Abstract
Deubiquitylating enzymes or DUBs are a class of enzymes that selectively remove the polypeptide posttranslational modification ubiquitin from a number of substrates. Approximately 100 DUBs exist in human cells and are involved in key regulatory cellular processes, which drive many disease states, making them attractive therapeutic targets. Several aspects of DUB biology have been studied through genetic knock-out or knock-down, genomic, or proteomic studies. However, investigation of enzyme activation and regulation requires additional tools to monitor cellular and physiological dynamics. A comparison between genetic ablation and dominant-negative target validation with pharmacological inhibition often leads to striking discrepancies. Activity probes have been used to profile classes of enzymes, including DUBs, and allow functional and dynamic properties to be assigned to individual proteins. The ability to directly monitor DUB activity within a native biological system is essential for understanding the physiological and pathological role of individual DUBs. We will discuss the evolution of DUB activity probes, from in vitro assay development to their use in monitoring DUB activity in cells and in animal tissues, as well as recent progress and prospects for assessing DUB inhibition in vivo.
Key words: ABP, Activity-based probe, Acyloxymethyl ketone, DUB, Deubiquitylating enzyme, Hemagglutinin, JAMM, JAB1/Mov34/Mpr1 Pad1 N-terminal + protease, MJD, MACHADO-Josephin domain proteas, MP, Mpr1/Pad1 N-terminal+, OTU, Ovarian tumor protease, PA, Propargy, SENP, Sentrin-specific protease, Ubl, Ubiquitin-like protein, UCH, Ubiquitin C-terminal hydrolase, USP, Ubiquitin-specific protease, VMS, Vinyl methyl sulfone, VME, Vinyl methyl ester
Ubiquitin–Proteasome System
Protein homeostasis is essential for most cellular processes. The ubiquitin–proteasome system is responsible for much of the regulated proteolysis in the cell, as well as many other regulatory processes such as transcriptional regulation, DNA damage, quality control, trafficking, inflammation, and autophagy. Ubiquitin is a small 76-amino acid protein that can be reversibly attached to protein substrates. Several ubiquitin-like proteins (Ubls) have also been identified including ISG15, NEDD8, and SUMO, which share a characteristic three-dimensional fold with ubiquitin but are otherwise distinct. The ubiquitin–proteasome system has multiple essential biological roles, and thus its function and dysfunction, are important factors in various human diseases, including cancer, infection, inflammation, and neurodegeneration [1–4].
Ubiquitylation of substrate proteins first involves an ATP-dependant activation of the ubiquitin polypeptide by the activating enzyme E1. Activation involves covalent linkage between the carboxy terminus of ubiquitin and a cysteine residue present on the E1, forming a thioester bond. The activated ubiquitin is then transferred to an E2 ubiquitin-conjugating enzyme forming a thioester linkage. In the final step, an E3 ligase transfers the ubiquitin from the E2 to the substrate protein. The majority of E3 ligases are classified as RING finger E3s and act by bringing the substrate and E2 enzyme in close proximity. The RING finger E3s directly transfer ubiquitin from the E2 to the substrate. The HECT domain E3s act by forming an intermediate thioester linkage with ubiquitin before transfer to the substrate (reviewed in [5]). More recently, a third class of E3 ligases with an intermediate mechanism of action has been identified. The RING-in-between-RING (RBR) E3s are an unusual family of ubiquitin E3-ligases composed of a dozen proteins. Their activities are autoinhibited, causing a requirement for activation by protein–protein interactions or posttranslational modifications. They catalyze ubiquitin conjugation by a concerted RING/HECT-like mechanism in which the RING1 domain facilitates E2-discharge to directly form a thioester intermediate with a cysteine in RING2. This short-lived, HECT-like intermediate then modifies the target [6, 7].
Following monoubiquitylation of a substrate, the process can either stop, forming monoadducts of ubiquitin, or be repeated forming an elongated chain of ubiquitin residues. Polyubiquitin chains can be formed using the N-terminus (linear) or any of the seven internal lysine residues found in ubiquitin, and these various chain topologies lead to different functional outcomes. Of the most well studied linkages, K63-linked polyubiquitin chains are often involved in nonproteolytic signal transduction while K48-linked chains generally target substrates for proteasomal degradation. A number of additional linkages such as Met1, K6, K11, K27, K29, and K33 have been identified and their nondegradative cellular signaling roles are still subject to a number of investigations. The complexity of ubiquitin chain signaling is further enhanced by the existence of mixed-lineage chains [8, 9].
Proteins destined for degradation via the ubiquitin–proteasome system include proteins that are damaged, improperly folded, or that have short half-lives [10]. Proteins that have been appropriately polyubiquitylated are recognized and degraded by the 26S macromolecular proteasome complex [11]. The 26S complex consists of a 20S catalytic core particle that is capped at both ends by 19S regulatory particles. The 19S regulatory particle can be further subdivided into lid and base components. Following recruitment to the proteasome, polyubiquitylated proteins undergo deubiquitylation and unfolding. The removal of ubiquitin is accomplished by deubiquitylating enzymes (DUBs) associated with the 19S lid. Ubiquitin polypeptides that are removed from substrate proteins can be directly recycled by the cell. The 19S base component plays a key role in the unfolding of the substrate protein and delivery of the deubiquitylated, unfolded protein into the 20S catalytic core particle. The 20S consists of four layers of ring-like structures [12]. The outer rings are composed of seven α subunits with the inner rings composed of seven β subunits. The β1 subunits exhibit caspase-like activity, the β2 subunits trypsin-like activity, and β5 subunits chymotrypsin-like activity, collectively degrading proteins into short oligopeptides as well as recycling amino acids [5, 13].
DUBs
Ubiquitin is covalently linked to many cellular proteins and regulates their activity, stability, localization, or interactions. Ubiquitylation is a reversible process carried out by the opposing activities of ubiquitin ligases and DUBs. The human genome encodes approximately 100 DUBs [14–16]. Of the five families of DUBs, four ( UCH, USP, MJD, and OTU) belong to the cysteine peptidase class, while one (JAMM) belongs to the metallopeptidase class. As DUBs have been shown to play critical roles in many pathological processes, particularly cancer, infectious disease, and neurodegeneration, they have begun to attract significant attention from the pharmaceutical industry [17–20]. Unlike most posttranslational modifications, ubiquitin is able to form polymeric chains [21]: the ubiquitin linkage in the chain as well as the length of the chain will impact on the fate of the protein modified by the polymer of ubiquitin [9, 22].
