Abstract
An in-situ hybridisation (ISH) technique for the detection of rabbit haemorrhagic disease virus (RHDV) was developed. Thirteen seronegative adult rabbits were infected oro-nasally using the BS89 RHDV strain. Liver and spleen samples were collected from 4 h post infection (p.i.) and repeated every 4 h till 44 h p.i. Each sample was tested immunohistochemically, by sandwich ELISA and by ISH. A 2.482-kb RNA probe, matching the genomic fragment coding for the VP60 structural protein of RHDV, was arranged. Two RNA probes (sense and antisense) were transcribed in vitro and UTP-digoxigenin-labelled. The antisense probe clearly detected positivity in the cytoplasm of the hepatocytes at 8 h p.i. Labelled hepatocytes were scattered throughout the sections until 24 h p.i. followed by a more diffuse perilobular positive reaction. A much weaker signal of similar distribution was detected up to 24 h p.i. using the sense RNA probe. All spleen samples tested negative for both probes. Liver samples were positive at 32 h p.i. using both ELISA and the immunoperoxidase test. Spleen samples were positive using only the ELISA at 32 h p.i. This study showed that RHDV replication occurred almost immediately after inoculation and that the liver appears to be the main site of replication.
Keywords: Rabbit hemorrhagic disease virus (RHDV), In situ hybridisation (ISH), RNA probe
1. Introduction
Rabbit haemorrhagic disease (RHD) is a highly acute and fatal disease of domestic and wild rabbits (Oryctolagus cuniculus) caused by a positive-stranded RNA virus which belongs to the Caliciviridae family (Capucci et al., 1990, Ohlinger et al., 1990). The disease is characterised by a short course (2–3 days), high levels of morbidity (100%) and mortality (80–90%), few but severe signs (dyspnoea, anorexia, depression, convulsions and epistaxis) and by a severe necrotising hepatitis (Marcato et al., 1991). RHD is endemic in many countries in Europe, East Asia and North Africa. Although the disease and its etiological agent have been investigated for the past decade, several aspects of pathogenesis are still to be clarified. These include: the site where virus replication occurs initially; how the virus reaches the hepatocytes; and the role of Kuppfer cells in viral distribution in the liver. Clarification of these aspects could provide a better understanding of the mechanisms which lead to the viral degradation associated with a chronic form of the disease (Barbieri et al., 1997) and why young animals below the age of 45 days are susceptible to RHDV infection but do not develop any sign of the disease.
Routine virological diagnosis is carried out commonly by ELISA (Capucci et al., 1991), and other diagnostic methods (Western blot, PCR) have been employed successfully for investigative studies. The use of in situ hybridisation (ISH) to detect viral genomes has not been a common practice in veterinary medicine. The application of ISH to RNA viruses of animals has been reported for foot and mouth disease virus (Brown et al., 1992), rabies virus (Jackson, 1992), African horse sickness virus (Brown et al., 1994), swine enteric and respiratory coronavirus (Sirinarumitr et al., 1996), and bovine syncytial virus (Viuff et al., 1996).
The availability of clones covering the entire RHDV cDNA sequence, prompted us to investigate the use of ISH with RNA probes to study the pathogenesis of RHDV. This paper describes the use of in situ hybridisation to study the presence of RHDV in livers and spleens of experimentally infected rabbits during the initial stages of infection.
2. Materials and methods
2.1. Experimental design
A group of 13 New Zealand adult rabbits, without anti-RHDV antibodies, was infected oro-nasally with 0.5 ml of a liver homogenate (10% w/v in PBS). This contained 2×103 LD50/ml and was obtained from a rabbit infected with RHDV reference strain (BS89). Following infection, one rabbit was killed humanely and necropsied each 4 h, until 44 h p.i. Two infected rabbits were kept as positive controls. They were observed constantly during the period between infection and death, and then necropsied. Two other rabbits were inoculated with a placebo and kept as uninfected controls in a separate unit. They were killed humanely at 24 and 44 h p.i. respectively and necropsied.
