Abstract
Accurate diagnosis of viral infections enhances the ability of the clinician to make decisions on appropriate treatment of patients, evaluate disease progression and prevent misuse of antibiotics. Knowledge of the pathogen involved also allow implementation of infection control and monitoring of success of antiviral treatments that may affect the prognosis of patients. Epidemiological data collected through accurate diagnostics play an important role in public health through identification and control of outbreaks, implementation of appropriate diagnostic tests, vaccination programs and treatment but also to recognize common and emerging pathogens in a community. It is key that the clinician have an understanding of appropriate specimens to send to the laboratory and the value of specific nucleic acid and serological testing for different viral pathogens. Molecular techniques have revolutionized viral diagnoses over the past decade and enhanced both the sensitivity and specificity of tests and the speed by which a diagnosis can be made and new tests be developed. The continued use of serology for viruses with a short viremia, or for chronic infections should however complement these tests. This chapter aims to provide an overview of the available tests, the principles of testing and appropriate tests to select for different viruses and syndromes. Also provided is a glimpse of new developments in diagnostics that may further enhance the capacity to make a conclusive diagnosis in the near future.
Keywords: Respiratory Syncytial Virus, West Nile Virus, Respiratory Virus, Severe Acute Respiratory Syndrome, Neutralization Assay
Introduction
Human virus infections may affect all ages and may impact morbidity and mortality through acute, chronic, recurrent or lifelong infections. This may depend on the immune status of the patient and their ability to clear virus infection as well as the characteristics of the pathogen. The development of sensitive and specific methods for both the detection of viral nucleic acids and antiviral antibodies has greatly advanced our ability to make accurate diagnoses at different stages of the disease. Previously the extended periods needed for identification of viral etiologies; which greatly depended upon virus isolation techniques, meant that most viral diagnoses were of epidemiological value only [1–4].
Advances made in diagnostic techniques over the past decade have significantly improved the accuracy and timeliness of a viral diagnosis, which in turn can aid in patient management, disease control and positively impact the disease outcome [1–4]. Since the development of antiviral drugs and treatment options available for viral infections, clinicians are encouraged to seek viral laboratory diagnosis that can provide clinically useful information in diagnosis and management of patients. This required the focus of laboratories to shift to providing better, faster diagnosis, which has driven the development of new approaches to monitor viral infections and to support antiviral treatment through: quantitative viral loads, antiviral susceptibility testing, viral genotyping and, point-of-care testing. Despite the massive impact that molecular diagnostics has had on viral diagnosis, significant strides have been made in antigen detection and serological tests, in development of “rapid tests”, for the direct detection of viral antigen in clinical specimens and detection of antibodies in convalescent or chronic infections. Laboratory controlled molecular and serological tests continue to have the advantage of superior sensitivity, specificity and differential diagnostic options in a controlled environment [1–4]. With the increase in sensitivity, specificity and diversity of virological diagnostic assays available, the clinician should work in collaboration with the virology laboratory to maximize the diagnostic potential of an appropriate clinical specimen. Understanding the relevance of the diagnostic test requested for specific viruses, at different ages and interpretation of a positive test, remains key in the clinical management of a patient [1–4].
The aim of this chapter is to provide an overview of diagnostic methodologies available for viral diagnosis rather than extensive technical details of each of the assays. It aims to provide an overview of options available for the clinician, from common assays to recent developments; the rationale for using each and how they could be successfully employed for better clinical management of patients.
Collecting and Sending Clinical Samples to the Laboratory
The most important factor influencing the accuracy of viral diagnostic results is the specimen. Whichever method is used in the laboratory, the results are largely dependent upon the right specimen type, taken at the right time and stored and transported correctly [5].
An understanding of the pathogenesis and epidemiology of the virus involved will help to identify the correct test and specimen type to collect. For acute viruses it is crucial to take the time since infection into consideration and whether the virus circulates commonly in the population and causes reinfections. Figure 6.1 depicts the typical period that acute viruses can be detected in blood and the time before IgM becomes visible and later IgG [4, 6]. For many acute viruses the viremia varies but may be relatively short and virus can only be detected in the first 10 days from the time that clinical symptoms became apparent, either in the blood or urine if it causes a systemic infection, such as arboviruses or measles; in the stool for enteric viruses such as rotavirus or poliovirus; in the site of infection such as the respiratory tract for respiratory viruses; or central nervous system for neuroinvasive infections. During this time virus specific tests such as virus isolation, antigen tests or molecular tests such as reverse transcription (RT) polymerase chain reaction (PCR) are appropriate. For viruses that are less common in the environment such as the arboviruses and childhood diseases prior to vaccination, IgM antibody tests can be requested after 7–10 days, but may not be detected in early specimens and are not appropriate for common viruses such as the respiratory viruses that may cause frequent reinfections. IgG antibody would only be used to diagnose acute infections if a paired serum is available 10–14 days apart and is not used for common viruses that may cause frequent reinfections. IgG testing may also be used to determine immune status following vaccines. Maternal antibody will interfere with IgG testing the first 4–6 months of life and therefore, for chronic diseases such as HIV transmission in children from HIV-infected mothers, DNA PCR testing is more suitable [4, 7, 8].
Fig. 6.1.
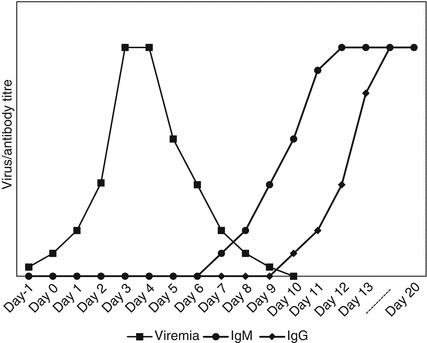
Diagram indicating a typical acute virus infection showing the period that virus, IgM and IgG antibody can be detected (compiled from [4, 6, 8])
Type of Specimen
Specimens to be used for virus isolation and RTPCR should be kept below 4 °C (39.2 °F) and reach the laboratory within 72 hours to keep RNA intact. For enveloped single stranded RNA viruses such as RSV the success rate declines from 48 hours and all effort should be made to keep the specimen on ice from the time it is collected until it reaches the laboratory.
Blood: the usual required volume is between 2 and 10 ml depending on the patient’s age, with the appropriate tube determined by the test required, and the appropriate blood component for the test (whole blood, plasma, serum). Anticoagulants such as heparin may inhibit PCR and EDTA tubes are preferred for molecular testing. For serology, clotted blood may be collected in SST (serum separation tubes) tubes that allow separation of red blood cells and serum through centrifugation. Virus isolation may be preferred from whole blood (EDTA) or serum (clotted blood) depending on the virus. Swabs: Swabs with a Dacron or rayon tip are preferred to ensure cells are collected and should be placed in viral transport medium that will preserve labile viruses for viral isolation and RTPCR. Washes or aspirates such as nasopharyngeal aspirates and other fluids such as saliva and urine should be placed in viral transport medium, although CSF is usually preferred undiluted. Stool: Obtain at least 4 g of stool and place in a sterile container. Tissue: Place in a sterile container with small amount of viral transport medium, for viral diagnosis. Specimens other than clotted blood must be kept at 4 °C (39.2 °F) and transported on ice to retain viability of the viruses and keep nucleic acids intact [7]. Table 6.1 summarizes the type of specimen and relevant tests available for viruses associated with different syndromes that may affect children (and adults).
Table 6.1.
| General syndrome | Agent | Specimen required | Diagnostic test options | Comments | |||||
|---|---|---|---|---|---|---|---|---|---|
| Commercial Rapid antigen detection test (RADT) and IFA available | Virus Isolation period in days | Diagnostic ELISA | PCR: Commercially or in-house assays | ||||||
| IgM | IgG | ||||||||
| Respiratory (pharyngitis, croup, bronchitis, pneumonia) | Adenovirus | Combination NP/OP swab; NPA, BAL | Yes | 21 or Shell-vial | N/A | N/A | Yes [10, 11] | Virus has no specific seasonality and is detected all year round [10, 12, 13]. Shell vial tests followed by antibody staining allows detection in cells after 48–96 hours | |
| Coronavirus | Combination NP/OP swab; NPA, BAL | No | RL | N/A | N/A | Yes [14–16] | This includes emerging viruses SARS-CoV and MERS-CoV [17] | ||
| Cytomegalovirus | Combination NP/OP swab; NPA, BAL, Blood | Yes | 28 | Yes | N/A | Yes [18] | CMV pneumonia in severely immunosuppressed HIV-positive patients and congenital infections [18] | ||
| Enterovirus | Combination NP/OP swab; NPA, BAL | No | 14 | N/A | N/A | Yes [10, 12, 13, 19–21] | Virus has no specific seasonality and is detected all year round [10, 12, 13] | ||
| Herpes simplex virus (HSV) | Combination NP/OP swab; NPA, BAL | No | 1–7 | Yes | N/A | Yes | HSV-1 has been associated with severe acute respiratory disease in severely immunocompromised patients [22] | ||
| Human metapneumovirus | Combination NP/OP swab; NPA, BAL | No | 2–21 | N/A | N/A | Yes [10, 12, 15, 16, 23–25] | Virus not routinely cultured and IFA not available. Seasonality similar to that of RSV [10, 12, 13, 25]. | ||
| Influenza virus | Combination NP/OP swab; NPA, BAL | Yes | 2–14 | N/A | N/A | Yes [10, 24, 26, 27] | Antigen detection 40–90% sensitive. Seasonal, test for Influenza A and B. Subtyping for seasonal subtypes (H1N1pdm09/H3N2) by specialist laboratories | ||
| Parainfluenza virus (PIV) | Combination NP/OP swab; NPA, BAL | Yes | 2–14 | N/A | N/A | Yes | Of all the PIVs, PIV3 is the main contributor to respiratory disease with a seasonality in the spring and summer months, PIV1, 2 and 4 less common [12, 13, 28]. | ||
| Respiratory syncytial virus | Combination NP/OP Swab in VTM; NPA, BAL | Yes | 2–21 | N/A | N/A | Yes [10, 15, 27, 29–34] | Rapid antigen detection variable specificity; 90% sensitive [35, 36]; Strong seasonal trends in autumn-winter months in South Africa; winter in temperate climates in Northern hemisphere, rainy season in tropics [10, 12, 13]. | ||
| Rhinovirus | Nasopharyngeal (NP) aspirate (NPA); NP wash; or NP swab, Oropharyngeal swab (OP), Combination NP/OP swab | No | 2–7 | No | No | Yes [19, 37, 38] | Too many strains to type serologically. Virus has no specific seasonality and is detected all year round [10, 12, 13]. | ||
| Exanthem | Maculopapular | Arboviruses | CSF if neurological; plasma/serum for febrile, VHF | No | RL | Yes | Rise in antibody levels: paired sera 10–14 days apart | Yes [39–44] | Viremia short, RTPCR in first 10 days only making acute serum for IgM serology important. Neurological cases detected in CSF. Specific to geographic region, mostly dengue, Zika. Cross reactivity for flaviviruses complicate confirmation by serum neutralization assays required |
