of a hydrophobic region, called fusion peptide, hRSV_F is cleaved twice at two polybasic sites separated by a 27 amino acid peptide (Gonzalez-Reyes et al., 2001, Zimmer et al., 2001) (Fig. 1 ). The cleavage site of hRSV_F immediately preceding the hydrophobic fusion peptide is equivalent to the cleavage site of other paramyxovirus F proteins. Once cleavage of hRSV_F is completed, the intervening 27 amino acid peptide is released from the mature protein (Begona Ruiz-Arguello et al., 2002). While hRSV_F is cleaved by furin mainly during transport to the cell membrane, probably in the trans-Golgi network (Collins and Mottet, 1991), hMPV_F is cleaved at the cell surface or in the virus particle by extracellular proteases (Shirogane et al., 2008). In both cases, cleavage generates two chains (F2 end terminal to F1) that remain covalently linked by two disulfide bridges. Hence, the mature Pneumovirinae F protein is a homotrimer of F1 + F2 subunits.
Fig. 1.
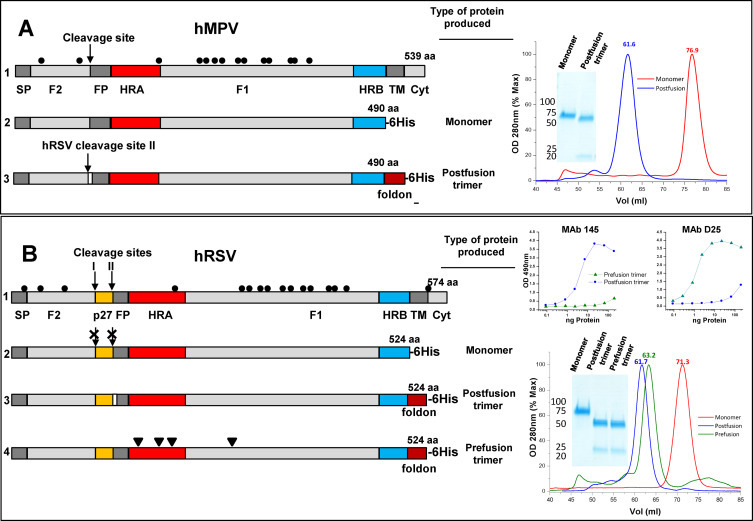
F proteins used in this study: Diagrams of the hMPV_F and hRSV_F primary structures are shown in line 1 of panels A and B, respectively. The signal peptide (SP), F2 subunit, cleavage sites, fusion peptide (FP), F1 subunit, heptad repeats A (HRA) and B (HRB), transmembrane region (TM), and cytoplasmic tail (Cyt) are denoted, as well as the 27 amino acid peptide (p27), unique to hRSV_F. Cysteines are represented by dots. All other lines of panels A and B show the soluble proteins that were produced and purified as described in Section 2, indicating only the structural features that are not represented in the corresponding full-length polypeptide. Panel (A) hMPV_F of line 2 was generated by introducing a 6-His tag after aa 490 to express a monomeric form of the F protein ectodomain. The hMPV_F of line 3 was produced as a soluble post-fusion F protein trimer by adding the foldon trimerization domain at the C-terminus of the protein ectodomain, replacing the hMPV cleavage site by the polybasic cleavage site II of hRSV_F and deleting the first eight amino acids of the fusion peptide (white rectangle). Panel (B) Line 2 shows the hRSV_F ectodomain, expressed as an uncleaved monomer by changing all basic residues of the furin cleavage sites to Ans. The hRSV_F of line 3 is produced as a soluble postfusion trimer (with the first eight FP amino acids deleted) and that of line 4 as a prefusion trimer, stabilized with the mutations (arrowheads) described by McLellan et al. to stabilize their DS-Cav1 variant (McLellan et al., 2013a). Gel filtration profiles and Coomassie stained SDS-PAGE gels of the purified proteins are shown in the right hand of panels A and B. The upper right hand part of panel B shows sandwich ELISAs of the indicated proteins with MAbs specific for either the postfusion (R145, Palomo et al., 2014) or prefusion (D25, McLellan et al., 2013b) conformations of hRSV_F.
The F trimer is incorporated into the virus particle in a prefusion metastable conformation. Once the incoming virus is bound to the surface of the target cell, the F protein is activated by still ill-defined mechanisms to initiate a series of conformational changes, including the formation and refolding of a pre-hairpin intermediate by which the viral and cell membranes are brought into proximity. Final refolding of F in a stable postfusion conformation leads to merging of the two membranes and formation of the fusion pore (Lamb and Jardetzky, 2007). Three-dimensional structures of soluble hRSV_F ectodomains stabilized in either the prefusion (McLellan et al., 2013a, McLellan et al., 2013b) or postfusion conformations (McLellan et al., 2011, Swanson et al., 2011) have been recently solved, providing crucial information to understand the conformational changes that the F protein experiences during membrane fusion. One of these changes involves refolding of HRA and HRB sequences of each F1 subunit in a highly stable coiled-coil structure, dubbed the 6-helix bundle motif (6HB).