2.2. Sampling and tissue preparation
Liver and spleen samples were collected from each animal, fixed in 4% PBS DEPC-treated sterile paraformaldehyde, dehydrated, and paraffin embedded, and 4-μm serial sections were then cut.
Before the ELISA, liver and spleen fragments were homogenised in 10% (w/v) distilled water and centrifuged at 3900×g for 15 min to eliminate gross debris.
2.3. RNA probe
The probe consisted of 2482 bases matching the RHDV genome from nucleotide 4955 to 7437, corresponding to the gene of the VP60 structural protein. The probe was transcribed in vitro from 1 μg of pMB11 plasmid (Boniotti et al., 1994) using the Dig RNA Labelling Kit (Boehringer Mannheim). The probe in the sense orientation was obtained through transcription from the T3 promoter after linearisation of the plasmid with XbaI. For the antisense probe, the plasmid was cut by XhoI and transcribed from the T7 promoter. Approximately 10 μg of each digoxygenin-labelled probe was produced and then diluted in 100 μl of DEPC-treated (Sigma) sterile distilled water and stored at −70°C. Just before use, the probes were partially fragmented by brief incubation in NaOH followed by neutralisation.
2.4. Immunohistochemical assay
Four-micrometre thick paraffin liver sections were dewaxed in two changes of xylene for 15 min each and hydrated through graded alcohols. All sections were treated with 3% hydrogen peroxide in methanol for 60 min at room temperature followed by several washes in Tris buffered saline (TBS). Sections were incubated overnight at +4°C with the anti-VP60 1H8 monoclonal antibody (mAb) (Capucci et al., 1995) which was diluted at 1:4000 in TBS. After incubation, the sections were washed in TBS and incubated sequentially with biotinylated horse anti-mouse IgG (Vector Laboratories) diluted at 1:200, and streptavidin peroxidase (Vector Laboratories) diluted at 1:500 for 30 min each, with rinses in TBS between incubations. 3-3 Diaminobenzidine (DAB) (Sigma) was used as a substrate for the peroxidase reaction. After developing, the sections were counterstained with Mayer's haematoxylin.
Negative controls consisted of a non-immune primary antibody (normal mouse serum diluted at 1:20) applied to RHDV-infected rabbit liver sections and mAb 1H8 applied to RHDV negative rabbit liver sections.
2.5. In situ hybridisation
To avoid RNA degradation due to RNAse, gloves, disposable or sterilised laboratory equipment, sterile DEPC-treated buffers and fixative solutions were used. Before hybridisation, histological sections were: dewaxed and rehydrated through graded alcohols to PBS; treated with proteinase K (1 mg/ml in 50 mM Tris, pH 8.0, 5 mM EDTA) at room temperature for 8 min; post-fixed in 4% paraformaldheyde; acetylated in 0.25% acetic anhydride containing 0.1 M triethanolamine for 10 min; dehydrated and air-dried. The sections were hybridised by: adding 60 μl of hybridisation buffer (50% formamide, 0.3 M NaCl, 20 mM Tris HCl pH 8.0, 5 mM EDTA pH 8.0, 10 mM Na2HPO4, 10% dextran sulphate, Denhardt's solution 1×, 0.5 mg/ml yeast RNA) containing 0.9 μg/ml of the labelled sense or antisense probe; sealed by coverslip and incubated overnight in a humidified chamber at 55°C; washed in decreasing concentration of standard sodium citrate (SSC; 150 mM NaCl and 15 mM sodium citrate, pH 7.0) after removing the coverslip; treated with RNAse (2 μg/ml SSC 2×) at 37°C for 30 min; incubated with anti-digoxigenin alkaline-phosphatase conjugated antibody (1:500 in 0.5 M Tris) at 37°C for 1 h, followed by incubation with NTB/BCIP (Boehringer Mannheim) chromogen substrate solution at 37°C for 90 min; rinsed in distilled water, counterstained with methyl-green stain, dehydrated and mounted in Eukitt (Kindler GmbH).