Pharmacological modulation of DUBs using a multitude of approaches in the last decade has seen limited success to date; however, recent progress is beginning to identify DUB inhibitors with the potential for drug development [23–27]. A number of conceptual and technological obstacles need to be overcome in order to progress genuine DUB therapies. A major challenge in characterization of DUB inhibitors is the development of high throughput assays monitoring “on-target” inhibition in cells and in vivo. Monitoring DUB target engagement by small molecule inhibitors in vivo has a number of implications. Firstly, as a biomarker readout of inhibition and for understanding the physiological implication of inhibiting a class of enzymes for which there is usually no known unique ubiquitylated substrate. Secondly, for assessment of the selectivity of compounds as well as understanding the mechanism of action of the inhibition, including duration, reversibility, and pharmacodynamic parameters.
Activity Probes
Activity-based probes (ABPs) rely on the design of chemical warheads which selectively react with the active site of an enzyme. ABPs are usually composed of a reactive electrophile, to covalently modify an active-site residue, and a reporter group to allow detection of the labeled enzyme [28], see Fig. 1a, b. Activity probes have been designed for a number of enzyme classes such as serine hydrolases [29], metalloproteases [30, 31], proteasomes [32], and oxidoreductases [33]. Epitope-tagged ubiquitin and ubiquitin-like derivatives have been utilized in a variety of assays to identify or monitor active DUBs in biological samples [34, 35] (Fig. 1c). Ubiquitin ABPs have been instrumental in the identification of a number of new DUBs [36] including a novel class of DUBs: OTUs [37]. Unlike other proteolytic enzymes, for optimal recognition, DUBs require not only an electrophilic trap but also a very large portion of ubiquitin or chains of ubiquitin for binding and recognition in the enzyme active site: truncated portions of ubiquitin are usually not sufficient to trap DUBs. In addition, the isopeptide nature of the covalent linkage of ubiquitin to the target protein imposes a restricted number of choices of electrophilic warheads. Monitoring the activity of endogenous enzymes such as DUBs in their native, full-length status as well as under all possible naturally occurring posttranslational modifications or interference/allosteric regulation from binding partners is a major advantage of ABPs. The irreversible covalent nature of ABPs toward their enzyme targets has a number of advantages when compared to many other analytical technologies that rely on weak, naturally transient and difficult to capture interactions between an enzyme and its substrate. Various warheads (Fig. 2) have been employed including alkyl halides (chloroethyl, bromoethyl, bromopropyl), Michael acceptors ( vinyl methyl ester (VME), vinyl methyl sulfone (VMS), vinyl phenyl sulfone, vinyl cyanide) and more recently propargyl (PA) [36, 38, 39].
Fig. 1.
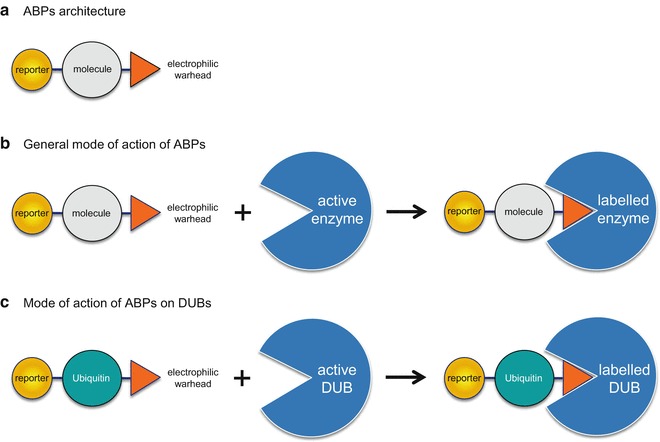
(a) General structure of an ABP consisting of a reporter (tag), specific molecule (protein), and warhead. (b) General mechanism of action of ABPs. Catalytically competent enzymes react with the electrophilic warhead resulting in a covalently labeled protein. (c) Mechanism of action for labeling DUBs by ubiquitin ABPs
Fig. 2.
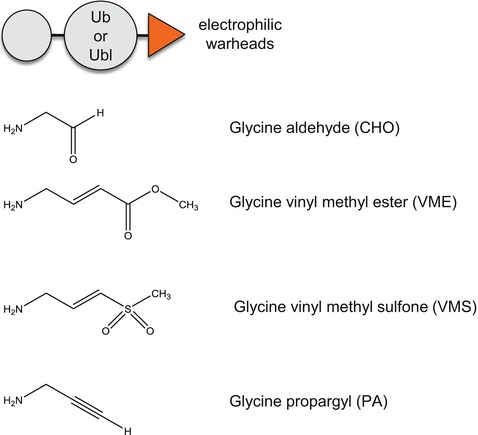
Common warheads used for ubiquitin ABPs
Activity Probes for MonitoringDUB Activity in Cells
The first attempt at generating activity probes to label DUBs on their catalytic site thiol group was described by Hidde Ploegh and colleagues [35]. Using a trypsin catalyzed transpeptidation to modify ubiquitin at its carboxy terminus with a vinyl sulfone group, they were able to demonstrate that ubiquitin vinyl sulfone labeled not only recombinant purified DUBs but also a number of yeast DUBs in a crude lysate. The identity of each labeled band was verified using individual yeast DUB mutant strains. The initial version of the ubiquitin vinyl sulfone probe was labeled with iodine125and allowed for detection of a number of DUBs in mouse tissues as well as in mouse cell lysates. In the same study, Borodovsky et al. described the use of unlabeled ubiquitin vinyl sulfone to detect a specific DUB by monitoring a shift in the apparent molecular weight in SDS-PAGE followed by immunoblotting: USP7 was labeled efficiently in mammalian cell lysates. Finally, the authors were also able to identify USP14 as a novel DUB associated with the proteasome thanks to the use of ubiquitin vinyl sulfone in fractionation and immune-purification assays.