| Enterovirus | CSF when clinically relevant; OP or rectal swab | No | 14 | N/A | N/A | Yes [10, 12, 13, 19–21] | |||
| Human herpes virus 6 and 7 | Serum | No | RL | Yes | No | Yes | Roseola agent | ||
| Measles virus | Serum, urine, respiratory secreta or CSF depending on syndrome | Yes | RL | Yes | Yes | Yes | Difficult to grow; RTPCR during acute, IgM serology later in disease for diagnostic purposes with paired sera [1, 9] | ||
| Parvovirus B19 | Serum | No | No | Yes | ND | Yes [45–50] | Erythema infectious agent; IgM serology is often diagnostic, but may be positive for a prolonged period [1] | ||
| Rubella virus | CSF when clinically relevant, serum, urine | No | >10 | Yes | Yes | Yes | Recommended that paired sera be tested simultaneously for diagnostic purposes [1, 9] | ||
| Vesicular | Herpes simplex virus | CSF when clinically relevant; Vesicle fluid, serum, EDTA | Yes | 21 | Yes | Yes | Yes [51, 52] | Serology rarely used for herpes simplex; IgM antibody used in selected cases [1]. Vesicle scrapings for direct IFA test [9] | |
| Varicella-zoster virus | CSF when clinically relevant; Vesicle fluid, serum, EDTA | Yes | 21 | Yes | Yes | Yes [52] | Vesicle scrapings for direct IFA test | ||
| CNS (Aseptic meningitis and encephalitis) | Arboviruses | CSF when clinically relevant and serum | No | No | Yes | Yes | Yes | Viremia brief, negative molecular test should be followed up with IgM serology with acute serum | |
| Dengue | CSF when clinically relevant, serum | Yes | RL | Yes [53] | Yes | Yes [54–57] | According to CDC; 80% seropositive at 6 days. Viremia brief, negative molecular test should be followed up with IgM serology with acute serum | ||
| Cytomegalovirus | CSF when clinically relevant | No | RL | Yes | Yes | Yes [51] | Immunocompromised patients; newborns | ||
| Enterovirus | CSF when clinically relevant | No | 3 | No | No | Yes [58–60] | Most common viral cause of meningoencephalitis | ||
| Epstein-Barr virus | CSF | No | No | Yes | Yes | Yes [51] | Neurological cases confirmed on CSF, positive on serum need to be interpreted with caution due to reactivation | ||
| Hantavirus | Serum, CSF when clinically relevant | No | RL | Yes | ND | RL | Diagnosis by presence of IgM antibody, Dependant on geographic location (Americas/Europe) | ||
| Herpes simplex virus | CSF when clinically relevant, serum | Yes | 21 | Yes | Yes | Yes [51, 61] | Culture of CSF has very low sensitivity; RTPCR on CSF most reliable | ||
| Measles virus | CSF when clinically relevant, serum | Yes | 21 | Yes | Yes | Yes | Culture of virus usually very successful. IgM serology diagnostic and RTPCR | ||
| Mumps virus | CSF when clinically relevant, urine | No | 21 | Yes [62] | Yes | Yes [63, 64] | RTPCR/IgM ELISA may allow diagnosis | ||
| Rabies virus | Saliva, nuchal skin biopsy, CSF (pre-mortem); brain biopsy (postmortem) | Not WHO endorsed | 2–3 | N/A | Neutralizing antibodies on CSF only | (WHO), no commercial tests | Antigen detection (FAT) on tissue (brain); RTPCR tissue; CSF, saliva; Serology only on unvaccinated individual | ||
| Varicella-zoster virus | CSF when clinically relevant, vesicular/skin swab | Yes | 3–21 days [1] | Yes | Yes | Yes [51, 52] | |||
| West Nile virus | CSF when clinically relevant, EDTA blood; serum | Yes | RL | Yes | Yes | Yes [39, 65–67] | Viremia brief, RTPCR on CSF or EDTA blood; if negative follow up with IgM serology with acute serum. Cross reaction of flaviviruses require confirmation by serum neutralization assays | ||
| Infectious mononucleosis | Epstein-Barr virus | Serum | No | No | Yes | Yes | Yes [51] | PCR on blood most common but correlate with clinical presentation. Serology for infection status only | |
| Hepatitis | Hepatitis A virus | Serum | No | No | Yes | ND | Yes [68–70] | Diagnosis by presence of IgM antibody/PCR | |
| Hepatitis B virus | Serum | Yes | No | Yes | ND | Yes | IgM to surface or core antigen confirms diagnosis; HBV viral load by PCR used for disease progression and clinical management | ||
| Hepatitis C virus | Serum | No | No | Yes | ND | Yes | Serology and PCR confirms infection; viral load by PCR used for management | ||
| Hepatitis D virus | Serum | No | No | Yes | No | No | Virus not routinely cultured | ||
| Hepatitis E virus | Serum, stool | No | No | Yes [71] | Yes | Yes [72, 73] | Virus not routinely cultured | ||
| Immunodeficiency virus | HIV1/2 | EDTA and serum | Yes | 15 | Yes 5–28 | ND? | Yes |
Antibody confirms infection in adults; DNA PCR for children <18 months due to passively acquired maternal antibody. Viral load by realtime PCR used for disease progression and clinical management |
|
| Gastroenteritis | Adenovirus | Rectal swab, stool | Yes | 10 | No | No | Yes [74–77] | Adenovirus 40 and 41 implicated in pediatric gastroenteritis | |
| Astrovirus | Stool | Yes | RL | No | No | Yes | Diagnosis by electron microscopy | ||
| Norovirus or Norwalk | Stool | No | RL | No | No | Yes [78–82] | One of the major causes of acute gastroenteritis in communities or cruise ships | ||
| Rotavirus | Stool | Yes | RL | No | No | Yes [76, 77, 83, 84] | Rapid assay are usually reliable, seasonal | ||
| Congenital infections | Cytomegalovirus | EDTA, serum, amniotic fluid, cord blood | Yes | 2 | Yes | N/A | Yes [51] | Presence of CMV IgM in cord blood is indicative of congenital infections; PCR and culture on dried blood spots, blood, saliva or urine in newborn < 3 weeks; retrospective, should be done on stored dried blood spots taken after birth [8] | |
| Enterovirus | CSF, serum, cord blood | No | 3 | Yes | N/A | Yes [58–60] | Cord blood most appropriate specimen in congenital infections | ||
| Herpes simplex virus | CSF, dermal, vesicle swab, tissue biopsy, amniotic fluid, cord blood | Yes | 1 | Yes | N/A | Yes [51] | Presence to HSV IgM in cord blood indicative of congenital infections | ||
| Parvovirus B19 | Synovial fluid, amniotic fluid, plasma | No | No | Yes | ND | Yes [45–50] | |||
| Rubella virus | CSF, serum | No | >10 | Yes | N/A | Yes | IgM for Rubella should be assayed using serum from infants up to 6 months of age IgG; could be false positive due to maternal antibodies | ||
| Zika virus | CSF, EDTA blood, serum. Pregnant mother/infant | No | RL | Yes | N/A | Yes | RTPCR recommended tests; IgM commercial tests need to be confirmed by PRNT by a reference laboratory for arboviruses due to cross reaction with dengue and other flaviviruses, depending on geographic relevance or travel history (CDC testing regiment [85, 86]) | ||
CSF cerebrospinal fluid, ELISA enzyme-linked immunosorbent assay, PCR polymerase chain reaction, EDTA ethylenediaminetetraacetic acid (purple top tube), NP nasopharyngeal, OP oropharyngeal, NPA nasopharyngeal aspirate, BAL bronchoalveolar lavage, IFA immunofluorescent antibody, RL indicates specialist or research laboratory only, ND not done, N/A Not applicable
Methods Used in Diagnostic Virology
Electron Microscopy
Although this is one of the oldest techniques it is not routinely used in diagnostic laboratories anymore. Electron microscopy (EM) is the only method available for directly visualizing the virus, and therefore has many applications beyond being purely diagnostic. The visualization of viruses with EM involves negative staining of the clinical specimen. Negative staining of the clinical sample is a relatively straightforward; inexpensive technique that would represent a “catch all” method of viral identification. EM could be particularly useful in identifying fastidious [87] or non-cultivable [88–90] virus in specimens, providing they have a high virus concentration with a sensitivity limit of approximately 106 viral particles per milliliter of specimen, making a negative result difficult to interpret [2]. While the sensitivity could be increased by ultracentrifugation or antibody-induced clumping, a further limitation is the lack of specificity, as EM can only identify up to the family level whereafter, other methods would have to be applied for a specific diagnosis [3]. Although the major advantage of EM is the speed with which a result could be obtained (30 min), the high cost of the instrument and specialized training and expertize needed, coupled with the lack of sensitivity and specificity, does not make this a viable option for routine diagnostics [4, 8].
Histology/Cytology
Direct microscopy of stained histology or cytology specimens may, in some instances, give the first indication of viral involvement that involves cellular changes. For viruses such as CMV, VZV, HPV, BK and B19, specific cytological changes can be confirmed through staining for specific antigen or genome sequences, using antibody or nucleic acid probes. Specific PCR amplification techniques may outperform these techniques in sensitivity, although detection of antigen in tissue is highly specific [4, 8].
Virus Isolation
Viral tissue culture was traditionally the “gold standard” used for diagnosing virus infections [91]. However, in the last 10 years molecular techniques have become routine. Virus isolation needs to remain an important part of viral diagnostics in order to maintain a source for analyzing, not only genotypic changes, but also phenotypic changes in virus populations for vaccine relevance and epidemiology. This allows identification of changes in antigenicity, pathogenicity and viral characteristics to update vaccines, such as the influenza vaccine, to match circulating strains [92]. Quality of the specimen, the time that it takes to reach the laboratory and transport under cold chain will determine the success of virus isolation. Detection of viruses in cell culture requires a considerable expertise and is performed by microscope examination, looking for degenerative morphological changes in the cell monolayer. This is called the cytopathic effect (CPE). Not all viruses grow in all cell types or produce CPE and further antigen or nucleic detection methods are required to correctly identify the specific virus involved. Clinical specimens are usually inoculated onto several cell lines to provide an optimum environment for a range of viruses (Table 6.2) [93].
Table 6.2.
List of viruses commonly isolated in clinical laboratories; compiled from [4]
| Virus | Rate of growth [1, 94]a | Type of CPE that can be detected [94] | Most permissive cell lineb |
|---|---|---|---|
| RNA viruses | |||
| Enteroviruses | 2–8 | Retractile angular or tear-shaped cells | PMK |
| Rhinoviruses | 4–10 | Retractile rounding of cells | HDF |
| Influenza viruses | 2–14 | Swollen vacuolated cells | PMK |
| RSV | 2–21 | Syncytia seen only in Hep-2 cells | HEp-2 |
| DNA viruses | |||
| Adenoviruses | 1–21 | Aggregation and rounding of cells in grape-like structures | HEp-2 |
| HSV | 1–7 | Retractile rounded cells | A549 |
| VZV | 5–10 | Foci of enlarged cells | HDF |
| CMV | 5–28 | Small foci of enlarged cells | HDF |
aTime in days needed for CPE to develop, depending on the initial viral load in the sample, the higher the viral load the quicker CPE will be detected
b PMK Primary monkey kidney, HDF human dermal fibroblasts, HEp-2 human epithelial type-2, A549 adenocarcinoma human alveolar basal epithelial cells
An adaption of traditional viral culture formats has been developed, which allows for more rapid detection of viruses, especially for viruses which are known to grow slowly in conventional cell culture. This is achieved by inoculating the specimen onto a microscope slide and centrifugation of the culture to enhance the infection rate (Shell vial assays). The enhanced detection rate may result from better contact between cells in the specimen and the cell culture, thus allowing for earlier and more extensive infection of the cell lines, as well as through the use of fluorescent-labeled (e.g. FITC) monoclonal antibodies directed to the viral antigen [93]. Nevertheless, most culture methods lack sensitivity and specificity relative to PCR. It remains, however, a catchall method of choice if the virus in question can be cultured [92].
