Abstract
A TaqMan® fluorogenic reverse transcriptase-polymerase chain reaction (RT-PCR) assay was developed for the detection and quantitation of canine coronavirus (CCoV) RNA in the faeces of naturally or experimentally infected dogs. The CCoV fluorogenic RT-PCR assay, which targeted the ORF5 (M gene), was more sensitive than a conventional RT-PCR assay targeting the same gene, showing a detection limit of 10 copies of CCoV standard RNA, and was linear from 10 to 108 copies, allowing quantitation of samples with a wide range of CCoV RNA loads. A total of 78 faecal samples of diarrhoeic dogs were tested simultaneously by conventional and fluorogenic RT-PCR: 29 were negative by both techniques, whereas 27 tested positive by conventional RT-PCR and 48 by the established CCoV fluorogenic assay. One sample, which was positive by conventional RT-PCR, gave no signal in the fluorogenic assay. In addition, by the fluorogenic assay CCoV shedding in the faecal samples of an experimentally infected dog was monitored for 28 days. The high sensitivity, simplicity and reproducibility of the CCoV fluorogenic RT-PCR assay, combined with its wide dynamic range and high throughput, make this method especially suitable for efficacy trials on CCoV vaccines.
Keywords: Canine coronavirus RNA, Quantitation, Fluorogenic RT-PCR
1. Introduction
Canine coronavirus (CCoV), a member of the family Coronaviridae, is an enveloped, single-stranded, positive-sense RNA virus, responsible for mild to severe enteritis in pups. CCoV belongs to the group I coronaviruses, which also include the transmissible gastroenteritis virus of swine (TGEV), the porcine epidemic diarrhoea virus (PEDV), the porcine respiratory coronavirus (PRCoV), the feline coronaviruses (FCoVs) and the human coronavirus 229E (HCoV 229E).
About two-thirds of the CCoV genomic RNA is occupied by two large, partially overlapping open reading frames (ORFs), ORF1a and ORF1b, which encode two polyproteins leading to the viral replicase formation. The 3’ one third of the genome consists of other ORFs encoding the structural proteins and the other non-structural ones. The structural proteins comprise the S, E, M and N proteins encoded by ORF2, ORF4, ORF5 and ORF6, respectively (Enjuanes et al., 2000).
In young pups, sometimes in combination with other pathogens, CCoV infection may cause severe diarrhoea, vomiting, dehydration, loss of appetite, and, occasionally, death. CCoV shedding in faeces occurs for 6–9 days post-infection (Keenan et al., 1976). Nevertheless long term viral shedding has been detected by PCR in CCoV infected pups (Pratelli et al., 2001b, Pratelli et al., 2002b).
Traditionally, diagnosis of CCoV infection was made using virus isolation or electron microscopy, but these methods have been demonstrated to be poorly sensitive or specific. Recently, the establishment of reverse transcriptase-polymerase chain reaction (RT-PCR) assays has led to an increase of both sensitivity and specificity, so that PCR has become the “gold standard” technique for the CCoV diagnosis (Pratelli et al., 1999, Pratelli et al., 2000). Several RT-PCR based methods have been developed for detecting CCoV RNA in the faeces of dogs, but none of these were designed to be quantitative (Bandai et al., 1999, Naylor et al., 2001, Pratelli et al., 1999, Pratelli et al., 2002c). Moreover, conventional RT-PCR assays are time consuming and contain a certain risk of carryover contamination due to the post-PCR manipulations and a second amplification step in nested PCR systems, especially when a high sample throughput is required. Finally, those methods are limited in sensitivity and allow only relatively few samples to be processed at one time. Conversely, it is well-known that real-time TaqMan RT-PCR enables a reproducible, sensitive and specific quantitation of viral RNA (Budevi and Weinstock, 2001, Gut et al., 1999, Martell et al., 1999, Callahan et al., 2001, Balasuriya et al., 2002, Reid et al., 2002, Smith et al., 2002, Spackman et al., 2002).
