Abstract
A minor groove binder (MGB) probe assay was developed to discriminate between type 2-based vaccines and field strains of canine parvovirus (CPV). Considering that most of the CPV vaccines contain the old type 2, no longer circulating in canine population, two MGB probes specific for CPV-2 and the antigenic variants (types 2a, 2b and 2c), respectively, were labeled with different fluorophores. The MGB probe assay was able to discriminate correctly between the old type and the variants, with a detection limit of 101 DNA copies and a good reproducibility. Quantitation of the viral DNA loads was accurate, as demonstrated by comparing the CPV DNA titres to those calculated by means of the TaqMan assay recognising all CPV types. This assay will ensure resolution of most diagnostic problems in dogs showing CPV disease shortly after CPV vaccination, although it does not discriminate between field strains and type 2b-based vaccines, recently licensed to market in some countries.
Keywords: Dog, Parvovirus, Type 2 vaccine, Field strains, MGB probe assay
1. Introduction
Canine parvovirus type 2 (CPV-2) is responsible for acute, sometimes fatal, gastroenteritis in dogs (Carmichael and Binn, 1981). CPV-2 emerged in the late 1970s (Kelly, 1978, Appel et al., 1979, Burtonboy et al., 1979, Johnson and Spreadbrow, 1979), but it was replaced in a few years by its antigenic variants (Parrish et al., 1985, Parrish et al., 1988, Parrish et al., 1991). Currently, three main antigenic variants of CPV-2 are known, named type 2a, 2b and 2c and variously distributed in dog population worldwide (Mochizuki et al., 1993, De Ybanez et al., 1995, Greenwood et al., 1996, Truyen et al., 1996, Truyen et al., 2000, Steinel et al., 1998, Sagazio et al., 1998, Buonavoglia et al., 2000, Pereira et al., 2000, Buonavoglia et al., 2001, Martella et al., 2004, Desario et al., 2005, Decaro et al., 2005b, Decaro et al., 2005c, Decaro et al., 2006). CPV-2c, designated previously as CPV Glu-426 mutant, emerged in Italy in 2000 (Buonavoglia et al., 2001) and has been detected in other countries (Nakamura et al., 2004). Moreover, its pathogenicity has been investigated (Decaro et al., 2005a). The original type 2, although disappeared from the field, is still present in the CPV-2 vaccines available on the market (Parrish et al., 1988).
Traditionally, identification of the CPV-2 variants is carried out by means of time-consuming techniques, such as haemagglutination inhibition (HI) test with monoclonal antibodies (MAbs) (Parrish and Carmichael, 1983, Nakamura et al., 2004), PCR-RFLP with enzyme MboII (Buonavoglia et al., 2001), PCR-based methods (Pereira et al., 2000), or sequence analysis, often requiring the use of combined methods for the definitive prediction of antigen specificity. Recently, two real-time PCR assays using minor groove binder (MGB) probes have been developed for rapid and unambiguous characterisation of CPV-2 (Decaro et al., 2005b, Decaro et al., 2006). The MGB probe assays are able to recognise the single nucleotide polymorphisms (SNPs) existing between types 2a/2b (A4062G) and between types 2b/2c (T4064A), which determine the presence at residue 426 of the capsid protein of amino acids Asn, Asp and Glu in types 2a, 2b and 2c, respectively (Parrish et al., 1991, Buonavoglia et al., 2001 and Table 1 ). Both type 2a/2b and type 2b/2c assays were found highly sensitive and specific, although the type 2a-specific probe was not able to discriminate type 2a CPVs from the original type 2. In fact, both types present nucleotide A at position 4062 of the viral genome, so that the old type is characterised erroneously as type 2a using the MGB strategy (Decaro et al., 2005b, Decaro et al., 2006). This makes the type 2a/2b MGB probe assay inadequate for discrimination between dogs vaccinated with the original type 2 and dogs infected with type 2a, although it has been noted that the vaccine virus is shed in the faeces at low titres and for a shorter time period than field strains and that usually the diagnostic tests are carried out on the faeces of sick animals (Decaro et al., 2006). Some problems may occur especially when testing faecal samples collected from dogs displaying diarrhoea few days after administration of a type 2-based vaccine, when it is crucial to be able to detect differentially the vaccine virus and the field strains of CPV-2.
Table 1.
