Abstract
An outbreak of highly virulent Chinese-type of Porcine Reproductive and Respiratory Syndrome Virus (H-PRRSV) in most areas of China recently has led to huge economic losses and drawn great attention to its diagnosis and disease control. To facilitate rapid identification of H-PRRSV, a fluorogenic-probe hydrolysis (TaqMan)-reverse transcriptase PCR for H-PRRSV has been developed. Primers and probe specificity were evaluated with RNA extracted from 5 strains of H-PRRSV and 24 strains of other viruses, the results showed 100% specificity for the selected panel. The assay met the sensitivity of 1 50% tissue culture infective dose (TCID50) per ml of samples from infected pigs. Analysis with 105–1 TCID50/ml H-PRRSV samples demonstrated high reproducibility with a coefficient of variation (CV) of 0.5–2.5%. More than two hundred samples from lung, spleen, blood serum specimens obtained from 22 outbreaks of suspected H-PRRS from March to June in 2007 were verified using this assay. The results showed that 68.5% (146 out of 213) of these samples were positive which is 100% consistent with that of the sequencing method. The assay can be performed in less than 3 h and thus provide a rapid method for the diagnosis of H-PRRSV as well as for elucidation of the epidemiology of H-PRRSV infections.
Keywords: Highly virulent, PRRS, Real-time PCR, TaqMan, Detection
1. Introduction
Porcine Reproductive and Respiratory Syndrome (PRRS) is considered one of the most economically important diseases in China as well as in other swine-producing countries throughout the world (Albina, 1997, Meredith, 1995). This disease was first reported in the USA in 1987 and named “blue-ear disease” or “mystery swine disease” (Keffaber, 1989). The causative agent of the disease is Porcine Reproductive and Respiratory Syndrome Virus (PRRSV), which is a small-enveloped RNA virus belonging to the family Arterivirida in the order Nidovirales (Cavanagh, 1997, Zimmerman et al., 1997). PRRS results in both reproductive failure in sows and respiratory disease (pneumonia) in nursery and grower/finishing pigs (Rossow, 1998, Stevenson et al., 1993, Snijder and Meulenberg, 1998).
PRRS was described initially in China in 1996 by the Harbin Veterinary Research Institute. Because of its economic significance, a great deal of resource has been invested in developing effective prevention and control strategies. But protocols providing consistent success have been elusive due to the high rate of genetic change and antigenic variability (Christopher-Hennings et al., 1995, Kapur et al., 1996, Halbur et al., 1996, Meng, 2000). A diagnosis of PRRSV infection is based mainly on typical clinical signs, seroconversion, characteristic light microscopic lesions and the demonstration of PRRSV by virus isolation, fluorescent antibody (FA) examination, immunohistochemistry, in situ hybridization, or PCR (Rossow et al., 1996, Rossow, 1998).
In June of 2006, outbreaks of highly pathogenic (acute, atypical) PRRS in most areas of China have been suggested to be caused by highly virulent Chinese-type PRRSV (H-PRRSV) strains. From January to July 2007, 39455 morbid pigs died among 143,221 infected pigs according to the administrative files. Rapid spread of the viruses and their persistence in some environments have made the control of outbreaks difficult and, at times, even impossible. A rapid, sensitive, specific diagnostic method would be extremely useful for the diagnosis and control of the highly virulent PRRSV in China. The present highly virulent PRRSV of Chinese-type isolate which lead to widespread epidemic in China has 87 bps deleted in the fixed region of its Nsp2 gene as compared to normal strains (Tian et al., 2007). The current immunoassay is not capable of differentiating this mutant strain from the others, furthermore, the traditional RT-PCR method relies upon electrophoresis of the nucleic acids in the presence of ethidium bromide and visual or densitometric analysis of the resulting bands after irradiation by ultraviolet light (Kidd et al., 2000). Real-time RT-PCR use specific fluorescence-labeled probes or primers to monitor the accumulating amplicon in real time, real time and automatic testing saves time. Real-time RT-PCR is valuable for detecting various pathogens around the globe, built on the data generated by conventional RT-PCR (Matthews and Kricka, 1990, Lomeli et al., 1989, Bustin, 2000).
