Abstract
The aim of this study was to investigate the effects of a porcine reproductive and respiratory syndrome virus (PRRSV) infection on the development of the immune response after pseudorabies virus (PRV) vaccination in pigs. Pigs were intranasally inoculated with the European PRRSV strain, Lelystad virus ter Huurne, and were vaccinated intramuscularly with PRV 2 weeks later (LV-PRV group). Control pigs were vaccinated with PRV only (PRV group). Eight weeks after PRV vaccination, pigs from both groups were challenged intranasally with wild-type PRV. We measured the lymphoproliferative, and the cytolytic responses to PRV of peripheral blood mononuclear cells (PBMC), isolated from blood samples. In addition, serum samples were examined for antibodies against PRV and LV. One week after PRV vaccination, PBMC proliferated abundantly to PRV in both groups. However, in the LV-PRV group the lymphoproliferative response declined after 1 week, whereas, in the PRV group, the lymphoproliferative response was high for 3 weeks and declined thereafter (P<0.05). After challenge, the lymphoproliferative response was 1 week earlier and was consistently and significantly higher in the PRV group than in the LV-PRV group. The PRV-specific killing was higher at 3 weeks after PRV vaccination and 5 weeks after PRV challenge 19±3 and 24±6%, respectively, in the PRV group, compared to 7±4 and 6±9%, respectively, in the LV-PRV group (P<0.05). However, later after vaccination and challenge the cytolytic response was identical in both groups. The antibody titre against PRV developed equally in both groups. After challenge, no PRV virus was isolated from both groups. From these results we conclude that, although PRRSV infection did cause changes in the time course of the T-lymphocyte response after PRV vaccination, PRRSV infection did not inhibit the development of vaccine-induced protection after PRV.
Abbreviations: PRV, pseudorabies virus; LV-TH, Lelystad virus ter Huurne; PRRSV, porcine reproductive and respiratory syndrome virus; PBMC, peripheral blood mononuclear cells; MAb, monoclonal antibody; PBS, phosphate-buffered saline; PFU, plaque-forming unit; VN, virus neutralisation; SLA, swine–leucocyte–antigen complex; IPMA, immunoperoxidase monolayer assay; OPF, oropharyngeal fluid; MRDG, mean relative daily gain; CPM, counts per minute
Keywords: Pig, Lymphoproliferative response, Cytolytic response, Killing, Porcine reproductive and respiratory virus, Pseudorabies virus, PBMC
1. Introduction
Porcine reproductive and respiratory syndrome virus (PRRSV) is a positive stranded RNA virus that causes reproductive failure in late term gestation sows and respiratory disease in pigs of all ages. The virus primarily infects and destroys alveolar macrophages, which are important in the host defence in the lungs. Therefore, PRRSV-infected pigs may be more susceptible to secondary infections. Since PRRSV occurs endemically in many countries and may influence the efficacy of host defence mechanisms, it is possible that a PRRSV infection interferes with the vaccination efficacy against PRV.
Previous studies on the effect of PRRSV infection on secondary infections under experimental conditions are contradictory. Galina et al. (1994) described more clinical signs, meningitis and bacteraemia after a Streptococcus suis infection only when pigs were pre-inoculated with PRRSV. Thacker et al. (1999) described more severe and persistent pneumonic lesions and clinical respiratory disease when a Mycoplasma hyopneumoniae infection was followed by a PRRSV infection. van Reeth et al., 1994, van Reeth et al., 1996 described more severe disease after a dual infection with PRRSV and porcine respiratory coronavirus or influenza virus compared with the single infections. In contrast, others described no influence of a PRRSV infection on the clinical signs of disease after infection with Pasteurella multocida, Haemophilus parasuis, S. suis or Salmonella cholerasuis (Cooper et al., 1995). In addition, Albina et al. (1998) could not demonstrate an immunosuppressive effect of PRRSV on a pseudorabies virus (PRV) glycoprotein immunisation and described an enhanced antibody response after challenge with PRV.
More illness and a more severe disease outcome after infection with other micro-organisms due to an earlier PRRSV infection have been described. These studies focussed on increased illness and pathological alterations by the secondary infection in PRRSV-infected pigs. However, the effect of PRRSV on the cellular immune response against secondary infections is not clear. Therefore, we examined the effects of a PRRSV infection on the development of the cellular immune response against a PRV infection in a well-defined PRV vaccination-challenge model.