The present study describes a real-time fluorogenic RT-PCR assay for CCoV. The method is based on the TaqMan® technology, which uses a dual-labeled fluorogenic probe combined with the 5′→3′ exonuclease activity of Taq polymerase, resulting in an increase of the reporter dye’s fluorescence released in the course of the PCR amplification (Holland et al., 1991, Heid et al., 1996). The analytical performance of the CCoV fluorogenic RT-PCR was evaluated in comparison to that of a conventional qualitative RT-PCR assay. The fluorogenic assay was then applied to detect and quantify viral load in CCoV naturally infected dogs and to trace the course of CCoV infection in the faeces of a dog experimentally infected with a field CCoV strain.
2. Materials and methods
2.1. Samples
A total of 78 faecal samples, collected from diarrhoeic pups in different geographical areas of Italy, were processed in order to detect CCoV and quantify viral RNA amounts in the faeces.
In addition, one dog, 3 months of age, which had been tested negative for CCoV antigen in the faeces by RT-PCR (Pratelli et al., 1999) and for CCoV antibodies by ELISA (Pratelli et al., 2002a), received 4 ml (2 ml intranasally and 2 ml orally) of a cell culture medium containing 105 TCID50/50 μl of a CCoV field strain, as previously described (Pratelli et al., 2003b, Pratelli et al., 2004). Faecal samples of the infected dog were collected daily for 28 days and subjected to both CCoV conventional RT-PCR and real-time testing.
2.2. RNA extraction
Total RNA was extracted from each faecal sample with QIAamp® RNeasy Mini Kit (Qiagen GmbH, Hilden, Germany) in accordance with the manufacturer’s protocol. The starting material consisted of 10 mg of faeces for each sample. Template RNAs were eluted in 50 μl of RNase-free water and stored at −70 °C until their use.
2.3. RNA standard for quantitation
To obtain a standard for the fluorogenic RT-PCR, the ORF5 (M-protein gene, 805 bp) of CCoV strain 45/93 (Buonavoglia et al., 1994) was cloned into pCR® 2.1-TOPO vector (TOPO TA Cloning®, Invitrogen, Milan, Italy) and transcribed with RiboMAX™ Large Scale RNA Production System-T7 (Promega Italia, Milan, Italy) from the T7 promoter, according to the manufacturer’s guidelines. After a DNase treatment to remove all DNA, the transcripts were purified using a commercial column (QIAamp® RNeasy Mini Kit, Qiagen GmbH) and quantified by spectrofotometrical analysis. Ten-fold dilutions of the RNA transcripts were carried out in TE (Tris–HCl, EDTA, pH 8.0) buffer containing 30 μg carrier RNA (tRNA from Escherichia coli, Sigma–Aldrich Srl, Milan, Italy) per ml. Aliquots of each dilution were frozen at −70 °C and used only once.
2.4. Primer and probe design
The ORF5 nucleotide sequences of several CCoV strains (Pratelli et al., 2003a) were aligned using the BioEdit software package (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Primers and TaqMan probe were designed using Beacon Designer software, version 2.0 (Premier Biosoft International, Palo Alto, CA, USA) to amplify a conserved 99-bp fragment within the aligned ORF5 sequences. Primers and probe were synthesized by MWG Biotech AG (Ebersberg, Germany). The TaqMan probe was labeled with 6-carboxyfluorescein (FAM) at the 5′ end and with 6-carboxytetramethylrhodamine (TAMRA) at the 3′ end. The position and sequence of the primers and probe used for TaqMan RT-PCR amplification are reported in Table 1 .
Table 1.