The development and the validation of a real-time RT-PCR for highly virulent Chinese-type isolate of PRRSV based on its characteristics of Nsp2 gene fragment are described. The established method was shown to be sensitive and reliable, and will be critical for the prevention and control of the highly virulent PRRS.
2. Materials and methods
2.1. Virus strains and cell
Five strains of highly virulent Chinese-type PRRSVs (Table 2) used in this experiment which were identified by virus isolation and sequencing were derived originally from the serum of clinically affected pigs. The virus strains were propagated twice in MARC-145 cell line at 37 °C and preserved at −80 °C. Marc-145 cells were grown in Eagle's minimal essential medium (EMEM) with 5% fetal calf serum at 37 °C in a humidified 5% CO2 atmosphere. Twenty four strains of other viruses used to confirm the specificity of the developed real-time RT-PCR assay were attained from different Animal Disease Control and Prevention Center (Table 2).
Table 2.
Specificity of real-time RT-PCR for highly virulent Chinese-type PRRSVa
| Genus | Strainb (source, No.) | Original source | Result c |
|---|---|---|---|
| Arterivirus | Highly virulent Chinese-type PRRSV ADCPC 101 | ChongQing, 2006 | + |
| Highly virulent Chinese-type PRRSV ADCPC 123 | ChongQing, 2006 | + | |
| Highly virulent Chinese-type PRRSV ADCPC 131 | ChongQing, 2006 | + | |
| Highly virulent Chinese-type PRRSV ADCPC 145 | ChongQing, 2006 | + | |
| Highly virulent Chinese-type PRRSV ADCPC 152 | ChongQing, 2006 | + | |
| Normal PRRSV ADCPC 1001 | ChongQing, 2001 | − | |
| Normal PRRSV ADCPC 1120 | ChongQing, 2003 | − | |
| Normal PRRSV ADCPC 1321 | ChongQing, 2004 | − | |
| Normal PRRSV ADCPC 1542 | ChongQing, 2005 | − | |
| Normal PRRSV LNCIQ b53 | ShengYang, 2004 | − | |
| Normal PRRSV LNCIQ b55 | ShengYang, 2005 | − | |
| Normal PRRSV SZCIQ 0131 | ShenZheng, 2005 | − | |
| Pestivirus | Classical Swine Fever Virus (CSFV) HNCIQ 3–125 | HuNan, 2004 | − |
| Classical Swine Fever Virus (CSFV) HNCIQ 3–121 | HuNan, 2005 | − | |
| Bovine viral diarrhea virus (BVDV) LNCIQm31 | LiaoNing, 2005 | − | |
| Bovine viral diarrhea virus (BVDV) SHCDC 0260 | Shanghai, 2005 | − | |
| Bovine viral diarrhea virus (BVDV) SHCDC 0317 | Shanghai, 2005 | − | |
| Parvovirus | Porcine parvovirus (PPV) SZCIQ 0138 | ShenZheng, 2004 | − |
| Porcine parvovirus (PPV) SZCIQ 0219 | ShengZheng, 2004 | − | |
| Varicellovirus | Porcine pseudorabies virus (PRV) GZCDC 0564 | Guangdong, 2004 | − |
| Porcine pseudorabies virus (PRV) GZCDC 0958 | Guangdong, 2004 | − | |
| Porcine pseudorabies virus (PRV) ADCPC 1983 | Sichuan, 2005 | − | |
| Coronavirus | Transmissible gastroenteritis virus (TGEV) GZCDC 0874 | Guangzhong, 2006 | − |
| Transmissible gastroenteritis virus (TGEV) GZCDC 0368 | Guangzhong, 2006 | − | |
| Transmissible gastroenteritis virus (TGEV) HNCIQ 7-303 | HuNan, 2005 | − | |
| Circovirus | Porcine circovirus 2 (PCV-2) SZCIQ 0165 | ShenZheng, 2005 | − |
| Porcine circovirus 2 (PCV-2) LNCIQ404 | LiaoNing, 2005 | − | |
| Enterovirus | Swine Vesicular Disease Virus (SVDV) SZCDC 0358 | ShengZheng, 2004 | − |
| Swine Vesicular Disease Virus (SVDV) ADCPC397 | Chongqing, 2005 | − | |
ADCPC, Chongqing Animal Disease Control and Prevention Center, Chongqing, China; LNCIQ, Liaoning Entry-Exit Inspection and Quarantine Bureau, Dalian, China; SZCIQ, Shenzhen Entry-Exit Inspection and Quarantine Bureau, Shenzhen, China; HNCIQ, Hunan Entry-Exit Inspection and Quarantine Bureau, Changsha, China; SHCDC, Shanghai Center for Disease Control and Prevention, Shanghai, China. GZCDC, Guangzhou Center for Disease Control and Prevention, Guangzhou, China; SZCDC, Shenzhen Center for Disease Control and Prevention, Shenzhen, China.