Nucleic Acid Detection Methods
Viruses can be detected directly in clinical samples using highly specific nucleic acid primers and probes that are complementary in sequence to RNA viruses, using RT-PCR or for DNA viruses, directly by PCR. Over the past 10 years, nucleic acid amplification tests have been developed for the major viruses of public health concern and have become the new benchmark for viral diagnoses. The published sensitivities and specificities are usually nearly 100% when compared with cell culture or antigen assays [92, 95–97]. In fact studies that have compared molecular assays, with tissue culture assays, have demonstrated significantly increased sensitivity, of up to 30% [92, 95, 96, 98, 99].
The development of real-time PCR, that incorporates the use of specific florescent labeled probes, has created the ability to monitor the DNA amplification process as it happens, or in “real-time” on a dedicated instrument that is capable of collecting the fluorescent data from every PCR cycle. The accumulation of the measured fluorescence at the end of every PCR cycle is plotted and displayed as a sigmoidal curve and when the data is analyzed a cycle threshold (Ct) value is assigned to each target’s amplification when it is first detected. The Ct is the point at which the amplicons’ fluorescence exceeds that of the background and this is indirectly proportionate to the initial concentration of the target DNA in the sample i.e. the higher the concentration in the initial sample the lower the Ct value will be [100–102].
Comparative studies have revealed that the detection of respiratory viruses using real-time reverse transcriptase polymerase chain reaction (rRT-PCR) assays is substantially more sensitive than using conventional methods such as viral culture and immunofluorescence assays (IFA) [92, 103, 104]. Furthermore, compared to conventional PCR and other real-time methods, multiplex rRT-PCR has the advantage of permitting simultaneous amplification of several viruses in a single reaction [16, 103, 104]. This facilitates cost-effective diagnosis, enabling the detection of multiple viruses in a single clinical specimen. Amplification of several viruses together may however sacrifice sensitivity of individual assays and much effort has gone into identifying ways of increasing sensitivity.
Multiplex PCR Assays
Multiplex PCR assays are now frequently used to detect the presence of a range of viruses involved in specific syndromes such as respiratory infections e.g. influenza virus (INF) A and B; [20, 95, 105–107], parainfluenza viruses (PIV) types 1, 2, 3 and 4 [19, 20, 95, 105, 107, 108]; human respiratory syncytial virus (RSV) [20, 105–108], human metapneumovirus (hMPV) [20, 105, 107], human rhinoviruses (RV) [19, 20, 105, 107], human coronaviruses (hCoV-229E, hCoV-OC43) and Severe Acute Respiratory Syndrome coronavirus (SARS-CoV) [20, 107], human enteroviruses (EV) [19, 20] and adenoviruses (AdV) [108]. Disadvantages, include higher start-up costs, higher reagent costs, and extensive and specific training for specialist laboratories and specialized equipment to run them [92].
The diversity, fastidious nature, short viremia periods of some pathogens that may cause the infection and lack of available diagnostic tests, can severely hamper the ability to identify etiologies to different clinical syndromes [81]. Several multiplex platforms have been developed either as a set of duplex assays, in-house and commercial [20, 104, 109] and multiplex systems that require specialized equipment to read and are now available for a range of syndromes [20, 104, 109]. The limiting factor has been sensitivity and most assays require PCR amplification before detecting products through a number of platforms. These include Mass-Tag PCR [110], microarray platforms [111], macro arrays [39], microbead based methods [20] and Taq-Man array cards [112–115]. Currently TaqMan array cards are increasing in popularity due to the ease with which these can be adapted for specific purposes. These assays have application in diagnosis of single cases or as part of epidemiological studies to describe the etiologies of specific syndromes. Taq-Man array card (TAC; Life Technologies, Foster City, CA, USA) assays have been developed and used with success for several syndromes such as respiratory disease [63], enteric disease [70] and neonatal sepsis [81]. Once developed, TaqMan array cards are stable at 4 °C (39.2 °F) for 2 years and can be shipped at ambient temperature [116]. The TAC assay is a 384-well microfluidic array which consists of identical arrays in eight individual microfluidic channels, each of which can be loaded with nucleic acid extract from a clinical specimen or positive control [63, 70, 81, 116, 117]. The individual channels consist of 48 wells, each of which contains singleplex qPCR reactions targeting a different pathogen. Thus eight specimens are assayed per TAC card, and can simultaneously detect up to 48 pathogens per specimen. This makes the TAC assay popular for the following reasons: (1) minimal specimen volume required; (2) reduction in cross contamination of specimens due to the closed system format; (3) the ability to tailor the panel of pathogens detected as required; (4) proven efficacy of this technology in pathogen detection for similar studies; (5) and simple to use format [63, 70, 81, 116, 117].
Future Trends
Multiplex methods are becoming more common in routine diagnostic laboratories. However, most of the large scale methods described above are predominantly research based and used in epidemiological studies or in specialist laboratories, rather than routinely. Next generation sequencing methods, that make use of deep sequencing of all nucleic acids present in a sample, are currently mostly used for pathogen discovery or in specialist laboratories to detect outbreaks. In general amplification steps are still needed before this can be used on clinical specimens. In addition these techniques are too expensive to run on a large scale, in routine diagnostic laboratories. Even though these techniques are becoming more affordable they generate significant amounts of data that require both trained bioinformaticians to interpret the outputs and large computational systems. Nevertheless, development of automated systems for identifying viruses directly in clinical specimens, may in future make these techniques more accessible for routine diagnostics [118–120].
Application of Molecular Virology Diagnostics in Clinical Management of Patients
Qualitative vs. Quantitative PCR
The increased sensitivity that the development of molecular assays have highlighted, is that while there are advantages in identifying new viruses associated with disease, such as the respiratory viruses described in the last decade (including hMPV [23], hCoV NL63 [14] and HKU1) [121], it has also revealed flaws that make interpreting such a positive PCR result problematic. Recent literature has shown that specifically in the case of RV [27, 37, 38] that the virus was detected in asymptomatic as well as symptomatic patients. Due to the increased sensitivity of molecular assays it is possible to detect the presence of a virus at a low genome copy number, which may represent the pre- or post-syndromic phase of a viral infection, redefining the nature of viral disease and the clinical interpretation thereof [27, 38]. Interpretation of qualitative and quantitative PCR results as well as the application of the appropriate choice would require a close liaison with the virological laboratory.
Qualitative detection in specimens that are normally virus free: A good example of this is the diagnosis of viral or aseptic encephalitis, in which testing CSF for HSV, CMV, VZV or enteroviruses are diagnostic [122]. Qualitative PCR offers significant advantages in terms of speed, especially with the development of the point-of-care testing. Early diagnosis and treatment of CNS infections has been proven to improve the prognosis [117] and reduce unnecessary treatment and hospitalization [123]. Viruses that only exhibit low-levels of virus shedding in the absence of symptoms such as viral gastroenteritis, caused by rotavirus or norovirus could be detected in stool samples [81, 82].
Quantitative viral loads: Assays that can quantify the amount of virus in infected patients have proven to be the most valuable tool in the management of chronic viral infections. For many persistent viral infections, with transient low-level viremia, the onset of symptoms is associated with a spike in viral replication and thus a higher viral load, allowing the prediction of disease onset [8]. This allows for better clinical management of the patient, as the clinician can monitor the progression of the disease, the success of treatment, the emergence of drug resistance and understanding the pathogenesis of a particular virus of which HIV-1 and 2 [124, 125], CMV [116, 126], EBV [127], HBV [128, 129] and HCV [124] are but a few.
Antiviral Resistance
As the availability of antiviral drugs increases, more emphasis is placed on assays to determine the causes of treatment failure, of which antiviral resistance is one possible outcome [8]. The emergence of antiviral resistance has been documented for virtually all antiviral compounds, with the specific viral mutations associated with resistance becoming better understood [8, 125, 130–132]. Laboratory assays to determine drug resistance fall into two major categories:
Phenotypic assays: Phenotypic assays have largely been replaced by molecular based genotypic assays, however, they remain the gold standard for determining drug efficacy and susceptibility, as the concentration of the drug required to inhibit viral replication can be calculated. HIV is the best example [130, 131, 133, 134]. Phenotypic assays have the added advantage of giving a complete overview of all mutations observed. However, it is an expensive and laborious technique, of which the success will greatly depend on the level of training of staff and whether or not the specific virus culture-adapted strains are available [8].
Genotypic assays: While the development of RT-PCR as a genotypic assay, focusing on specific areas on the virus genome, have the added advantage of being rapid, relatively inexpensive and semi-quantitative (single point mutation assays, and allelic discrimination assays), it is difficult to interpret a single point mutation without all the required information that a phenotypic assay would provide [8]. The development of new automated sequencing methods have enabled the study of the genetic basis of drug resistance and made the assessment of virus isolates, with reduced drug susceptibility, more accessible [8]. The use of sequence based methods for testing for antiviral resistance have also have become routine in viral diagnostics, especially for HIV [132, 135–137] and HBV [138–140]. The biggest drawback of this technique, other than the expense, is the downstream analysis of sequencing data that is generated. Sequencing editing and interpretation is required, and in the case of HIV, the identification of resistance is dependent upon the recognition of specific sequence patterns on the software system used [141].
Serology
Serological techniques can either be targeted at the antigen, during the acute phase of infection, or to virus specific antibody later in infection. While virus specific antigen may only be detected in the first 10 days of acute infections, IgM antibody is detected within 7–14 days following infection and may remain for a month or more in the patient’s blood after the infection was cleared. Therefore, IgM can be used to detect a recent infection. IgG antibody is detected after 10–14 days of infection and can be present for life. Sero-conversion to IgG is measured with paired sera taken during the acute and the convalescent phase, 10–14 days apart. A significant rise in antibody of fourfold increase is seen as a positive reaction and new infection, while a single IgG positive test may reflect infection any time in the past. In pediatric infections, maternal immunity needs to be taken into consideration in the first 6 months of life and may only be cleared by 18 months, and may therefore, interfere with serological diagnosis in infants. Assays for detection of IgM or IgG are usually qualitative, since the presence or absence of antibody is enough to make a diagnosis. However, when a rise in antibody has to be detected, the test needs to be quantitative in order to detect an increase in antibody from the first to the second specimen. Antibody titre is measured as the reciprocal of the highest serum dilution where a positive reaction can still be detected. For example, a titre of 32 indicates that positive antibody binding could be detected in serum diluted up to 1 in 32, but not beyond that. Serological techniques are easily automated and play an important part in routine diagnostic laboratories. They have an important role in diagnosing acute and chronic infections and are useful for development of rapid tests and should, in addition, complement molecular techniques where clinically relevant [8, 142].