Conformation specific antibodies would be very useful reagents to monitor the transition of Pneumovirinae F from the prefusion to postfusion conformation in structural or functional studies. We have reported previously the generation of rabbit antibodies specific for the 6-HB of hRSV_F (Palomo et al., 2014) and the serendipitous isolation of a rabbit monoclonal antibody (MAb) specific of this structural motif. We now describe a systematic approach for the isolation of murine MAbs specific of the 6HB which recognize hMPV_F or hRSV_F folded in their respective postfusion conformations. We propose that this approach could be extended to isolate postfusion specific MAbs directed against the so-called type I fusion proteins of several virus families.
2. Materials and methods
2.1. Ethics statement
Animal work complied with Spanish and European legislation concerning vivisection and the use of genetically modified organisms. Protocols were approved by the “Comité de Ética de la Investigación y del Bienestar Animal” of “Instituto de Salud Carlos III” (CBA PA 19_2012).
2.2. Cloning and expression of HRA, HRB and HRA-L-HRB sequences of hMPV_F fused to glutathione-S-transferase
Sequences encoding amino acids 130–176 of hMPV_F (NL/1/00 strain, subtype A1, GenBank accession number AAK62968, cloned in the pRB21plasmid), corresponding to the HRA domain plus a C-terminal linker GSSGGV (see Fig. 2A) were amplified by PCR using the following primers: forward 5′-GGCGGATCCTTGAAAGTGAAGTAACAGCA-3′ (underlined is the Bam HI site used for cloning) and reverse 5′-GGCGTC GACGCCCCCCGACGACCCTGCACGTGTTAGATTCTTGCTCAC-3′ (the Sal I site is underlined and the sequence encoding the GSSGGV linker is shown in italics). Similarly, F gene sequences encoding amino acids 479–520 of the HRB domain were amplified using the primers: forward 5′-GGCGGATCCTTGACCCAGTCAAGTTTCCT-3′ (underlined is the Bam HI site) and reverse 5′-GGCGCGGCCGCTCAGTTTCCTTTCTCTGCACTGCT-3′ (underlined is the Not I site). The amplified DNAs were digested with the indicated enzymes and cloned into the pGEX-5X-3 plasmid (GE Healthcare). The resulting constructs had the heptad repeat sequences linked through a factor Xa cleavage site and the tripeptide GIL to the C-terminus of the glutathione-S-transferase (GST, see Fig. 2A). The plasmid encoding GST-HRA-L-HRB (Fig. 2A) was obtained by inserting amplified HRB sequences, flanked by Sal I and Not I sites into pGEX-5X-3-HRA-L.
Fig. 2.
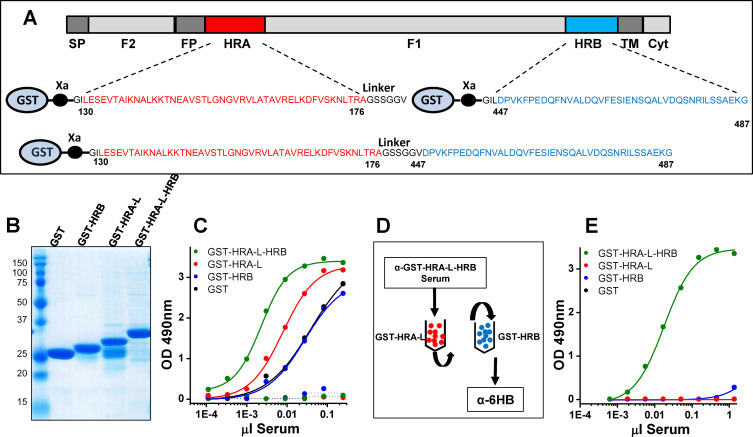
Generation of α-6HB polyclonal antisera against hMPV_F: (A) Scheme of the hMPV_F protein primary structure (as shown in Fig. 1A). The HRA (red), HRB (blue) and HRA-L-HRB sequences that were fused to GST are indicated below. Note that HRA is followed by a linker sequence (L, black) encoded by one of the primers used in the amplification. (B) Coomassie blue stained SDS-PAGE gel of the purified proteins indicated in each lane. (C) Serial dilutions of a rabbit serum (abscissa) raised against purified GST-HRA-L-HRB were tested for binding by direct ELISA to either GST or the indicated chimeric proteins. Dotted lines represent preimmune serum binding. (D) Diagram of the depletion protocol used to obtain α-GST-HRA-L-HRB specific antibodies. Rabbit serum was first loaded onto a column of GST-HRA-L bound to Sepharose (red beads). The unbound material was loaded onto a second column of GST-HRB (blue beads) and unbound antibodies (tentatively named α-6HB) were collected and saved. (E) Direct ELISA binding of α-6HB antibodies to either GST or the indicated proteins. ELISA results of this and other Figures are representative of at least three independent assays.
The different GST-fusion proteins were expressed in E. coli BL21 grown at 37 °C in autoinducible Overnight Express Instant TB Medium (Novagen). Bacteria were collected by sedimentation, resuspended in PBS with 5 mM dithiothreitol (DTT) and lysed by sonication. After clarification, the bacterial extracts were added to Glutathione-Sepharose 4 Fast Flow (GE Healthcare) and incubated overnight at 4 °C in a mixing rotor. After washing with PBS, the bound GST chimeric proteins were eluted with 50 mM Tris–HCl, pH 8.0, 10 mM glutathione.
