Highlights
► A simple and rapid measure for generation of a scFv library against pAPN using T7Select phage display. ► The scFv library possesses a sufficiently large capacity. ► The scFv library has a sufficient diversity.
Keywords: pAPN, scFv, Phage library, TGEV, PEDV
Abstract
Porcine aminopeptidase N (pAPN) is a common cellular receptor for swine transmissible gastroenteritis virus (TGEV) and porcine epidemic diarrhea virus (PEDV). To investigate single-chain fragment variable (scFv) repertoire against pAPN, the genes encoding the immunoglobulin light chain variable region (VL) and heavy chain variable region (VH) were amplified by reverse transcript polymerase chain reaction (RT-PCR) using a series of degenerate primers from the spleen of BABL/c mice immunized with native pAPN. The VL and VH amplicons were combined randomly by a 12 amino acid flexible linker by splicing by overlap extension PCR (SOE-PCR), which produced the scFv gene repertoire. After ligation of the scFv gene repertoire into the T7Select10-3b vector, a mouse scFv phage library specific for pAPN was produced through in vitro packaging. The primary scFv library against pAPN contained 2.0 × 107 recombinant phage clones, and the titer of the amplified library was 3.6 × 109 pfu/mL. BstNI restriction analysis and DNA sequencing revealed that 28 phage clones from the primary pAPN scFv library showed excellent diversity. The effectiveness of the scFv library against pAPN was verified further by phage ELISA using the recombinant protein of the pAPN C subunit as coating antigen. The construction and evaluation of a murine scFv library against the common receptor pAPN of porcine coronaviruses TGEV and PEDV using the T7 phage display system are described.
1. Introduction
Porcine aminopeptidase N (pAPN) is a type II transmembrane glycoprotein belonging to the membrane-bound metallopeptidase family; it is composed of 963 amino acids and has a molecular weight of 150–160 kDa (Delmas et al., 1994, Hooper, 1994). The pAPN is distributed mainly on the surface of porcine intestinal epithelial cells, where some pAPN proteins are cleaved into a B subunit (aa 1–572, 95 kDa) and a C subunit (aa 573–963, 50 kDa) by trypsin (Kenny and Maroux, 1982, Semenza, 1986, Jongeneel et al., 1989). pAPN has been revealed to be a common cellular receptor for swine transmissible gastroenteritis virus (TGEV) and porcine epidemic diarrhea virus (PEDV) to allow their entry into cells (Delmas et al., 1992, Li et al., 2007).
TGEV and PEDV belong to group I of the genus Coronavirus, family Coronaviridae, order Nidovirales, and cause severe diarrhea and high mortality rates of up to 100% in piglets (Pensaert and Debouck, 1978, Reynolds and Garwes, 1979). In the past 30 years, disease caused by TGEV and PEDV has broken out frequently in many swine-raising countries, and has led to heavy economic losses (Jung and Chae, 2005, Kim et al., 2007, Vemulapalli et al., 2009, Chen et al., 2010). Many studies of these viruses have been reported over the past decades, but TGEV and PEDV still exist and cause frequent occurrences of piglet diarrhea in various countries, notably in China (Sun et al., 2007, Sun et al., 2008, Elia et al., 2010, Ribes et al., 2011, Nogales et al., 2011). Most researchers have focused mainly on how to kill or control the pathogens TGEV and PEDV. Only a little information has been reported regarding anti-viral measures against TGEV and PEDV that are based on the cellular receptor pAPN (Liu et al., 2009, Ren et al., 2010).
The single-chain fragment variable (scFv) is a small engineered antibody in which the variable light chain (VL) and heavy chain (VH) of the antibody are connected by a short polypeptide linker. The scFv holds important potential for receptor blockade, pathogen neutralization and therapeutic antigen targeting in vivo (Haidaris et al., 2001, Pansri et al., 2009). At present, isolation and identification of interesting scFvs are carried out through a powerful phage display library. In the current study, the T7 phage display system was use to construct a mouse scFv library specific for pAPN, the common receptor for TGEV and PEDV. The scFv library against pAPN was generated, and the potential utility of the scFv library was confirmed by restriction analysis, sequencing, and specific antigen binding. The pAPN scFv library and its preparation will provide useful information and a basis for further research into anti-viral agents against TGEV and PEDV, and other coronaviruses that use APN as a cellular receptor.