Sections from the following samples were used as controls: RHDV-positive liver incubated with hybridisation buffer only; RHDV-positive liver incubated with either the antisense or the sense probe; and RHDV-negative liver incubated with either the antisense or the sense probe.
2.6. ELISA
A sandwich ELISA for RHDV detection was carried out as described previously (Capucci et al., 1995). Briefly, ELISA plates (96 wells, Nunc Maxisorb) were coated overnight at 4°C with 50 μl of 20 μg/ml anti-RHDV hyperimmune rabbit serum in carbonate buffer pH 9.6. The organs were extracted at 10% w/v in PBS pH 7.4 and, for the test, diluted 1/10 and 1/50 in PBS containing 1% yeast extract and 0.1% Tween 20. They were incubated for 60 min at 37°C with gentle shaking. The final detection of RHDV linked to the solid phase was carried out using horseradish peroxidase (HRP)-conjugated mAb 1H8 and mAb 6G2 in separate wells (Capucci et al., 1995). After each incubation the plates were washed 3×5 min with PBS containing 0.05% Tween 20. HRP activity was quantified using o-phenylenediamine (OPD) (Sigma) at 0.5 mg/ml in phosphate citrate buffer pH 5.0.
3. Results
3.1. Clinical signs and gross lesions
All the rabbits killed between 4 and 36 h p.i. showed no clinical signs or lesions specific to RHD. The last two rabbits to be killed (40 and 44 h p.i.) showed anorexia and mild prostration. In the two positive controls more severe signs (anorexia, apathy, prostration and neurological signs) were observed during the last 24 h before death, which occurred spontaneously at between 55 and 60 h p.i.
At necropsy these last four rabbits showed the most severe and typical RHD lesions, i.e. friable, discoloured and degenerate livers, and diffuse parenchymal and serosal haemorrhages.
3.2. Histological and immunohistochemical examination
The histological lesions observed in liver samples were similar to those reported previously (Marcato et al., 1991, Gelmetti et al., 1992) (Table 1 ). There were no specific lesions, but mild leucocyte infiltration and binucleated hepatocytes were observed up to 20 h p.i. Small and scattered necrotic foci were observed at 24 h p.i. The number of necrotic foci increased between 28 and 36 h p.i. From 40 h p.i. characteristic necrotising hepatitis (diffuse necrosis mainly in perilobular areas) was evident. Similar but more severe lesions were evident in liver sections from the positive controls. None of the spleen samples examined showed specific lesions.
Table 1.