2.3. Generation of polyclonal anti-six-helix bundle (α-6HB) antibodies
New Zealand white rabbits were inoculated intradermally at multiple sites with 100 μg of purified GST-HRA-L-HRB from hMPV_F, emulsified with an equal volume of Freund's complete adjuvant. After three weeks, rabbits were bled repeatedly.
To deplete the antibodies able to recognize either GST or HRA-L or HRB, two affinity columns were made with 10 mg each of purified GST-HRA-L or GST-HRB covalently bound to 1 g of CNBr-activated Sepharose beads following the manufacturer's instructions (GE Healthcare). The beads were washed and equilibrated in PBS. Sera of rabbits inoculated with GST-HRA-L-HRB were loaded first onto the GST-HRA-L-Sepharose column. The unbound material was collected and loaded onto the GST-HRB-Sepharose column (Fig. 2D). Unbound antibodies (dubbed α-6HB) were collected and stored at −20 °C until used.
2.4. Purification of soluble F proteins
The recombinant vaccinia virus Vac/FTM- which encodes a soluble form of the hRSV_F protein ectodomain (Long strain), generated by the change Ile525Stop (ATC to TAA) has been described (Bembridge et al., 1999). Vaccinia viruses expressing the hMPV_F and hRSV_F variants depicted in Fig. 1 were obtained by the method of Blasco and Moss (Blasco and Moss, 1995) as described before in detail (Bembridge et al., 1998). The different proteins were purified from supernatants of CV1 cells infected for 48 h with the different vaccinia recombinants (0.1 pfu/cell). Culture supernatants were concentrated and buffer exchanged using Vivaflow membranes (Sartorius); then, loaded onto Ni2+ columns in 50 mM Na2HPO4 pH 8.0, 300 mM NaCl, 10 mM Imidazole buffer and, after washing, proteins were eluted with the same buffer containing 250 mM imidazole, Finally, the proteins were concentrated with Vivaspin and exchanged to buffer without imidazole before being loaded onto a HiLoad 16/600 Superdex 200 pg gel filtration column (GE Healthcare) equilibrated and eluted with the same buffer.
2.5. Mouse immunization and isolation of α-6HB monoclonal antibodies (MAbs)
Six-eight week old female BALB/c mice were inoculated intramuscularly (im) with 20 μg of either hMPV_F or hRSV_F, purified and stabilized as soluble post-fusion proteins, as described in the previous section. Four weeks later, mice were boosted im with 20 μg of purified GST-HRA-L-HRB. Four days later, mice were sacrificed by ISOVA inhalation and their splenocytes fused to Sp2-0 myeloma cells with PEG4000, as described (Sanchez-Fauquier et al., 1987). Hybridomas were selected in Clonacell medium supplemented with HAT (hypoxanthine, aminopterin and thymidine) components and after five days transferred to the same medium supplemented with HT. Antibody production was tested by ELISA of hybridoma supernatants as indicated below. Positive cultures were recloned at least twice by limiting dilution.
2.6. Enzyme linked immunosorbent assay
Direct ELISA: 96 well plates were coated overnight at 4 °C with a predetermined amount of antigen, as indicated in the Figure legends. Nonspecific binding was blocked for 1 h with 0.5% serum albumin (BSA) in PBS. Antibody or serum serial dilutions were then added to the plates for 1 h at room temperature and unbound antibodies were removed by washing five times with water. Antibody binding was revealed by incubation with peroxidase-conjugated anti-rabbit or anti-mouse secondary antibodies (GE Healthcare) and subsequent addition of OPD (Sigma) as substrate as per manufacturer's instructions.
Sandwich ELISA: Wells were coated overnight with purified MAbs and unspecific binding blocked as indicated before. Subsequently, serial dilutions of His-tag proteins were added to the wells. After washing, bound proteins were revealed with an excess of anti-His-Biotin, Streptavidin-horseradish peroxidase antibody and OPD.
2.7. Electron microscopy
Purified proteins were applied to glow-discharged carbon-coated grids and negatively stained with 1% aqueous uranyl formate. Micrographs were recorded with a Gatan ERLANGHEN 1000 W CCD camera in a Jeol JEM-1011 electron microscope operated at 100 kV at a nominal magnification of 20,400×.
3. Results
3.1. Purification of soluble F proteins stabilized in different conformations
Fig. 1 shows a diagram of the primary structures of hMPV_F (panel A, line 1) and hRSV_F (panel B, line 1). The different proteins used in this study were produced by using vaccinia virus recombinants, made as described (Blasco and Moss, 1995;Bembridge et al., 1998). A monomeric form of hMPV_F (Fig. 1A, line 2) was obtained by expressing the protein ectodomain followed by a 6-His tag, added for purification purposes. Stabilization of a trimeric postfusion soluble form of the same protein required insertion of the foldon trimerization domain (Meier et al., 2004) before the 6-His tag. In addition, the hMPV_F cleavage site was replaced with the polybasic cleavage site II of hRSV to facilitate proteolytic processing without added trypsin and the first eight amino acids of the fusion peptide were deleted to avoid aggregation of the trimeric postfusion hMPV_F (Fig. 1A, line 3).