PRV is an alphaherpesvirus that causes Aujeszky’s disease in pigs. Both humoral and cellular immunity appear to be involved in the development of a protective immune response to PRV. Previous reports have described the time course of the lymphoproliferative and cytolytic responses after a PRV infection and challenge in detail (Kimman et al., 1995b, De Bruin et al., 1998). Therefore, we used this PRV model to investigate the immunosuppressive effect of PRRSV on a PRV vaccination and challenge in pigs. We investigated the development of the humoral, lymphoproliferative and cytolytic responses against PRV in pigs that were inoculated with PRRSV prior to PRV vaccination.
2. Materials and methods
2.1. Viruses
PRRSV stocks for inoculation were prepared on alveolar macrophages as described by Wensvoort et al. (1991). Pigs were inoculated with PRRSV strain Lelystad virus ter Huurne (LV-TH). PRV stocks for vaccination and challenge were prepared on SK-6 cells (Kasza et al., 1971) and secondary porcine kidney cells, respectively, as described by Kimman et al. (1995b). Pigs were vaccinated with an avirulent mutant strain M141 (gE−) of PRV (De Wind et al., 1990, Kimman et al., 1992). The pigs were then challenged with wild-type PRV strain NIA-3 (McFerran and Dow, 1975). This strain was also used to infect target cells for the cytolytic assay, as described by Kimman et al. (1995a).
2.2. Animals and experimental design
Minnesota miniature pigs from our institute, which are inbred for swine–leukocyte–antigen complex (SLA) (haplotype d/d) (Sachs et al., 1976), were kept under specific pathogen-free conditions. The pigs were born from unvaccinated sows and had no antibodies against PRV or PRRSV. Pigs were randomly allocated to two groups and housed separately. Six pigs were intranasally inoculated with 0.5 ml of PRRSV strain LV-TH at a concentration of 105 TCID50/ml per nostril. Five control SPF pigs were left uninoculated. After 2 weeks both groups were vaccinated intramuscularly with 105 plaque-forming units (PFU)/ml PRV strain M141. These pigs were subsequently challenged intranasally with 105 PFU/ml NIA-3 virus, 8 weeks after PRV vaccination. Blood samples were taken weekly, starting 1 week before and then 2 weeks after the LV inoculation. Rectal temperatures were recorded daily for 25 days after LV inoculation and 25 days after PRV challenge. Body weights were recorded at weekly intervals for 3 months. Pigs were considered to have fever when body temperatures were above or equal to 40°C. Growth performance was assessed by calculating the mean relative daily gain (MRDG) in body weight according to Stellman et al. (1989). The ethical committee for animal experiments of ID-Lelystad approved the experiments (Table 1 ).
Table 1.
Experimental design
| LV inoculation | M141 vaccination | NIA-3 challenge | |
| PRV (n=5) | ND | 2 weeks | 10 weeks |
| LV-PRV (n=6) | Day 0 | 2 weeks | 10 weeks |
2.3. Isolation and culture of PBMC
PBMC isolation and culturing was done as previously described by Kimman et al. (1995b). Briefly, PBMC were stimulated in vitro for 4 days with negative cell-lysate of SK-6 cells, or infectious NIA-3 (PRV) virus. For cytolytic assays, PBMC were used directly (direct killing) or they were stimulated for 6 days in vitro with NIA-3 (indirect killing) as described by De Bruin et al. (1998).
2.4. Lymphocyte proliferation assay
Proliferation of PBMC was measured by 3H-thymidine incorporation as described by De Bruin et al. (1998) and expressed as delta counts, i.e. the number of counts of virus-stimulated PBMC minus the number of counts of negative cell-lysate-stimulated PBMC. Proliferation was investigated starting 1 week before LV inoculation and then at weekly intervals from 2 weeks after LV inoculation and was measured after subsequent PRV inoculations.