Summarised results of histological, immunoperoxidase, ISH and ELISA examinations of liver samples
| Hours p.i. | Histology | Immunoperoxidase | ISH | ELISA | |
|---|---|---|---|---|---|
| Antisense | Sense | ||||
| 4 | No lesions | − | − | − | − |
| 8 | Rare etherophils | − | + | + | − |
| 12 | No lesions | − | + | + | − |
| 16 | Rare neutrophils | − | − | − | − |
| 20 | Scattered foci of neutrophils | − | + | + | − |
| 24 | Few necrotic foci | − | + | + | − |
| 28 | Several necrotic foci | − | + | − | − |
| 32 | Several necrotic foci | + | + | − | + |
| 36 | Focal necrotising hepatitis | + | + | − | + |
| 40 | Focal necrotising hepatitis | + | + | − | + |
| 44 | Diffuse necrotising hepatitis | + | + | − | + |
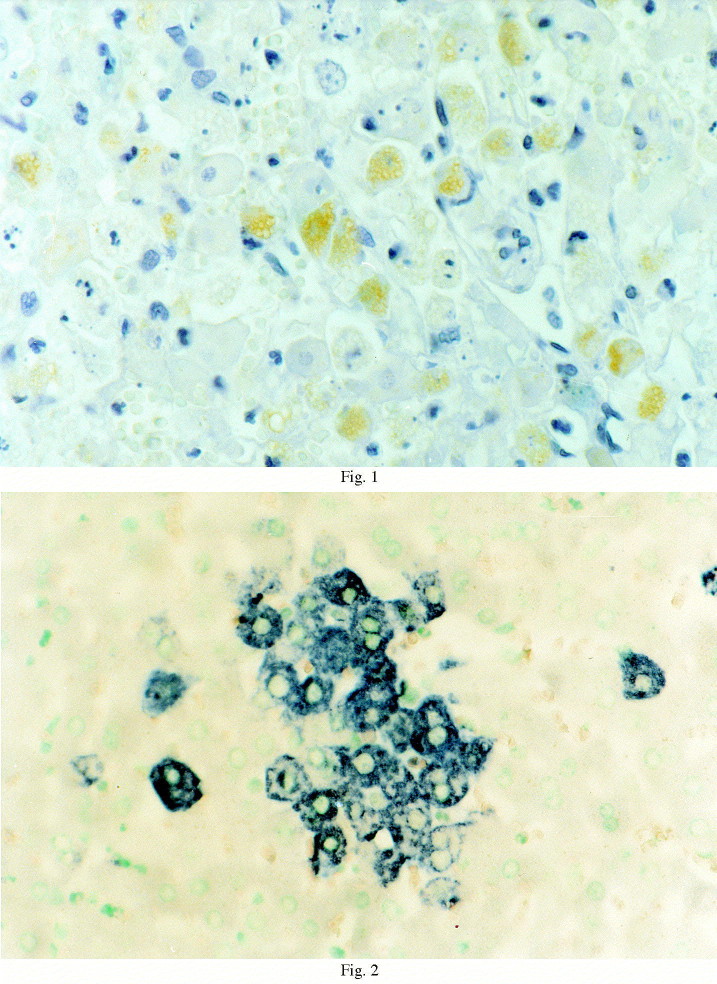
The immunoperoxidase test on the liver was positive from 32 h p.i. onwards. The positive signal appeared as small, brownish, vacuolated and granular structures, and was confined to the cytoplasm of apparently normal as well as necrotic hepatocytes (Fig. 1 ). After this, positive cells were detected mainly in the perilobular areas.
Fig. 1.
Streptavidin-biotin immunoperoxidase, liver, 32 h p.i. Small, brownish, vacuolated and granular structures are evident in the cytoplasm of hepatocytes (1000×).
No positive reactions were detected in the spleen samples at any time point.
3.3. ISH examination
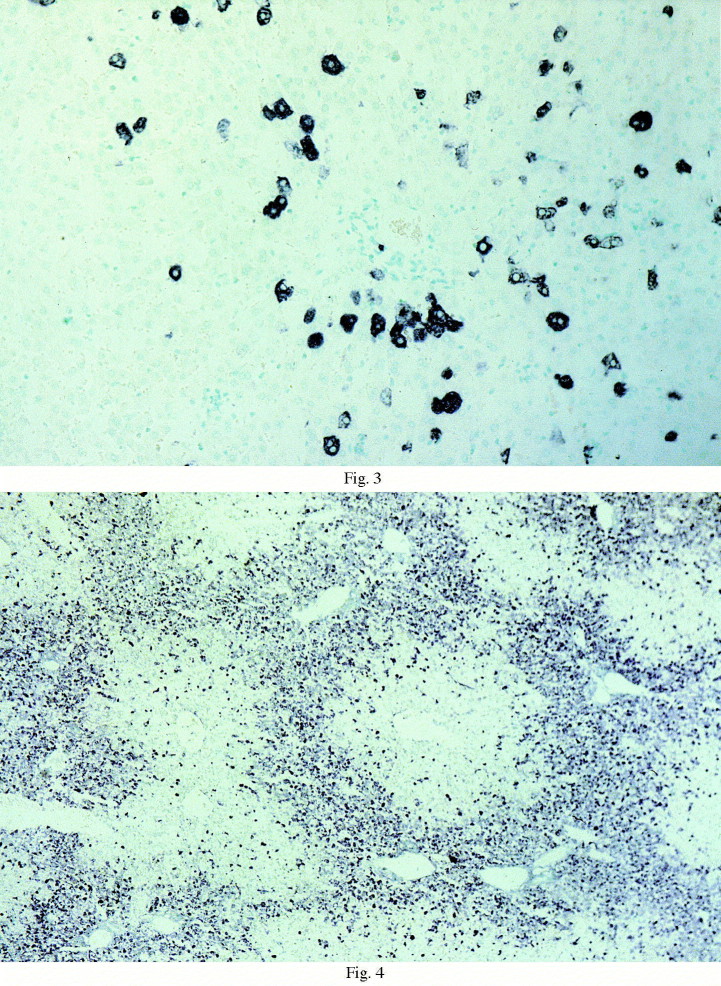
The antisense RNA probe clearly showed positivity in the liver from 8 h p.i. The positive signal was blue and granular, and confined to the hepatocyte cytoplasm (Fig. 2 ). Nuclei showed no reactivity. Sometimes, the signal was so intense as to mask the nucleus and cover the whole cellular surface. Labelled hepatocytes were scattered throughout the sections until 24 h p.i. (Fig. 3 ). Their number constantly and significantly increased with time, reaching a maximum at 44 h p.i. (Fig. 4 ). The liver and spleen samples of one rabbit, killed at 16 h p.i., was negative with both probes.
Fig. 2.