Immunofluorescence Assay (IFA)
These assays allow for the rapid detection of antigen and can be applied directly on clinical samples such as nasopharyngeal aspirates or on tissue culture or tissue from biopsy specimens, such as brain tissue for rabies virus. IFA is quick and convenient for individual specimens but requires a skilled operator and is not as easy to scale up and is not as sensitive as molecular techniques for viral detection. It is, however, relatively cheap and a popular choice, for this reason, in identification of viral infections.
Direct IFA detects virus in infected specimens or tissue using commercially available antibodies labelled with a florescent marker, while indirect IFA detects antibody in the patient sera by binding to the antigen in virus infected tissue cultured cells. A secondary anti-human IgG or IgM antibody is then used to detect the patient’s bound antibodies. Direct IFA is frequently used for respiratory virus antigen detection in respiratory secretions (RSV, influenza, PIV1–3) while indirect IFA for IgG or IgM antibody detection is used to detect infections such as EBV or VZV, amongst others [4, 8, 142, 143].
Enzyme-Linked Immunoassay for Antibody Detection (ELISA)
ELISAs are the most commonly used antibody or antigen detection assays since they have a high throughput, are rapid, are easily automated and are objective since the output can be read using a spectrophotometer. ELISA works on the principle of detecting antibody in patient sera through a reaction where antigen is bound to the surface of a micro-titre plate, the patient serum added to bind to the antigen and any bound antibody is then detected through addition of a secondary anti-human antibody coupled to an enzyme. Addition of a substrate to the enzyme linked antigen antibody complex results in a colour change which will induce a positive reaction. The assay can be adjusted for IgG or IgM through addition of the anti-human antibody. Antigen detection ELISA is performed by coating the solid phase with antibody to detect the antigen in the patient sera in order to reveal the detection antibody complex.
ELISA is frequently used for detection of IgG or IgM antibodies to rubella, measles, mumps, HIV, Hepatitis A and arboviruses such as West Nile virus, Zika virus, dengue or JEV. Although ELISA can have a very high sensitivity and specificity, some viral families may cross react and for exact identification of viruses such as the flaviviruses, neutralization assays are needed for confirmation [6, 142].
Neutralization Assays
Virus neutralization assays are highly specific assays testing for neutralizing antibodies and are also used to confirm results of other serological assays, such as ELISA, which are known to cross-react between different viruses of the same family e.g. the Flaviviruses (Zika virus, dengue and West Nile virus). It can also be used to determine if a vaccine would provide protection e.g. to detect antigenic drift in the neutralizing epitopes of the annual influenza vaccine. Antigen is mixed with dilutions of antibody and the inhibition of CPE observed through inoculation on a tissue monolayer. The inhibition effect can either be read through observation of CPE, or through overlay of agar which allows plaque formation for plaque reduction neutralization assays (PRNT). Micro-neutralization assays can also be read through ELISA methods, which help to automate the process and reduce the test run time before infected cells can be detected. These techniques are labour intensive and not routinely done by diagnostic laboratories, but rather by reference or specialist laboratories [6, 142, 144].
Other Serological Techniques
Several further formats of serological techniques exist that are used for different purposes. The hemagglutination inhibition assay (HAI) test detects antibodies to viruses that have a hemagglutinin antigen. These include rubella, Influenza and the flaviviruses. The test is still routinely used in reference laboratories to identify especially, cross reactive viruses such as the flaviviruses, before confirming specific viruses by neutralization assays or to investigate influenza antigenic variation relative to sera raised against the vaccine. Due to cross reactivity and requirement for fresh red blood cells, it is less commonly employed in routine laboratories. The Western blot technique is still used for confirmation of HIV and HCV, and it is based on the principle of transferring specific viral proteins separated on a gel or blotting paper, followed by binding to patient serum and detection with an anti-human enzyme labeled antibody and substrate. It is very specific but sensitivity may vary. Antigen detection methods allow for the development of rapid antigen or antibody tests and allow for bedside diagnosis. However, variable sensitivity and specificity determine the value of these tests [142].
Future Trends in Serology
Following the trend in molecular diagnostics, development of multiplex serological assays, that cover a range of viral antigens associated with specific syndromes, will significantly improve the diagnostic capacity of laboratories. Methods that use multiplex microsphere-based suspension immuno assays (SIAs) for the simultaneous detection of IgG antibodies against a range of viruses, enables development of syndrome or application specific tests. An example would be a B19, CMV and T. gondii combination SIAs multiplex for rapid antibody screening during pregnancy [145]. These assays bind a number of antigens through antibody to a microsphere. They are then incubated with the patient sera before being visualized with a labeled anti-human IgM antibody. Similar tests have been described for arbovirus screening in the Northern hemisphere [146]. Multiplex formats, based on protein arrays, have also been developed to detect a range of viruses. For these assays the antigens are fixed to a solid phase microchip slide and tested against patient anti-serum and fluorescent labeled anti-human antibodies used for detection. The position on the chip identifies the pathogen involved. These arrays may be based on peptides synthesized from pathogen sequence [147] recombinant proteins, [148] or inactivated virus antigens [149]. These techniques are not yet widely available in routine diagnostic laboratories but developed in specialist and research laboratories. However, the automation possibilities will very likely improve their accessible in the future.
Quality Assurance and Control
The ability of the laboratory to provide accurate diagnostic results is essential for effective clinical management of patients, solving of outbreaks and for responsible decision making [150]. Therefore monitoring ongoing quality assurance (QA) and improvement in all aspects of the laboratory is crucial. This involves the managing and monitoring of all services and processes related to releasing a diagnostic results [4, 150]. Processes that should be monitored relate to the pre-analytical phase i.e. specimen transport, collection and storage; the analytical phase i.e. testing and monitoring of the laboratory procedures and environment as well as the post analytical phase i.e. of result reporting and result interpretation [4, 150]. QA ensures annual assessment of staff competency, calibration and servicing of equipment as well as the quality of diagnostic tests. Clinical laboratories are strictly regulated by appointed agencies and are audited according to specific standards set forth by the International Organization for Standardization (ISO) and the International Electrotechnical Commission (IEC) [4, 150].
The primary quality control (QC) concern in a molecular laboratory is the specimen and nucleic acid quality or integrity, assay sensitivity and specificity, as well as the false positive tests because of PCR contamination. The RNA and DNA integrity can be insured through use of RNase and DNase free reagents and consumables, in addition to handling specimens on ice. While, PCR contamination can be avoided through physical organization of the laboratory and workflow, separating work areas and equipment; relevant PCR controls should be included in each run to ensure correct interpretation of the results [4, 8, 150]. The use of uracil N-glycosylase (UNG) in PCR reactions provides for chemical control for carry over contamination [151].
It is vital that when PCR diagnostics are undertaken, every effort is made to minimize contamination and that these assays are tested in a laboratory environment in which staff are well trained and competent for this type of work [8]. Using an accredited laboratory ensures the diagnostic findings are reliable.
Conclusion
Molecular and serological techniques should be used in a complimentary fashion for the diagnoses of virus infections. Knowledge of the stage of infection, viral pathogenesis and epidemiology help to make decisions regarding the correct test to choose for appropriate diagnosis. Advances in specificity and sensitivity and capacity to test for a range of viruses through multiple platforms make accurate diagnosis and rapid identification of circulating viruses for real-time clinical relevant data more feasible today. Virus isolation and collaboration with specialist laboratories make newer techniques for identification of emerging and re-emerging viruses possible. Quality of specimens, patient clinical history and presentation and close collaboration between the clinician, pathologist and laboratory remain key for useful diagnostic data for improved patient management.
Contributor Information
Robin J. Green, Phone: +272712 354 5277, Email: Robin.Green@up.ac.za
Marietjie Venter, Email: marietjie.venter.up@gmail.com, Email: marietjie.venter@up.ac.za
References
- 1.Levin M, Asturias E, Weinberg A. Infections: viral & rickettsial. New York, NY: McGraw-Hill; 2016. [Google Scholar]
- 2.Miller SE. Electron microscopy of viral infections. In: Lennette EH, Smith TF, editors. Laboratory diagnosis of viral infections. 3rd. New York, NY: Marcel Dekker, Inc.; 1999. pp. 45–70. [Google Scholar]
- 3.Petric M, Szymanski M. Electron microscopy and immunoelectron microscopy. In: Specter S, Hodinka RL, Young SA, editors. Clinical virology manual. 3rd. Washington, DC: American Society of Microbiology Press; 2000. pp. 54–65. [Google Scholar]
- 4.Specter S, Hodinka RL, Wiedbrauk DL, Young SA. Diagnosis of viral infections. In: Richman DD, Whitley RJ, Hayden FG, editors. Clinical virology. 2nd. Washington, DC: American Society for Microbiology Press; 2002. pp. 243–272. [Google Scholar]
- 5.Nutting PA, Main DS, Fischer PM, Stull TM, Pontious M, Seifert M, Jr, Boone DJ, Holcomb S. Toward optimal laboratory use. Problems in laboratory testing in primary care. JAMA. 1996;275:635–639. doi: 10.1001/jama.1996.03530320059035. [DOI] [PubMed] [Google Scholar]
- 6.Lambert AJ, Lanciotti RS. Laboratory diagnosis of arboviruses. Norfolk, UK: Caister Academic press; 2016. [Google Scholar]
- 7.Storch G. Diagnostic virology. In: Knipe DM, Howley PM, editors. Fields virology. 5. Philadelphia, PA: Lippincott Williams & Wilkins; 2007. pp. 565–604. [Google Scholar]
- 8.Jeffery K, Aarons E. Diagnostic approaches. In: Zuckerman AJ, Banatvala JE, Pattison JR, editors. Principles and practice of clinical virology. New York: Wiley; 2009. pp. 1–27. [Google Scholar]
- 9.Grys TE, Smith TF. Specimen requirements: selection, collection, transport and processing. In: Steven S, Hodinka RL, Young SA, Wiedbrauk DL, editors. Clinical virology manual. 4. Washington, DC: ASM Press; 2009. pp. 18–35. [Google Scholar]
- 10.Pretorius MA, Madhi SA, Cohen C, Naidoo D, Groome M, Moyes J, Buys A, Walaza S, Dawood H, Chhagan M, Haffjee S, Kahn K, Puren A, Venter M. Respiratory viral coinfections identified by a 10-plex real-time reverse-transcription polymerase chain reaction assay in patients hospitalized with severe acute respiratory illness--South Africa, 2009-2010. J Infect Dis. 2012;206(Suppl 1):S159–S165. doi: 10.1093/infdis/jis538. [DOI] [PubMed] [Google Scholar]
- 11.Heim A, Ebnet C, Harste G, Pring-Akerblom P. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J Med Virol. 2003;70:228–239. doi: 10.1002/jmv.10382. [DOI] [PubMed] [Google Scholar]
- 12.Cohen C, Walaza S, Moyes J, Groome M, Tempia S, Pretorius M, Hellferscee O, Dawood H, Chhagan M, Naby F, Haffejee S, Variava E, Kahn K, Nzenze S, Tshangela A, von Gottberg A, Wolter N, Cohen AL, Kgokong B, Venter M, Madhi SA. Epidemiology of viral-associated acute lower respiratory tract infection among children <5 years of age in a high HIV prevalence setting, South Africa, 2009-2012. Pediatr Infect Dis J. 2015;34:66–72. doi: 10.1097/INF.0000000000000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen C, Walaza S, Moyes J, Groome M, Tempia S, Pretorius M, Hellferscee O, Dawood H, Haffejee S, Variava E, Kahn K, Tshangela A, von Gottberg A, Wolter N, Cohen AL, Kgokong B, Venter M, Madhi SA. Epidemiology of severe acute respiratory illness (SARI) among adults and children aged >/=5 years in a high HIV-prevalence setting, 2009-2012. PLoS One. 2015;10:e0117716. doi: 10.1371/journal.pone.0117716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJ, Wolthers KC, Wertheim-van Dillen PM, Kaandorp J, Spaargaren J, Berkhout B. Identification of a new human coronavirus. Nat Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lassauniere R, Kresfelder T, Venter M. A novel multiplex real-time RT-PCR assay with FRET hybridization probes for the detection and quantitation of 13 respiratory viruses. J Virol Methods. 2010;165:254–260. doi: 10.1016/j.jviromet.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venter M, Lassauniere R, Kresfelder TL, Westerberg Y, Visser A. Contribution of common and recently described respiratory viruses to annual hospitalizations in children in South Africa. J Med Virol. 2011;83:1458–1468. doi: 10.1002/jmv.22120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sisk JM, Frieman MB. Emerging coronaviruses: severe acute respiratory syndrome (SARS) and middle east respiratory syndrome (MERS), eLS 1–12. New York: John Wiley & Sons, Ltd; 2001.