Amino acid variations in the VP2 protein of different CPV types
| Amino acid variations at residuea |
||||||||
|---|---|---|---|---|---|---|---|---|
| 87 | 101 | 297 | 300 | 305 | 375 | 426 | 555 | |
| 3045–3047b | 3087–3089b | 3675–3677b | 3684–3686b | 3699–3701b | 3909–3911b | 4062–4064b | 4449–4451b | |
| ATG (Met)c | ATT (Ile)c,d | TCT (Ser)c | GCT (Ala)c | GAT (Asp)c | AAT (Asn)c | AAT (Asn)c | GTA (Val)c | |
| TTG (Leu) | ACT (Thr)d | GCT (Ala) | GGT (Gly) | TAT (Tyr) | GAT (Asp) | GAT (Asp), GAA (Glu) | ATA (Ile) | |
| CPV-2 | Met | Ile | Ser | Ala | Asp | Asn | Asn | Val |
| CPV-2a | Leu | Thr | Ser | Gly | Tyr | Asp | Asn | Ile |
| CPV-2b | Leu | Thr | Ser | Gly | Tyr | Asp | Asp | Val |
| New CPV-2b | Leu | Thr | Ala | Gly | Tyr | Asp | Asp | Val |
| New CPV-2a | Leu | Thr | Ala | Gly | Tyr | Asp | Asn | Val |
| Asp-300 (CPV-2a/CPV-2b) | Leu | Thr | Ala | Asp | Tyr | Asp | Asn Asp | Val |
| CPV-2c | Leu | Thr | Ala | Gly | Tyr | Asp | Glu | Val |
Positions are referred to the amino acid and nucleotide sequences of strain CPV-b (accession no. M38245).
Nucleotide position.
Codon observed.
Codon affected by the SNP used to design the type-specific probes CPV2-Pb (type 2) and CPVv-Pb (variants).
In such an effort, we have developed an MGB probe assay for rapid discrimination between vaccine (old type) and field (types 2a, 2b, 2c) strains of CPV-2.
2. Materials and methods
2.1. Samples
A total of 56 samples were tested, consisting of 11 samples positive for the original type 2 and 45 samples positive for the antigenic variants (type 2a, n = 15; type 2b, n = 15; type 2c, n = 15). Prediction of CPV specificity was carried out by HI test with MAbs (Parrish and Carmichael, 1983, Nakamura et al., 2004), PCR-RFLP with enzyme MboII (Buonavoglia et al., 2001), or sequence analysis, as described by Desario et al. (2005). Samples containing the antigenic variants were recruited from a previous study (Desario et al., 2005). Since the old type no longer circulates in dog population, samples containing CPV-2 included only the following vaccine formulations licensed in Italy: Vanguard 7, Vanguard CPV (Pfizer Inc., NY, USA), Tetradog-CHPL, Parvodog-P, Primodog, Eurican-CHPPI2-L (Merial Italia S.p.A., Milan, Italy), Nobivac® CEPPi, Nobivac® PARVO-c, Nobivac® PUPPY CP (Intervet Italia S.r.l., Milan, Italy), Canigen CEPPi/L (Virbac s.r.l., Milan, Italy). The experimental type 2-based vaccine 17/80-ISS (Buonavoglia et al., 1983) was also tested. For each commercial vaccine, three doses from different batches were tested in separate runs.
2.2. Template preparation
Faecal samples were homogenised (10%, w/v) in phosphate buffered saline (PBS, pH 7.2) and subsequently clarified by centrifuging at 1500 × g for 15 min. DNA was extracted from the supernatants by boiling the faecal homogenates for 10 min and chilling on ice (Schunck et al., 1995, Uwatoko et al., 1995). Lyophilised vaccines were resuspended in 1 ml of PBS and then processed as the faecal homogenates. To reduce residual inhibitors of DNA polymerase activity to ineffective concentrations, the DNA extracts were diluted 1:10 in distilled water (Decaro et al., 2005c, Decaro et al., 2006).
2.3. Standard DNAs
Standard DNAs for the old type and field strains (types 2a, 2b and 2c) were obtained from vaccine Vanguard CPV (Pfizer Italia srl, Rome, Italy) and from a field faecal sample characterised as type 2a (Decaro et al., 2006), respectively. In order to increase the viral DNA titre, the vaccine strain was passaged three times on A-72 canine cell line. DNA loads for vaccine and field viruses were calculated using a real-time PCR assay able to recognise all CPV-2 strains (Decaro et al., 2005c). Ten-fold dilutions of the standard DNAs were carried out in a CPV-negative faecal suspension. Aliquots of each dilution were frozen at −70 °C and used only once.