The type of these virus strains had been determined by serological assay or virus isolation, especially highly virulent Chinese-type PRRSV were identified by sequencing.
Results of real-time RT-PCR: +, positive result; −, negative result.
2.2. Specimens collection
Two hundred and thirteen lung, spleen and blood serum specimens were obtained from 22 outbreaks of suspected highly virulent Chinese-type PRRS from March to June in 2007 (Table 4).
Table 4.
Detection results of 213 suspected positive samples
| Real-time PCR/sequencing/virus isolation | Lung (76) | Spleen (54) | Serum (83) |
|---|---|---|---|
| Positive/positive/positive | 47 | 31 | 63 |
| Negative/negative/positive | 6 | 8 | 3 |
| Positive/negative/positive | 0 | 0 | 0 |
| Negative/positive/positive | 0 | 0 | 0 |
| Positive/positive/negative | 2 | 1 | 2 |
| Negative/negative/negative | 11 | 14 | 15 |
| Positive/negative/negative | 0 | 0 | 0 |
| Negative/positive/negative | 0 | 0 | 0 |
| Positive ratio | 64.47% | 59.26% | 78.31% |
2.3. RNA extraction
Viral RNA extracted from samples submitted for diagnostic assays was performed by using either Trizol (Invitrogen, Carlsban, CA) or the Qiagen viral RNA kit (Qiagen, Inc.) according to the manufacturer's instructions. Extractions of RNA from clinical tissue specimens were performed with the Trizol reagent, for the virus-infected cell culture and blood serum RNA was extracted by the Qiagen viral RNA kit. RNAs were eluted with 40 μl of diethyl pyrocarbonate-treated water and kept at −80 °C until used for real-time RT-PCR.
2.4. Primers and probe design
The difference of genome sequence between the highly virulent Chinese-type PRRSV and normal PRRSV was the 87 base deletion in the fixed site in Nsp2 gene (Tian et al., 2007). Nsp2 gene sequences of normal PRRSV strains and H-PRRSV strains were analyzed to generate similarity plot (Fig. 1 ), from which a pair of primers and a TaqMan fluorogenic probe were designed and shown in Table 1 . The TaqMan probe, was labeled with a reporter fluorescent dye 6-carboxyfluorescein (FAM) in 5′ end, and with a quencher fluorescent dye, 6-carboxy-tetramethyl-rhodamine (TAMRA) in 3′ end. The primers and probe were synthesized by ShangHai Chaoshi Biotechnology Co.
Fig. 1.