- 18.Salomon N, Gomez T, Perlman DC, Laya L, Eber C, Mildvan D. Clinical features and outcome of HIV-related cytomegalovirus pneumonia. AIDS. 1997;11:319–324. doi: 10.1097/00002030-199703110-00009. [DOI] [PubMed] [Google Scholar]
- 19.Coiras MT, Aguilar JC, Garcia ML, Casas I, Perez-Brena P. Simultaneous detection of fourteen respiratory viruses in clinical specimens by two multiplex reverse transcription nested-PCR assays. J Med Virol. 2004;72:484–495. doi: 10.1002/jmv.20008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahony J, Chong S, Merante F, Yaghoubian S, Sinha T, Lisle C, Janeczko R. Development of a respiratory virus panel test for detection of twenty human respiratory viruses by use of multiplex PCR and a fluid microbead-based assay. J Clin Microbiol. 2007;45:2965–2970. doi: 10.1128/JCM.02436-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahony JB. Detection of respiratory viruses by molecular methods. Clin Microbiol Rev. 2008;21:716–747. doi: 10.1128/CMR.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simoons-Smit AM, Kraan EM, Beishuizen A, van Schijndel RJS, Vandenbroucke-Grauls CM. Herpes simplex virus type 1 and respiratory disease in critically-ill patients: real pathogen or innocent bystander? Clin Microbiol Infect. 2006;12:1050–1059. doi: 10.1111/j.1469-0691.2006.01475.x. [DOI] [PubMed] [Google Scholar]
- 23.van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, Osterhaus AD. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7:719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen C, Moyes J, Tempia S, Groome M, Walaza S, Pretorius M, Dawood H, Chhagan M, Haffejee S, Variava E, Kahn K, von Gottberg A, Wolter N, Cohen AL, Malope-Kgokong B, Venter M, Madhi SA. Mortality amongst patients with influenza-associated severe acute respiratory illness, South Africa, 2009-2013. PLoS One. 2015;10:e0118884. doi: 10.1371/journal.pone.0118884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Groome MJ, Moyes J, Cohen C, Walaza S, Tempia S, Pretorius M, Hellferscee O, Chhagan M, Haffejee S, Dawood H, Kahn K, Variava E, Cohen AL, Gottberg A, Wolter N, Venter M, Madhi SA. Human metapneumovirus-associated severe acute respiratory illness hospitalisation in HIV-infected and HIV-uninfected South African children and adults. J Clin Virol. 2015;69:125–132. doi: 10.1016/j.jcv.2015.06.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen AL, Hellferscee O, Pretorius M, Treurnicht F, Walaza S, Madhi S, Groome M, Dawood H, Variava E, Kahn K, Wolter N, von Gottberg A, Tempia S, Venter M, Cohen C. Epidemiology of influenza virus types and subtypes in South Africa, 2009-2012. Emerg Infect Dis. 2014;20:1162–1169. doi: 10.3201/eid2007.131869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pretorius MA, Tempia S, Walaza S, Cohen AL, Moyes J, Variava E, Dawood H, Seleka M, Hellferscee O, Treurnicht F, Cohen C, Venter M. The role of influenza, RSV and other common respiratory viruses in severe acute respiratory infections and influenza-like illness in a population with a high HIV sero-prevalence, South Africa 2012–2015. J Clin Virol. 2016;75:21–26. doi: 10.1016/j.jcv.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen AL, Sahr PK, Treurnicht F, Walaza S, Groome MJ, Kahn K, Dawood H, Variava E, Tempia S, Pretorius M, Moyes J, Olorunju SA, Malope-Kgokong B, Kuonza L, Wolter N, von Gottberg A, Madhi SA, Venter M, Cohen C. Parainfluenza virus infection among human immunodeficiency virus (HIV)-infected and HIV-uninfected children and adults hospitalized for severe acute respiratory illness in South Africa, 2009-2014. Open Forum Infect Dis. 2015;2:ofv139. doi: 10.1093/ofid/ofv139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haynes AK, Manangan AP, Iwane MK, Sturm-Ramirez K, Homaira N, Brooks WA, Luby S, Rahman M, Klena JD, Zhang Y, Yu H, Zhan F, Dueger E, Mansour AM, Azazzy N, McCracken JP, Bryan JP, Lopez MR, Burton DC, Bigogo G, Breiman RF, Feikin DR, Njenga K, Montgomery J, Cohen AL, Moyes J, Pretorius M, Cohen C, Venter M, Chittaganpitch M, Thamthitiwat S, Sawatwong P, Baggett HC, Luber G, Gerber SI. Respiratory syncytial virus circulation in seven countries with Global Disease Detection Regional Centers. J Infect Dis. 2013;208(Suppl 3):S246–S254. doi: 10.1093/infdis/jit515. [DOI] [PubMed] [Google Scholar]
- 30.Moyes J, Cohen C, Pretorius M, Groome M, von Gottberg A, Wolter N, Walaza S, Haffejee S, Chhagan M, Naby F, Cohen AL, Tempia S, Kahn K, Dawood H, Venter M, Madhi SA. Epidemiology of respiratory syncytial virus-associated acute lower respiratory tract infection hospitalizations among HIV-infected and HIV-uninfected South African children, 2010-2011. J Infect Dis. 2013;208(Suppl 3):S217–S226. doi: 10.1093/infdis/jit479. [DOI] [PubMed] [Google Scholar]
- 31.Nair H, Brooks WA, Katz M, Roca A, Berkley JA, Madhi SA, Simmerman JM, Gordon A, Sato M, Howie S, Krishnan A, Ope M, Lindblade KA, Carosone-Link P, Lucero M, Ochieng W, Kamimoto L, Dueger E, Bhat N, Vong S, Theodoratou E, Chittaganpitch M, Chimah O, Balmaseda A, Buchy P, Harris E, Evans V, Katayose M, Gaur B, O’Callaghan-Gordo C, Goswami D, Arvelo W, Venter M, Briese T, Tokarz R, Widdowson M-A, Mounts AW, Breiman RF, Feikin DR, Klugman KP, Olsen SJ, Gessner BD, Wright PF, Rudan I, Broor S, SimÃμes EAF, Campbell H. Global burden of respiratory infections due to seasonal influenza in young children: a systematic review and meta-analysis. Lancet. 2011;378:1917–1930. doi: 10.1016/S0140-6736(11)61051-9. [DOI] [PubMed] [Google Scholar]
- 32.Pretorius MA, van Niekerk S, Tempia S, Moyes J, Cohen C, Madhi SA, Venter M. Replacement and positive evolution of subtype a and B respiratory syncytial virus G-protein genotypes from 1997-2012 in South Africa. J Infect Dis. 2013;208(Suppl 3):S227–S237. doi: 10.1093/infdis/jit477. [DOI] [PubMed] [Google Scholar]
- 33.Tempia S, Walaza S, Viboud C, Cohen AL, Madhi SA, Venter M, McAnerney JM, Cohen C. Mortality associated with seasonal and pandemic influenza and respiratory syncytial virus among children <5 years of age in a high HIV prevalence setting—South Africa, 1998–2009. Clin Infect Dis. 2014;58:1241–1249. doi: 10.1093/cid/ciu095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venter M, Collinson M, Schoub BD. Molecular epidemiological analysis of community circulating respiratory syncytial virus in rural South Africa: comparison of viruses and genotypes responsible for different disease manifestations. J Med Virol. 2002;68:452–461. doi: 10.1002/jmv.10225. [DOI] [PubMed] [Google Scholar]
- 35.Slinger R, Milk R, Gaboury I, Diaz-Mitoma F. Evaluation of the QuickLab RSV Test, a new rapid lateral-flow immunoassay for detection of respiratory syncytial virus antigen. J Clin Microbiol. 2004;42:3731–3733. doi: 10.1128/JCM.42.8.3731-3733.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Englund JA, Piedra PA, Jewell A, Patel K, Baxter BB, Whimbey E. Rapid diagnosis of respiratory syncytial virus infections in immunocompromised adults. J Clin Microbiol. 1996;34:1649–1653. doi: 10.1128/jcm.34.7.1649-1653.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fry AM, Lu X, Olsen SJ, Chittaganpitch M, Sawatwong P, Chantra S, Baggett HC, Erdman D. Human rhinovirus infections in rural Thailand: epidemiological evidence for rhinovirus as both pathogen and bystander. PLoS One. 2011;6:e17780. doi: 10.1371/journal.pone.0017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pretorius MA, Tempia S, Treurnicht FK, Walaza S, Cohen AL, Moyes J, Hellferscee O, Variava E, Dawood H, Chhagan M, Haffjee S, Madhi SA, Cohen C, Venter M. Genetic diversity and molecular epidemiology of human rhinoviruses in South Africa. Influenza Other Respi Viruses. 2014;8:567–573. doi: 10.1111/irv.12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaayman D, Human S, Venter M. A highly sensitive method for the detection and genotyping of West Nile virus by real-time PCR. J Virol Methods. 2009;157:155–160. doi: 10.1016/j.jviromet.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 40.Garcia S, Crance JM, Billecocq A, Peinnequin A, Jouan A, Bouloy M, Garin D. Quantitative real-time PCR detection of rift valley fever virus and its application to evaluation of antiviral compounds. J Clin Microbiol. 2001;39:4456–4461. doi: 10.1128/JCM.39.12.4456-4461.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moureau G, Temmam S, Gonzalez JP, Charrel RN, Grard G, de Lamballerie X. A real-time RT-PCR method for the universal detection and identification of flaviviruses. Vector Borne Zoonotic Dis. 2013;7(4):467–478. doi: 10.1089/vbz.2007.0206. [DOI] [PubMed] [Google Scholar]
- 42.Patel P, Landt O, Kaiser M, Faye O, Koppe T, Lass U, Sall A, Niedrig M. Development of one-step quantitative reverse transcription PCR for the rapid detection of flaviviruses. Virol J. 2013;10:58. doi: 10.1186/1743-422X-10-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Eeden C, Zaayman D, Venter M. A sensitive nested real-time RT-PCR for the detection of Shuni virus. J Virol Methods. 2014;195:100–105. doi: 10.1016/j.jviromet.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 44.Forrester NL, Palacios G, Tesh RB, Savji N, Guzman H, Sherman M, Weaver SC, Lipkin WI. Genome-scale phylogeny of the alphavirus genus suggests a marine origin. J Virol. 2012;86:2729–2738. doi: 10.1128/JVI.05591-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zaaijer HL, Koppelman MH, Farrington CP. Parvovirus B19 viraemia in Dutch blood donors. Epidemiol Infect. 2004;132:1161–1166. doi: 10.1017/S0950268804002730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koppelman MH, Cuypers HT, Emrich T, Zaaijer HL. Quantitative real-time detection of parvovirus B19 DNA in plasma. Transfusion. 2004;44:97–103. doi: 10.1046/j.0041-1132.2004.00610.x. [DOI] [PubMed] [Google Scholar]