2.5. Target cells
The following target cells were used in the cytolytic assays: PRV-infected and uninfected L14 cells (a retrovirus immortalised B-lymphoblastoid cell line of the haplotype d/d SLA) (Kaeffer et al., 1990) and K562 cells (a cell line from human erythroleukemia cells) (ATCC, Rockville, MD). The K562 cells were used to assess the killing by porcine NK cells (Pescovitz et al., 1988). Infected L14 cells were obtained by infecting the cells, 24 h before the start of the cytolytic assay, with NIA-3 virus at a multiplicity of infection of 10. The cells were labelled by incubation of various numbers of cells in a volume of 50 μl serum-free medium which contained 400 μCi 51Cr (Amersham CJS4, Den Bosch, The Netherlands) for 2 h at 37°C on a Rock’n Roller (Labinco, Breda, The Netherlands). After being labelled, the cells were washed three times in RPMI complete medium. Volumes of 50 μl medium containing 104 cells were added to the wells of 96-well V-bottomed microtitre plates (Nunc, Life Technology, Breda, The Netherlands).
2.6. Cytolytic assay
The cytolytic activity of the effector cells was measured by 51Cr release. Volumes of 50 μl medium containing effector cells and 50 μl medium containing 104 51Cr-labelled target cells were mixed in order to obtain effector:target ratios of 50:1 to 6:1. The plates were then centrifuged for 5 min at 200×g. Maximal release of 51Cr was determined by adding 50 μl 20% Triton X-100. Spontaneous release was determined in wells that did not contain effector cells. Killing of the target cells was determined by measuring the release of 51Cr in the supernatant after an incubation period of 5 h. Volumes of 50 μl supernatant were mixed with 100 μl optiphase supermix liquid scintillation fluid (EG&G Instruments, Nieuwegein, The Netherlands). Radioactivity was then measured in a Wallac Microbetaplus 1450 (EG&G Instruments, Nieuwegein, The Netherlands). The percentage of specific 51Cr release was calculated as
| cpm experimental release−cpm spontaneous releasecpm maximal release−cpm spontaneous release×100 |
Direct killing was investigated at 10 and 18 days after PRV vaccination. Indirect killing was investigated 1 week before the LV inoculation and then 2 weeks after LV inoculation and was measured at weekly intervals after subsequent PRV inoculations.
2.7. Serological examinations
Blood samples were collected from all pigs 1 day before LV inoculation and at weekly intervals after PRV inoculations. Serum samples were stored at −20°C. Sera were tested for the presence of virus neutralising antibodies directed against PRV in a virus neutralisation test (VN titres) as described by Kimman et al. (1992). Sera were tested for the presence of virus antibodies directed against PRRSV in an immunoperoxidase monolayer assay (IPMA) and for the presence of PRRSV by virus isolation as described by Wensvoort et al. (1991).
2.8. PRV isolation
PRV virus excretion was monitored by collecting swab specimens of oropharyngeal fluid (OPF) from the day before challenge until day 10 after challenge. Swab specimens were extracted with 4 ml of Dulbecco’s minimal essential medium supplemented with 2% foetal bovine serum and antibiotics. To determine the virus content per gram OPF, we measured the weight of the collected fluid after centrifugation of the swabs (Kimman et al., 1992). The amount of PRV excretion was quantitated by titration of the virus on SK-6 monolayers as described by Kimman et al. (1992).
2.9. Statistical analysis
Differences in lymphoproliferative responses, virus-specific killing activity, VN antibody titres and MRDG were tested for statistical significance by analysis of variance (ANOVA) after testing for homogeneity of variances within groups (Barletts’ test). The significance level was set at 5%.
3. Results
3.1. Time course of the humoral response against LV and PRV
LV specific antibodies were detected from 1 week after LV inoculation in IPMA. This response peaked on 7 weeks after LV inoculation (titre log 4.6) and declined thereafter. After 15 weeks post-LV inoculation, antibodies against LV (titre log 3.4) were still detected.
PRV VN antibodies were detected from 1 week after PRV vaccination with M141 (gE− PRV-strain), in the LV-PRV and the PRV group, with a virus neutralisation assay (titres log 2.7−3.3). After challenge with PRV, no increase of the antibody-titres was observed and no difference was observed in the development of the humoral response between both groups. In contrast to the antibody-titres against LV, the antibody-titres after PRV vaccination and challenge did not decrease during the experiment (15 weeks).