In a second generation of activity probes, the thiol-reactive group was added to ubiquitin using an intein-based chemical ligation method [36]. The reactivity of the DUBs depends on the type of electrophilic warhead fused to ubiquitin. The second generation of probes were additionally used for the identification of bound DUBs by affinity purification/ mass spectrometry [34]. More recently, ABPs using a fluorescent reporter tag have been generated to replace the initial tags (e.g., HA) to allow replacement of the immunoblot procedure with fluorescent imaging [39–41].
While the historical production of ubiquitin ABPs was based on a trypsin catalyzed transpeptidation to modify ubiquitin at its carboxy terminus with a vinyl sulfone group or based on the addition of the electrophilic warhead via intein-based chemical ligation methods, recent approaches have moved toward the full-chemical synthesis of ubiquitin ABPs [41, 42]. This latest improvement has the added advantage of allowing the incorporation of modified amino acid residues at any position in the ABPs, whether natural or not.
In addition their major role in monitoring or identifying active DUBs in biological samples, ubiquitin-based probes are useful tools for structural analysis of DUBs. A number of cocrystals of DUBs with ubiquitin have been solved [34, 39] and in some cases, the structure of the apo-DUB was not achieved in the absence of the modified ubiquitin ABP [43]. ABPs are sometimes the only option available for cocrystallizing DUBs with ubiquitin substrates or ubiquitin chains.
Activity Probes for Monitoring DUB Activity in Tissues, Viruses, or Parasites
A limited number of studies have demonstrated the utility of activity probes for monitoring DUB activity in normal or diseased animal tissues. In earlier DUB activity probe publications, mouse tissues were examined and significant differences in the profile of active DUBs in tissues was observed [35]. More recently, in a very detailed study, Altun et al. investigated the activity of DUBs in models of aging and dietary restriction [44]. Dramatic differences in the levels of active DUBs in cell lines derived from various tissues as well as in primary tissues has been observed [45]. For a small number of DUBs, the activity as monitored by activity probe binding can be correlated with the malignant status of the cell line or tissue, suggesting a possible therapeutic window for DUB inhibitors [45]. Given the sensitivity and significance of such techniques, we can expect an increase in the number of studies taking full advantage of ubiquitin ABPs to monitor differential DUB activity in pathological versus normal conditions in the near future.
ABPs have been used to identify and monitor the activity of bacterial, viral, or parasitic DUBs including Herpes viridae, Chlamydia trachomatis, Toxoplasma gondii, and Plasmodium falciparum: ABPs are invaluable tools to identify functionally active DUBs in complex in sometimes relatively poor or difficult to annotate organisms [46–50]. While viruses or bacteria do not encode a full complement of ubiquitin proteasome enzyme systems, they express DUBs to evade the detection of their proteins by the immune system or otherwise enhance virulence [51]. In addition, since DUBs are essential for viral proliferation, viral DUBs have been considered as possible therapeutic strategies for the treatment of certain viral infections such as SARS or MERS [52].
Chemical Proteomics-Activity Probes for Characterizing DUB Inhibitors
Mass spectrometry has emerged as an important tool for characterizing the various forms of ubiquitin. Initial global characterization of the ubiquitin-modified proteome has been made possible in proteomic studies taking advantage of a monoclonal antibody that recognizes (di-Gly)-containing isopeptides following trypsin digestion of complex cellular lysates [53, 54]. In the Ubiquitin-AQUA approach, synthetic isotopically labeled internal standard peptides are used to quantify branched peptides and the branched -GG signature peptides generated by trypsin digestion of ubiquitin signals [55]. Proteomic studies looking at DUB interaction partners have also generated a great deal of information about their substrates, regulation, and function [56]. Additional studies have evaluated the functional role of DUBs using RNAi libraries [57, 58] or GFP-DUB fusions [59, 60], and have linked DUBs to specific cellular pathways. While such studies are very informative and have generated a wealth of data on the biological roles of DUBs, they provide only limited information regarding the dynamic activity profile of DUBs, and are not able to distinguish the catalytic state (active versus inactive) of DUBs. As the cellular activity of DUBs can be controlled by multiple factors including protein interactions [61], stoichiometric changes to the structure of the protein [62, 63], and posttranslational modifications [64, 65], the advantage of activity probes is their specific reactivity with catalytically active DUBs.
One of the benefits of ABPs for the characterization of DUB inhibitors is the ability to monitor compound selectivity. A chemical activity-based proteomic approach using HA-tagged ubiquitin labeled with electrophilic warheads (HA-UbBr2 or HA-Ub-VME) was undertaken to characterize the selectivity of two USP7 inhibitors either in immunoblots or by quantitative mass spectrometry following treatment of cells or cell lysates with compounds [66]. An independent study using another USP7 inhibitor displaying selectivity in a panel of biochemical DUB assays, was also subjected to cellular selectivity profiling using HA-Ub-VMS followed by immunoblotting [24]. In a more targeted approach, an active-site ubiquitin probe (HA-Ub-VMS) has been used to demonstrate that USP14/UCHL5 inhibition by a small molecule (b-AP15) inhibits the 19S proteasome in a reconstituted biochemical assay. A similar probe approach was also used to demonstrate that b-AP15 is not a general inhibitor of DUBs in a cell lysate probed with an anti-HA antibody detecting the conjugated ubiquitin species [67]. While the studies mentioned above are paving the way for elucidating DUB selectivity profiles in a cellular context, coverage of the “ DUBome” is still limited. Technological improvements are still required to increase sensitivity and accurately monitor DUBs in a given cell or tissue experiment.
While DUB proteomic studies using activity probes have mainly been used for monitoring the selectivity of first generation DUB inhibitors, the potential for ubiquitin ABPs is much broader. Indeed, it is possible to determine the dynamic nature of DUB inhibitors by using ABPs to monitor the reversibility or the duration of DUB inhibition. Furthermore, most of the work so far on DUBs using ABPs has been restricted to cellular studies. Recent progress in developing DUB inhibitors with in vivo preclinical potential is currently driving the tools for pharmacodynamic as well as mode-of-action understanding of DUB inhibitors in vivo. Activity probes based on selective inhibitors of peptidases have already been developed such as probes targeting proteasomes [68], cathepsins [69] or caspases [70] and are proving their usefulness for in vivo imaging studies as well as for diagnostic purposes [71].