2.4. Design of primers and MGB probes
Primers and MGB probes specific for the vaccine and wild types were designed and synthesised by Applied Biosystems (Foster City, CA) taking into account the SNP T3088C encountered in the capsid protein gene between the original type 2 and its variants (2a, 2b and 2c), which is responsible for the change Ile to Thr at residue 101 of the capsid protein (Martella et al., in press and Table 1). Probes specific for vaccine and field strains were labeled with FAM and VIC fluorophores, respectively. Specificity, sequence and position of real-time PCR primers and MGB probes are reported in Table 2 .
Table 2.
Sequence, position and specificity of the oligonucleotides used in the study
| Assay | Primer/probe | Sequence 5′ to 3′ | Polarity | Specificity | Position | Amplicon size |
|---|---|---|---|---|---|---|
| TaqMan assaya | CPV-For | AAACAGGAATTAACTATACTAATATATTTA | + | All types | 4104–4135c | 93 bp |
| CPV-Rev | AAATTTGACCATTTGGATAAACT | − | 4176–4198c | |||
| CPV-Pb | FAM-TGGTCCTTTAACTGCATTAAATAATGTACC-TAMRA | + | 4143–4172c | |||
| Type 2/variants MGB probe assayb | CPV2/v-For | GCAGTTAACGGAAACATGGCTTTAG | + | All types | 3057–3081c | 68 bp |
| 772–796d | ||||||
| CPV2/v-Rev | TCAACCAATGACCAAGGTGTTACAA | − | 3100–3124c | |||
| 815–839d | ||||||
| CPV2-Pb | FAM-TGTGCATGAATATCAT-MGB | + | Type 2 (old type) | 3082–3097c | ||
| CPVv-Pb | VIC-TTTGTGCATGAGTATCAT-MGB | + | Antigenic variants | 797–814d | ||
2.5. MGB probe assay
Real-time PCR was conducted in an i-Cycler iQ™ Real-Time Detection System (Bio-Rad Laboratories Srl, Milan, Italy). The reactions (25 μl) contained 10 μl of template or standard DNA, 12.5 μl of IQ™ Supermix (Bio-Rad Laboratories Srl), 900 nM of primers CPV2/v-For and CPV2/v-Rev, 200 nM of probes CPV2-Pb and CPVv-Pb. Two different wells were used for each test sample and each dilution of standard DNA. After activation of iTaq DNA polymerase at 95 °C for 10 min, 45 cycles of two-step PCR were performed, consisting of denaturation at 95 °C for 30 s and primer annealing-extension at 60 °C for 1 min. The increase in fluorescent signal was registered during the annealing-extension step of the reaction and the data were analysed with the appropriate sequence detector software (version 3.0).
2.6. Detectability, linearity, specificity and reproducibility
Serial log 10 dilutions of each standard DNA (type 2 and type 2a) were carried out in a CPV-negative faecal suspension and used to determine the detectability and the linearity of the assay. Since vaccine virus was found to have DNA titres lower with respect to field CPVs, standard DNA concentrations ranged from 108 to 100 and from 109 to 100 DNA copies/10 μl of template for type 2 and 2a, respectively. C T values were measured in triplicate and were plot against the log of the input DNA copy number.
The specificity of the assay was evaluated by processing high and low concentrations of DNA from the original type 2 and types 2a, 2b and 2c, as well as DNA preparations from four other unrelated DNA viruses of dogs, including minute virus of canine (Decaro et al., 2002a), canid herpesvirus 1 (Decaro et al., 2002b), canine adenovirus type 1 (Pratelli et al., 2001) and type 2 (Decaro et al., 2004), or sterile water.
The intra-assay and interassay reproducibilities were evaluated using high, intermediate and low input of the standard DNAs of the old type and CPV-2a (dilutions in a CPV-negative faecal suspension). The coefficients of variation for the absolute copy number obtained for each dilution were calculated as described previously (Decaro et al., 2005c, Decaro et al., 2006).
2.7. Simultaneous detection of vaccine and field strains
In order to determine the ability of quantifying correctly and discriminating between vaccinal and field strains, artificially generated DNA mixtures composed of DNAs from vaccines and from field strains (2a, 2b or 2c) in various concentrations were tested by the MGB probe assay and the absolute copy numbers for each type were calculated.