- 47.Weimer T, Streichert S, Watson C, Groner A. High-titer screening PCR: a successful strategy for reducing the parvovirus B19 load in plasma pools for fractionation. Transfusion. 2001;41:1500–1504. doi: 10.1046/j.1537-2995.2001.41121500.x. [DOI] [PubMed] [Google Scholar]
- 48.Schmidt I, Blumel J, Seitz H, Willkommen H, Lower J. Parvovirus B19 DNA in plasma pools and plasma derivatives. Vox Sang. 2001;81:228–235. doi: 10.1046/j.1423-0410.2001.00120.x. [DOI] [PubMed] [Google Scholar]
- 49.Harder TC, Hufnagel M, Zahn K, Beutel K, Schmitt HJ, Ullmann U, Rautenberg P. New LightCycler PCR for rapid and sensitive quantification of parvovirus B19 DNA guides therapeutic decision-making in relapsing infections. J Clin Microbiol. 2001;39:4413–4419. doi: 10.1128/JCM.39.12.4413-4419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aberham C, Pendl C, Gross P, Zerlauth G, Gessner M. A quantitative, internally controlled real-time PCR assay for the detection of parvovirus B19 DNA. J Virol Methods. 2001;92:183–191. doi: 10.1016/S0166-0934(00)00292-5. [DOI] [PubMed] [Google Scholar]
- 51.Espy MJ, Uhl JR, Sloan LM, Buckwalter SP, Jones MF, Vetter EA, Yao JD, Wengenack NL, Rosenblatt JE, Cockerill FR, 3rd, Smith TF. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin Microbiol Rev. 2006;19:165–256. doi: 10.1128/CMR.19.1.165-256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Neill HJ, Wyatt DE, Coyle PV, McCaughey C, Mitchell F. Real-time nested multiplex PCR for the detection of Herpes simplex virus types 1 and 2 and Varicella zoster virus. J Med Virol. 2003;71:557–560. doi: 10.1002/jmv.10516. [DOI] [PubMed] [Google Scholar]
- 53.Shu PY, Chen LK, Chang SF, Yueh YY, Chow L, Chien LJ, Chin C, Lin TH, Huang JH. Comparison of capture immunoglobulin M (IgM) and IgG enzyme-linked immunosorbent assay (ELISA) and nonstructural protein NS1 serotype-specific IgG ELISA for differentiation of primary and secondary dengue virus infections. Clin Diagn Lab Immunol. 2003;10:622–630. doi: 10.1128/CDLI.10.4.622-630.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shu PY, Chang SF, Kuo YC, Yueh YY, Chien LJ, Sue CL, Lin TH, Huang JH. Development of group- and serotype-specific one-step SYBR green I-based real-time reverse transcription-PCR assay for dengue virus. J Clin Microbiol. 2003;41:2408–2416. doi: 10.1128/JCM.41.6.2408-2416.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Houng HS, Chung-Ming Chen R, Vaughn DW, Kanesa-thasan N. Development of a fluorogenic RT-PCR system for quantitative identification of dengue virus serotypes 1-4 using conserved and serotype-specific 3′ noncoding sequences. J Virol Methods. 2001;95:19–32. doi: 10.1016/S0166-0934(01)00280-4. [DOI] [PubMed] [Google Scholar]
- 56.Callahan JD, Wu SJ, Dion-Schultz A, Mangold BE, Peruski LF, Watts DM, Porter KR, Murphy GR, Suharyono W, King CC, Hayes CG, Temenak JJ. Development and evaluation of serotype- and group-specific fluorogenic reverse transcriptase PCR (TaqMan) assays for dengue virus. J Clin Microbiol. 2001;39:4119–4124. doi: 10.1128/JCM.39.11.4119-4124.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laue T, Emmerich P, Schmitz H. Detection of dengue virus RNA in patients after primary or secondary dengue infection by using the TaqMan automated amplification system. J Clin Microbiol. 1999;37:2543–2547. doi: 10.1128/jcm.37.8.2543-2547.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Monpoeho S, Coste-Burel M, Costa-Mattioli M, Besse B, Chomel JJ, Billaudel S, Ferre V. Application of a real-time polymerase chain reaction with internal positive control for detection and quantification of enterovirus in cerebrospinal fluid. Eur J Clin Microbiol Infect Dis. 2002;21:532–536. doi: 10.1007/s10096-002-0766-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Corless CE, Guiver M, Borrow R, Edwards-Jones V, Fox AJ, Kaczmarski EB, Mutton KJ. Development and evaluation of a ‘real-time’ RT-PCR for the detection of enterovirus and parechovirus RNA in CSF and throat swab samples. J Med Virol. 2002;67:555–562. doi: 10.1002/jmv.10138. [DOI] [PubMed] [Google Scholar]
- 60.Verstrepen WA, Kuhn S, Kockx MM, Van De Vyvere ME, Mertens AH. Rapid detection of enterovirus RNA in cerebrospinal fluid specimens with a novel single-tube real-time reverse transcription-PCR assay. J Clin Microbiol. 2001;39:4093–4096. doi: 10.1128/JCM.39.11.4093-4096.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aberle SW, Puchhammer-Stockl E. Diagnosis of herpesvirus infections of the central nervous system. J Clin Virol. 2002;25(Suppl 1):S79–S85. doi: 10.1016/S1386-6532(02)00037-9. [DOI] [PubMed] [Google Scholar]
- 62.Krause CH, Molyneaux PJ, Ho-Yen DO, McIntyre P, Carman WF, Templeton KE. Comparison of mumps-IgM ELISAs in acute infection. J Clin Virol. 2007;38:153–156. doi: 10.1016/j.jcv.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 63.Krause CH, Eastick K, Ogilvie MM. Real-time PCR for mumps diagnosis on clinical specimens--comparison with results of conventional methods of virus detection and nested PCR. J Clin Virol. 2006;37:184–189. doi: 10.1016/j.jcv.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 64.Uchida K, Shinohara M, Shimada S, Segawa Y, Doi R, Gotoh A, Hondo R. Rapid and sensitive detection of mumps virus RNA directly from clinical samples by real-time PCR. J Med Virol. 2005;75:470–474. doi: 10.1002/jmv.20291. [DOI] [PubMed] [Google Scholar]
- 65.Lanciotti R. Molecular amplification assays for the detection of flaviviruses. Adv Virus Res. 2003;61:67–99. doi: 10.1016/S0065-3527(03)61002-X. [DOI] [PubMed] [Google Scholar]
- 66.Lanciotti RS, Ebel GD, Deubel V, Kerst AJ, Murri S, Meyer R, Bowen M, McKinney N, Morrill WE, Crabtree MB, Kramer LD, Roehrig JT. Complete genome sequences and phylogenetic analysis of West Nile virus strains isolated from the United States, Europe, and the Middle East. Virology. 2002;298:96–105. doi: 10.1006/viro.2002.1449. [DOI] [PubMed] [Google Scholar]
- 67.Zaayman D, Venter M. West Nile Virus neurologic disease in humans, South Africa, September 2008–May 2009. Emerg Infect Dis. 2012;18:2051–2054. doi: 10.3201/eid1812.111208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sanchez G, Populaire S, Butot S, Putallaz T, Joosten H. Detection and differentiation of human hepatitis A strains by commercial quantitative real-time RT-PCR tests. J Virol Methods. 2006;132:160–165. doi: 10.1016/j.jviromet.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 69.Brooks HA, Gersberg RM, Dhar AK. Detection and quantification of hepatitis A virus in seawater via real-time RT-PCR. J Virol Methods. 2005;127:109–118. doi: 10.1016/j.jviromet.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 70.Costa-Mattioli M, Di Napoli A, Ferre V, Billaudel S, Perez-Bercoff R, Cristina J. Genetic variability of hepatitis A virus. J Gen Virol. 2003;84:3191–3201. doi: 10.1099/vir.0.19532-0. [DOI] [PubMed] [Google Scholar]
- 71.Mansuy JM, Peron JM, Bureau C, Alric L, Vinel JP, Izopet J. Immunologically silent autochthonous acute hepatitis E virus infection in France. J Clin Microbiol. 2004;42:912–913. doi: 10.1128/JCM.42.2.912-913.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Orru G, Masia G, Orru G, Romano L, Piras V, Coppola RC. Detection and quantitation of hepatitis E virus in human faeces by real-time quantitative PCR. J Virol Methods. 2004;118:77–82. doi: 10.1016/j.jviromet.2004.01.025. [DOI] [PubMed] [Google Scholar]
- 73.Mansuy JM, Peron JM, Abravanel F, Poirson H, Dubois M, Miedouge M, Vischi F, Alric L, Vinel JP, Izopet J. Hepatitis E in the south west of France in individuals who have never visited an endemic area. J Med Virol. 2004;74:419–424. doi: 10.1002/jmv.20206. [DOI] [PubMed] [Google Scholar]
- 74.Banerjee A, De P, Manna B, Chawla-Sarkar M. Molecular characterization of enteric adenovirus genotypes 40 and 41 identified in children with acute gastroenteritis in Kolkata, India during 2013–2014. J Med Virol. 2016;89:606–14. [DOI] [PubMed]
- 75.Sanaei Dashti A, Ghahremani P, Hashempoor T, Karimi A. Molecular epidemiology of enteric adenovirus gastroenteritis in under-five-year-old children in Iran. Gastroenterol Res Pract. 2016;2016:2045697. doi: 10.1155/2016/2045697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ozsari T, Bora G, Kaya B, Yakut K. The prevalence of rotavirus and adenovirus in the childhood gastroenteritis. Jundishapur J Microbiol. 2016;9:e34867. doi: 10.5812/jjm.34867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu L, Qian Y, Zhang Y, Zhao L, Jia L, Dong H. Epidemiological aspects of rotavirus and adenovirus in hospitalized children with diarrhea: a 5-year survey in Beijing. BMC Infect Dis. 2016;16:1–7. doi: 10.1186/s12879-016-1829-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Qiao N, Wang X-Y, Liu L. Temporal evolutionary dynamics of norovirus GII.4 variants in China between 2004 and 2015. PLoS One. 2016;11:e0163166. doi: 10.1371/journal.pone.0163166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kabue JP, Meader E, Hunter PR, Potgieter N. Norovirus prevalence and estimated viral load in symptomatic and asymptomatic children from rural communities of Vhembe district, South Africa. J Clin Virol. 2016;84:12–18. doi: 10.1016/j.jcv.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brown JR, Shah D, Breuer J. Viral gastrointestinal infections and norovirus genotypes in a paediatric UK hospital, 2014–2015. J Clin Virol. 2016;84:1–6. doi: 10.1016/j.jcv.2016.08.298. [DOI] [PubMed] [Google Scholar]