Activity Probes for Ubiquitin-Like Deconjugating Enzymes
The utility of ubiquitin activity probes to identify and characterize DUBs in a number of conditions is not limited to ubiquitin. Indeed, probes for enzymes that remove Ubls have been generated. The exquisite selectivity of DUBs for their cognate substrates suggested that specific probes are also required for Ubl peptidases. An initial approach based on the synthesis of peptide vinyl sulfones harboring various portions of the ubiquitin-like carboxy terminus has suggested that truncated Ubls are able to bind Ubl-specific proteases in a manner similar to the ubiquitin-based vinyl sulfone polypeptides [72]. Ubl-based probes for Nedd8, SUMO-1, ISG15, GATE-16, MAP1-LC3, GABARAP, and Apg8L have been successfully synthesized [73–75].
An alternative to classical activity probes containing a full ubiquitin or ubiquitin-like polypeptide is based on the use of small molecule inhibitors to label the catalytic site of desumoylating enzymes (sentrin-specific proteases, SENPs). A peptide acyloxymethyl ketone (AOMK) containing a large aromatic O-acyl group are selective covalent inhibitors of SENPs and can be modified using fluorescent labels to detect SENPs activity in biological samples [76]. A similar approach has been described using a different family of proteins: glycine fluoromethylketones, which serve as probes to selectively target SENPs [77]. A more conventional derivatization of the carboxy-terminal end of Ubls with electrophilic warheads has also been pursued and a general derivatization procedure to produce any Ubl domain chemically activated at its C-terminus by formation of a thiol ester. Reaction of the thiol with a nucleophile produces the desired derivatives taking advantage of the intein fusion technology [78]. There is no technical challenges preventing the development of fully synthetic Ubl ABPs and indeed, a number of such reagents are already commercially available from various sources.
As the mechanism for the removal of Ubls by specific enzymes has not yet been fully characterized, ABPs will certainly play a key role in the elucidation of such understanding. Similarly, the biological or mechanistic functions of a number of DUBs or SENPs remains poorly understood, and existing ABPs or novel more selective ABPs can serve as tools for extending our knowledge.
Activity Probes Using Ubiquitin Chains or Modified Ubiquitin
In parallel with the development of monoubiquitin ABPs, a number of groups have also achieved total (semi)-synthesis of di-ubiquitin [42, 79–81] or even tetra-ubiquitin chains [82, 83]. However, incorporation of electrophilic warheads into polyubiquitin chains remains problematic. An intermediate approach to the generation of polyubiquitin ABPs was elaborated on the basis of the synthesis of branched-peptides incorporating an isopeptide-linked ubiquitin and an electrophilic warhead [84]. In addition, the synthesis and characterization of K48- or K63-linked di-ubiquitin probes bearing dehydroalanine as a warhead near the isopeptide bond has been described [85]. Finally, ABPs engineered for di-ubiquitin chains incorporating the 8 known ubiquitin linkages have been successful and now allow DUB ubiquitin-linkage specificity in a cellular context to be addressed[86–88]. Structural studies of DUBs with di-ubiquitin have demonstrated that in addition to the peptide flanking the ubiquitylated residues, more extensive interactions between DUBs and the proximal ubiquitin in the chain also contribute to the recognition by DUBs. Probing DUB selectivity with the latest generation of probes not only generates a distinct pattern from that obtained using mono-ubiquitin ABPs, but also suggests that the promiscuity of some DUBs for their substrates is probably much less pronounced than initially anticipated.
In the last couple of years, posttranslational modifications of ubiquitin, especially phosphorylation of ubiquitin at specific residues (e.g., Ser57 and Ser65) have been shown to play important roles in a number of cellular processes [89, 90]. Ubiquitin ABPs bearing the phosphorylated variants of ubiquitin have been generated and used to probe the selectivity of the modifications for conjugating and deconjugating enzymes. E1 and E2 enzymes are usually able to tolerate phosphorylated ubiquitin, however, a number of DUBs have difficulty recognizing the modified substrates [91, 92]. Studies evaluating additional posttranslational modifications of ubiquitin such as methylation, acetylation, hydroxylation, or other phosphorylation will certainly be unraveled in the near future: the corresponding ABPs will again serve as useful tools to understand the mechanistic and physiological role of novel variants of ubiquitin.
Activity Probes to Measure Target Engagement
A key issue facing researchers involved in deciphering the roles of DUBs in a cellular context is the lack of understanding of the most direct or relevant substrate of specific DUBs in a given cellular pathway. Some DUBs have very well characterized substrates (e.g., USP1 or USP7) [15] that are clearly linked to the function of the DUBs, however, the known substrate specificity is still relatively poor or partial at best for most DUBs. In certain cases, it is quite clear that unique substrates do not exist: e.g., USP14 or UCHL5 are DUBs that indiscriminately recognize any ubiquitylated substrates which is targeted to the proteasome [93]. Ubiquitin ABPs can play a critical role as tools to monitor the dynamics of the activation or inhibition of DUBs under specific physiological or pharmacological pathway alterations. The problem is especially acute for the monitoring of DUB activity upon inhibition with specific inhibitors: the pharmaceutical development of DUB inhibitors requires a good understanding of the pharmacokinetic modulation of the target upon treatment with compounds. The development of ABPs for proteomic evaluation of target engagement is currently being investigated by a number of groups. In addition, higher throughput ABP-based strategies are also under development for the determination of DUB target engagement in cellular contexts as well as in tissues or eventually for clinical sample evaluation (Fig. 3).
Fig. 3.