2. Materials and methods
2.1. Primers for mouse scFv gene repertoire
Primers for PCR amplification of the immunized mouse VL and VH genes were designed according to the degenerate primers described by Okamoto et al. (2004). The 5′ ends of the mouse VL forward primers and VH reverse primers were modified to include EcoRI and HindIII sites, respectively. In order to splice the VL and VH amplicons, a complementary overlapping sequence encoding a flexible linker of 12 amino acids (GGGGSGGGGSGG) was added to the 5′ ends of the mouse VL reverse primers and VH forward primers, respectively. The primers MscFv-F and MscFv-R were designed for generation of the scFv genes by splicing by overlap extension PCR (SOE-PCR). All primers are shown in Table 1 .
Table 1.
Primers for PCR amplification of the immunized mouse VH and VL genes.
| Mouse VL forward primers | |
| MVL-F1 | 5′-cggcatttcgtcGAATTCcGAYATTGTDHTVWCHCAGTC-3′ |
| MVL-F2 | 5′-cggcatttcgtcGAATTCcGAYATTNWKMTVAHDCAGTC-3′ |
| MVL-F3 | 5′-cggcatttcgtcGAATTCcGAYRTYBWRMTSACMCARWC-3′ |
| MVL-F4 | 5′-cggcatttcgtcGAATTCcGAYATYSWGMTGACNCARBC-3′ |
| MVL-F5 | 5′-cggcatttcgtcGAATTCc GAYRYTGTKRTRMYYMRGDW-3′ |
| Mouse VL reverse primers | |
| MVL-R1 | 5′-tcctcctgagccgccgccgccagaaccaccaccaccCCGTTYNAKYTCCARCTTDG-3′ |
| MVL-R1 | 5′-tcctcctgagccgccgccgccagaaccaccaccaccMCSTWBNABHKYCAVYYTDG-3′ |
| Mouse VH forward primers | |
| MVH-F1 | 5′-ggtggtggtggttctggcggcggcggctcaggaggaSAKGTBMAGCTBMAGSASTC-3′ |
| MVH-F2 | 5′-ggtggtggtggttctggcggcggcggctcaggaggaSAGGTYCARCTBCARCARTC-3′ |
| MVH-F3 | 5′-ggtggtggtggttctggcggcggcggctcaggaggaSAVGTSMWSBTGRWGSARTC-3′ |
| MVH-F4 | 5′-ggtggtggtggttctggcggcggcggctcaggaggaGAKGTGMAVSKGRTGGARTC-3′ |
| MVH-F5 | 5′-ggtggtggtggttctggcggcggcggctcaggaggaGARGTRMARSTTSWBGAGTC-3′ |
| MVH-F6 | 5′-ggtggtggtggttctggcggcggcggctcaggaggaSAKGTBMMNYTVVWVSWRYS-3′ |
| Mouse VH reverse primers | |
| MVH-R1 | 5′-ttctatgcgcAAGCTTttaYGARGARACDSTGASMRKRGT-3′ |
| MVH-R2 | 5′-ttctatgcgcAAGCTTttaYGARGARRMSKKKASWGWGRT-3′ |
| MVH-R3 | 5′-ttctatgcgcAAGCTTttaYGAGGAGACKGTGASHGDGGH-3′ |
| Primers for generation of scFv genes in SOE-PCR | |
| MscFv-F | 5′-cggcatttcgtcGAATTCc-3′ |
| MscFv-R | 5′-ttctatgcgcAAGCTTtta-3′ |
S = G/C, R = G/A, K = G/T, M = A/C, Y = C/T, W = A/T, H = A/C/T, B = C/G/T, V = A/C/G, D = A/G/T, and N = A/T/G/C.
2.2. Immunization of mice with pAPN
Female BALB/c mice, 6–8 weeks old (Harbin Veterinary Research Institute Experimental Animal Center, Harbin, China), were injected subcutaneously with 50 μg of native pAPN from porcine kidney (Sigma–Aldrich, St. Louis, MO, USA) emulsified in an equal amount of complete Freund's adjuvant (Sigma, St. Louis, MO, USA). At 3-week intervals two boosters of 50 μg of the native pAPN emulsified in an equal amount of incomplete Freund's adjuvant (Sigma, St. Louis, MO, USA) were administered. The spleens of the immunized mice were isolated 3 days after the last booster inoculation, and stored at −80 °C.