ISH, anti-sense probe, liver, positive control rabbit. Blue and granular positive signal was confined to the cytoplasm of the hepatocytes (1000×).
Fig. 3.
ISH, anti-sense probe, liver, 28 h p.i. Labelled hepatocytes were scattered throughout the section (600×).
Fig. 4.
ISH, anti-sense probe, liver, 44 h p.i. Labelled hepatocytes with perilobular distribution (150×).
Using the sense RNA probe, a similar but weak positivity was detected until 24 h p.i. None of the liver samples tested positive from either rabbits killed later or the infected controls.
All spleen samples were negative with either probe.
RHDV-positive liver sections incubated with hybridisation buffer only and liver sections of both non-infected rabbits, incubated with the antisense and sense probes, showed negative results. No specific signals were detected in hepatocytes and only a low or no background was visible.
3.4. ELISA
The ELISA on liver (Table 1) and spleen samples was positive from 32 h p.i.
4. Discussion
A non-isotopic RNA probe was developed to detect RHDV in fixed tissue samples by ISH. The use of a digoxigenin-labelled probe was preferred to isotopic marker detection, because it is considered more suitable for application in diagnostic laboratories, and it allows better localisation of the signal by histological examination of the sections.
The ISH signal was restricted to the cytoplasm of the hepatocytes. The number of labelled cells and their distribution were directly correlated to the progress of the infection. In particular, the maximum signal intensity was observed at 44 h p.i., mainly in the hepatocytes of periportal areas.
The ISH method employed above yielded reliable and reproducible results. Both positive and negative controls behaved as expected. Serial sections of the same paraffin block always gave similar results (data not shown). RNA probes constantly gave a very low background in all the sections examined. This may have been due to their single-strandedness (Jackson, 1992) and high specificity.