2.8. Quantitation of viral DNA by TaqMan assay
All the DNA extracts were processed in parallel by a TaqMan assay able to recognise both the old type and the antigenic variants (Decaro et al., 2005c). The assay is internally controlled by using as exogenous DNA the nucleic acid extracted from ovine herpesvirus type 2 (Decaro et al., 2003). Real-time PCR was carried out in a 25 μl reaction containing 12.5 μl of master mix (Bio-Rad Laboratories Srl), 600 nM of primers CPV-For and CPV-Rev, 200 nM of probe CPV-Pb (Table 2) and 10 μl of DNA. The following thermal protocol was used: activation of iTaq DNA polymerase at 95 °C for 10 min and 40 cycles consisting of denaturation at 95 °C for 15 s, primer annealing at 52 °C for 30 s and extension at 60 °C for 1 min.
3. Results
3.1. Analytical performances
The detection limits of the MGB probe assay were shown to be 101 DNA copies for both types 2 and 2a. Standard curves demonstrated a strong linear correlation between 101 and 108 copies (type 2) or 109 copies (type 2a) (Fig. 1 ).
Fig. 1.
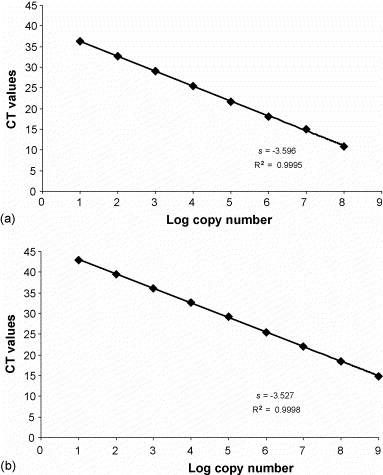
Standard curves obtained for types 2 (a) and 2a (b) by the type 2/variants MGB probe assay. The dilutions of standard DNA are indicated on the x-axis, whereas the corresponding cycle threshold (CT) values are presented on the y-axis. Each dot represents the result of duplicate amplifications of each dilution. The coefficient of determination (R2) and the slope value (s) of the regression curve were calculated and are indicated.
No cross-reactions were observed between the old type and the variants. FAM fluorescence signals were generated only by DNA templates from vaccines (original type 2), whereas VIC fluorescence was registered when DNA preparations from the wild types were tested. Furthermore, none of the other four canine viruses showed cross-reactivity, as well as FAM or VIC fluorescence was not obtained from sterile water.
CVs intra-assay ranged between 8.88 and 18.96% for the old type and between 7.80 and 27.86% for CPV-2a. CVs interassay were slightly higher, with values ranging between 14.26 and 35.44% for the original type 2 and between 12.76 and 33.54% for type 2a (Fig. 2 ).
Fig. 2.
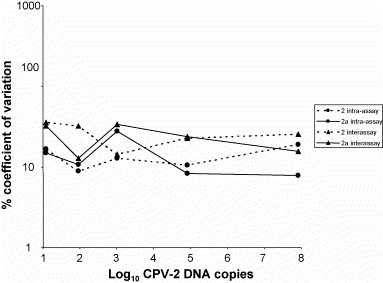
Coefficients of variation (%) intra-assay and interassay over the dynamic range of the type 2/variants MGB probe assay.
3.2. Simultaneous detection of vaccine and field strains
Samples spiked with low (103 copies) and high (108 copies) concentrations of DNA from the old type and the variants showed no interference during detection and quantitation of vaccine and field strains contained in the same sample, with DNA titres calculated correctly for all CPV types (data not shown).
3.3. Quantitation of viral DNA by TaqMan and MGB probe assays
CPV DNA loads in vaccines and field samples were calculated in parallel by the developed type2/variants MGB probe assay and by the TaqMan assay established previously (Decaro et al., 2005c). As shown in Fig. 3 , the two assays were found to correlate well over the shared 8 log10 dynamic ranges, calculating similar DNA titres for the same samples. In contrast, a poor correlation was found between the vaccine titres reported by manufacturers and expressed by CCID50 and the DNA copy numbers calculated by the MGB probe assay (Table 3 ).
Fig. 3.