3.2. Time course of the lymphoproliferative response against PRV
One week after PRV vaccination, PBMC from PRV-infected pigs proliferated abundantly in vitro in both groups on stimulation with PRV (Fig. 1 ). The proliferation of PBMC tended to be higher (but not statistically significantly) in the PRV group (50,587±36,565 CPM) than in the LV-PRV group (18,042±19,888 CPM) (P<0.09). The response peaked 2 weeks after PRV vaccination in both groups. In the LV-PRV group the lymphoproliferative response declined 1 week later, whereas in the PRV group the lymphoproliferative response remained high for 3 weeks and declined thereafter. The proliferation was significantly higher (P<0.05) in the PRV group than in the LV-PRV group in weeks 4 and 5 after PRV vaccination. In contrast to the VN antibodies, the lymphoproliferative response increased after challenge with wild-type NIA-3 virus (Fig. 1) in both groups. This secondary lymphoproliferative response started 1 week after challenge in the PRV group and 2 weeks after challenge in the LV-PRV group (P<0.05). After PRV challenge, the specific proliferation of PBMC to PRV stimulation was consistently and significantly lower, in the LV-PRV group than in the PRV group (P<0.05) (Fig. 1).
Fig. 1.
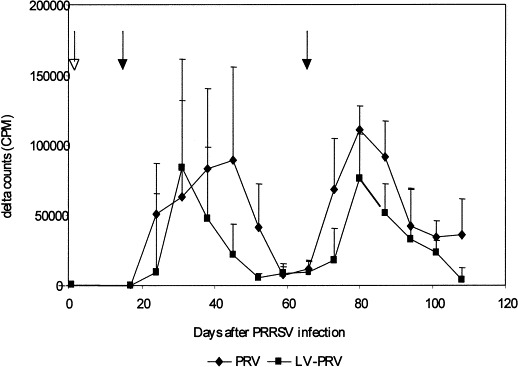
Time course of PRV-specific lymphoproliferation after inoculation with PRRSV and PRV. PBMC isolated from inoculated SLA d/d minipigs were stimulated in vitro with PRV for 4 days. Presented are delta counts, i.e. 3H-thymidine incorporation (CPM) of PRV-stimulated PBMC minus 3H-thymidine incorporation (CPM) of negative-cell-lysate-stimulated PBMC. The open arrow indicates inoculation with PRRSV. Closed arrows indicate PRV vaccination and challenge. Significant differences are indicated with an asterisk (P<0.05).
3.3. Time course of cytolytic activity
3.3.1. Indirect killing after LV and PRV inoculations:
When PBMC from the PRV and LV-PRV group were stimulated in vitro with NIA-3 virus, PRV-specific killing (i.e. killing of PRV-infected L14 cells and killing of uninfected L14 cells) increased between weeks 3 and 6 after PRV vaccination (up to 30% of PRV-infected target cells were killed compared with up to 15% of uninfected target cells). In both groups, PRV-specific killing by these cells was detected for a period of at least 6 weeks after challenge; (up to 35% of PRV-infected target cells were killed compared with up to 15% of uninfected target cells). K562 cells were only poorly killed (10% after PRV vaccination). The PRV-specific killing was higher at 3 weeks after PRV vaccination and 5 weeks after PRV challenge 19±3 and 24±6%, respectively, in the PRV group, compared to 7±4 and 6±9%, respectively, in the LV-PRV group (P<0.05) (Fig. 2 ). However, later after PRV vaccination the killing was comparable in both groups. K562 cells were poorly killed in both groups (up to 20% in the PRV and LV-PRV group 1 week after challenge).
Fig. 2.
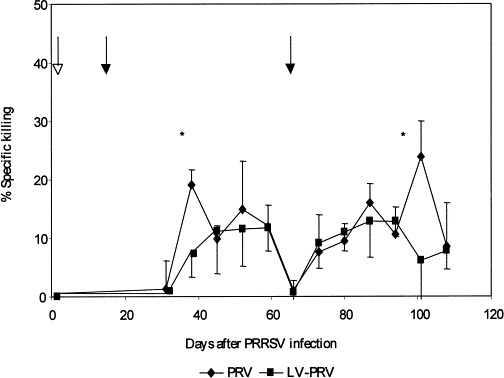
Time course of PRV-specific killing, i.e. killing of PRV-infected target cells minus killing of uninfected target cells. PBMC from inoculated SLA d/d minipigs were stimulated in vitro with PRV for 6 days and incubated for 5 h with PRV-infected or uninfected 51Cr-labelled target L14 cells. The open arrow indicates inoculation with PRRSV. Closed arrows indicate PRV vaccination and challenge. Significant differences are indicated with an asterisk (P<0.05).