Specific oligonucleotides used in CCoV fluorogenic assay and conventional RT-PCR
| Primer/probe | Sequence 5′→3′ | Sense | Positiona | Amplicon size |
|---|---|---|---|---|
| CcoV1b | TCCAGATATGTAATGTTCGG | + | 6729–6748 | 409 bp |
| CcoV2b | TCTGTTGAGTAATCACCAGCT | − | 7118–7138 | |
| CcoV-Forc | TTGATCGTTTTTATAACGGTTCTACAA | + | 6585–6611 | 99 bp |
| CcoV-Revc | AATGGGCCATAATAGCCACATAAT | − | 6660–6683 | |
| CcoV-Pbc | FAMd-ACCTCAATTTAGCTGGTTCGTGTATGGCATT-TAMRAe | + | 6620–6650 |
Oligonucleotide position is referred to the sequence of CCoV strain Insavc-1 (accession no.: D13096).
Conventional RT-PCR (Pratelli et al., 1999).
Fluorogenic assay.
FAM, 6-carboxyfluorescein.
TAMRA, 6-carboxytetramethylrhodamine.
2.5. Reverse transcription
Triplicates of the standard dilutions and RNA templates were subjected simultaneously to reverse transcription (RT) with GeneAmp® RNA PCR (Applied Biosystems, Applera Italia, Monza, Italy). One microliter of each triplicate of standard dilutions or template RNA was reverse transcribed in a reaction volume of 20 μl containing PCR buffer 1× (KCl 50 mM, Tris–HCl 10 mM, pH 8,3), MgCl2 5 mM, 1 mM of each deoxynucleotide (dATP, dCTP, dGTP, dTTP), RNase Inhibitor 1 U, MuLV reverse transcriptase 2.5 U, random hexamers 2.5 U. Synthesis of c-DNA was carried out at 42 °C for 30 min, followed by a denaturation step at 99 °C for 5 min.
2.6. Fluorogenic PCR
The 50-μl PCR mixture for one reaction contained 25 μl of IQ™ Supermix (Bio-Rad Laboratories Srl, Milan, Italy), 300 nM of each primer (CCoV-For and CCoV-Rev), 200 nM of probe CCoV-Pb and 20 μl of c-DNA. The thermal cycle protocol used was the following: activation of iTaq DNA polymerase at 95 °C for 10 min and 45 cycles consisting of denaturation at 95 °C for 15 s and primer annealing-extension at 60 °C for 1 min. Fluorogenic PCR was performed in an i-Cycler iQ™ Real-Time Detection System (Bio-Rad Laboratories Srl) and the data were analysed with the appropriate sequence detector software (version 3.0). The accumulation of the PCR products was detected by monitoring the increase in fluorescence of the reporter dye. Signals were regarded as positive if the fluorescence intensity exceeded 10 times the standard deviation of the baseline fluorescence (threshold cycle [C T]).
2.7. Conventional RT-PCR
Conventional RT-PCR, amplifying a 409 bp fragment of the ORF5 of CCoV, was carried out as previously described (Pratelli et al., 1999). Briefly, PCR amplification was carried out GeneAmp® RNA PCR (Applied Biosystems, Applera Italia) and the following thermal conditions: reverse transcription at 42 °C for 30 min, inactivation of MuLV reverse transcriptase at 99 °C for 4 min, 45 cycles of 94 °C for 1 min, 55 °C for 1 min, 72 °C for 1 min, with a final extension at 72 °C for 10 min. The PCR products were detected by electrophoresis through a 1.5% agarose gel and visualization under UV light after bromide ethidium staining.
The position and sequence of the primers used for conventional amplification are reported in Table 1.
3. Results
3.1. Analytical performance of the CCoV fluorogenic RT-PCR assay
To compare the analytical sensitivity, 10-fold dilutions of the standard RNA, ranging from 108 to 100 copies/μl, were tested by both fluorogenic and conventional RT-PCR. In addition, 10-fold dilutions in Dulbecco’s Minimal Essential Medium (D-MEM) of the CCoV vaccinal strain 257/98-3c (Pratelli et al., 2004), starting from 105.50 TCID50/50 μl, were processed. Each standard or virus dilution was quantified three times separately. As shown in Table 2 , the detection limit of the TaqMan RT-PCR was 1–2 log higher than that of conventional RT-PCR, ranging around 101 copies/μl and 10−1.50 TCID50/50 μl for standard RNA and CCoV strain, respectively, with a detection rate of 100% for each positive dilution.