The equivalent monomeric form of hRSV_F (Fig. 1B, line 2) was similarly produced, except that cleavage of the monomer was avoided by changing the basic residues of the two furin cleavage sites to Asn (Begona Ruiz-Arguello et al., 2002). However, in the case of hRSV_F -and in contrast to hMPV_F- about half of the protein eluted from the gel filtration chromatography as a trimer. The monomeric peak was collected and loaded again onto a second gel filtration column to obtain the profile shown at right of Fig. 1B. To increase the yield of trimeric postfusion hRSV_F, the foldon domain was added at the C-terminus of the ectodomain, before the 6-His tag (Fig. 1B, line 3). Finally, a stabilized prefusion form of hRSV_F (Fig. 1B, line 4) was made by incorporating the mutations described by McLellan et al. for their DS-Cav1 protein (McLellan et al., 2013a). Prefusion hMPV_F could not be made since equivalent stabilizing mutations have not been described yet. All proteins were purified by using Ni2+ columns followed by gel filtration chromatography. They were quality checked by SDS-PAGE (Fig. 1) and electron microscopy (Fig. 5) and in the case of hRSV_F by reactivity with conformation specific MAbs (Fig. 1B). MAb D25 has been reported to be specific of the prefusion conformation of hRSV_F (McLellan et al., 2013b) whereas R145 was found to be specific of the postfusion conformation of the same molecule (Palomo et al., 2014).
Fig. 5.
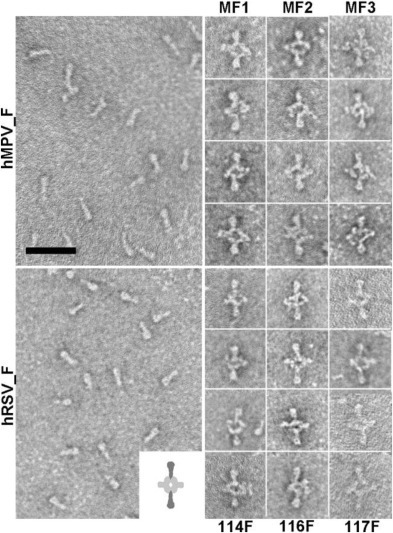
Electron microscopy of proteins and immune complexes: The left large panels show negatively stained preparations of soluble hMPV_F (upper) and hRSV_F (lower) folded in their respective postfusion conformations. Scale bar: 50 nm. The right hand columns are galleries of selected F proteins bound to the indicated MAbs, forming complexes as diagrammed in the lower large panel (inset).
3.2. Isolation and characterization of α-6HB polyclonal antibodies specific of hMPV_F
Based on a previous report describing cloning and expression in E. coli of hRSV_F heptad repeat sequences fused to GST (Palomo et al., 2014), equivalent constructs were made with sequences of hMPV_F (strain NL/1/00, subtype A1), as shown in Fig. 2A. The corresponding proteins, purified by affinity chromatography showed a major band of the expected size after Coomassie staining of SDS-PAGE gels (Fig. 2B), except GST-HRA-L which contained an additional band co-migrating with GST, probably generated by spontaneous cleavage at the GST-HRA linkage during the purification process.
New Zealand white rabbits were then immunized with purified GST-HRA-L-HRB and the immune serum was tested by ELISA for reactivity with either the immunogen or with its purified moieties; i.e., GST-HRA-L, GST-HRB or GST. The results obtained (Fig. 2C) demonstrated the presence of antibodies that reacted with each of the antigens included in the ELISA.
To enrich for antibodies that required both heptad repeats for binding, the anti-GST-HRA-L-HRB antiserum was passed successively through Sepharose columns that contained covalently bound either GST-HRA-L or GST-HRB (Fig. 2D). The antibodies that did not bind to either column were collected and tested again in ELISA with the same antigens. Although the antibodies so depleted lost binding to GST, GST-HRA-L or GST-HRB, they still maintained most of the reactivity with GST-HRA-L-HRB (Fig. 2E) and were tentatively dubbed anti-six-helix bundle (α-6HB) antibodies, by analogy to equivalent antibodies previously described for hRSV_F (Palomo et al., 2014).
3.3. Isolation and characterization of murine MAbs specific for the α-6HB motif of hMPV_F
Once it was demonstrated that the GST-HRA-L-HRB protein could induce antibodies that required the two F protein heptad repeats for binding, mice were first immunized with purified hMPV_F stabilized in the postfusion conformation and then with GST-HRA-L-HRB to boost production of antibodies specific for the 6-HB motif. Hybridoma supernatants derived from mouse splenocytes were screened in parallel by ELISA for antibodies binding to both postfusion F and GST-HRA-L-HRB. Three hybridomas that produced such antibodies were selected and cloned twice by limiting dilution. The specificity of the corresponding antibodies was finally confirmed as shown in Fig. 3 . Fig. 3A demonstrates that the three MAbs MF1, MF2 and MF3 bound efficiently to GST-HRA-L-HRB but not to any of the individual moieties, GST, GST-HRA-L or GST-HRB. Furthermore, the three MAbs efficiently reacted with postfusion soluble trimers of hMPV_F derived from either NL/1/00 (A1 subtype) or NL/1/99 (B1 subtype) strains but not with the monomeric form of the latest protein. As controls, a polyclonal anti-GST (α-GST) antiserum and an anti-hMPV_F MAb (MF14) were included in Fig. 3 to demonstrate reactivity of GST and monomeric hMPV_F, respectively. MF14 antibody (to be described elsewhere) behaves with hMPV_F as palivizumab (Johnson et al., 1997) does with hRSV_F (see Fig. 4B); i.e., both antibodies recognize epitopes that are shared by their respective monomeric and trimeric proteins.