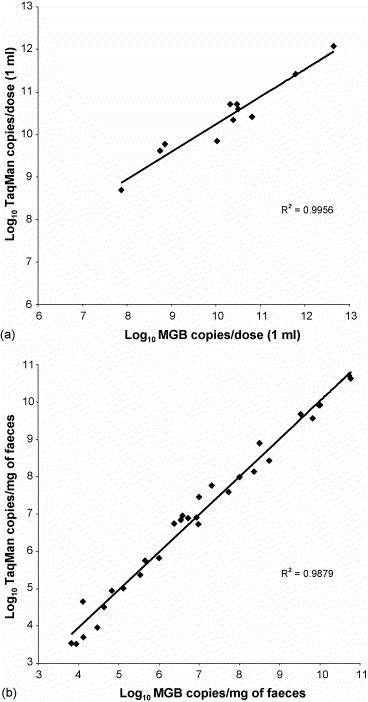
CPV-2 DNA titres measured by the TaqMan assay and the type 2/variants MGB probe assay. The type 2-based vaccines marketed in Italy (a) and 30 field samples tested positive to types 2a (n = 10), 2b (n = 10) or 2c (n = 10) (b) and covering the shared log10 dynamic ranges of the assays were analysed. DNA titres are expressed as copy numbers per dose and per mg of faeces for vaccines and field samples, respectively.
Table 3.
CPV DNA titres calculated by the MGB probe assay in commercial and experimental vaccines
| Vaccine | Company/reference | Specificity | Reported titre (CCID50)/dose | Viral DNA titre/dose |
|---|---|---|---|---|
| Vanguard 7 | Pfizer Inc. | Type 2 | ≥107a | 4.43 × 1012 |
| Vanguard CPV | Pfizer Inc. | Type 2 | ≥107a | 6.09 × 1011 |
| Tetradog-CHPL | Merial Italia S.p.A. | Type 2 | ≥103b | 7.26 × 108 |
| Parvodog-P | Merial Italia S.p.A. | Type 2 | ≥103b | 5.52 × 108 |
| Primodog | Merial Italia S.p.A. | Type 2 | ≥105.5b | 1.90 × 108 |
| Eurican-CHPPI2-L | MERIAL Italia S.p.A. | Type 2 | ≥104.9b | 2.92 × 1010 |
| Duramune DA2LP + Pv | Fort Dodge Animal Health S.p.A. | Type 2 | >104.5a | 7.45 × 107 |
| Nobivac® CEPPi | Intervet Italia S.r.l. | Type 2 | ≥107a | 3.17 × 1010 |
| Nobivac® PARVO-c | Intervet Italia S.r.l. | Type 2 | ≥107a | 2.08 × 1010 |
| Nobivac® PUPPY CP | Intervet Italia S.r.l. | Type 2 | ≥107a | 2.44 × 1010 |
| Canigen CEPPi/L | Virbac S.r.l | Type 2 | >103a | 1.07 × 1010 |
| 17/80-ISS | Buonavoglia et al. (1983) | Type 2 | 107.8c | 6.58 × 1010 |
Cell lines used for determination of viral titres are not reported.
Titres calculated on feline cells.
Titres calculated on canine A-72 cells.
4. Discussion
Diagnosis of CPV infection may be ambiguous when carried out on faecal samples from dogs presenting with diarrhoea few days after vaccination. In fact, the modified-live virus contained in the vaccines is able to replicate in the intestinal mucosa of vaccinated dogs (Carmichael et al., 1984, Buonavoglia et al., 1983), despite the unnatural route of administration (intramuscular or subcutaneous instead of oronasal), and to be shed in the faeces albeit at low titres and for a shorter time period with respect to field strains, as noted in a previous work (Decaro et al., 2006). In such a circumstance, the detection of CPV-2 or its nucleic acid in the faeces of vaccinated dogs could provide false-positive results, leading to a misdiagnosis of the disease probably caused by other enteric pathogens of dogs, i.e., canine coronavirus, canine distemper virus, reoviruses, rotaviruses, Salmonella spp., etc. Moreover, it would be important to rule out vaccine-induced disease due to regaining of virulence of the vaccine (old type) virus. For this purpose, the characterisation of CPV using traditional techniques is often inconclusive, since a simultaneous infection by the type 2-based vaccine and wild-type virus may mislead the results of HI with MAbs, PCR-RFLP and sequence analysis.
A PCR-based approach has been proposed by Senda et al. (1995) to address this point, which takes advantage of two SNPs, A3045T and C3685G, that determine the replacement of Met by Leu at position 87 and of Ala by Gly at position 300, in old- and wild-type strains, respectively (Table 1). Two primers specific for the wild types (types 2a and 2b) were selected to have one such mutation at the very 3′ end, as nucleotide mismatches that occur at the 3′ end of a primer are highly detrimental to primer extension and strongly decrease PCR amplification. However, in our experience, such mutations were not sufficient to prevent completely amplification of the old type virus (vaccine) (V. Martella, N. Decaro and C. Buonavoglia, personal observation). Moreover, samples containing both vaccine and wild-type strains are amplified successfully, so that the PCR-based strategy will not be able to detect the simultaneous presence of the two viruses in the faeces.