Table 2.
Analytical sensitivity of CCoV fluorogenic assay and conventional RT-PCR
| Template | Amounta,b | Fluorogenic assay | RT-PCR | ||||
|---|---|---|---|---|---|---|---|
| Standard RNA | 105 | + | + | + | + | + | + |
| 104 | + | + | + | + | + | + | |
| 103 | + | + | + | + | + | + | |
| 102 | + | + | + | − | − | − | |
| 101 | + | + | + | − | − | − | |
| 100 | − | − | − | − | − | − | |
| CCoV 257/98 | 102.50 | + | + | + | + | + | + |
| 101.50 | + | + | + | + | + | + | |
| 100.50 | + | + | + | + | + | + | |
| 10−0.50 | + | + | + | + | − | + | |
| 10−1.50 | + | + | + | − | − | − | |
| 10−2.50 | − | − | − | − | − | − | |
The higher tested amounts were all positive and are not indicated in the table.
Standard RNA amounts are expressed as number of copies; CCoV 257/98 amounts are expressed as TCID50/50μl.
Serial 10-fold dilutions of standard RNA (from 10 to 108 copies/μl) were used to generate a standard curve to quantify CCoV RNA in faecal samples from naturally or experimentally infected dogs. The standard curve was created automatically by the i-Cycler IQ Optical System Software, version 3.0 (Bio-Rad Laboratories Srl), by plotting the C T values against each standard dilution of known concentration. The standard curve that was generated spans eight orders of magnitude and shows linearity over the entire quantitation range (slope=−3.338), providing an accurate measurement over a very large variety of starting target amounts. The coefficient of linear regression (R 2) was equal to 0.999 and the PCR efficiency ranged around 99% (Fig. 1 ).
Fig. 1.
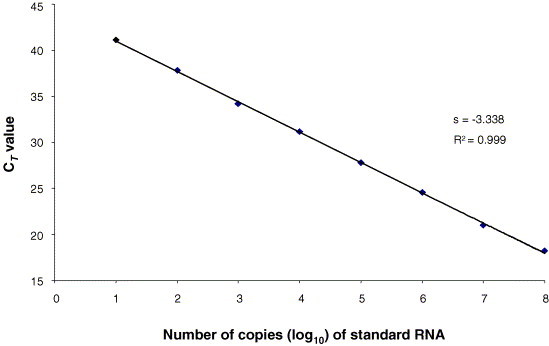
Standard curve of the CCoV fluorogenic RT-PCR assay. Ten-fold dilutions of standard RNA prior to amplification were used, as indicated on the x-axis, whereas the corresponding cycle threshold (CT) values are presented on the y-axis. Each dot represents the result of triplicate amplifications of each dilution. The coefficient of determination (R2) and the slope value (s) of the regression curve were calculated and are indicated.
The reproducibility of the method was established with the C T values obtained for the same standard dilution in different assays and within an assay, in order to calculate the interassay and intra-assay coefficient of variation (CV). The CV was obtained by dividing the standard deviation of the standard dilution by its mean and multiplying that result for 100. To estimate the interassay reproducibility, 105 copies of the standard RNA were submitted in triplicate to 20 consecutive runs. The intra-assay reproducibility was determined by pipetting the same standard copy number (105 molecules) 50 times on the same 96-well reaction plate. The CV between runs and within-run was 4.74 and 1.13%, respectively.