The results obtained using the antisense and sense probes were clearly different. The antisense probe recognised the genome of replicated virions and gave a strong positive signal, while the sense probe gave a weak signal. This observation is in line with the lower expected concentration of the target of the sense probe, i.e. the negative strand genome. This represents the intermediate of viral replication. In the picornaviridae it has been estimated that negative strand RNA amounts, on average, to less than 3% of total viral RNA in cells where the virus is actively replicating (Novak and Kirkegaard, 1991). Unfortunately these data cannot be confirmed for RHDV by in vitro studies because no suitable cell culture system has been identified so far (Xu and Chen, 1989).
The aim of this study was to assess the use of ISH for the detection of RHDV. However, we also wanted to check its applicability to study the pathogenesis of the infection. The results confirmed the data of a previous study (Gelmetti et al., 1992) suggesting that the liver is the most important site for RHDV replication. Viral replication appeared to begin almost immediately after inoculation, as indicated by the presence of scattered positive hepatocytes as early as 8 h p.i. Thereafter, with the exception of one animal (16 h p.i.), all liver samples were positive with the antisense probe and the signal gradually increased. The negative result at 16 h was best explained by individual differences in susceptibility to infection, as previously observed (Gelmetti et al., 1992).
Using ISH, all the spleen samples were negative. This result differs from previous data obtained using different methods (ELISA, Western blot and PCR). In particular, in a recent study on early stages of RHDV infection using PCR, it was reported that both the liver and the spleen contained viral RNA from 18 h p.i. (Guittré et al., 1996). However all these methods differ from ISH in terms of preparation of the sample since they involve organ homogenisation and therefore the contamination by virions present in the blood stream or associated with blood cells cannot be excluded. Passive transfer of virions from liver to spleen, and not RHDV replication in the latter, was previously suggested (Barbieri et al., 1997). The results obtained above further support this hypothesis. It may well be that mature virus particles pool in the spleen but do not replicate in spleen cells.
The ISH method had a higher sensitivity than immunohistochemistry for RHDV detection in hepatocytes. Even in rabbits infected intramuscularly, the earliest detection of virus using the immunoperoxidase test was at 20 h p.i. (Gelmetti et al., 1992).
The ISH method proved to be sensitive and specific enough to be used in diagnostic as well as investigative studies of RHDV. Since the probe employed spans highly conserved regions of the RHDV structural protein gene, ISH could be especially useful for diagnosis of new emerging mutated strains with different antigenicity. It seemed particularly suitable for pathogenetic studies and it will be applied to further investigation of virus distribution in tissue after oro-nasal infection.
Acknowledgements
We would like to thank Mrs. G. Botti, Mr. G. Bertocchi, Mr. G. Bonatti and Mr. M. Datteri for technical assistance. We would also like to thank Dr. R. Coates (Centro Linguistico della Università di Brescia) for his linguistic revision. This work was supported by the Veterinary Service of the Ministry of Health of Italy (30/93).
References
- Barbieri, I., Lavazza, A., Brocchi, E., König, M., Capucci, L., 1997. Morphological, structural and antigenic modifications of rabbit haemorrhagic disease virus in the course of the disease. In: Chasey, D., Gaskell, R.M., Clarke, I.N. (Eds.), Proceedings of the First International Symposium on Caliciviruses. ESVV, Reading, UK, 15–17 September 1996, pp. 182–193.