3.3.2. Direct killing after PRV vaccination:
On day 10 after PRV vaccination, directly used PBMC from the PRV group killed the PRV-infected L14 cells more efficiently (30±5%) than PBMC from the LV-PRV group (18±7%) (P<0.05). In contrast, on day 18 after PRV vaccination, PBMC from the PRV and LV-PRV group killed the PRV-infected L14 cells equally well (14±5%) and (11±4%). No killing of K562 cells was detected.
3.4. Daily body temperature, virus isolation, clinical signs and MRDG
Pigs from the LV-PRV group had fever after inoculation with LV from days 3 to 6 after LV inoculation and again after PRV vaccination on days 17 and 18 after LV infection. In contrast, pigs from the PRV group had no fever after vaccination (Fig. 3 ). Pigs from both groups had no fever after challenge. LV was isolated from sera from 4 pigs (n=6). LV was isolated on day 3 (2 pigs), day 10 (1 pig) and day 24 (1 pig) after LV inoculation. No PRV was isolated from OPF of both groups after challenge with PRV. The mean daily weight gain did not differ after LV and after both PRV inoculations between both groups. Pigs had no clinical signs of PRRSV disease after LV inoculation (except fever) and no clinical signs of Aujeszky’s disease after PRV inoculations.
Fig. 3.
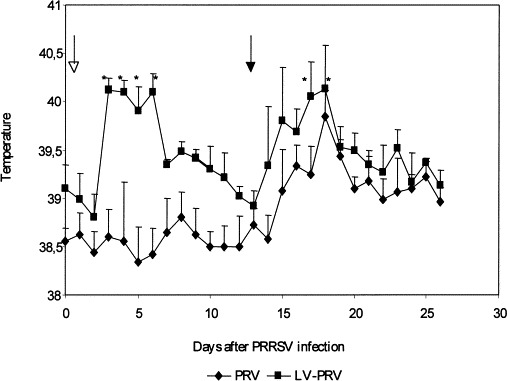
Body temperature curve of inoculated pigs during 25 days after LV inoculation and subsequent PRV vaccination. An open arrow indicates inoculation with PRRSV and a closed arrow indicates vaccination with PRV. Fever (>40°C) is indicated with an asterisk.
4. Discussion
Results showed that a PRRSV infection prior to a PRV vaccination caused differences in the level and time course of the cellular immune response against PRV. The lymphoproliferative response tended to be quicker (not significantly) and longer (2 weeks) (P<0.05) in the PRV group than in the LV-PRV group after PRV vaccination. This lymphoproliferative response was also quicker (1 week) and consistently higher in the PRV group than in the LV-PRV group after PRV challenge (P<0.05). Furthermore, when we used in vitro PRV-stimulated PBMC, the cytolytic response against PRV reached a maximum in week 3 after PRV vaccination and week 5 after PRV challenge in the PRV-group, whereas the LV-PRV group did not (P<0.05). Later after vaccination and challenge, the cytolytic response was comparable in both groups. The same kinetics of the cytolytic response against PRV vaccination with M141 was described in an other study (De Bruin et al., 1998), which indicates that the LV-PRV group in this study did not reach a maximum level in the cytolytic response, whereas the PRV group did. When PBMC were used directly, we also observed a significantly higher cytolytic response against PRV on day 10 after PRV vaccination in the PRV group than in the LV-PRV group (P<0.05). These results indicate a mild immunosuppressive effect by the PRRSV infection on the development of a cellular immune response after PRV vaccination and challenge. An explanation for this lower T-lymphocyte response may be that PRRSV infects monocytes and macrophages, which may hamper efficient antigen-presentation and thereby suppressing the ability of PBMC to proliferate.
Although PRRSV clearly affected the time course of the cellular immune response against a PRV infection, no effect on the humoral response against PRV was detected after vaccination and challenge in the LV-PRV group. This finding is in contrast to Brun et al. (1994), who observed an enhanced secondary humoral response after an influenza challenge, and Albina et al. (1998) and Molitor et al. (1992), who also observed an enhanced humoral response after challenge with PRV, after an earlier PRRSV infection. It is possible that PRRSV infection stimulates the humoral response. However, an explanation for this discrepancy is that we used a very potent vaccine strain, inducing protection and no increase of antibody-titres (Kimman et al., 1992, Kimman et al., 1994, De Bruin et al., 1998). We could not detect differences in protection against a PRV challenge between both groups, but a negative effect of a PRRSV infection on protection after PRV vaccination can not be excluded when less potent vaccine strains are used. However, the M141 vaccine strain was used, because this strain induced a high cellular immune response (De Bruin et al., 1998) and eventually negative effects of a PRRSV infection on the development of the cellular immune response could be measured very well.