3.2. Examination of faecal samples of dogs naturally infected with CCoV
Twenty-nine of the 78 faecal samples examined were negative for CCoV by both conventional and fluorogenic RT-PCR; in 13/29 CCoV negative samples, canine parvovirus type 2 was detected by a specific haemagglutination assay (data not shown). As shown in Fig. 2 , by conventional RT-PCR 27 samples were found to be positive and 51 negative for CCoV. Conversely, 48 samples tested positive and 30 negative by real-time RT-PCR. Totally, 55 faecal samples were in agreement by both tests (26 positive and 29 negative samples). Twenty-two samples, CCoV negative by conventional amplification, resulted positive by the fluorogenic RT-PCR assay. One sample tested positive by conventional amplification and negative by real-time analysis. Four samples which gave a positive signal in the fluorogenic RT-PCR assay could not be quantified, since their viral titre was below the sensitivity limit of the assay (10 copies). Regarding the CCoV RNA loads assessed by real-time analysis, it was demonstrated that the faeces of the naturally infected dogs contained a wide range of CCoV RNA amounts, from 10 to 7.5×107/μl of template, with a median titre of about 103/μl of template.
Fig. 2.
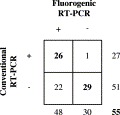
Detection of CCoV in faecal samples of naturally infected dogs: comparison between conventional and fluorogenic RT-PCR. Numbers indicate the samples positive (+) or negative (−) for CCoV. Results according to both techniques are shown in bold.
3.3. Examination of faecal samples of the dog experimentally infected with CCoV
The results of the conventional amplification and real-time analysis carried out on the faecal samples of the CCoV experimentally infected dog are summarized in Fig. 3 . The dog tested positive for CCoV by conventional RT-PCR for 24 days, from day 1 to 24 post-infection (dpi). In contrast, by quantitative fluorogenic RT-PCR, CCoV shedding was demonstrated during the entire observation period (28 days), reaching a peak at dpi 4 (1.7×105 RNA copies/μl of template). The infected dog showed mild diarrhoea from dpi 3 to 6 (Pratelli et al., 2003b, Pratelli et al., 2004), concomitantly with the faecal shedding of the highest CCoV titres.
Fig. 3.
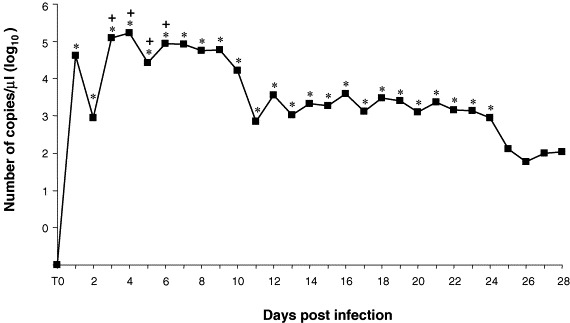
Number of copies of CCoV genomic RNA in the faecal samples of the experimentally infected dog by fluorogenic RT-PCR. Asterisks indicate the CCoV positive samples by conventional RT-PCR. Symbol (+) indicates days post-infection in which a mild diarrhoea was observed.
4. Discussion
The development is described of a quantitative, simple, rapid and reproducible method for the detection and quantitation of CCoV RNA in faecal samples of dogs infected experimentally or naturally with CCoV. The minimum copy number which could be detected by the CCoV fluorogenic assay was approximately 10 copies of standard RNA. On the other hand, this assay was able to quantify correctly samples with more than 107 copies/μl of template, since the linearity of the generated standard curve persisted up to the highest titre of in vitro-transcribed RNA analysed. In contrast, the detection limit of conventional RT-PCR was about 103 molecules. This considerably wide range of linearity allows the use of this system to analyse samples with a wide range of CCoV loads.
In comparison with conventional RT-PCR, the fluorogenic assay presents many advantages. In addition to its greater sensitivity, this technique is rapid, allowing several samples to be processed in few hours, with a large increase in throughput. The assay is a closed system in which the tube is never opened post-amplification, and this eliminates the possibility of cross-contamination of new samples with products amplified previously. A limited carryover may occur due to the separation between RT and fluorogenic PCR, but we preferred a two-step assay, since one-tube methods are less cheap and sensitive than a two-step RT-PCR procedure (Nakamura et al., 1993). In order to reduce the risk of contamination, we have separated strictly the different working steps and carried out pipetting in different laminar flow hoods.