Fig. 3.
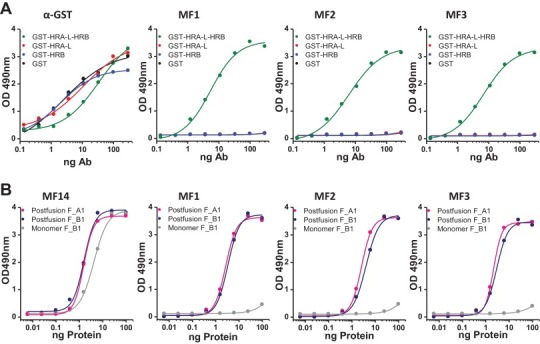
ELISA binding of anti-6HB MAbs specific of hMPV_F: (A) Direct ELISA of polyclonal (α-GST) and monoclonal (MF1, MF2 and MF3) antibodies with either GST or the indicated chimeric proteins. (B) Sandwich ELISA of either a neutralizing MAb (MF14, Olmedillas et al., under preparation) or the three MAbs shown in panel A with the trimeric postfusion forms of hMPV_F derived from strains NL/1/00 (sublineage A1) or NL/1/99 (sublineage B1) or a monomeric form of the latest protein.
Fig. 4.
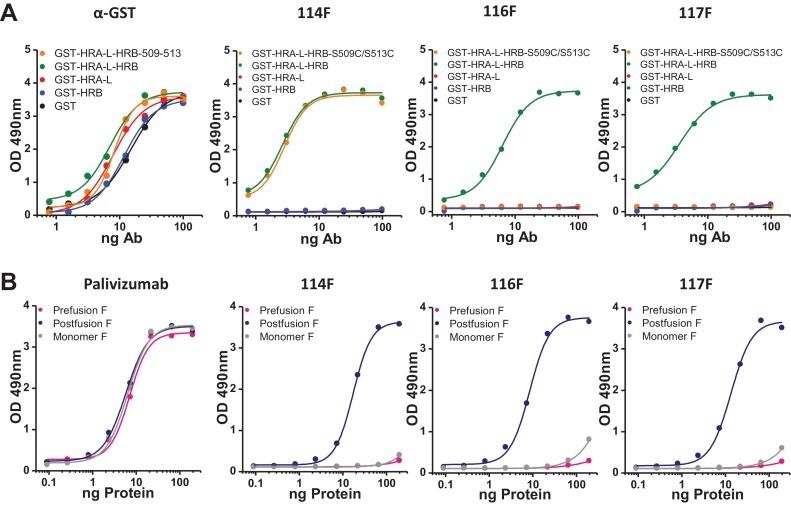
ELISA binding of anti-6HB MAbs specific of hRSV_F: (A) Direct ELISA of polyclonal (α-GST) and monoclonal (114F, 116F and 117F) antibodies with either GST or the indicated chimeric proteins. The GST-HRA-L-HRB double mutant S509C/S513C has been previously described (Palomo et al., 2014). (B) Sandwich ELISA of either the neutralizing MAb palivizumab (Johnson et al., 1997) or the three MAbs shown in panel A with either the postfusion or prefusion soluble trimers of hRSV_F (Long strain) or a monomeric form of the same molecule (Swanson et al., 2014).
The results of Fig. 3 hence demonstrate that the MAbs MF1, MF2 and MF3 bind specifically to the HRA-L-HRB motif which is reproduced in the post-fusion F trimer of the two strains tested but not in the protein monomer.
3.4. Isolation and characterization of MAbs specific for the α-6HB motif of hRSV_F
We have previously shown that GST-chimeric proteins with heptad repeat sequences from hRSV_F were capable of inducing rabbit polyclonal antibodies equivalent to those shown in Fig. 2 for hMPV_F (Palomo et al., 2014). Hence, a similar strategy to that described in the previous section was used to obtain murine MAbs that reacted with both post-fusion hRSV_F and GST-HRA-L-HRB. Three hybridomas were thus isolated which produced MAbs 114F, 116F and 117F. These three MAbs specifically bound GST-HRA-L-HRB but not any of its moieties (GST, GST-HRA-L or GST-HRB) (Fig. 4A). Interestingly, MAb 114F was able to bind a previously described double mutant of GST-HRA-L-HRB (S509C/S513C) which could not bind rabbit MAb R145 nor the new murine MAbs 116F and 117F. Therefore, the three murine MAbs described in this study recognize at least two different, although probably highly overlapping epitopes. Additionally, the three MAbs bound soluble hRSV_F stabilized in the post-fusion conformation but they did not react with either the pre-fusion conformation or monomeric F (Fig. 4B). Hence, binding of MAbs 114F, 116F and 117F to hRSV_F is fully dependent on folding of the protein trimer into the postfusion conformation.
3.5. Electron microscopy
To visualize binding of the MAbs described in the two previous sections to their corresponding F protein trimers, they were incubated with either soluble hMPV_F or hRSV_F stabilized in their respective post-fusion conformations and negatively stained before being observed by electron microscopy. Fig. 5 shows representative images of the purified proteins with the characteristic cone shape of post-fusion trimers and selected pictures of protein-antibody complexes. The MAbs bound to the protein stem end (i.e., distal from the protein head) where the 6-HB motif is located, confirming the MAb specificity inferred from the ELISA binding results.