No weight loss or differences in mean daily weight gain were observed between both groups. Furthermore, no PRV was excreted after challenge indicating optimal protection. The only observed clinical signs were fever for 4 days after LV inoculation and again 2 days after the PRV vaccination in LV-PRV group. van Reeth et al. (1994) also described fever and more clinical signs after dual infections of PRRSV with porcine respiratory coronavirus and swine influenza virus. Although we measured fever after LV inoculation and PRV vaccination, we could not detect clinical signs of Aujeszky’s disease after vaccination with M141 (a PRV mutant strain that causes mild clinical signs (Kimman et al., 1994, De Bruin et al., 1998)). Obviously, even though pigs were recovering from the earlier PRRSV infection, the PRV mutant strain M141 could not cause Aujeszky’s disease. Thus, although T-lymphocyte responses were diminished against PRV, and pigs had some fever in the LV-PRV group, pigs were clinically protected against a PRV challenge.
In conclusion, we demonstrated that a PRRSV infection prior to a PRV vaccination mildly affects the level and time course of the T-lymphocyte response against PRV. Despite this difference, the PRRSV infection did not influence the capacity of the gE− PRV strain to induce protection against a wild-type PRV challenge.
References
- Albina E, Piriou L, Hutet E, Cariolet R, L’Hsopitalier R. Immune responses in pigs infected with porcine reproductive and respiratory syndrome virus (PRRSV) Vet. Immunol. Immunopathol. 1998;61:49–66. doi: 10.1016/S0165-2427(97)00134-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun, A., Charreyre, C., Vaganay, A., Reynaud, G., 1994. Porcine reproductive respiratory syndrome role of the togavirus and other infectious agents in the respiratory disease of swine. In: Proceedings of the 13th International Pig Veterinary Congress, Bangkok, Thailand, 26–30 June 1994, p. 52.
- Cooper V.L, Doster A.R, Hesse R.A, Harris N.B. Porcine reproductive and respiratory syndrome: NEB-I PRRSV infection did not potentiate bacterial pathogens. J. Vet. Diagn. Invest. 1995;7:313–320. doi: 10.1177/104063879500700303. [DOI] [PubMed] [Google Scholar]
- De Bruin M.G.M, De Visser Y.E, Kimman T.G, Bianchi A.T.J. Time course of the porcine cellular and humoral immune responses in vivo against pseudorabies virus after inoculation and challenge: significance of in vitro antigenic restimulation. Vet. Immunol. Immunopathol. 1998;65:75–87. doi: 10.1016/s0165-2427(98)00175-5. [DOI] [PubMed] [Google Scholar]
- De Wind N, Zijderveld A, Glazenburg K, Gielkens A, Berns A. Linker insertion mutagenesis of herpesviruses: inactivation of single genes within the US region of pseudorabies virus. J. Virol. 1990;64:4691–4696. doi: 10.1128/jvi.64.10.4691-4696.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galina L, Pijoan C, Sitjar M, Christianson W.T, Rossow K, Collins J.E. Interaction between Streptococcus suis serotype-2 and porcine reproductive and respiratory syndrome virus in specific pathogen-free piglets. Vet. Rec. 1994;134:60–64. doi: 10.1136/vr.134.3.60. [DOI] [PubMed] [Google Scholar]
- Kaeffer B, Bottreau E, Phan Thanh L, Olivier M, Salmon H. Histocompatible miniature boar model: selection of transformed cell lines of B and T lineages producing retrovirus. Int. J. Cancer. 1990;46:481–488. doi: 10.1002/ijc.2910460326. [DOI] [PubMed] [Google Scholar]
- Kasza L, Shadduck J.A, Christofinis G.J. Establishment, viral susceptibility, and biological characteristics of a swine kidney cell line SK-6. Res. Vet. Sci. 1971;13:46–51. [PubMed] [Google Scholar]