2.3. RNA extraction and cDNA synthesis
Total RNA was prepared from spleen tissue of the immunized mice using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The mRNA was purified from extracted total RNA using a Poly AT tract mRNA isolation kit (Promega, Madison, WI, USA). The first strand cDNA was generated using an OrientExpress Oligo (dT) cDNA Synthesis Kit (Novagen, San Diego, CA, USA) according to the manufacturer's instructions.
2.4. Generation of mouse scFv gene repertoire
The PCR amplification of the mouse VL and VH genes was carried out in a 50 μL mixture containing 10 nmol of each forward and reverse primer, 4 μL cDNA extracted from spleen tissue of the immunized mice, 0.25 mmol each dNTP mixture (Takara, Dalian, China), 10× buffer, and 0.5 U ExTaq DNA polymerase (Takara, Dalian, China). The amplification conditions comprised an initial denaturation at 95 °C for 5 min, followed by 35 PCR cycles of 95 °C for 1 min, 55 °C for 1 min, and 72 °C for 1 min, and a final extension step of 72 °C for 10 min. The amplified products of the mouse VL and VH genes were purified with a Qiaquick Gel Extraction Kit (Qiagen, Hilden, Germany). A SOE-PCR was performed for generation of the mouse scFv gene. Briefly, the SOE-PCR reaction contained 100 ng of purified VL products and 100 ng of purified VH products, 10 nmol of MscFv-F and MscFv-R primers, 0.25 mmol each dNTP mixture (Takara, Dalian, China), 10× buffer, and 0.5 U ExTaq DNA polymerase (Takara, Dalian, China). The cycling conditions for the SOE-PCR were the same as mentioned previously.
2.5. Cloning of mouse scFv gene repertoire into T7Select10-3b vector
The mouse scFv gene products were digested with restriction enzymes EcoRI and HindIII. After purification of the digested products, cohesive ligation of the scFv gene products (0.12 pmol) with T7Select 10-3b EcoRI/HindIII Vector Arms (0.04 pmol) was carried out in a working volume of 5 μL using T4 DNA Ligase (Novagen, San Diego, CA, USA) at 16 °C for 12 h. The ligation products were added directly to 25 μL T7 Packaging Extracts (Novagen, San Diego, CA, USA), and the mixtures were incubated for 2 h at 22 °C for in vitro packaging. After the reaction had been stopped by the addition of 270 μL sterile LB medium, the mouse scFv library against pAPN was obtained from the packaged phages. The primary pAPN scFv library was amplified by liquid lysate amplification according to the T7Select System Manual. The titers of the primary and amplified library were determined by plaque assay.
2.6. BstNI restriction analysis and DNA sequencing
Thirty phage clones were selected randomly from the primary scFv library against pAPN, and they were cultured by liquid lysate amplification. After purification by the PEG/NaCl method, the genomic DNA of each phage clone was extracted by simple heat treatment. The inserted scFv gene of each recombinant phage was amplified by PCR using the primers T7SelectUP (5′-GGAGCTGTCGTATTCCAGTC-3′) and T7SelectDOWN (5′-ACCCCTCAAGACCCGTTTA-3′), which were specific for amplification of the region surrounding the multiple cloning site of the vector T7Select 10-3b. The amplified product of each phage clone was digested with the frequent cutting restriction enzyme BstNI (New England Biolabs Inc., Beverly, MA, USA). Furthermore, the nucleotide sequences of three scFv clones with different restriction patterns were obtained. The framework region (FR) and complementarity determining region (CDR) of each sequenced mouse VL and VH gene were analyzed using the IMGT/V-QUEST program version 3.2.20 of the international immunogenetics information system (http://www.imgt.org./IMGT_Vquest/share/textes/).