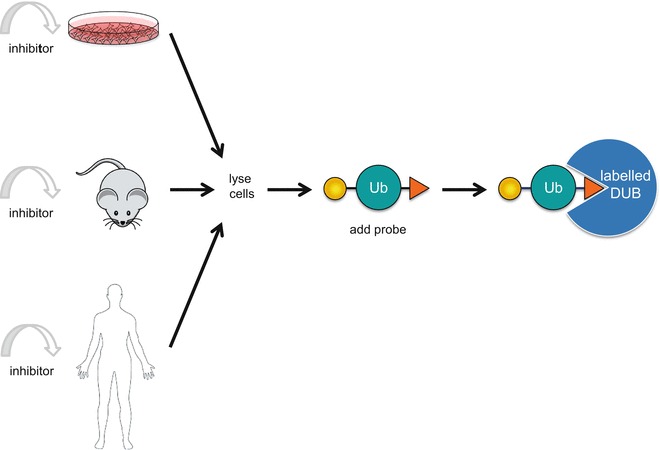
High-throughput assay design to monitor DUB target engagement using ubiquitin ABPs in cells, animals, or patients tissues: (1) treatment of cells, animals, or patients with DUB inhibitor; (2) generation of protein lysates; (3) incubation of lysates with ubiquitin ABPs; (4) visualization of DUB activity or inhibition
What Is Next for Chemical Probes Targeting DUBs?
The development of activity probes for DUBs has lagged behind the development of probes for more classical proteases. Indeed, the complexity of the recognition site of DUBs, which requires the binding of full-length ubiquitin in the catalytic site as well as the challenges in the characterization of potent and selective DUB inhibitors, has hindered production of ABPs for DUBs. However, following on from the ground-breaking evolution of cell-permeable and in vivo-compatible activity-based imaging probes developed for other proteases such as caspases or cathepsins [69, 70], the next generation of probes for DUBs will certainly be agents that enable direct visualization and quantification of DUB activity in vivo. Such noninvasive agents have great potential for early diagnosis as well as pharmacodynamic evaluation of DUB inhibition in preclinical as well as clinical settings. One attractive avenue to explore for the development of selective DUB activity probes is based on the design of copper-catalyzed click-labeled DUB inhibitors with quenchable or nonfluorescent labels [94]. Click-labeled ABPs allows for selective labeling, visualization, and enrichment of active enzymes in a complex proteome. Another approach will likely be based on the generation of noninvasive substrate probes that do not bind covalently to the enzyme. The advantage of this approach is based on the theoretically higher signal that can be generated, in contrast to covalent activity probes which are limited by the stoichiometric labeling of the enzyme (the signal being proportional to the amount of enzyme in various tissues). So far a very limited number of reporter substrates are available, none being cell permeable, or suitable for in vivo applications. Again, noninvasive permeable substrates will likely be derived from selective inhibitors of individual DUBs or knowledge around selectively ubiquitylated sites on DUB substrates. Probably one of the most promising avenues for developing cell- and tissue-permeable selective ubiquitin ABPs for DUBs will rely on the modification of selective small molecule inhibitors of DUBs. Similar approaches have already achieved some preliminary success for other enzymes of the UPS such as E1 enzymes [95] and proteasome probes [96]. The limiting step in developing such probes for DUBs is currently a lack of potent, specific and selective DUB inhibitors available, however, the community is successfully designing novel generations of selective DUB inhibitors.
While a number of ABPs have been successfully designed for monitoring the activity of cysteine peptidase DUBs, there is still a gap in the development of ABPs for DUBs of the metalloenzyme class (MPN+/JAMMs) . A number of approaches are currently being investigated for the design of ABPs for metallo-DUBs and will certainly aid the characterization of inhibitors for that class of enzymes which is showing great promise as therapeutic targets [97–99].
Summary
Protein ubiquitylation is critical for the control of protein half-life, localization, and function. Deregulation of this process is a causative factor of many diseases. The development of ABPs has allowed for major advancement in the identification and characterization of cysteine DUBs. Significant progress has been made in terms of probe design and preparation. For example, five papers have been published in the past 3 years describing di-ubiquitin ABPs, underscoring the importance of these tools for DUB research. The JAMM family remains difficult to target using ABPs due to the catalytic mechanism which does not involve a covalent DUB-substrate intermediate. Hopefully new approaches and novel probe designs will yield better tools to investigate this class of metalloproteases. ABPs will ultimately shed light on the function and relevance of DUBs involved in various chain-specific ubiquitin signaling, and will continue to advance our knowledge of DUB regulation and function in a cellular context. Furthermore, ABPs will aid the development and characterization of DUB inhibitors, allowing the monitoring of target engagement as well as selectivity in vivo. Finally, while ubiquitin ABPs have not yet been as broadly used as one might expect to monitor DUBs in developmental or pathological evaluations, they can provide a unique dynamic assessment of the activity of DUBs, and will undoubtedly become a more familiar option for many researchers.
Acknowledgements
The authors would like to acknowledge networking support by the Proteostasis COST Action (BM1307).
Contributor Information
Rune Matthiesen, Phone: +35135100351 217508145, Email: runem2009@gmail.com.
Xavier Jacq, Email: xjacq@missiontherapeutics.com.