- Kimman T.G, De Wind N, Oei-Lie N, Pol J.M.A, Berns A.J.M, Gielkens A.L.J. Contribution of single genes within the unique short region of Aujeszky’s disease virus (suid herpesvirus type 1) to virulence pathogenesis and immunogenicity. J. Gen. Virol. 1992;73:243–251. doi: 10.1099/0022-1317-73-2-243. [DOI] [PubMed] [Google Scholar]
- Kimman T.G, De Wind N, De Bruin T, De Visser Y, Voermans J. Inactivation of glycoprotein gE and thymidine kinase or the US3-encoded protein kinase synergistically decreases in vivo replication of pseudorabies virus and the induction of protective immunity. Virology. 1994;205:511–518. doi: 10.1006/viro.1994.1672. [DOI] [PubMed] [Google Scholar]
- Kimman T.G, Bianchi A.T.J, De Bruin T.G.M, Mulder W.A.M, Priem J, Voermans J.J.M. Interaction of pseudorabies virus with immortalized porcine B-cells: influence on surface class I and II major histocompatibility complex and immunoglobulin M expression. Vet. Immunol. Immunopathol. 1995;67:253–263. doi: 10.1016/0165-2427(94)05344-r. [DOI] [PubMed] [Google Scholar]
- Kimman T.G, De Bruin M.G.M, Voermans J.J.M, Peeters B.P.H, Bianchi A.T.J. Development and antigen specificity of the lymphoproliferation response of pigs to pseudorabies virus. Dichotomy between secondary B and T cell responses. Immunology. 1995;86:372–378. [PMC free article] [PubMed] [Google Scholar]
- McFerran J.B, Dow C. Studies on immunization of pigs with the Bartha strain of Aujeszky’s disease virus. Res. Vet. Sci. 1975;19:17–22. [PubMed] [Google Scholar]
- Molitor T, Leitner G, Choi C, Risdahl J, Rossow K, Collins J. Modulation of host immune responses by SIRS virus. Am. Assoc. Swine Practitioners Newslett. 1992;4(4):27–28. [Google Scholar]
- Pescovitz M.D, Lowman M.A, Sachs D.H. Expression of T-cell associated antigens by porcine natural killer cells. Immunology. 1988;65:267–271. [PMC free article] [PubMed] [Google Scholar]
- Sachs D.H, Leight G, Cone J, Schwarz S, Stuart L. Transplantation in miniature swine. I. Fixation of the major histocompatibility complex. Transplantation. 1976;22:559–566. doi: 10.1097/00007890-197612000-00004. [DOI] [PubMed] [Google Scholar]
- Stellman C, Vannier P, Chappuis G, Brun A, Dauvergne M, Fargeaud D, Bugand M, Colson X. The potency testing of pseudorabies vaccines in pigs. A proposal for a quantitative criterion and a minimal requirement. J. Biol. Stand. 1989;17:17–27. doi: 10.1016/0092-1157(89)90024-3. [DOI] [PubMed] [Google Scholar]
- Thacker E.L, Halbur P.G, Ross R.F, Thanawongnuwech R, Thacker B.J. Mycoplasma hyopneumoniae potentation of porcine reproductive and respiratory syndrome virus-induced pneumonia. J. Clin. Microbiol. 1999;37:620–627. doi: 10.1128/jcm.37.3.620-627.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Reeth, K., Koyen, A., Pensaert, M., 1994. Clinical effects of dual infections with porcine epidemic abortion and respiratory syndrome virus, porcine respiratory coronavirus and swine influenza virus. In: Proceedings of the 13th International Pig Veterinary Congress, Bangkok, Thailand, 26–30 June 1994, p. 51.
- van Reeth K, Nauwynck H, Pensaert M. Dual infections of feeder pigs with porcine reproductive and respiratory syndrome virus followed by porcine respiratory coronavirus or swine influenza virus: a clinical and virological study. Vet. Microbiol. 1996;48:325–335. doi: 10.1016/0378-1135(95)00145-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensvoort G, Terpstra C, Pol J.M, ter Laak E.A, Bloemraad M, de Kluyver E.P, Kragten C, van Buiten L, den Besten A, Wagenaar F, Broekhuijsen J.M, Moonen P.L.J.M, Zetstra T, de Boer E.A, Tibben H.J, de Jong M.F, van’t Veld P, Groenland G.J.R, van Gennep J.A, Voets M.Th, Verheijden J.H.M, Braamskamp J. Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet. Q. 1991;13:121–130. doi: 10.1080/01652176.1991.9694296. [DOI] [PubMed] [Google Scholar]