- 81.O’Neill HJ, McCaughey C, Coyle PV, Wyatt DE, Mitchell F. Clinical utility of nested multiplex RT-PCR for group F adenovirus, rotavirus and norwalk-like viruses in acute viral gastroenteritis in children and adults. J Clin Virol. 2002;25:335–343. doi: 10.1016/S1386-6532(02)00124-5. [DOI] [PubMed] [Google Scholar]
- 82.O’Neill HJ, McCaughey C, Wyatt DE, Mitchell F, Coyle PV. Gastroenteritis outbreaks associated with Norwalk-like viruses and their investigation by nested RT-PCR. BMC Microbiol. 2001;1:14. doi: 10.1186/1471-2180-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Do LP, Kaneko M, Nakagomi T, Gauchan P, Agbemabiese CA, Dang AD, Nakagomi O. Molecular epidemiology of Rotavirus A, causing acute gastroenteritis hospitalisations among children in Nha Trang, Vietnam, 2007–2008: Identification of rare G9P[19] and G10P[14] strains. J Med Virol. 2016;89:621–31. [DOI] [PubMed]
- 84.Wandera EA, Mohammad S, Komoto S, Maeno Y, Nyangao J, Ide T, Kathiiko C, Odoyo E, Tsuji T, Taniguchi K, Ichinose Y. Molecular epidemiology of rotavirus gastroenteritis in Central Kenya before vaccine introduction, 2009–2014. J Med Virol. 2016;89:809–17. [DOI] [PubMed]
- 85.CDC . Guidance for U.S. laboratories testing for Zika virus infection. Atlanta, GA: Centres for Disease Control and Prevention; 2016. [Google Scholar]
- 86.Petersen EE, Polen KND, Meaney-Delman D, Ellington SR, Oduyebo T, Cohn A, Oster AM, Russell K, Kawwass JF, Karwowski MP, Powers AM, Bertolli J, Brooks JT, Kissin D, Villanueva J, Muñoz-Jordan J, Kuehnert M, Olson CK, Honein MA, Rivera M, Jamieson DJ, Rasmussen SA. Update: interim guidance for health care providers caring for women of reproductive age with possible Zika virus exposure — United States. MMWR Morb Mortal Wkly Rep. 2016;65:315–322. doi: 10.15585/mmwr.mm6512e2. [DOI] [PubMed] [Google Scholar]
- 87.Chow VT, Tambyah PA, Yeo WM, Phoon MC, Howe J. Diagnosis of nipah virus encephalitis by electron microscopy of cerebrospinal fluid. J Clin Virol. 2000;19:143–147. doi: 10.1016/S1386-6532(00)00094-9. [DOI] [PubMed] [Google Scholar]
- 88.Itoh Y, Takahashi M, Fukuda M, Shibayama T, Ishikawa T, Tsuda F, Tanaka T, Nishizawa T, Okamoto H. Visualization of TT virus particles recovered from the sera and feces of infected humans. Biochem Biophys Res Commun. 2000;279:718–724. doi: 10.1006/bbrc.2000.4013. [DOI] [PubMed] [Google Scholar]
- 89.Khaskhely NM, Uezato H, Kamiyama T, Maruno M, Kariya KI, Oshiro M, Nonaka S. Association of human papillomavirus type 6 with a verruciform xanthoma. Am J Dermatopathol. 2000;22:447–452. doi: 10.1097/00000372-200010000-00012. [DOI] [PubMed] [Google Scholar]
- 90.Otsu R, Ishikawa A, Mukae K. Detection of small round structured viruses in stool specimens from outbreaks of gastroenteritis by electron microscopy and reverse transcription-polymerase chain reaction. Acta Virol. 2000;44:53–55. [PubMed] [Google Scholar]
- 91.Fenner FJ, White DO. Laboratory diagnosis of viral disease. 4. New York: Academic Press; 1994. [Google Scholar]
- 92.Hendrickson KJ. Advances in the laboratory diagnosis of viral respiratory disease. Pediatr Infect Dis J. 2004;23:S6–10. doi: 10.1097/01.inf.0000108187.63151.ea. [DOI] [PubMed] [Google Scholar]
- 93.Kesson AM. Respiratory virus infections. Paediatr Respir Rev. 2007;8:240–248. doi: 10.1016/j.prrv.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Drew WL, Stevens GR. How your laboratory should perform viral studies (continued): isolation and identification of commonly encountered viruses. Laboratory Med. 1980;11:14–23. doi: 10.1093/labmed/11.1.14. [DOI] [Google Scholar]
- 95.Fan J, Hendrickson KJ, Slavatski LL. Rapid simultaneous diagnosis of infections with respiratory syncytial viruses A and B influenza viruses A and B, and human parainfluenza viruses types 1, 2 and 3 by multiplex quantitiative reverse trancription-polymerase chain reaction-enzyme hybridization assay. Clin Infect Dis. 1998;26:1397–1402. doi: 10.1086/516357. [DOI] [PubMed] [Google Scholar]
- 96.Fan J, Henrickson KJ. Rapid diagnosis of human parainfluenza virus type 1 infection by quantitative reverse transcription-PCR-enzyme hybridization assay. J Clin Microbiol. 1996;34:1914–1917. doi: 10.1128/jcm.34.8.1914-1917.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Woo PC, Chiu SS, Seto WH, Peiris M. Cost-effectiveness of rapid diagnosis of viral respiratory tract infections in pediatric patients. J Clin Microbiol. 1997;35:1579–1581. doi: 10.1128/jcm.35.6.1579-1581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Falsey AR, Formica MA, Treanor JJ, Walsh EE. Comparison of quantitative reverse transcription-PCR to viral culture for assessment of respiratory syncytial virus shedding. J Clin Microbiol. 2003;41:4160–4165. doi: 10.1128/JCM.41.9.4160-4165.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Falsey AR, Formica MA, Walsh EE. Diagnosis of respiratory syncytial virus infection: comparison of reverse transcription-PCR to viral culture and serology in adults with respiratory illness. J Clin Microbiol. 2002;40:817–820. doi: 10.1128/JCM.40.3.817-820.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 101.Higuchi R, Fockler C, Dollinger G, Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology (N Y) 1993;11:1026–1030. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- 102.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 103.Paranhos-Baccala G, Komuruan-Pradel F, Richard N, Vernet G, Lina B, Floret D. Mixed Respiratory Virus Infections. J Clin Virol. 2008;43:407–410. doi: 10.1016/j.jcv.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lassauniere R, Kresfelder T, Venter M. A novel multiplex real0time RT-PCR assay with FRET hybridizations probes for the detection and quantitation of 13 respiratory viruses. J Virol Methods. 2010;165:254–60. [DOI] [PMC free article] [PubMed]
- 105.Bellau-Pujol S, Vabret A, Legrand L, Dina J, Gouarin S, Petitjean-Lecherbonnier J, Pozzetto B, Ginevra C, Freymuth F. Development of three multiplex RT-PCR assays for the detection of 12 respiratory RNA viruses. J Virol Methods. 2005;126:53–63. doi: 10.1016/j.jviromet.2005.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kaye M, Skidmore S, Osman H, Weinbren M, Warren R. Surveillance of respiratory virus infections in adult hospital admissions using rapid methods. Epidemiol Infect. 2006;137:792–798. doi: 10.1017/S0950268805005364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lam WY, Yeung AC, Tang JW, Ip M, Chan EW, Hui M, Chan PK. Rapid multiplex nested PCR for detection of respiratory viruses. J Clin Microbiol. 2007;45:3631–3640. doi: 10.1128/JCM.00280-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Osiowy C. Direct detection of respiratory syncytial virua, prainfluenza virus and adenovirus in clinical specimens by a multiplex reverse trancription-PCR assay. J Clin Microbiol. 1998;36:3419–3154. doi: 10.1128/jcm.36.11.3149-3154.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sakthivel SK, Whitaker B, Lu X, Oliveira DBL, Stockman LJ, Kamili S, Oberste MS, Erdman DD. Comparison of fast-track diagnostics respiratory pathogens multiplex real-time RT-PCR assay with in-house singleplex assays for comprehensive detection of human respiratory viruses. J Virol Methods. 2012;185:259–266. doi: 10.1016/j.jviromet.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Palacios G, Briese T, Kapoor V, Jabado O, Liu Z, Venter M, Zhai J, Renwick N, Grolla A, Geisbert TW, Drosten C, Towner J, Ju J, Paweska J, Nichol ST, Swanepoel R, Feldmann H, Jahrling PB, Lipkin WI. MassTag polymerase chain reaction for differential diagnosis of viral hemorrhagic fevers. Emerg Infect Dis. 2006;12:692–695. doi: 10.3201/eid1204.051515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Petrik J. Microarray blood testing: Pros & cons. Biologicals. 2010;38:2–8. doi: 10.1016/j.biologicals.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 112.Liu J, Ochieng C, Wiersma S, Stroher U, Towner JS, Whitmer S, Nichol ST, Moore CC, Kersh GJ, Kato C, Sexton C, Petersen J, Massung R, Hercik C, Crump JA, Kibiki G, Maro A, Mujaga B, Gratz J, Jacob ST, Banura P, Scheld WM, Juma B, Onyango CO, Montgomery JM, Houpt E, Fields B. Development of a TaqMan Array Card for acute-febrile-illness outbreak investigation and surveillance of emerging pathogens, including ebola virus. J Clin Microbiol. 2016;54:49–58. doi: 10.1128/JCM.02257-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Harvey JJ, Chester S, Burke SA, Ansbro M, Aden T, Gose R, Sciulli R, Bai J, DesJardin L, Benfer JL, Hall J, Smole S, Doan K, Popowich MD, St George K, Quinlan T, Halse TA, Li Z, Perez-Osorio AC, Glover WA, Russell D, Reisdorf E, Whyte T, Jr, Whitaker B, Hatcher C, Srinivasan V, Tatti K, Tondella ML, Wang X, Winchell JM, Mayer LW, Jernigan D, Mawle AC. Comparative analytical evaluation of the respiratory TaqMan Array Card with real-time PCR and commercial multi-pathogen assays. J Virol Methods. 2016;228:151–157. doi: 10.1016/j.jviromet.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Driscoll AJ, Karron RA, Bhat N, Thumar B, Kodani M, Fields BS, Whitney CG, Levine OS, O’Brien KL, Murdoch DR. Evaluation of fast-track diagnostics and TaqMan array card real-time PCR assays for the detection of respiratory pathogens. J Microbiol Methods. 2014;107:222–226. doi: 10.1016/j.mimet.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu J, Gratz J, Amour C, Kibiki G, Becker S, Janaki L, Verweij JJ, Taniuchi M, Sobuz SU, Haque R, Haverstick DM, Houpt ER. A laboratory-developed TaqMan Array Card for simultaneous detection of 19 enteropathogens. J Clin Microbiol. 2013;51:472–480. doi: 10.1128/JCM.02658-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Boeckh M, Gooley TA, Myerson D, Cunningham T, Schoch G, Bowden RA. Cytomegalovirus pp65 antigenemia-guided early treatment with ganciclovir versus ganciclovir at engraftment after allogeneic marrow transplantation: a randomized double-blind study. Blood. 1996;88:4063–4071. [PubMed] [Google Scholar]