4. Discussion
As reported in our previous publication (Palomo et al., 2014), immunization of rabbits with GST chimeras expressing HRA-L-HRB sequences derived from either hMPV_F or hRSV_F induced high levels of polyclonal antibodies that require both heptad repeats for binding. Although HRA can assemble by itself in a coiled-coil helix trimer, the α-6HB antibodies required incorporation of HRB to the inner core for binding, reflecting the structure acquired when the two heptad repeats are mixed together (Lawless-Delmedico et al., 2000, Matthews et al., 2000). These binding requirements are reproduced in the murine MAbs described here (Fig. 3, Fig. 4) and are likely responsible of the restricted specificity of these antibodies for the post-fusion forms of hMPV_F and HRSV_F.
Our strategy for raising MAbs specific of the post-fusion conformation originates from reports demonstrating that the 6-HB motif can be assembled in the test tube when HRA and HRB peptides are properly mixed or are linked together by a short spacer (Zhao et al., 2000, Lawless-Delmedico et al., 2000, Matthews et al., 2000). Hence, we expected that immunizations with soluble forms of post-fusion Pneumovirinae F followed by a boost with the corresponding GST-HRA-L-HRB proteins should preferentially induce antibodies of the desired specificity. This strategy indeed made isolation of MAbs that bound both GST-HRA-L-HRB and postfusion F protein a straightforward process. In our experience, screening of hybridoma supernatants with the chimeric GST-HRA-L-HRB is the most critical step for identification of the desired α-6HB MAbs, even with alternative immunization protocols. The MAbs thus isolated were unable to bind either monomeric F or prefusion stabilized F protein (in the case of hRSV). Although a few conformation dependent MAbs specific for the F proteins of PIV5 (Bissonnette et al., 2006), hRSV (Palomo et al., 2014) and Newcastle Disease Virus (NDV) (Umino et al., 1990) have been serendipitously found, the procedure used in this study provides a reliable approach to generate post-fusion specific MAbs.
The 6-HB motif characteristic of the post-fusion Pneumovirinae F protein is shared with the F proteins of all viruses of the Paramyoxividae family (Lamb and Jardetzky, 2007). Similar structure core motifs are also found in the so-called type I fusion proteins which mediate fusion of viral and cell membranes for entry of viruses of the following families: Filoviridae, Retroviridae, Arenaviridae, Orthomyxoviridae and Coronaviridae (Igonet and Rey, 2012). Partial core helical structures have been reported for at least the PIV5 fusion protein (Baker et al., 1999), the coronavirus spike S glycoprotein (Bosch et al., 2003, Tripet et al., 2004, Xu et al., 2004), the HIV1 gp41 envelope subunit (Chan et al., 1997), the filovirus (Malashkevich et al., 1999) and the arenavirus (Igonet et al., 2011) glycoproteins. These core structures could hence be used to generate MAbs specific of the post-fusion conformations of the related proteins, as done here for the post-fusion Pneumovirinae F proteins. Those antibodies would be valuable tools to follow the structural transitions that type I fusion glycoproteins experience during the membrane fusion process.
Acknowledgements
We would like to thank members of the Genomics and Animal House Core Facilities for technical help. This work was supported in part by grants SAF2012-31217 from Plan Nacional I + D + I (JAM, MINECO) and BFU2013-43149-R (DL, MINECO) and PI10/00895 from AES/FIS (DL, ISCIII).
References
- Baker K.A., Dutch R.E., Lamb R.A., Jardetzky T.S. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell. 1999;3:309–319. doi: 10.1016/s1097-2765(00)80458-x. [DOI] [PubMed] [Google Scholar]
- Begona Ruiz-Arguello M., Gonzalez-Reyes L., Calder L.J., Palomo C., Martin D., Saiz M.J., Garcia-Barreno B., Skehel J.J., Melero J.A. Effect of proteolytic processing at two distinct sites on shape and aggregation of an anchorless fusion protein of human respiratory syncytial virus and fate of the intervening segment. Virology. 2002;298:317–326. doi: 10.1006/viro.2002.1497. [DOI] [PubMed] [Google Scholar]
- Bembridge G.P., Lopez J.A., Bustos R., Melero J.A., Cook R., Mason H., Taylor G. Priming with a secreted form of the fusion protein of respiratory syncytial virus (RSV) promotes interleukin-4 (IL-4) and IL-5 production but not pulmonary eosinophilia following RSV challenge. J Virol. 1999;73:10086–10094. doi: 10.1128/jvi.73.12.10086-10094.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bembridge G.P., Lopez J.A., Cook R., Melero J.A., Taylor G. Recombinant vaccinia virus coexpressing the F protein of respiratory syncytial virus (RSV) and interleukin-4 (IL-4) does not inhibit the development of RSV-specific memory cytotoxic T lymphocytes, whereas priming is diminished in the presence of high levels of IL-2 or gamma interferon. J Virol. 1998;72:4080–4087. doi: 10.1128/jvi.72.5.4080-4087.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissonnette M.L., Connolly S.A., Young D.F., Randall R.E., Paterson R.G., Lamb R.A. Analysis of the pH requirement for membrane fusion of different isolates of the paramyxovirus parainfluenza virus 5. J Virol. 2006;80:3071–3077. doi: 10.1128/JVI.80.6.3071-3077.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco R., Moss B. Selection of recombinant vaccinia viruses on the basis of plaque formation. Gene. 1995;158:157–162. doi: 10.1016/0378-1119(95)00149-z. [DOI] [PubMed] [Google Scholar]