However, the main advantage of the fluorogenic dye system consists of quantifying CCoV RNA amounts in faecal samples with a high degree of reproducibility and precision (CV between runs=4.74%, CV within-run=1.13%). Quantitative gel-based PCR assays have been established for measuring several cellular and viral RNAs, but they required time-consuming and potentially contaminating post-amplification steps and showed a lower precision (Wang et al., 1989, Kinoshita et al., 1992, Nagano and Kelly, 1994, Hua et al., 1999, Martell et al., 1999). Nevertheless, quantitation of CCoV in the faeces is essential to trace the course of natural as well as experimental infection in dogs. Among the faecal samples collected from the CCoV experimentally infected dog, four were only found to be positive by the fluorogenic assay, probably due to their low viral load combined with the higher sensitivity of the TaqMan assay. Conversely, one of the faecal samples of dogs infected naturally were only found to be positive by conventional RT-PCR. A possible explanation for this ambiguous result may be that the M gene, which is the target for both the conventional and fluorogenic assay, presents a certain degree of variability, so that mismatches in the binding site of primers and probe may reduce and, eventually, prevent an efficient amplification (Pratelli et al., 2001a, Pratelli et al., 2002b).
Finally, CCoV quantitation by real-time analysis could be a useful and complementary method to evaluate the efficacy of vaccines in challenged dogs. Usually, dogs experimentally inoculated with field CCoV strains do not develop considerable clinical signs, impairing any comparison between vaccinated and unvaccinated dogs. Thus, the efficacy of CCoV vaccines may be evaluated, as described previously (Pratelli et al., 2003b, Pratelli et al., 2004), by monitoring viral shedding in the faeces of vaccinated dogs after CCoV challenge, using virus isolation and conventional RT-PCR. Theoretically, CCoV amounts in the faeces could be assessed by virus titration on cell cultures. However, the low sensitivity of CCoV isolation and, mainly, the appearance of CCoV specific antibodies in the faeces after the challenge may affect this virological assay (personal observation). Conversely, the use of the CCoV fluorogenic RT-PCR assay could give a quantitation of viral loads, potentially showing differences in CCoV shedding.
References
- Balasuriya U.B.R, Leutenegger C.M, Topol J.B, McCollum W.H, Timoney P.J, MacLachlan N.J. Detection of equine arteritis virus by real-time TaqMan® reverse transcription-PCR assay. J. Virol. Methods. 2002;101:21–28. doi: 10.1016/s0166-0934(01)00416-5. [DOI] [PubMed] [Google Scholar]
- Bandai C, Ishiguro S, Masuya N, Hohdatsu T, Mochizuki M. Canine coronavirus infections in Japan: virological and epidemiological aspects. J. Vet. Med. Sci. 1999;61:731–736. doi: 10.1292/jvms.61.731. [DOI] [PubMed] [Google Scholar]
- Budevi B, Weinstock D. Fluorogenic RT-PCR assay (TaqMan) for detection and classification of bovine viral diarrhea virus. Vet. Microbiol. 2001;83:1–10. doi: 10.1016/S0378-1135(01)00390-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonavoglia, C., Marsilio, F., Cavalli, A., Tiscar, P.G., 1994. L’infezione da coronavirus del cane: indagine sulla presenza del virus in Italia, Notiziario Farmaceutico Veterinario, Nr. 2/94, ed. SCIVAC.