References
- 1.Bedford L, et al. Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat Rev Drug Discov. 2011;10(1):29–46. doi: 10.1038/nrd3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldberg M, et al. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421(6926):952–956. doi: 10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- 3.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol. 2008;9(9):679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt M, Finley D. Regulation of proteasome activity in health and disease. Biochim Biophys Acta. 2014;1843(1):13–25. doi: 10.1016/j.bbamcr.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson DE. The ubiquitin-proteasome system: opportunities for therapeutic intervention in solid tumors. Endocr Relat Cancer. 2015;22(1):T1–T17. doi: 10.1530/ERC-14-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wenzel DM, Klevit RE. Following Ariadne’s thread: a new perspective on RBR ubiquitin ligases. BMC Biol. 2012;10:24. doi: 10.1186/1741-7007-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smit JJ, Sixma TK. RBR E3-ligases at work. EMBO Rep. 2014;15(2):142–154. doi: 10.1002/embr.201338166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rahighi S, Dikic I. Selectivity of the ubiquitin-binding modules. FEBS Lett. 2012;586(17):2705–2710. doi: 10.1016/j.febslet.2012.04.053. [DOI] [PubMed] [Google Scholar]
- 9.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 10.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6(1):79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 11.Gallastegui N, Groll M. The 26S proteasome: assembly and function of a destructive machine. Trends Biochem Sci. 2010;35(11):634–642. doi: 10.1016/j.tibs.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Groll M, et al. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386(6624):463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 13.Suraweera A, et al. Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol Cell. 2012;48(2):242–253. doi: 10.1016/j.molcel.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10(8):550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 15.Clague MJ, et al. Deubiquitylases from genes to organism. Physiol Rev. 2013;93(3):1289–1315. doi: 10.1152/physrev.00002.2013. [DOI] [PubMed] [Google Scholar]
- 16.Eletr ZM, Wilkinson KD. Regulation of proteolysis by human deubiquitinating enzymes. Biochim Biophys Acta. 2014;1843(1):114–128. doi: 10.1016/j.bbamcr.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mattern MR, Wu J, Nicholson B. Ubiquitin-based anticancer therapy: carpet bombing with proteasome inhibitors vs surgical strikes with E1, E2, E3, or DUB inhibitors. Biochim Biophys Acta. 2012;1823(11):2014–2021. doi: 10.1016/j.bbamcr.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sippl W, Collura V, Colland F. Ubiquitin-specific proteases as cancer drug targets. Future Oncol. 2011;7(5):619–632. doi: 10.2217/fon.11.39. [DOI] [PubMed] [Google Scholar]
- 19.Lill JR, Wertz IE. Toward understanding ubiquitin-modifying enzymes: from pharmacological targeting to proteomics. Trends Pharmacol Sci. 2014;35(4):187–207. doi: 10.1016/j.tips.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 20.Jacq X, et al. Deubiquitylating enzymes and DNA damage response pathways. Cell Biochem Biophys. 2013;67(1):25–43. doi: 10.1007/s12013-013-9635-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye Y, et al. Ubiquitin chain conformation regulates recognition and activity of interacting proteins. Nature. 2012;492(7428):266–270. doi: 10.1038/nature11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heideker J, Wertz IE. DUBs, the regulation of cell identity and disease. Biochem J. 2015;467(1):191. doi: 10.1042/bj4670191. [DOI] [PubMed] [Google Scholar]
- 23.Colland F, et al. Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Mol Cancer Ther. 2009;8(8):2286–2295. doi: 10.1158/1535-7163.MCT-09-0097. [DOI] [PubMed] [Google Scholar]
- 24.Reverdy C, et al. Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chem Biol. 2012;19(4):467–477. doi: 10.1016/j.chembiol.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 25.Chauhan D, et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell. 2012;22(3):345–358. doi: 10.1016/j.ccr.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian Z, et al. A novel small molecule inhibitor of deubiquitylating enzyme USP14 and UCHL5 induces apoptosis in multiple myeloma and overcomes bortezomib resistance. Blood. 2014;123(5):706–716. doi: 10.1182/blood-2013-05-500033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peterson LF, et al. Targeting deubiquitinase activity with a novel small molecule inhibitor as therapy for B-cell malignancies. Blood. 2015;125(23):3588–3597. doi: 10.1182/blood-2014-10-605584. [DOI] [PubMed] [Google Scholar]
- 28.Cravatt BF, Wright AT, Kozarich JW. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu Rev Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci U S A. 1999;96(26):14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan EW, et al. Developing photoactive affinity probes for proteomic profiling: hydroxamate-based probes for metalloproteases. J Am Chem Soc. 2004;126(44):14435–14446. doi: 10.1021/ja047044i. [DOI] [PubMed] [Google Scholar]
- 31.Saghatelian A, et al. Activity-based probes for the proteomic profiling of metalloproteases. Proc Natl Acad Sci U S A. 2004;101(27):10000–10005. doi: 10.1073/pnas.0402784101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berkers CR, et al. Activity probe for in vivo profiling of the specificity of proteasome inhibitor bortezomib. Nat Methods. 2005;2(5):357–362. doi: 10.1038/nmeth759. [DOI] [PubMed] [Google Scholar]
- 33.Adam GC, Sorensen EJ, Cravatt BF. Proteomic profiling of mechanistically distinct enzyme classes using a common chemotype. Nat Biotechnol. 2002;20(8):805–809. doi: 10.1038/nbt714. [DOI] [PubMed] [Google Scholar]
- 34.Galardy P, Ploegh HL, Ovaa H. Mechanism-based proteomics tools based on ubiquitin and ubiquitin-like proteins: crystallography, activity profiling, and protease identification. Methods Enzymol. 2005;399:120–131. doi: 10.1016/S0076-6879(05)99008-3. [DOI] [PubMed] [Google Scholar]
- 35.Borodovsky A, et al. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 2001;20(18):5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borodovsky A, et al. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem Biol. 2002;9(10):1149–1159. doi: 10.1016/S1074-5521(02)00248-X. [DOI] [PubMed] [Google Scholar]