- Bosch B.J., van der Zee R., de Haan C.A., Rottier P.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan D.C., Fass D., Berger J.M., Kim P.S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- Collins P.L., Karron R.A. Respiratory syncytial virus and metapneumovirus. In: Knipe D.M., Howley P.M., editors. Fields Virology. Wolters Kluwer/Lippincott Wlliams & Wilkins; Piladelphia: 2013. pp. 1086–1123. [Google Scholar]
- Collins P.L., Melero J.A. Progress in understanding and controlling respiratory syncytial virus: still crazy after all these years. Virus Res. 2011;162:80–99. doi: 10.1016/j.virusres.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins P.L., Mottet G. Post-translational processing and oligomerization of the fusion glycoprotein of human respiratory syncytial virus. J. Gen. Virol. 1991;72(Pt 12):3095–3101. doi: 10.1099/0022-1317-72-12-3095. [DOI] [PubMed] [Google Scholar]
- Falsey A.R., Hennessey P.A., Formica M.A., Cox C., Walsh E.E. Respiratory syncytial virus infection in elderly and high-risk adults. N Engl J Med. 2005;352:1749–1759. doi: 10.1056/NEJMoa043951. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Reyes L., Ruiz-Arguello M.B., Garcia-Barreno B., Calder L., Lopez J.A., Albar J.P., Skehel J.J., Wiley D.C., Melero J.A. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc. Natl Acad. Sci U.S.A. 2001;98:9859–9864. doi: 10.1073/pnas.151098198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C.B., Weinberg G.A., Iwane M.K., Blumkin A.K., Edwards K.M., Staat M.A., Auinger P., Griffin M.R., Poehling K.A., Erdman D., Grijalva C.G., Zhu Y., Szilagyi P. The burden of respiratory syncytial virus infection in young children. N. Engl. J. Med. 2009;360:588–598. doi: 10.1056/NEJMoa0804877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igonet S., Rey F.A. SnapShot: Viral and eukaryotic protein fusogens. Cell. 2012;151:1634. doi: 10.1016/j.cell.2012.11.041. [DOI] [PubMed] [Google Scholar]
- Igonet S., Vaney M.C., Vonrhein C., Bricogne G., Stura E.A., Hengartner H., Eschli B., Rey F.A. X-ray structure of the arenavirus glycoprotein GP2 in its postfusion hairpin conformation. Proc. Natl. Acad. Sci. U.S.A. 2011;108:19967–19972. doi: 10.1073/pnas.1108910108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S., Oliver C., Prince G.A., Hemming V.G., Pfarr D.S., Wang S.C., Dormitzer M., O’Grady J., Koenig S., Tamura J.K., Woods R., Bansal G., Couchenour D., Tsao E., Hall W.C., Young J.F. Development of a humanized monoclonal antibody (MEDI-493) with potent in vitro and in vivo activity against respiratory syncytial virus. J. Infect. Dis. 1997;176:1215–1224. doi: 10.1086/514115. [DOI] [PubMed] [Google Scholar]
- Lamb R.A., Jardetzky T.S. Structural basis of viral invasion: lessons from paramyxovirus F. Curr. Opin. Struct. Biol. 2007;17:427–436. doi: 10.1016/j.sbi.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawless-Delmedico M.K., Sista P., Sen R., Moore N.C., Antczak J.B., White J.M., Greene R.J., Leanza K.C., Matthews T.J., Lambert D.M. Heptad-repeat regions of respiratory syncytial virus F1 protein form a six-membered coiled-coil complex. Biochemistry. 2000;39:11684–11695. doi: 10.1021/bi000471y. [DOI] [PubMed] [Google Scholar]
- Malashkevich V.N., Schneider B.J., McNally M.L., Milhollen M.A., Pang J.X., Kim P.S. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-A resolution. Proc. Natl. Acad. Sci. U.S.A. 1999;96:2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews J.M., Young T.F., Tucker S.P., Mackay J.P. The core of the respiratory syncytial virus fusion protein is a trimeric coiled coil. J. Virol. 2000;74:5911–5920. doi: 10.1128/jvi.74.13.5911-5920.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan J.S., Chen M., Joyce M.G., Sastry M., Stewart-Jones G.B., Yang Y., Zhang B., Chen L., Srivatsan S., Zheng A., Zhou T., Graepel K.W., Kumar A., Moin S., Boyington J.C., Chuang G.Y., Soto C., Baxa U., Bakker A.Q., Spits H., Beaumont T., Zheng Z., Xia N., Ko S.Y., Todd J.P., Rao S., Graham B.S., Kwong P.D. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013;342:592–598. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan J.S., Chen M., Leung S., Graepel K.W., Du X., Yang Y., Zhou T., Baxa U., Yasuda E., Beaumont T., Kumar A., Modjarrad K., Zheng Z., Zhao M., Xia N., Kwong P.D., Graham B.S. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science. 2013;340:1113–1117. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan J.S., Yang Y., Graham B.S., Kwong P.D. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J. Virol. 2011;85:7788–7796. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier S., Guthe S., Kiefhaber T., Grzesiek S. Foldon, the natural trimerization domain of T4 fibritin, dissociates into a monomeric A-state form containing a stable beta-hairpin: atomic details of trimer dissociation and local beta-hairpin stability from residual dipolar couplings. J. Mol. Biol. 2004;344:1051–1069. doi: 10.1016/j.jmb.2004.09.079. [DOI] [PubMed] [Google Scholar]