2.7. scFv library validation
To determine the presence of antigen-specific scFv that bound to the recombinant C subunit of pAPN (pAPN-C) expressed in Escherichia coli in a previous study, a biopanning procedure was run for enrichment of the specific scFv phages according to the T7Select System Manual. Twenty-five single phage clones were selected from output phages of the third round of biopanning, and their reactivity with pAPN-C was analyzed by phage ELISA. Briefly, the ELISA plate wells were coated using the purified pAPN-C protein at 4 °C for 12 h. After blocking with skimmed milk, 1.0 × 108 pfu of selected phages were added to the well, and incubated at 37 °C for 1 h. Phages that bound to the immobilized pAPN-C protein were detected by incubation with HRP-conjugated anti-T7 tag antibody (EMD Chemicals Inc., San Diego, CA, USA), followed by a coloration reaction using the substrate TMB solution (Amresco, USA). The absorbance at 450 nm was measured. The phages generated by the T7Select Control Insert were used as negative controls. The sample OD450 value/negative control OD450 value (S/N) > 2 was determined as the positive standard.
3. Results
3.1. Amplification of the VL and VH genes from immunized mice
The cDNA was prepared using the extracted total RNA from spleen tissue of the immunized mice by reverse transcription (RT). The VL and VH genes were amplified clearly at about 400 and 340 bp by PCR using the synthesized cDNA as a template. Furthermore the amplified products of the VL and VH genes were combined randomly by the 12 amino acid flexible linker, using splicing, by SOE-PCR, resulting in the scFv gene repertoire (Fig. 1 ).
Fig. 1.
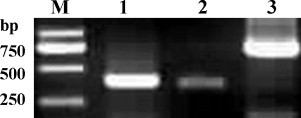
Amplification of the VL and VH genes from immunized mice. Lane M: DNA Marker DL2000; Lane 1: PCR products of the VH gene; Lane 2: PCR products of the VL gene; Lane 3: Amplified products of the scFv gene by SOE-PCR.
3.2. Diversity analysis of the scFv library
The primary scFv library specific for pAPN was generated by cloning of the scFv gene repertoire into the T7Select10-3b vector and in vitro packaging; subsequently the primary library generated was amplified by the liquid lysate method. The titers of the primary and amplified library were 2.0 × 107 pfu/mL and 3.6 × 109 pfu/mL, respectively. Diversity analysis of the primary library was carried out by BstNI fingerprinting of the 30 random phage clones. The BstNI digestion pattern indicated that 28 phage clones had a different digestion pattern, with the exception of two unamplified scFv clones (Fig. 2 ). Sequence analysis revealed that the framework region (FR) and complementarity determining region (CDR) of the three random scFv clones showed the greatest difference in amino acid sequences (Table 2 ).
Fig. 2.
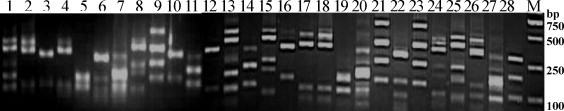
Fingerprint analysis of the scFv phage clones by the frequent-cutting enzyme BstNI. Lane 1–25: The restriction patterns of each scFv clone; Lane M: DNA Marker DL2000.
Table 2.
The VL and VH amino acid sequences of scFv clones selected from primary library.
| Phage clones | Chain type | FRI | CDRI | FR2 | CDR2 | FR3 | CDR3 | FR4 |
|---|---|---|---|---|---|---|---|---|
| mouC-1 | VL | DIVLTQTTLTLSVTIGQPASISCKSS | QSLLDSDGKTY | LNWLLQRPGQSPKRLIY | LVS | KLDSGVPDRFTGSGSGTDFTLKISRVEAEDLGVYYC | WQGTHFPQT | FGGGTKLESNNS |
| mouC-15 | DIVMTQSTSSLAMSVGQKVTMSC | KSSQSLLNSSNQKNYLA | WYQQKPGQSPKLLVY | FASTRES | GVPDRFIGSGSGTDFTLTISSVQAEDLADYFC | QQHYSTPWT | FGGGTKLEIK | |
| mouC-21 | DIVLTQTTAIMSASPGEKVTMTCSA | SSSVSY | MHWYQQKPGSSPKLWIY | STS | NLASGVPARFSGSGSGTSYSLTISSMEAEDAATYYC | HQYHRSPYT | FGGGTKLEIK | |
| mouC-1 | VH | QVQLQESGPGLVAPSQSLSITCTVS | GFSLTGYG | VNWVRQPPGKGLEWLGM | IWGDGST | DYNSALKSRLSISKDNSKSQVFLKMNSLQTDDTARYYC | ARQGNYFDY | WGRAATLIV |
| mouC-15 | QVQLPESGPGLVAPSQSLSITCTVS | GFSLTGYG | VNWVRQPPGKGLEWLGM | IWGDGST | DYNSALKSRLSISKDNSKSQVFLKMNSLQTDDTARYYC | ARGGNYFDY | WGQGTTLIV | |
| mouC-21 | RGEAAESGPGLVAPSQSLSITCTVS | GFSLTDYG | VSWIRQPPGKGLEWLGV | IWGGGST | YYNSALKSRLSISKDNSKSQVFLKMNSLQTDDTARYYC | ARDRGILRYFDY | WGQGTTLIVSS |
3.3. Validation of the scFv library by phage ELISA
After performing three rounds of biopanning for the primary scFv library, the reactivity of 25 scFv phage clones with the recombinant protein pAPN-C was evaluated by phage ELISA. Among the 25 phage clones, 10 phage clones showed positive reactions (S/N value > 2) with the recombinant protein pAPN-C in the phage ELISA (Fig. 3 ).