- Callahan J.D, Wu S.-J.L, Dion-Schultz A, Mangold B.E, Peruski L.F, Watts D.M, Porter K.R, Murphy G.R, Suharyono W, King C.-C, Hayes C.G, Temenak J.J. Development and evaluation of serotype- and group-specific fluorogenic reverse transcriptase PCR (TaqMan) assay for dengue virus. J. Clin. Microbiol. 2001;39:4119–4124. doi: 10.1128/JCM.39.11.4119-4124.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuanes, L., Brian, D., Cavanagh, D., Holmes, K., Lai, M.M.C., Laude, H., Masters, P., Rottier, P., Siddell, S., Spaan, W.J.M., Taguchi, F., Talbot, P., 2000. Coronaviridae. In: van Regenmortel, MH.V., Fauquet, C.M., Bishop, D.H.L., Carstens, E.B., Estes, M.K., Lemon, S.M., Maniloff, J., Mayo, M.A., McGeoch, D.J., Pringle, C.R., Wickner, R.B. (Eds.), Virus Taxonomy, Classification and Nomenclature of Viruses. Academic Press, New York, pp. 835–849.
- Gut M, Leutenegger C.M, Huder J.B, Pedersen N.C, Lutz H. One-tube fluorogenic reverse transcription-polymerase chain reaction for the quantitation of feline coronaviruses. J. Virol. Methods. 1999;77:37–46. doi: 10.1016/S0166-0934(98)00129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid C, Stevens J, Livak K, Williams P.M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- Holland P.M, Abramson R.D, Watson R, Gelfand D.H. Detection of specific polymerase chain reaction product by utilizing the 5′-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Li H, Crawford T.B. Quantitation of sheep-associated malignant catarrhal fever viral DNA by competitive polymerase chain reaction. J. Vet. Diagn. Invest. 1999;11:117–121. doi: 10.1177/104063879901100202. [DOI] [PubMed] [Google Scholar]
- Keenan K.P, Jervis H.R, Marchwicki R.H, Binn L.N. Intestinal infection of neonatal dogs with canine coronavirus 1-71: studies by virologic, histologic, histochemical and immunofluorescent techniques. Am. J. Vet. Res. 1976;37:247–256. [PubMed] [Google Scholar]
- Kinoshita T, Imamura J, Nagai H, Shimotohno K. Quantification of gene expression over a wide range by the polymerase chain reaction. Anal. Biochem. 1992;206:231–235. doi: 10.1016/0003-2697(92)90358-e. [DOI] [PubMed] [Google Scholar]
- Martell M, Gómez J, Esteban J.I, Sauleda S, Quer J, Cabot B, Esteban R, Guardia J. High-throughput real-time reverse transcription-PCR quantitation of hepatitis C virus RNA. J. Clin. Microbiol. 1999;37:327–332. doi: 10.1128/jcm.37.2.327-332.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano M, Kelly P.A. Tissue distribution and regulation of rat prolactin receptor gene expression quantitative analysis by polymerase chain reaction. J. Biol. Chem. 1994;269:13337–13345. [PubMed] [Google Scholar]
- Nakamura S, Katamine S, Yamamoto T, Foung S, Kurata T, Hirabayashi Y, Shimada K, Hino S, Miyamoto T. Amplification and detection of a single molecule of human immunodeficiency virus RNA. Virus Genes. 1993;4:325–338. doi: 10.1007/BF01703389. [DOI] [PubMed] [Google Scholar]
- Naylor M.J, Harrison G.A, Monckton R.P, McOrist S, Lehrbach P.R, Deane E.M. Identification of canine coronavirus strains from faeces by S gene nested PCR and molecular characterization of a new Australian isolate. J. Clin. Microbiol. 2001;39:1036–1041. doi: 10.1128/JCM.39.3.1036-1041.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A, Tempesta M, Greco G, Martella V, Buonavoglia C. Development of a nested PCR assay for the detection of canine coronavirus. J. Virol. Methods. 1999;80:11–15. doi: 10.1016/S0166-0934(99)00017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A, Buonavoglia D, Martella V, Tempesta M, Lavazza A, Buonavoglia C. Diagnosis of canine coronavirus infection using nested-PCR. J. Virol. Methods. 