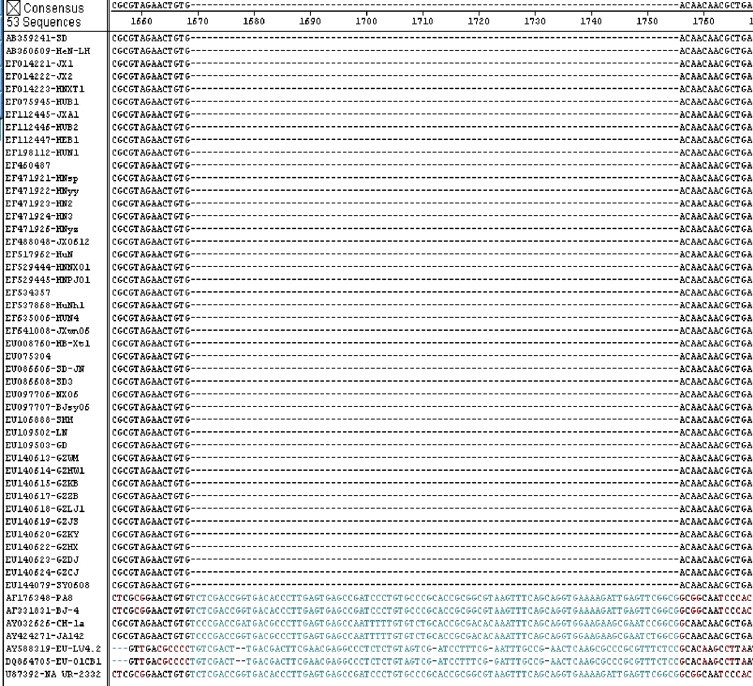
Alignment of NSP2 gene sequence. All the PRRSV sequences were derived from GenBank. The deleted sequence of H-PRRSV is shown in blue, normal PRRSV strains include NA origin (U87392, VR-2332), Asian origin (AF331831, BJ-4; AY032626, CH-1a) and EU origin (AY588319, LV4.2.1; DQ854705, 01CB1).
Table 1.
Nucleotide sequences of primers and fluorogenic probe
| Primer or probea | Sequence (5′ → 3′)b | Tm (°C)c | Locationd within the genome | Size (bp) | GC content (%) |
|---|---|---|---|---|---|
| Forward primer | CCCAAGCTGATGACACCTTTG | 59.2 | 2869–2889 | 21 | 52.38% |
| Reverse primer | AATCCAGAGGCTCATCCTGGT | 58.6 | 2945–2965 | 21 | 52.38% |
| Probe | FAM-CGCGTAGAACTGTGACA ACAACGCTGA-TAMRA | 68.8 | 2915–2941 | 27 | 51.85% |
Primers and probe were designed by using Primer Express V2.0 software.
FAM fluorescent reporter dye and with TAMRA the quencher dye.
Melting temperature estimated by Primer Express 2.0 primer test document, dyes are not accounted for in the calculations.
Nucleotide positions are based on GenBank EF641008.
2.5. Virus isolation
Virus isolation was performed on monolayers of 1–3-day-old MARC-145 cells in 24-well plates. 213 serum and tissue samples were obtained from 22 outbreaks of suspected highly virulent Chinese-type PRRS from March to June in 2007. 400 μl of serum or 250 μl of tissue homogenate plus 150 μl of MEM with 50 μg of gentamicin per ml were inoculated into the cells and incubated for 1 h at 37 °C with 5% CO2. Specific procedures referred to the method (Wills et al., 2003).
2.6. Real-time RT-PCR
Reagents from one step RT-PCR kit (TAKARA Bio Inc.) were used to prepare master mixture recipes according to the guidelines of the manufacturer for individual component concentrations, and both RT and DNA polymerization were carried out in the same tube without subsequent addition of enzymes or buffer. The RT-PCR master mixes of 25 μl-volume consisted of 2.5 μl 10× buffer, 0.5 μl Taq polymerase (5 U/μl), 0.5 μl reverse transcriptase (5 U/μl), 0.5 μl RNase inhibitor (40 U/μl), 5 μl RNA template, 2.5 μl dNTP (10 mM), forward and reverse primers, probe, Mgcl2 (25 mM) and nuclease-free water was used to give a total volume of 25 μl for each reaction tube, master mixes were maintained on ice at all times prior to real-time RT-PCR. In preliminary studies, concentrations of 0.8–1.5 μl forward primer (10 μM), 1.0–2.0 μl reverse primer (10 μM), 0.5–1.2 μl probe (10 μM), 2–5 μl MgCl2 (25 mM) were, respectively, evaluated to give the optimal results with the highest amplification efficiency, meanwhile, cycling parameters including the temperatures for the annealing, extension, and denaturation steps were also optimized.