The MGB probe assays developed for the rapid characterisation of the CPV strains (Decaro et al., 2005b, Decaro et al., 2006) do not discriminate between the old type and type 2a. Thus, all samples collected from vaccinated dogs and characterised as type 2a should be tested by the novel MGB probe assay in order to assess whether they are true type 2a (field) strains or vaccine (old type) virus. A correct discrimination will help resolution of the diagnostic dilemma arising when dogs develop gastroenteric signs few days after CPV vaccination with the old virus.
Recently, type 2b vaccines have been licensed and are available on the market. Although at the moment such vaccines are not used widely, some problems may arise when testing samples collected from dogs administered a type 2b vaccine. In fact, vaccine virus shed in the faeces would be recognised by the probe specific for the CPV variants, being characterised as a field strain instead of a vaccine strain. Consequently, an additional assay should be developed which is able to discriminate the type 2b vaccines from type 2b field CPVs, in order to obtain a correct discrimination between dogs vaccinated and infected with CPV-2b. In conclusion, the test we have developed does not allow the discrimination of all vaccine viruses from field strains, but it could help a correct diagnosis when dogs display enteritis shortly after the administration of type 2-based vaccines, that are mostly used worldwide.
Acknowledgements
We thank Donato Narcisi and Carlo Armenise for their excellent technical assistance and student Anna Morea for her assistance with part of the experimental work. This work was supported by grants from University of Bari, Italy: project ex 60% 2006 “Caratterizzazione delle varianti di campo del parvovirus del cane mediante real-time PCR con sonde minor groove binding (MGB)”.
References
- Appel M.J.G., Scott W.F., Carmichael L.E. Isolation and immunization studies of canine parvo-like virus from dogs with haemorrhagic enteritis. Vet. Rec. 1979;105:156–159. doi: 10.1136/vr.105.8.156. [DOI] [PubMed] [Google Scholar]
- Buonavoglia C., Compagnucci M., Orfei Z. Dog response to plaque variant of canine parvovirus. Zbl. Vet. Med. B. 1983;30:526. doi: 10.1111/j.1439-0450.1983.tb01876.x. [DOI] [PubMed] [Google Scholar]
- Buonavoglia C., Martella V., Pratelli A., Tempesta M., Cavalli A., Buonavoglia D., Bozzo G., Elia G., Decaro N., Carmichael L.E. Evidence for evolution of canine parvovirus type-2 in Italy. J. Gen. Virol. 2001;82:1555–1560. doi: 10.1099/0022-1317-82-12-3021. [DOI] [PubMed] [Google Scholar]
- Buonavoglia D., Cavalli A., Pratelli A., Martella V., Greco G., Tempesta M., Buonavoglia C. Antigenic analysis of canine parvovirus strains isolated in Italy. New Microbiol. 2000;23:93–96. [PubMed] [Google Scholar]
- Burtonboy G., Coignoul F., Pastoret P.P., Delferriere N. Canine hemorrhagic enteritis detection of viral particles by electron microscopy. Arch. Virol. 1979;61:1–11. doi: 10.1007/BF01320586. [DOI] [PubMed] [Google Scholar]
- Carmichael L.E., Binn L.N. New enteric viruses in the dog. Adv. Vet. Sci. Comp. Med. 1981;25:1–37. [PubMed] [Google Scholar]
- Carmichael L.E., Pollock R.V., Joubert J.C. Response of puppies to canine-origin parvovirus vaccines. Mod. Vet. Pract. 1984;65:99–102. [PubMed] [Google Scholar]
- Decaro N., Altamura M., Pratelli A., Pepe M., Tinelli A., Casale D., Martella V., Tafaro A., Camero M., Elia G., Tempesta M., Jirillo E., Buonavoglia C. Evaluation of the innate immune response in pups during canine parvovirus type 1 infection. New Microbiol. 2002;25:291–298. [PubMed] [Google Scholar]