2000;84:91–94. doi: 10.1016/S0166-0934(99)00134-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A, Martella V, Elia G, Decaro N, Aliberti A, Buonavoglia D, Tempesta M, Buonavoglia C. Variation of the sequence in the gene encoding for transmembrane protein M of canine coronavirus (CCV) Mol. Cell. Probes. 2001;15:229–233. doi: 10.1006/mcpr.2001.0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A, Martella V, Elia G, Tempesta M, Guarda F, Capucchio M.T, Carmichael L.E, Buonavoglia C. Severe enteric disease in an animal shelter associated with dual infections by canine adenovirus type 1 and canine coronavirus. J. Vet. Med. B. 2001;48:385–392. doi: 10.1046/j.1439-0450.2001.00466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A, Elia G, Martella V, Palmieri A, Cirone F, Tinelli A, Corrente M, Buonavoglia C. Prevalence of canine coronavirus (CCoV) antibodies in dogs in Bari, Italy, by an enzyme-linked immunosorbent assay. J. Virol. Methods. 2002;102:67–71. doi: 10.1016/S0166-0934(01)00450-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A, Elia G, Martella V, Tinelli A, Decaro N, Marsilio F, Buonavoglia D, Tempesta M, Buonavoglia C. M gene evolution of canine coronavirus in naturally infected dogs. Vet. Rec. 2002;151:758–761. [PubMed] [Google Scholar]
- Pratelli A, Tinelli A, Decaro N, Camero M, Elia G, Gentile A, Buonavoglia C. PCR assay for the detection and the identification of atypical canine coronavirus in dogs. J. Virol. Methods. 2002;106:209–213. doi: 10.1016/S0166-0934(02)00165-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A, Martella V, Pistello M, Elia G, Decaro N, Buonavoglia D, Camero M, Tempesta M, Buonavoglia C. Identification of coronaviruses in dogs that segregate separately from the canine coronavirus genotype. J. Virol. Methods. 2003;107:213–222. doi: 10.1016/S0166-0934(02)00246-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A, Tinelli A, Decaro N, Cirone F, Elia G, Roperto S, Tempesta M, Buonavoglia C. Efficacy of an inactivated canine coronavirus vaccine in pups. New Microbiol. 2003;26:151–155. [PubMed] [Google Scholar]
- Pratelli A, Tinelli A, Decaro N, Martella V, Camero M, Tempesta M, Martini M, Carmichael L.E, Buonavoglia C. Safety and efficacy of a modified-live canine coronavirus vaccine in dogs. Vet. Microbiol. 2004;99:43–49. doi: 10.1016/j.vetmic.2003.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid S.M, Ferris N.P, Hutchings G.H, Zhang Z, Belsham G.J, Alexandersen S. Detection of all seven serotypes of foot-and-mouth disease virus by real-time, fluorogenic reverse transcription polymerase chain reaction assay. J. Virol. Methods. 2002;105:67–80. doi: 10.1016/s0166-0934(02)00081-2. [DOI] [PubMed] [Google Scholar]
- Smith I.L, Northill J.A, Harrower B.J, Smith G.A. Detection of Australian bat lyssavirus using a fluorogenic probe. J. Clin. Virol. 2002;25:285–291. doi: 10.1016/s1386-6532(02)00083-5. [DOI] [PubMed] [Google Scholar]
- Spackman E, Senne D.A, Myers T.J, Bulaga L.L, Garber L.P, Perdue M.L, Lohman K, Daum L.T, Suarez D.L. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 2002;40:3256–3260. doi: 10.1128/JCM.40.9.3256-3260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A.M, Doyle M.V, Mark D.F. Quantitation of mRNA by the polymerase chain reaction (published erratum appears in Proc. Natl. Acad. Sci. U.S.A. 87 (1990) 2865) Proc. Natl. Acad. Sci. U.S.A. 1989;86:9717–9721. doi: 10.1073/pnas.86.24.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]