Fig. 3.
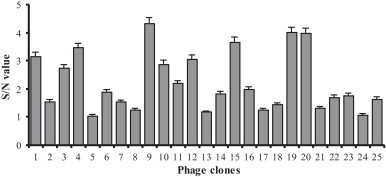
Phage ELISA of the phage clones generated from the third round of biopanning.
4. Discussion
As a basic functional unit of the antibody, the single-chain fragment variable (scFv) region maintains antigen specificity, and has a wide range of biomedical applications (Intorasoot et al., 2007, Bhatia et al., 2010). The phage library in which the scFv gene repertoires are expressed on the surface of phages becomes a powerful tool for isolation and identification of scFv molecules of interest (Cabezas et al., 2008, Tang et al., 2009). Here we describe a simple and rapid method for the generation and evaluation of a murine scFv library specific for porcine aminopeptidase N (pAPN), a common cellular receptor for TGEV and PEDV, using the T7Select Phage Display System. Compared with other similar technologies, the T7Select Phage Display System is easy to use and has the capacity to display peptides of up to about 50 amino acids or 1200 amino acids in size using the T7Select phage display vectors of different copy numbers.
In the current study the optimized degenerate primer sets reported by Okamoto et al. (2004) were chosen to amplify the immunoglobulin light chain variable region (VL) and heavy chain variable region (VH) genes. It has been revealed that these primer sets showed good coverage for amplification of the whole VL and VH gene repertoire by RT-PCR. The VL and VH amplicons were connected by a flexible linker of 12 amino acids by SOE-PCR, to generate the scFv gene repertoire. A number of studies have shown that the length of the linker region between the VH and VL genes affects the activity of scFv molecules, and a linker 12 amino acids in length is suitable to maintain the affinity of the scFv molecules (Wang et al., 2008). In this study the mid-copy number vector T7Select10-3b with EcoRI/HindIII arms for directional cloning of appropriately prepared inserts was used to construct the scFv phage library against pAPN. Generally, each recombinant T7 phage produced by the T7Select10-3b vector is capable of displaying 5–15 copy scFv molecules, and is suitable for the stable expression of the scFv gene and other genes up to about 750 bp in size. The size of the primary scFv library and the amplified scFv library specific for pAPN was 2.0 × 107 pfu/mL and 3.6 × 109 pfu/mL, respectively. Although this is considered to represent a sufficiently large scFv library, the size of both the primary library and the amplified library was slightly lower than the recommended size (6 × 108 pfu/mL and 1010 pfu/mL, respectively). It is suggested that the inserted scFv genes may affect the growth of the recombinant phages to a certain extent.
Using BstNI restriction analysis and DNA sequencing we have shown that the scFv phage library against pAPN has a sufficient diversity. In addition, among the 30 random phage clones from the primary scFv library against pAPN, the inserted scFv genes of two phages were not amplified. This demonstrates that there may have been some non-recombinant phages in the scFv phage library constructed. The phage ELISA indicated that, in the output phages of the third round of biopanning, forty percent of phage clones demonstrated a positive reaction with the recombinant protein pAPN-C. This result suggests that a large number of mouse spleen lymphocytes were activated and had proliferated as a result of the repeated immunization using the well-characterized native pAPN as immunogen. The pAPN scFv phage library will have a potential use for further receptor antagonist screening and analysis of the function of the receptors of TGEV and PEDV.