- Decaro N., Tinelli A., Campolo M., Elia G., Ventriglia G., Terio V., Bozzo G., Guarda F. Infezione erpetica del cane e mortalità neonatale dei cuccioli: descrizione di un focolaio. Veterinaria Anno. 2002;16(3):77–82. [Google Scholar]
- Decaro N., Tinelli A., Pratelli A., Martella V., Tempesta M., Buonavoglia C. First two confirmed cases of malignant catarrhal fever in Italy. New Microbiol. 2003;26:339–344. [PubMed] [Google Scholar]
- Decaro N., Camero M., Greco G., Zizzo N., Elia G., Campolo M., Pratelli A., Buonavoglia C. Canine distemper and related diseases: report of a severe outbreak in a kennel. New Microbiol. 2004;27:177–181. [PubMed] [Google Scholar]
- Decaro N., Desario C., Campolo M., Elia G., Martella V., Ricci D., Lorusso E., Buonavoglia C. Clinical and virological findings in pups naturally infected by canine parvovirus type 2 Glu-426 mutant. J. Vet. Diagn. Invest. 2005;17:133–138. doi: 10.1177/104063870501700206. [DOI] [PubMed] [Google Scholar]
- Decaro N., Elia G., Campolo M., Desario C., Lucente M.S., Bellacicco A.L., Buonavoglia C. New approaches for the molecular characterization of canine parvovirus type 2 strains. J. Vet. Med. B. 2005;52:316–319. doi: 10.1111/j.1439-0450.2005.00869.x. [DOI] [PubMed] [Google Scholar]
- Decaro N., Elia G., Martella V., Desario C., Campolo M., Di Trani L., Tarsitano E., Tempesta M., Buonavoglia C. A real-time PCR assay for rapid detection and quantitation of canine parvovirus type 2 DNA in the feces of dogs. Vet. Microbiol. 2005;105:19–28. doi: 10.1016/j.vetmic.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Decaro N., Elia G., Martella V., Campolo M., Desario C., Camero M., Cirone F., Lorusso E., Lucente M.S., Narcisi D., Scalia P., Buonavoglia C. Characterisation of the canine parvovirus type 2 variants using minor groove binder probe technology. J. Virol. Methods. 2006;133:92–99. doi: 10.1016/j.jviromet.2005.10.026. [DOI] [PubMed] [Google Scholar]
- Desario C., Decaro N., Campolo M., Cavalli A., Cirone F., Elia G., Martella V., Lorusso E., Camero M., Buonavoglia C. Canine parvovirus infection: which diagnostic test for virus? J. Virol. Methods. 2005;121:179–185. doi: 10.1016/j.jviromet.2005.02.006. [DOI] [PubMed] [Google Scholar]
- De Ybanez R.R., Vela C., Cortes E., Simarro I., Casal J.I. Identification of types of canine parvovirus circulating in Spain. Vet. Rec. 1995;136:174–175. doi: 10.1136/vr.136.7.174. [DOI] [PubMed] [Google Scholar]
- Greenwood N.M., Chalmers W.S.K., Baxendale W., Thompson H. Comparison of isolates of canine parvovirus by monoclonal antibody and restriction-enzyme analysis. Vet. Rec. 1996;138:495–496. doi: 10.1136/vr.138.20.495. [DOI] [PubMed] [Google Scholar]
- Johnson R.H., Spreadbrow P.B. Isolation from dogs with severe enteritis of a parvovirus related to feline panleukopenia virus. Aust. Vet. J. 1979;55:151. [Google Scholar]
- Kelly W.R. An enteric disease of dogs resembling feline panleukopenia. Aust. Vet. J. 1978;54:593. doi: 10.1111/j.1751-0813.1978.tb02426.x. [DOI] [PubMed] [Google Scholar]
- Martella V., Cavalli A., Pratelli A., Bozzo G., Camero M., Buonavoglia D., Narcisi D., Tempesta M., Buonavoglia C. A canine parvovirus mutant is spreading in Italy. J. Clin. Microbiol. 2004;42:1333–1336. doi: 10.1128/JCM.42.3.1333-1336.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martella, V., Decaro, N., Buonavoglia, C., in press. Genetic and antigenic variation of CPV-2 and implicance in antigenic/genetic characterization. Virus Genes. [DOI] [PubMed]
- Mochizuki M., Harasawa R., Nakatami H. Antigenic and genomic variabilities among recently prevalent parvoviruses of canine and feline origin in Japan. Vet. Microbiol. 1993;38:1–10. doi: 10.1016/0378-1135(93)90070-n. [DOI] [PubMed] [Google Scholar]