- 117.Raschilas F, Wolff M, Delatour F, Chaffaut C, De Broucker T, Chevret S, Lebon P, Canton P, Rozenberg F. Outcome of and prognostic factors for herpes simplex encephalitis in adult patients: results of a multicenter study. Clin Infect Dis. 2002;35:254–260. doi: 10.1086/341405. [DOI] [PubMed] [Google Scholar]
- 118.Boonham N, Kreuze J, Winter S, van der Vlugt R, Bergervoet J, Tomlinson J, Mumford R. Methods in virus diagnostics: from ELISA to next generation sequencing. Virus Res. 2014;186:20–31. doi: 10.1016/j.virusres.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 119.Fischer N, Indenbirken D, Meyer T, Lutgehetmann M, Lellek H, Spohn M, Aepfelbacher M, Alawi M, Grundhoff A. Evaluation of unbiased next-generation sequencing of RNA (RNA-seq) as a diagnostic method in influenza virus-positive respiratory samples. J Clin Microbiol. 2015;53:2238–2250. doi: 10.1128/JCM.02495-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Datta S, Budhauliya R, Das B, Chatterjee S, Vanlalhmuaka VV. Next-generation sequencing in clinical virology: discovery of new viruses. World J Virol. 2015;4:265–276. doi: 10.5501/wjv.v4.i3.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Woo PC, Lau SK, Chu CM, Chan KH, Tsoi HW, Huang Y, Wong BH, Poon RW, Cai JJ, Luk WK, Poon LL, Wong SS, Guan Y, Peiris JS, Yuen KY. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jeffery KJ, Read SJ, Peto TE, Mayon-White RT, Bangham CR. Diagnosis of viral infections of the central nervous system: clinical interpretation of PCR results. Lancet. 1997;349:313–317. doi: 10.1016/S0140-6736(96)08107-X. [DOI] [PubMed] [Google Scholar]
- 123.Nigrovic LE, Chiang VW. Cost analysis of enteroviral polymerase chain reaction in infants with fever and cerebrospinal fluid pleocytosis. Arch Pediatr Adolesc Med. 2000;154:817–821. doi: 10.1001/archpedi.154.8.817. [DOI] [PubMed] [Google Scholar]
- 124.Hodinka RL. 5 - Antiviral susceptibility testing using DNA-DNA hybridization. In: Wiedbrauk DL, Farkas DH, editors. Molular Methods for Virus Detection. San Diego: Academic Press; 1995. pp. 103–130. [Google Scholar]
- 125.Erali M, Page S, Reimer LG, Hillyard DR. Human immunodeficiency virus type 1 drug resistance testing: a comparison of three sequence-based methods. J Clin Microbiol. 2001;39:2157–2165. doi: 10.1128/JCM.39.6.2157-2165.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Verkruyse LA, Storch GA, Devine SM, Dipersio JF, Vij R. Once daily ganciclovir as initial pre-emptive therapy delayed until threshold CMV load > or =10000 copies/ml: a safe and effective strategy for allogeneic stem cell transplant patients. Bone Marrow Transplant. 2006;37:51–56. doi: 10.1038/sj.bmt.1705213. [DOI] [PubMed] [Google Scholar]
- 127.Riddler SA, Breinig MC, McKnight JL. Increased levels of circulating Epstein-Barr virus (EBV)-infected lymphocytes and decreased EBV nuclear antigen antibody responses are associated with the development of posttransplant lymphoproliferative disease in solid-organ transplant recipients. Blood. 1994;84:972–984. [PubMed] [Google Scholar]
- 128.Iloeje UH, Yang HI, Su J, Jen CL, You SL, Chen CJ, Risk Evaluation of Viral Load E, Associated Liver Disease/Cancer-In HBVSG Predicting cirrhosis risk based on the level of circulating hepatitis B viral load. Gastroenterology. 2006;130:678–686. doi: 10.1053/j.gastro.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 129.Ohkubo K, Kato Y, Ichikawa T, Kajiya Y, Takeda Y, Higashi S, Hamasaki K, Nakao K, Nakata K, Eguchi K. Viral load is a significant prognostic factor for hepatitis B virus-associated hepatocellular carcinoma. Cancer. 2002;94:2663–2668. doi: 10.1002/cncr.10557. [DOI] [PubMed] [Google Scholar]
- 130.Hirsch MS, Brun-Vézinet F, D’Aquila RT, et al. Antiretroviral drug resistance testing in adult hiv-1 infection: recommendations of an international aids society–usa panel. JAMA. 2000;283:2417–2426. doi: 10.1001/jama.283.18.2417. [DOI] [PubMed] [Google Scholar]
- 131.Larder B, Kemp S. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT) Science. 1989;246:1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 132.J-f L, Linley L, Kline R, Ziebell R, Heneine W, Johnson JA. Sensitive sentinel mutation screening reveals differential underestimation of transmitted HIV drug resistance among demographic groups. AIDS. 2016;30:1439–1445. doi: 10.1097/QAD.0000000000001099. [DOI] [PubMed] [Google Scholar]
- 133.Petropoulos CJ, Parkin NT, Limoli KL, Lie YS, Wrin T, Huang W, Tian H, Smith D, Winslow GA, Capon DJ, Whitcomb JM. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob Agents Chemother. 2000;44:920–928. doi: 10.1128/AAC.44.4.920-928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Clavel F, Hance AJ. HIV drug resistance. N Engl J Med. 2004;350:1023–1035. doi: 10.1056/NEJMra025195. [DOI] [PubMed] [Google Scholar]
- 135.Dudley DM, Chin EN, Bimber BN, Sanabani SS, Tarosso LF, Costa PR, Sauer MM, Kallas EG, O’Connor DH. Low-cost ultra-wide genotyping using Roche/454 pyrosequencing for surveillance of HIV drug resistance. PLoS One. 2012;7:e36494. doi: 10.1371/journal.pone.0036494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Inzaule SC, Ondoa P, Peter T, Mugyenyi PN, Stevens WS, de Wit TFR, Hamers RL. Affordable HIV drug-resistance testing for monitoring of antiretroviral therapy in sub-Saharan Africa. Lancet Infect Dis. 2016;16:267–75. [DOI] [PubMed]
- 137.Chen X, Zou X, He J, Zheng J, Chiarella J, Kozal MJ. HIV drug resistance mutations (DRMs) detected by deep sequencing in virologic failure subjects on therapy from hunan province, China. PLoS One. 2016;11:e0149215. doi: 10.1371/journal.pone.0149215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lowe CF, Merrick L, Harrigan PR, Mazzulli T, Sherlock CH, Ritchie G. Implementation of next-generation sequencing for hepatitis B virus resistance testing and genotyping in a clinical microbiology laboratory. J Clin Microbiol. 2016;54:127–133. doi: 10.1128/JCM.02229-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang Q, Liao Y, Chen J, Cai B, Su Z, Ying B, Lu X, Tao C, Wang L. Corrigendum: epidemiology study of HBV genotypes and antiviral drug resistance in multi-ethnic regions from Western China. Sci Rep. 2016;6:20451. doi: 10.1038/srep20451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Archampong TN, Boyce CL, Lartey M, Sagoe KW, Obo-Akwa A, Kenu E, Blackard JT, Kwara A. HBV genotypes and drug resistance mutations in antiretroviral treatment-naive and treatment-experienced HBV-HIV-coinfected patients. Antivir Ther. 2016;22:13–20. [DOI] [PMC free article] [PubMed]
- 141.Woods CK, Brumme CJ, Liu TF, Chui CKS, Chu AL, Wynhoven B, Hall TA, Trevino C, Shafer RW, Harrigan PR. Automating HIV drug resistance genotyping with RECall, a freely accessible sequence analysis tool. J Clin Microbiol. 2012;50:1936–1942. doi: 10.1128/JCM.06689-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kudesia G, Wreghitt T. Clinical and diagnostic virology. Cambridge: Cambridge University Press; 2009. [Google Scholar]
- 143.Korsman SNJ, van Zyl GU, Nutt L, Andersson MI, Preiser WP. Virology, an illustrated colour text. London: Elsevier; 2012. [Google Scholar]
- 144.World Health Organization . Manual for the laboratory diagnosis and virology surveillance of influenza. Geneva: WHO Press; 2011. [Google Scholar]
- 145.Wang Y, Hedman L, Perdomo MF, Elfaitouri A, Bolin-Wiener A, Kumar A, Lappalainen M, Soderlund-Venermo M, Blomberg J, Hedman K. Microsphere-based antibody assays for human parvovirus B19V, CMV and T. gondii. BMC Infect Dis. 2016;16:8. doi: 10.1186/s12879-015-1194-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Basile AJ, Horiuchi K, Panella AJ, Laven J, Kosoy O, Lanciotti RS, Venkateswaran N, Biggerstaff BJ. Multiplex microsphere immunoassays for the detection of IgM and IgG to arboviral diseases. PLoS One. 2013;8:e75670. doi: 10.1371/journal.pone.0075670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rizwan M, Ronnberg B, Cistjakovs M, Lundkvist A, Pipkorn R, Blomberg J. Serology in the digital age: using long synthetic peptides created from nucleic acid sequences as antigens in microarrays. Microarrays (Basel). 2016;5(3). pii: E22 [DOI] [PMC free article] [PubMed]
- 148.Cleton NB, Godeke GJ, Reimerink J, Beersma MF, Doorn HR, Franco L, Goeijenbier M, Jimenez-Clavero MA, Johnson BW, Niedrig M, Papa A, Sambri V, Tami A, Velasco-Salas ZI, Koopmans MP, Reusken CB. Spot the difference-development of a syndrome based protein microarray for specific serological detection of multiple flavivirus infections in travelers. PLoS Negl Trop Dis. 2015;9:e0003580. doi: 10.1371/journal.pntd.0003580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sivakumar PM, Moritsugu N, Obuse S, Isoshima T, Tashiro H, Ito Y. Novel microarrays for simultaneous serodiagnosis of multiple antiviral antibodies. PLoS One. 2013;8:e81726. doi: 10.1371/journal.pone.0081726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ginocchio CC. Quality assurance in clinical virology. In: Specter S, Hodinka R, Young S, Wiedbrauk D, editors. Clinical virology manual. 4th ed. Washington, DC: American Society of Microbiology; 2009; p.3–17.
- 151.Longo MC, Berninger MS, Hartley JL. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene. 1990;93:125–128. doi: 10.1016/0378-1119(90)90145-H. [DOI] [PubMed] [Google Scholar]