Acknowledgements
This work is supported by the State National Key Laboratory of Veterinary Biotechnology (Grant No. SKLVBF201104), the National Natural Science Foundation of China (Grant No. 31001081), and the Technological Innovation Team Building Program of University of Heilongjiang Province (Grant No. 2010td05).
Contributor Information
Dongbo Sun, Email: dongbosun@yahoo.com.cn.
Li Feng, Email: fl@hvri.ac.cn.
References
- Bhatia S., Gangil R., Gupta D.S., Sood R., Pradhan H.K., Dubey S.C. Single-chain fragment variable antibody against the capsid protein of bovine immunodeficiency virus and its use in ELISA. J. Virol. Methods. 2010;167:68–73. doi: 10.1016/j.jviromet.2010.03.012. [DOI] [PubMed] [Google Scholar]
- Cabezas S., Rojas G., Pavon A., Alvarez M., Pupo M., Guillen G., Guzman M.G. Selection of phage-displayed human antibody fragments on Dengue virus particles captured by a monoclonal antibody: application to the four serotypes. J. Virol. Methods. 2008;147:235–243. doi: 10.1016/j.jviromet.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Chen J., Wang C., Shi H., Qiu H., Liu S., Chen X., Zhang Z., Feng L. Molecular epidemiology of porcine epidemic diarrhea virus in China. Arch. Virol. 2010;155:1471–1476. doi: 10.1007/s00705-010-0720-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas B., Gelfi J., Haridon L.R., Vogel L.K., Sjöström H., Norén O., Laude H. Aminopeptidase N is a major receptor for the enteropathogenic coronavirus TGEV. Nature. 1992;357:417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas B., Gelfi J., Kut E., Sjöström H., Noren O., Laude H. Determinants essential for the transmissible gastroenteritis virus-receptor interaction reside within a domain of aminopeptidase N that is distinct from the enzymatic site. J. Virol. 1994;68:5216–5224. doi: 10.1128/jvi.68.8.5216-5224.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia G., Decaro N., Martella V., Lorusso E., Mari V., Maria S.L., Cordioli P., Buonavoglia C. An ELISA based on recombinant spike protein S for the detection of antibodies to transmissible gastroenteritis virus of swine-like canine coronaviruses. J. Virol. Methods. 2010;163:309–312. doi: 10.1016/j.jviromet.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haidaris C.G., Malone J., Sherrill L.A., Bliss J.M., Gaspari A.A., Insel R.A., Sullivan M.A. Recombinant human antibody single chain variable fragments reactive with Candida albicans surface antigens. J. Immunol. Methods. 2001;257:185–202. doi: 10.1016/s0022-1759(01)00463-x. [DOI] [PubMed] [Google Scholar]
- Hooper N.M. Families of zinc metalloproteases. FEBS Lett. 1994;354:1–6. doi: 10.1016/0014-5793(94)01079-x. [DOI] [PubMed] [Google Scholar]
- Intorasoot S., Tanaka H., Shoyama Y., Leelamanit W. Characterization and diagnostic use of a recombinant single-chain antibody specific for the gp116 envelop glycoprotein of Yellow head virus. J. Virol. Methods. 2007;143:186–193. doi: 10.1016/j.jviromet.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Jongeneel C.V., Bouvier J., Bairoch A. A unique signature identifies a family of zinc-dependent metallopeptidases. FEBS Lett. 1989;242:211–214. doi: 10.1016/0014-5793(89)80471-5. [DOI] [PubMed] [Google Scholar]
- Jung K., Chae C. RT-PCR-based dot blot hybridization for the detection and differentiation between porcine epidemic diarrhea virus and transmissible gastroenteritis virus in fecal samples using a non-radioactive digoxigenin cDNA probe. J. Virol. Methods. 2005;123:141–146. doi: 10.1016/j.jviromet.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Kenny A.J., Maroux S. Topology of microvillar membrane hydrolases of kidney and intestine. Physiol. Rev. 1982;62:91–128. doi: 10.1152/physrev.1982.62.1.91. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Kim I.J., Pyo H.M., Tark D.S., Song J.Y., Hyun B.H. Multiplex real-time RT-PCR for the simultaneous detection and quantification of transmissible gastroenteritis virus and porcine epidemic diarrhea virus. J. Virol. Methods. 