3. Results
3.1. Optimization of real-time RT-PCR
For optimal conditions of the real-time RT-PCR for the H-PRRSV, the final RT-PCR mixture for a 25 μl-volume assay of the following reaction components was prepared to the indicated end-concentration: 1.0 μl forward primer (10 μM), 1.5 μl reverse primer (10 μM), 0.8 μl probe (10 μM), 3 μl Mgcl2 (25 mM), 2.5 μl dNTP (10 mM), 2.5 μl 10× buffer, 0.5 μl Taq polymerase (5 U/μl), 0.5 μl reverse transcriptase (5 U/μl), 0.5 μl RNase inhibitor (40 U/μl), 5 μl RNA template, 7.2 μl water. Optimized experimental run protocol for ABI7500 Sequence Detector System was used as follows: reverse transcript program (50 °C for 30 min), denaturation program (95 °C for 3 min), amplification program repeated 40 times (95 °C for 5 s, 60 °C for 40 s) and a single fluorescence FAM measurement was set at 60 °C.
H-PRRSV RNA and a without template as negative control reaction mixture were performed with the above protocol, the amplification result was illustrated in Fig. 2 . Positive real-time RT-PCR result was obtained with the H-PRRSV sample.
Fig. 2.
Real-time RT-PCR assay for highly virulent Chinese-type PRRSV.
3.2. Specificity of real-time RT-PCR
Specificity of the real-time RT-PCR was determined by analyzing 29 different virus strains. H-PRRSV strains have been identified by sequencing and the other virus strains were verified by a serological method or a PCR method. Viral RNA extracted from H-PRRSV-infected cell supernatants with approximate viral titers of 105 TCID50/ml were tested with the real-time RT-PCR assay. All H-PRRSVs were detected with positive results (Table 2 ). Positive fluorogenic signals were observed when testing RNA obtained from tissue culture supernatant of Marc-145 cells infected with H-PRRSV; no positive fluorogenic signal was observed in the real-time RT-PCR for other viral RNA (Table 2), which showed 100% specificity for the selected panel; and no cross reactions were observed in this real-time RT-PCR.
3.3. Sensitivity of real-time RT-PCR
To assess the sensitivity of the assay developed, H-PRRSV titers in clinical samples (identified by sequencing) were determined using 96-well plates with Marc-145 cells, and titers were calculated by the method of Reed and Muench and expressed as TCID50/ml (Reed and Muench, 1938). The samples were diluted in 10-fold in Eagle's minimal essential medium; RNA of H-PRRSV samples with 0.1–105 TCID50/ml were extracted and tested to determine the end-point dilution at which a positive amplification signal could be obtained. The detection sensitivity was defined as the minimum TCID50/ml that can be detected; a PCR reaction was considered positive if the cycle threshold (Ct) level was obtained at <40 cycles. The results (Fig. 3 ) demonstrated that the sensitivity of the assay was 1 TCID50/ml. Standard curve (Fig. 4 ) was obtained which displays the cycle threshold value versus different dilutions of 1–105 TCID50/ml samples. The coefficient of determination (R 2) was >0.99 and the slope of the curve was −3.349 which indicate a high PCR efficiency of the experiment.
Fig. 3.
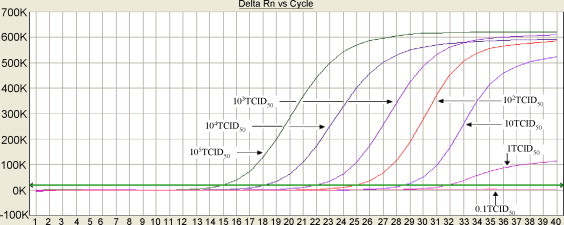
Sensitivity detection of real-time RT-PCR assay for highly virulent Chinese-type PRRSV.
Fig. 4.
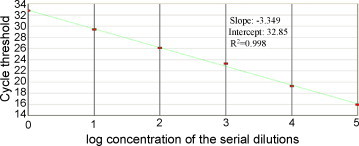
Standard curve was generated with the log10 TCID50/ml samples against the corresponding Ct value, the linear relationship was observed with 105–1 TCID50/ml H-PRRSV sample.
3.4. Intra- and inter-assay variability of real-time RT-PCR
Intra-assay reproducibility of the developed real-time RT-PCR was assessed with four replicates each of a 10-fold dilution series (105–100 TCID50/ml H-PRRSV samples), Ct values obtained from different dilution series were used for calculation; the intra-assay coefficient of variation (% CV) ranged from 0.5 to 2.3 (Table 3 ); the inter-assay reproducibility were determined with 10-fold dilution series (105–100 TCID50/ml H-PRRSV samples) on four separate days; the %CV of the inter-assay ranged from 0.9 to 2.5 (Table 3); the %CV of both the intra-assay and inter-assay indicated the repeatability of the assay developed and the sensitivity of the assay could be confirmed to 1 TCID50/ml.