- 37.Balakirev MY, et al. Otubains: a new family of cysteine proteases in the ubiquitin pathway. EMBO Rep. 2003;4(5):517–522. doi: 10.1038/sj.embor.embor824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hemelaar J, et al. Chemistry-based functional proteomics: mechanism-based activity-profiling tools for ubiquitin and ubiquitin-like specific proteases. J Proteome Res. 2004;3(2):268–276. doi: 10.1021/pr0341080. [DOI] [PubMed] [Google Scholar]
- 39.Ekkebus R, et al. On terminal alkynes that can react with active-site cysteine nucleophiles in proteases. J Am Chem Soc. 2013;135(8):2867–2870. doi: 10.1021/ja309802n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGouran JF, et al. Fluorescence-based active site probes for profiling deubiquitinating enzymes. Org Biomol Chem. 2012;10(17):3379–3383. doi: 10.1039/c2ob25258a. [DOI] [PubMed] [Google Scholar]
- 41.de Jong A, et al. Ubiquitin-based probes prepared by total synthesis to profile the activity of deubiquitinating enzymes. Chembiochem. 2012;13(15):2251–2258. doi: 10.1002/cbic.201200497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.El Oualid F, et al. Chemical synthesis of ubiquitin, ubiquitin-based probes, and diubiquitin. Angew Chem Int Ed Engl. 2010;49(52):10149–10153. doi: 10.1002/anie.201005995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Renatus M, et al. Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure. 2006;14(8):1293–1302. doi: 10.1016/j.str.2006.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altun M, et al. Muscle wasting in aged, sarcopenic rats is associated with enhanced activity of the ubiquitin proteasome pathway. J Biol Chem. 2010;285(51):39597–39608. doi: 10.1074/jbc.M110.129718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ovaa H, et al. Activity-based ubiquitin-specific protease (USP) profiling of virus-infected and malignant human cells. Proc Natl Acad Sci U S A. 2004;101(8):2253–2258. doi: 10.1073/pnas.0308411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kattenhorn LM, et al. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol Cell. 2005;19(4):547–557. doi: 10.1016/j.molcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 47.Schlieker C, et al. Structure of a herpesvirus-encoded cysteine protease reveals a unique class of deubiquitinating enzymes. Mol Cell. 2007;25(5):677–687. doi: 10.1016/j.molcel.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Misaghi S, et al. Chlamydia trachomatis-derived deubiquitinating enzymes in mammalian cells during infection. Mol Microbiol. 2006;61(1):142–150. doi: 10.1111/j.1365-2958.2006.05199.x. [DOI] [PubMed] [Google Scholar]
- 49.Frickel EM, et al. Apicomplexan UCHL3 retains dual specificity for ubiquitin and Nedd8 throughout evolution. Cell Microbiol. 2007;9(6):1601–1610. doi: 10.1111/j.1462-5822.2007.00896.x. [DOI] [PubMed] [Google Scholar]
- 50.Artavanis-Tsakonas K, et al. Identification by functional proteomics of a deubiquitinating/deNeddylating enzyme in Plasmodium falciparum. Mol Microbiol. 2006;61(5):1187–1195. doi: 10.1111/j.1365-2958.2006.05307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Le Negrate G, et al. ChlaDub1 of Chlamydia trachomatis suppresses NF-kappaB activation and inhibits IkappaBalpha ubiquitination and degradation. Cell Microbiol. 2008;10(9):1879–1892. doi: 10.1111/j.1462-5822.2008.01178.x. [DOI] [PubMed] [Google Scholar]
- 52.Baez-Santos YM, St John SE, Mesecar AD. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim W, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44(2):325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Phu Lilian, Izrael-Tomasevic Anita, Matsumoto Marissa L., Bustos Daisy, Dynek Jasmin N., Fedorova Anna V., Bakalarski Corey E., Arnott David, Deshayes Kurt, Dixit Vishva M., Kelley Robert F., Vucic Domagoj, Kirkpatrick Donald S. Improved Quantitative Mass Spectrometry Methods for Characterizing Complex Ubiquitin Signals. Molecular & Cellular Proteomics. 2010;10(5):M110.003756. doi: 10.1074/mcp.M110.003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bennett EJ, et al. Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell. 2010;143(6):951–965. doi: 10.1016/j.cell.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sowa ME, et al. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138(2):389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dirac AM, et al. Functional annotation of deubiquitinating enzymes using RNA interference. Methods Enzymol. 2005;398:554–567. doi: 10.1016/S0076-6879(05)98045-2. [DOI] [PubMed] [Google Scholar]
- 58.Buus R, et al. Deubiquitinase activities required for hepatocyte growth factor-induced scattering of epithelial cells. Curr Biol. 2009;19(17):1463–1466. doi: 10.1016/j.cub.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Urbe S, et al. Systematic survey of deubiquitinase localization identifies USP21 as a regulator of centrosome- and microtubule-associated functions. Mol Biol Cell. 2012;23(6):1095–1103. doi: 10.1091/mbc.E11-08-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nishi R, et al. Systematic characterization of deubiquitylating enzymes for roles in maintaining genome integrity. Nat Cell Biol. 2014;16(10):1016–1026. doi: 10.1038/ncb3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Faesen AC, et al. Mechanism of USP7/HAUSP activation by its C-terminal ubiquitin-like domain and allosteric regulation by GMP-synthetase. Mol Cell. 2011;44(1):147–159. doi: 10.1016/j.molcel.2011.06.034. [DOI] [PubMed] [Google Scholar]
- 62.Lander GC, et al. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482(7384):186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wiener R, et al. E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1. Nat Struct Mol Biol. 2013;20(9):1033–1039. doi: 10.1038/nsmb.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cotto-Rios XM, Jones MJ, Huang TT. Insights into phosphorylation-dependent mechanisms regulating USP1 protein stability during the cell cycle. Cell Cycle. 2011;10(23):4009–4016. doi: 10.4161/cc.10.23.18501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang OW, et al. Phosphorylation-dependent activity of the deubiquitinase DUBA. Nat Struct Mol Biol. 2012;19(2):171–175. doi: 10.1038/nsmb.2206. [DOI] [PubMed] [Google Scholar]
- 66.Altun M, et al. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem Biol. 2011;18(11):1401–1412. doi: 10.1016/j.chembiol.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 67.D’Arcy P, et al. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat Med. 2011;17(12):1636–1640. doi: 10.1038/nm.2536. [DOI] [PubMed] [Google Scholar]