- Boniotti M.B., Wirblich C., Sibilia M., Meyers G., Thiel H.J., Rossi C. Identification and characterization of 3C-like protease from rabbit hemorragic disease virus, a calicivirus. J. Virol. 1994;68:6487–6495. doi: 10.1128/jvi.68.10.6487-6495.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.C., Meyer R.F., Olander H.J., House C., Mebus A. A pathogenesis study of foot and mouth disease in cattle, using in situ hybridization. Can. J. Vet. Res. 1992;56:189–193. [PMC free article] [PubMed] [Google Scholar]
- Brown C.C., Meyer R.F., Grubman M.J. Presence of African horse sickness virus in equine tissues, as determined by in situ hybridization. Vet. Pathol. 1994;31:689–694. doi: 10.1177/030098589403100609. [DOI] [PubMed] [Google Scholar]
- Capucci L., Scicluna M.T., Lavazza A., Brocchi E. Purification and characterization of the causative agent of viral haemorrhagic disease of rabbit. Sel. Vet. 1990;31:301–312. [Google Scholar]
- Capucci L., Scicluna M.T., Lavazza A. Diagnosis of viral haemorrhagic disease of rabbits and European brown hare syndrome. Rev. Sci. Tech. Off. Int. Epiz. 1991;10:347–370. doi: 10.20506/rst.10.2.561. [DOI] [PubMed] [Google Scholar]
- Capucci L., Frigoli G., Ronsholt L., Lavazza A., Brocchi E., Rossi C. Antigenicity of the rabbit haemorrhagic disease virus studied by its reactivity with monoclonal antibodies. Vir. Res. 1995;37:221–238. doi: 10.1016/0168-1702(95)00033-m. [DOI] [PubMed] [Google Scholar]
- Gelmetti D., Colmegna S., Capucci L., Scicluna M.T., Guadagnini P.F., Gallazzi D., Mandelli G. Viral haemorrhagic disease (VHD): a pathogenetic study under experimental conditions. J. Appl. Rabbit Res. 1992;15:1453–1471. [Google Scholar]
- Guittré C., Ruvoen-Clouet N., Barraud L., Cherel Y., Baginski I., Prave M., Ganiere J.P., Trépo C., Cova L. Early stages of rabbit haemorrhagic disease virus infection monitored by polymerase chain reaction. J. Vet. Med. B. 1996;43:109–118. doi: 10.1111/j.1439-0450.1996.tb00294.x. [DOI] [PubMed] [Google Scholar]
- Jackson A.C. Detection of rabies virus mRNA in mouse brain by using in situ hybridization with digoxigenin-labelled RNA probes. Mol. Cell. Probes. 1992;6:131–136. doi: 10.1016/0890-8508(92)90057-5. [DOI] [PubMed] [Google Scholar]
- Marcato P.S., Benassi C., Vecchi G., Galeotti L., Della Salda L., Sarli G., Lucidi P. Clinical and pathological features of viral haemorrhagic disease of rabbits and the European brown hare syndrome. Rev. Sci. Tech. Off. Int. Epiz. 1991;10:371–390. doi: 10.20506/rst.10.2.560. [DOI] [PubMed] [Google Scholar]
- Novak J.E., Kirkegaard K. Improved method for detecting poliovirus negative strands used to demonstrate specificity of positive-strand encapsidation and the ratio of positive to negative strands in infected cells. J. Virol. 1991;65:3384–3387. doi: 10.1128/jvi.65.6.3384-3387.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlinger R.F., Haas B., Meyers G., Weiland F., Thiel H.J. Identification and characterization of the virus causing rabbit haemorrhagic disease. J. Virol. 1990;64:3331–3336. doi: 10.1128/jvi.64.7.3331-3336.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirinarumitr T., Paul P.S., Kluge J.T., Halbur P.G. In situ hybridization technique for the detection of swine enteric and respiratory coronaviruses, transmissible gastro-enteritis virus (TGEV) and porcine respiratory coronavirus (PRCV), in formalin-fixed paraffin-embedded tissues. J. Virol. Meth. 1996;56:149–160. doi: 10.1016/0166-0934(95)01901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viuff B., Uttenthal A., Tegtmeier C., Alexandersen S. Sites of replication of bovine respiratory syncytial virus in naturally infected calves as determined by in situ hybridization. Vet. Pathol. 1996;33:383–390. doi: 10.1177/030098589603300403. [DOI] [PubMed] [Google Scholar]
- Xu Z.J., Chen W.X. Viral haemorrhagic disease in rabbits: a review. Vet. Res. Commun. 1989;13:205–212. doi: 10.1007/BF00142046. [DOI] [PubMed] [Google Scholar]