Table 3.
Intra-and inter-assay reproducibility of real-time RT-PCR
| Concentration of specimens (TCID50/ml) | Replicate (Cta value) |
Ct mean (S.D.b) | %CVc | |||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |||
| Intra-assay | ||||||
| 105 | 15.68 | 15.54 | 15.41 | 15.38 | 15.50 (0.14) | 0.9 |
| 104 | 19.01 | 18.54 | 18.98 | 18.72 | 18.81 (0.23) | 1.2 |
| 103 | 21.23 | 22.43 | 22.12 | 21.94 | 21.93 (0.51) | 2.3 |
| 102 | 25.23 | 25.36 | 26.02 | 25.43 | 25.51 (0.35) | 1.4 |
| 101 | 28.56 | 28.39 | 28.12 | 28.40 | 28.37 (0.17) | 0.6 |
| 100 | 31.98 | 31.77 | 32.12 | 31.95 | 31.96 (0.16) | 0.5 |
| Inter-assay | ||||||
| 105 | 14.98 | 15.72 | 15.47 | 15.84 | 15.50 (0.39) | 2.5 |
| 104 | 18.89 | 18.62 | 18.63 | 18.33 | 18.62 (0.22) | 1.2 |
| 103 | 22.31 | 22.58 | 22.21 | 22.02 | 22.28 (0.22) | 1.0 |
| 102 | 25.13 | 25.46 | 25.71 | 24.98 | 25.32 (0.33) | 1.3 |
| 101 | 28.10 | 28.64 | 28.01 | 28.34 | 28.27 (0.25) | 0.9 |
| 100 | 31.25 | 31.76 | 32.53 | 31.89 | 31.86 (0.54) | 1.7 |
Cycle threshold value.
Standard deviation.
Coefficient of variation = S.D./Ct mean.
3.5. Analysis of clinical samples
Real-time RT-PCR was used to detect RNA from lung, spleen and serum specimens which were obtained from symptomatic individuals. H-PRRSV RNA was detected in 65 of 83 serum specimens, 49 of 76 lung specimens, 32 of 54 spleen specimens by real-time RT-PCR. The average Ct values from positive serum specimens, spleen specimens and lung specimens were 20.44, 24.87, 23.41, respectively. All these samples were detected using virus isolation and sequencing simultaneously. Briefly, virus was isolated with Marc-145 cells and identified with monoclonal antibody as described previously, H-PRRSV were determined by sequencing of partial Nsp2 gene fragment with the forward primer and reverse primer. The results (Table 4 ) obtained by real-time RT-PCR and sequencing method indicated further the accuracy of the developed method.
4. Discussion
Real-time PCR has been used widely in the field of developmental studies (Steuerwald et al., 2000), clinical diagnosis and the detection of very low copy numbers of gene or gene products (Fujimaki et al., 2000, Kreuzer et al., 2000, Pfitzner et al., 2000, Zerr et al., 2000). The nucleotide sequence characteristics of H-PRRSV is that it has 87 bps deleted in the fixed region of Nsp2 gene by comparison to that of normal PRRSV, Thus, it provides an ideal site for designing a probe for real-time RT-PCR assay for H-PRRSV. In this paper, the TaqMan probe is oligonucleotides designed to bind to complementary target sequences presented within the Nsp2 gene of highly virulent Chinese-type PRRSV. No probe binding to the Nsp2 gene sequence of normal PRRSV due to the addition of 87 bps in this region by comparison to that of H-PRRSV. There are no other reports of real-time RT-PCR techniques to differentiate H-PRRSV strains from other strains of PRRSV due to the similarity of the target sequence between H-PRRSV and normal PRRSV, the method developed could specifically detect the highly virulent Chinese-type PRRS virus that has 87 bps deleted in the fixed region of Nsp2 gene.