- Nakamura M., Tohya Y., Miyazawa T., Mochizuki M., Phung H.T., Nguyen N.H., Huynh L.M., Nguyen L.T., Nguyen P.N., Nguyen P.V., Nguyen N.P., Akashi H. A novel antigenic variant of canine parvovirus from a Vietnamese dog. Arch. Virol. 2004;149:2261–2269. doi: 10.1007/s00705-004-0367-y. [DOI] [PubMed] [Google Scholar]
- Parrish C.R., Carmichael L.E. Antigenic structure and variation of canine parvovirus type-2, feline panleukopenia virus, and mink enteritis virus. Virology. 1983;129:401–414. doi: 10.1016/0042-6822(83)90179-4. [DOI] [PubMed] [Google Scholar]
- Parrish C.R., O′Connel P.H., Evermann J.F., Carmichael L.E. Natural variation of canine parvovirus. Science. 1985;230:1046–1048. doi: 10.1126/science.4059921. [DOI] [PubMed] [Google Scholar]
- Parrish C.R., Have P., Foreyt W.J., Evermann J.F., Senda M., Carmichael L.E. The global spread and replacement of canine parvovirus strains. J. Gen. Virol. 1988;69:1111–1116. doi: 10.1099/0022-1317-69-5-1111. [DOI] [PubMed] [Google Scholar]
- Parrish C.R., Aquadro C.F., Strassheim M.L., Evermann J.F., Sgro J.-Y., Mohammed H.O. Rapid antigenic-type replacement and DNA sequence evolution of canine parvovirus. J. Virol. 1991;65:6544–6552. doi: 10.1128/jvi.65.12.6544-6552.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira C.A., Monezi T.A., Mehnert D.U., D′Angelo M., Durigon E.L. Molecular characterisation of canine parvovirus in Brazil by polymerase chain reaction assay. Vet. Microbiol. 2000;75:127–133. doi: 10.1016/s0378-1135(00)00214-5. [DOI] [PubMed] [Google Scholar]
- Pratelli A., Martella V., Elia G., Tempesta M., Guarda F., Capucchio M.T., Carmichael L.E., Buonavoglia C. Severe enteric disease in an animal shelter associated with dual infection by canine adenovirus type 1 and canine coronavirus. J. Vet. Med. B. 2001;48:385–392. doi: 10.1046/j.1439-0450.2001.00466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagazio P., Tempesta M., Buonavoglia D., Cirone F., Buonavoglia C. Antigenic characterization of canine parvovirus strains isolated in Italy. J. Virol. Methods. 1998;73:197–200. doi: 10.1016/s0166-0934(98)00055-x. [DOI] [PubMed] [Google Scholar]
- Schunck B., Kraft W., Truyen U. A simple touch-down polymerase chain reaction for the detection of canine parvovirus and feline panleukpenia virus in feces. J. Virol. Methods. 1995;55:427–433. doi: 10.1016/0166-0934(95)00069-3. [DOI] [PubMed] [Google Scholar]
- Senda M., Parrish C.R., Harasawa R., Gamoh K., Muramatsu M., Hirayama N., Itoh O. Detection by PCR of wild-type canine parvovirus which contaminates dog vaccines. J. Clin. Microbiol. 1995;33:110–113. doi: 10.1128/jcm.33.1.110-113.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinel A., Venter E.H., van Vuuren M., Truyen U. Antigenic and genetic analysis of canine parvoviruses in southern Africa. Ondersteeport J. Vet. Res. 1998;65:239–242. [PubMed] [Google Scholar]
- Truyen U., Platzer G., Parrish C.R. Antigenic type distribution among canine parvoviruses in dogs and cats in Germany. Vet. Rec. 1996;138:365–366. doi: 10.1136/vr.138.15.365. [DOI] [PubMed] [Google Scholar]
- Truyen U., Steinel A., Bruckner L., Lutz H., Mostl K. Distribution of antigenic types of canine parvovirus in Switzerland, Austria and Germany. Schweiz. Arch. Tierheilkd. 2000;142:115–119. [PubMed] [Google Scholar]
- Uwatoko K., Sunairi M., Nakajima M., Yamaura K. Rapid method utilizing the polymerase chain reaction for detection of canine parvovirus in feces of diarrhoeic dogs. Vet. Microbiol. 1995;43:315–323. doi: 10.1016/0378-1135(94)00102-3. [DOI] [PubMed] [Google Scholar]