- 68.Li H, et al. Assessing subunit dependency of the plasmodium proteasome using small molecule inhibitors and active site probes. ACS Chem Biol. 2014;9(8):1869–1876. doi: 10.1021/cb5001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Edgington LE, et al. Noninvasive optical imaging of apoptosis by caspase-targeted activity-based probes. Nat Med. 2009;15(8):967–973. doi: 10.1038/nm.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blum G, et al. Noninvasive optical imaging of cysteine protease activity using fluorescently quenched activity-based probes. Nat Chem Biol. 2007;3(10):668–677. doi: 10.1038/nchembio.2007.26. [DOI] [PubMed] [Google Scholar]
- 71.Sanman LE, Bogyo M. Activity-based profiling of proteases. Annu Rev Biochem. 2014;83:249–273. doi: 10.1146/annurev-biochem-060713-035352. [DOI] [PubMed] [Google Scholar]
- 72.Borodovsky A, et al. Small-molecule inhibitors and probes for ubiquitin- and ubiquitin-like-specific proteases. Chembiochem. 2005;6(2):287–291. doi: 10.1002/cbic.200400236. [DOI] [PubMed] [Google Scholar]
- 73.Ovaa H, Galardy PJ, Ploegh HL. Mechanism-based proteomics tools based on ubiquitin and ubiquitin-like proteins: synthesis of active site-directed probes. Methods Enzymol. 2005;399:468–478. doi: 10.1016/S0076-6879(05)99032-0. [DOI] [PubMed] [Google Scholar]
- 74.Hemelaar J, et al. A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1-LC3, GABARAP, and Apg8L. J Biol Chem. 2003;278(51):51841–51850. doi: 10.1074/jbc.M308762200. [DOI] [PubMed] [Google Scholar]
- 75.Hemelaar J, et al. Specific and covalent targeting of conjugating and deconjugating enzymes of ubiquitin-like proteins. Mol Cell Biol. 2004;24(1):84–95. doi: 10.1128/MCB.24.1.84-95.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Albrow VE, et al. Development of small molecule inhibitors and probes of human SUMO deconjugating proteases. Chem Biol. 2011;18(6):722–732. doi: 10.1016/j.chembiol.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dobrota C, et al. Glycine fluoromethylketones as SENP-specific activity based probes. Chembiochem. 2012;13(1):80–84. doi: 10.1002/cbic.201100645. [DOI] [PubMed] [Google Scholar]
- 78.Wilkinson KD, Gan-Erdene T, Kolli N. Derivitization of the C-terminus of ubiquitin and ubiquitin-like proteins using intein chemistry: methods and uses. Methods Enzymol. 2005;399:37–51. doi: 10.1016/S0076-6879(05)99003-4. [DOI] [PubMed] [Google Scholar]
- 79.Virdee S, et al. Engineered diubiquitin synthesis reveals Lys29-isopeptide specificity of an OTU deubiquitinase. Nat Chem Biol. 2010;6(10):750–757. doi: 10.1038/nchembio.426. [DOI] [PubMed] [Google Scholar]
- 80.Kumar KS, et al. Total chemical synthesis of di-ubiquitin chains. Angew Chem Int Ed Engl. 2010;49(48):9126–9131. doi: 10.1002/anie.201003763. [DOI] [PubMed] [Google Scholar]
- 81.Yang R, et al. Synthesis of K48-linked diubiquitin using dual native chemical ligation at lysine. Chem Commun (Camb) 2010;46(38):7199–7201. doi: 10.1039/c0cc01382j. [DOI] [PubMed] [Google Scholar]
- 82.Kumar KS, et al. Total chemical synthesis of a 304 amino acid K48-linked tetraubiquitin protein. Angew Chem Int Ed Engl. 2011;50(27):6137–6141. doi: 10.1002/anie.201101920. [DOI] [PubMed] [Google Scholar]
- 83.Bavikar SN, et al. Chemical synthesis of ubiquitinated peptides with varying lengths and types of ubiquitin chains to explore the activity of deubiquitinases. Angew Chem Int Ed Engl. 2012;51(3):758–763. doi: 10.1002/anie.201106430. [DOI] [PubMed] [Google Scholar]
- 84.Iphofer A, et al. Profiling ubiquitin linkage specificities of deubiquitinating enzymes with branched ubiquitin isopeptide probes. Chembiochem. 2012;13(10):1416–1420. doi: 10.1002/cbic.201200261. [DOI] [PubMed] [Google Scholar]
- 85.Haj-Yahya N, et al. Dehydroalanine-based diubiquitin activity probes. Org Lett. 2014;16(2):540–543. doi: 10.1021/ol403416w. [DOI] [PubMed] [Google Scholar]
- 86.Mulder MP, et al. A native chemical ligation handle that enables the synthesis of advanced activity-based probes: diubiquitin as a case study. Chembiochem. 2014;15(7):946–949. doi: 10.1002/cbic.201402012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McGouran JF, et al. Deubiquitinating enzyme specificity for ubiquitin chain topology profiled by di-ubiquitin activity probes. Chem Biol. 2013;20(12):1447–1455. doi: 10.1016/j.chembiol.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li G, et al. Activity-based diubiquitin probes for elucidating the linkage specificity of deubiquitinating enzymes. Chem Commun (Camb) 2014;50(2):216–218. doi: 10.1039/C3CC47382A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Koyano F, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510(7503):162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 90.Kane LA, et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205(2):143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wauer T, et al. Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis. EMBO J. 2015;34(3):307–325. doi: 10.15252/embj.201489847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bondalapati S, et al. Chemical synthesis of phosphorylated ubiquitin and diubiquitin exposes positional sensitivities of E1-E2 enzymes and deubiquitinases. Chemistry. 2015;21(20):7360–7364. doi: 10.1002/chem.201500540. [DOI] [PubMed] [Google Scholar]
- 93.Lander GC, Martin A, Nogales E. The proteasome under the microscope: the regulatory particle in focus. Curr Opin Struct Biol. 2013;23(2):243–251. doi: 10.1016/j.sbi.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Martell J, Weerapana E. Applications of copper-catalyzed click chemistry in activity-based protein profiling. Molecules. 2014;19(2):1378–1393. doi: 10.3390/molecules19021378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.An H, Statsyuk AV. Development of activity-based probes for ubiquitin and ubiquitin-like protein signaling pathways. J Am Chem Soc. 2013;135(45):16948–16962. doi: 10.1021/ja4099643. [DOI] [PubMed] [Google Scholar]
- 96.Carmony KC, Kim KB. Activity-based imaging probes of the proteasome. Cell Biochem Biophys. 2013;67(1):91–101. doi: 10.1007/s12013-013-9626-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Clague MJ, Urbe S. Endocytosis: the DUB version. Trends Cell Biol. 2006;16(11):551–559. doi: 10.1016/j.tcb.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 98.Ambroggio XI, Rees DC, Deshaies RJ. JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2004;2(1):E2. doi: 10.1371/journal.pbio.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hannss R, Dubiel W. COP9 signalosome function in the DDR. FEBS Lett. 2011;585(18):2845–2852. doi: 10.1016/j.febslet.2011.04.027. [DOI] [PubMed] [Google Scholar]