- Nair H., Nokes D.J., Gessner B.D., Dherani M., Madhi S.A., Singleton R.J., O’Brien K.L., Roca A., Wright P.F., Bruce N., Chandran A., Theodoratou E., Sutanto A., Sedyaningsih E.R., Ngama M., Munywoki P.K., Kartasasmita C., Simoes E.A., Rudan I., Weber M.W., Campbell H. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet. 2010;375:1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomo C., Mas V., Vazquez M., Cano O., Luque D., Terron M.C., Calder L.J., Melero J.A. Polyclonal and monoclonal antibodies specific for the six-helix bundle of the human respiratory syncytial virus fusion glycoprotein as probes of the protein post-fusion conformation. Virology. 2014;460–461C:119–127. doi: 10.1016/j.virol.2014.05.001. [DOI] [PubMed] [Google Scholar]
- Sanchez-Fauquier A., Villanueva N., Melero J.A. Isolation of cross-reactive, subtype-specific monoclonal antibodies against influenza virus HA1 and HA2 hemagglutinin subunits. Arch. Virol. 1987;97:251–265. doi: 10.1007/BF01314425. [DOI] [PubMed] [Google Scholar]
- Schuster J.E., Williams J.V. Human metapneumovirus. Pediatr. Rev. 2013;34:558–565. doi: 10.1542/pir.34-12-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirogane Y., Takeda M., Iwasaki M., Ishiguro N., Takeuchi H., Nakatsu Y., Tahara M., Kikuta H., Yanagi Y. Efficient multiplication of human metapneumovirus in Vero cells expressing the transmembrane serine protease TMPRSS2. J. Virol. 2008;82:8942–8946. doi: 10.1128/JVI.00676-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson K.A., Balabanis K., Xie Y., Aggarwal Y., Palomo C., Mas V., Metrick C., Yang H., Shaw C.A., Melero J.A., Dormitzer P.R., Carfi A. A monomeric uncleaved RSV F antigen retains pre-fusion-specific neutralizing epitopes. J. Virol. 2014 doi: 10.1128/JVI.01225-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson K.A., Settembre E.C., Shaw C.A., Dey A.K., Rappuoli R., Mandl C.W., Dormitzer P.R., Carfi A. Structural basis for immunization with postfusion respiratory syncytial virus fusion F glycoprotein (RSV F) to elicit high neutralizing antibody titers. Proc. Natl Acad. Sci U.S.A. 2011;108:9619–9624. doi: 10.1073/pnas.1106536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripet B., Howard M.W., Jobling M., Holmes R.K., Holmes K.V., Hodges R.S. Structural characterization of the SARS-coronavirus spike S fusion protein core. J. Biol. Chem. 2004;279:20836–20849. doi: 10.1074/jbc.M400759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umino Y., Kohama T., Sato T.A., Sugiura A., Klenk H.D., Rott R. Monoclonal antibodies to three structural proteins of Newcastle disease virus: biological characterization with particular reference to the conformational change of envelope glycoproteins associated with proteolytic cleavage. J. Gen. Virol. 1990;71(Pt 5):1189–1197. doi: 10.1099/0022-1317-71-5-1189. [DOI] [PubMed] [Google Scholar]
- van den Hoogen B.G., Herfst S., Sprong L., Cane P.A., Forleo-Neto E., de Swart R.L., Osterhaus A.D., Fouchier R.A. Antigenic and genetic variability of human metapneumoviruses. Emerg. Infect. Dis. 2004;10:658–666. doi: 10.3201/eid1004.030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh E.E., Hruska J. Monoclonal antibodies to respiratory syncytial virus proteins: identification of the fusion protein. J. Virol. 1983;47:171–177. doi: 10.1128/jvi.47.1.171-177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whimbey E., Ghosh S. Respiratory syncytial virus infections in immunocompromised adults. Curr. Clin Top. Infect. Dis. 2000;20:232–255. [PubMed] [Google Scholar]
- Xu Y., Lou Z., Liu Y., Pang H., Tien P., Gao G.F., Rao Z. Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core. J. Biol. Chem. 2004;279:49414–49419. doi: 10.1074/jbc.M408782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Singh M., Malashkevich V.N., Kim P.S. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc. Natl. Acad. Sci. U.S.A. 2000;97:14172–14177. doi: 10.1073/pnas.260499197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer G., Budz L., Herrler G. Proteolytic activation of respiratory syncytial virus fusion protein. Cleavage at two furin consensus sequences. J. Biol. Chem. 2001;276:31642–31650. doi: 10.1074/jbc.M102633200. [DOI] [PubMed] [Google Scholar]