2007;146:172–177. doi: 10.1016/j.jviromet.2007.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B.X., Ge J.W., Li Y.J. Porcine aminopeptidase N is a functional receptor for the PEDV coronavirus. Virology. 2007;365:166–172. doi: 10.1016/j.virol.2007.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Li G., Sui X., Yin J., Wang H., Ren X. Expression and functional analysis of porcine aminopeptidase N produced in prokaryotic expression system. J. Biotechnol. 2009;141:91–96. doi: 10.1016/j.jbiotec.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales A., Galán C., Márquez-Jurado S., García-Gallo M., Kremer L., Enjuanes L., Almazán F. Immunogenic characterization and epitope mapping of transmissible gastroenteritis virus RNA dependent RNA polymerase. J. Virol. Methods. 2011;175:7–13. doi: 10.1016/j.jviromet.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto T., Mukai Y., Yoshioka Y., Shibata H., Kawamura M., Yamamoto Y., Nakagawa S., Kamada H., Hayakawa T., Mayumi T., Tsutsumi Y. Optimal construction of non-immune scFv phage display libraries from mouse bone marrow and spleen established to select specific scFvs efficiently binding to antigen. Biochem. Biophys. Res. Commun. 2004;323:583–591. doi: 10.1016/j.bbrc.2004.08.131. [DOI] [PubMed] [Google Scholar]
- Pansri P., Jaruseranee N., Rangnoi K., Kristensen P., Yamabhai M. A compact phage display human scFv library for selection of antibodies to a wide variety of antigens. BMC Biotechnol. 2009;9:6. doi: 10.1186/1472-6750-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert M.B., Debouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Li G., Liu B. Binding characterization of determinants in porcine aminopeptidase N, the cellular receptor for transmissible gastroenteritis virus. J. Biotechnol. 2010;150:202–206. doi: 10.1016/j.jbiotec.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds D., Garwes D. Virus isolation and serum antibody-responses after infection of cats with transmissible gastroenteritis virus. Arch. Virol. 1979;60:161–166. doi: 10.1007/BF01348032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribes J.M., Ortego J., Ceriani J., Montava R., Enjuanes L., Buesa J. Transmissible gastroenteritis virus (TGEV)-based vectors with engineered murine tropism express the rotavirus VP7 protein and immunize mice against rotavirus. Virology. 2011;410:107–118. doi: 10.1016/j.virol.2010.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza G. Anchoring and biosynthesis of stalked brush border membrane proteins: glycosidases and peptidases of enterocytes and renal tubuli. Annu. Rev. Cell Biol. 1986;2:255–313. doi: 10.1146/annurev.cb.02.110186.001351. [DOI] [PubMed] [Google Scholar]
- Sun D.B., Feng L., Shi H.Y., Chen J.F., Cui X.C., Chen H.Y., Liu S.W., Tong Y.E., Wang Y.F., Tong G.Z. Identification of two novel B cell epitopes on porcine epidemic diarrhea virus spike protein. Vet. Microbiol. 2008;131:73–81. doi: 10.1016/j.vetmic.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D.B., Feng L., Shi H.Y., Chen J.F., Liu S.W., Chen H.Y., Wang Y.F. Spike protein region (aa 636–789) of porcine epidemic diarrhea virus is essential for induction of neutralizing antibodies. Acta Virol. 2007;51:149–156. [PubMed] [Google Scholar]
- Tang K.H., Yusoff K., Tan W.S. Display of hepatitis B virus PreS1 peptide on bacteriophage T7 and its potential in gene delivery into HepG2 cells. J. Virol. Methods. 2009;159:194–199. doi: 10.1016/j.jviromet.2009.03.015. [DOI] [PubMed] [Google Scholar]
- Vemulapalli R., Gulani J., Santrich C. A real-time TaqMan RT-PCR assay with an internal amplification control for rapid detection of transmissible gastroenteritis virus in swine fecal samples. J. Virol. Methods. 2009;162:231–235. doi: 10.1016/j.jviromet.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Zheng C., Liu Y., Zheng H., Wang Z. Construction of multiform scFv antibodies using linker peptide. J. Genet. Genomics. 2008;35:313–316. doi: 10.1016/S1673-8527(08)60045-4. [DOI] [PubMed] [Google Scholar]