Two hundred and thirteen specimens were detected by the real-time RT-PCR. Results are 100% consistent with that of sequencing, no false-positive samples were found, and the serum sample which obtained the lower Ct value but higher positive ratio as 78.31% may be due to the higher level of viral load in serum. The other reason may be attributed to the different extraction protocol between serum and tissues. Finally we can conclude that the lower Ct value and higher positive ratio clearly indicated that compared with tissue from lung and spleen, serum has a higher predicative value of detecting H-PRRSV infection during surveillance screening. In the comparison of the real-time RT-PCR and virus isolation for H-PRRSV, 5 strains of the H-PRRSV were positive by real-time RT-PCR but negative by virus isolation, it may due to that VI has a lower sensitivity or the infection of H-PRRSV is in its window period. Virus was isolated from seventeen samples were positive but real-time RT-PCR-negative, the final identification by RT-PCR for PRRSV showed that they belong to the typical PRRSV.
The sensitivity of the method developed is up to 1 TCID50/ml. The slope value of standard curve indicated the high amplification efficiency, and further, low coefficient of variation for inter and intra-assay also confirmed the repeatability of the assay. The high specificity and sensitivity of the method demonstrated the practicality of real-time RT-PCR for the detection of H-PRRSV. The present identification assay for H-PRRSV by virus isolation was tedious because of the cell culture and sequencing. While, the assay described in this paper, a fluorogenic-probe hydrolysis (TaqMan)-real-time RT-PCR for rapid detection of H-PRRSV is developed with high specificity, sensitivity and reproducibility. The simplicity and accuracy of the assay make it a powerful tool for the detection and control of highly virulent Chinese-type PRRSV.
Acknowledgements
This work was partly supported by Zeng zheng from Chongqing Animal Disease Control and Prevention Center and a grant from Shenzhen Taitai genomics, Inc.
References
- Albina E. Epidemiology of porcine reproductive and respiratory syndrome (PRRS): an overview. Vet. Microbiol. 1997;55:309–316. doi: 10.1016/s0378-1135(96)01322-3. [DOI] [PubMed] [Google Scholar]
- Bustin S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- Christopher-Hennings J., Nelson E.A., Nelson J.K., Hines R.J., Swenson S.L., Hill H.T., Zimmerman J.J., Katz J.B., Yaeger M.J., Chase C.C. Detection of porcine reproductive and respiratory syndrome virus in boar semen by PCR. J. Clin. Microbiol. 1995;33:1730–1734. doi: 10.1128/jcm.33.7.1730-1734.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimaki S., Funato T., Harigae H., Imaizumi M., Suzuki H., Kaneko Y., Miura Y., Sasaki T. A quantitative reverse transcriptase polymerase chain reaction method for the detection of leukaemic cells in peripheral blood. Eur. J. Haematol. 2000;64:252–258. doi: 10.1034/j.1600-0609.2000.90091.x. [DOI] [PubMed] [Google Scholar]
- Halbur P.G., Paul P.S., Meng X.J., Lum M.A., Andrews J.J., Rathje J.A. Comparative pathogenicity of nine US porcine reproductive and respiratory syndrome virus (PRRSV) isolates in a five-week-old cesarean-derived, colostrum-deprived pig model. J. Vet. Diagn. Invest. 1996;8:11–20. doi: 10.1177/104063879600800103. [DOI] [PubMed] [Google Scholar]
- Kapur V., Elam M.R., Pawlovich T.M., Murtaugh M.P. Genetic variation in porcine reproductive and respiratory syndrome virus isolates in the midwestern United States. J. Gen. Virol. 1996;77:1271–1276. doi: 10.1099/0022-1317-77-6-1271. [DOI] [PubMed] [Google Scholar]
- Keffaber K.K. Reproductive failure of unknown etiology. Am. Assoc. Swine Pratt News. 1989;11:1–9. [Google Scholar]
- Kidd I.M., Clark D.A., Emery V.C. A non-radioisotopic quantitative competitive polymerase chain reaction method: application in measurement of human herpesvirus 7 load. J. Virol. Methods. 2000;87:177–181. doi: 10.1016/s0166-0934(00)00164-6. [DOI] [PubMed] [Google Scholar]
- Kreuzer K.A., Lass U., Nagel S., Ellerbrok H., Pauli G., PawlaczykPeter B., Siegert W., Huhn D., Schmidt C.A. Applicability of an absolute quantitative procedure to monitor intra-individual bcr/abl transcript kinetics in clinical samples from chronic myelogenous leukemia patients. Int. J. Cancer. 2000;86:741–746. doi: 10.1002/(sici)1097-0215(20000601)86:5<741::aid-ijc22>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Lomeli H., Tyagi S., Pritchard C.G., Lizardi P.M., Kramer F.R. Quantitative assays based on the use of replicatable hybridization probes. Clin. Chem. 1989;35:1826–1831. [PubMed] [Google Scholar]
- Matthews J.A., Kricka L.J. Minimizing the time required for DNA amplification by efficient heat transfer to small samples. Anal. Biochem. 1990;169:1–25. doi: 10.1016/0003-2697(90)90090-v. [DOI] [PubMed] [Google Scholar]
- Meng X.J. Heterogeneity of porcine reproductive and respiratory syndrome virus: implications for current vaccine efficacy and future vaccine development. Vet. Microbiol. 2000;74:309–329. doi: 10.1016/S0378-1135(00)00196-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith M.S. European ed. Pig Disease and Information Centre; Cambridge: 1995. Porcine Reproductive and Respiratory Syndrome (PRRS) pp. 3–12. [Google Scholar]
- Pfitzner T., Engert A., Wittor H., Schinkothe T., Oberhauser F., Schulz H., Diehl V., Barth S. A real-time PCR assay for the quantification of residual malignant cells in B cell chronic lymphatic leukemia. Leukemia. 2000;14:754–766. doi: 10.1038/sj.leu.2401706. [DOI] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty percent end points. Am. J. Hyg. 1938;27:709–716. [Google Scholar]
- Rossow K.D., Benfield D.A., Goyal S.M., Nelson E.A., Christopher-Hennings J., Collins J.E. Chronological immunohistochemical detection and localization of porcine reproductive and respiratory syndrome virus in gnotobiotic pigs. Vet. Pathol. 1996;33:551–556. doi: 10.1177/030098589603300510. [DOI] [PubMed] [Google Scholar]
- Rossow K.D. Porcine reproductive and respiratory syndrome. Vet. Pathol. 1998;35:1–20. doi: 10.1177/030098589803500101. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Meulenberg J.J.M. The molecular biology of arteriviruses. J. Gen. Virol. 1998;79:961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- Steuerwald N., Cohen J., Herrera R.J., Brenner C.A. Quentification of mRNA in single oocytes and embroyos by real-time rapid cycle fluorescence monitored RT-PCR. Mol. Hum. Reprod. 2000;64:252–258. doi: 10.1093/molehr/6.5.448. [DOI] [PubMed] [Google Scholar]
- Stevenson G.W., Van Alstine W.G., Kanitz C.L., Keffaber K.K. Endemic porcine reproductive and respiratory syndrome virus infection of nursery pigs in two swine herds without current reproductive failure. J. Vet. Diagn. Invest. 1993;5:432–434. doi: 10.1177/104063879300500322. [DOI] [PubMed] [Google Scholar]
- Tian K., Yu X., Zhao T., Feng Y., Cao Z. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS ONE. 2007;2(6):e526. doi: 10.1371/journal.pone.0000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills R.W., Doster A.R., Galeota J.A., Sur J.-H., Osorio F.A. Duration of infection and proportion of pigs persistently infected with porcine reproductive and respiratory syndrome virus. Clin. Microbiol. 2003;41:58–62. doi: 10.1128/JCM.41.1.58-62.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerr D.M., Huang M.L., Corey L., Erickson M., Parker H.L., Frenkel L.M. Sensitive method for detection of human herpesviruses 6 and 7 in saliva collected in field studies. J. Clin. Microbiol. 2000;38:1981–1983. doi: 10.1128/jcm.38.5.1981-1983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman J.J., Yoon K.J., Wills R.W., Swenson S.L. General overview of PRRSV: a perspective from the United States. Vet. Microbiol. 1997;55:187–196. doi: 10.1016/s0378-1135(96)01330-2. [DOI] [PubMed] [Google Scholar]


