Abstract
The ability of an influenza virus to transmit efficiently from human-to-human is a major factor in determining the epidemiological impact of that strain. The use of a relevant animal model to identify viral determinants of transmission, as well as host and environmental factors affecting transmission efficiency, is therefore critical for public health. The characterization of newly emerging influenza viruses in terms of their potential to transmit in a mammalian host is furthermore an important part of pandemic risk assessment. For these reasons, a guinea pig model of influenza virus transmission was developed in 2006. The guinea pig provides an important alternative to preexisting models for influenza. Most influenza viruses do not readily transmit among mice. Ferrets, while highly relevant, are expensive and can be difficult to obtain in high numbers. Moreover, it is generally accepted that efforts to accurately model human disease are strengthened by the use of multiple animal species. Herein, we provide an overview of influenza virus infectivity, growth, and transmission in the guinea pig and highlight knowledge gained on the topic of influenza virus transmission using the guinea pig model.
Keywords: Influenza Virus, Sialic Acid, Avian Influenza Virus, Influenza Virus Infection, Swine Influenza Virus
Introduction
The guinea pig (Cavia porcellus) has been used as a model to study influenza virus infection and immune responses since the 1970s (Wetherbee 1973; Phair et al. 1979; Azoulay-Dupuis et al. 1984). The utility of this species for the study of influenza virus transmission was, however, first described in 2006 (Lowen et al. 2006). The development of the guinea pig transmission model offered an important alternative to the better-established ferret and mouse models. Unlike mice, guinea pigs support efficient transmission of influenza viruses adapted to human hosts and are highly susceptible to infection with a broad range of influenza A and B viruses—without prior adaptation of the viruses through serial passage. Compared to ferrets, guinea pigs are relatively small and inexpensive, and seronegative guinea pigs are easily obtained, facilitating the performance of experiments that are adequately powered to gain statistically significant results. The utility of the guinea pig transmission model is evidenced by the expansion of its use since 2006. Research performed in the model has contributed important insights into influenza virus transmission, including the effects of viral, host, and environmental factors. This chapter describes influenza virus infection, growth, and transmission in guinea pigs; highlights how these properties differ among influenza viruses adapted to human, swine, and avian hosts; and provides an overview of knowledge gained through the study of influenza virus transmission in the guinea pig model.
Influenza Virus Infection in the Guinea Pig
Susceptibility to Infection
Guinea pigs are highly susceptible to infection by the intranasal route with influenza viruses derived from human, avian, and swine hosts. Determinations of 50 % infectious dose (ID50) for a handful of human and avian isolates have yielded values ranging from 3 PFU to 66 PFU (Lowen et al. 2006; Bouvier et al. 2008; Steel et al. 2009; Gabbard et al. 2013). While precise ID50 values have not been reported for swine isolates, the success of low dose inoculations with isolates of both North American and Eurasian swine influenza virus lineages indicates that the ID50 is approximately 100 PFU or less in each case (J. Steel, unpublished data). These results for ID50 in guinea pigs are similar to what has been reported for humans and ferrets (Alford et al. 1966), indicating that guinea pigs and ferrets have comparable natural susceptibilities to infection with a broad range of influenza viruses (Gustin et al. 2011; Lakdawala et al. 2011). Indeed, a sero-survey of domestic guinea pigs in Ecuador, obtained either from farms or live animal markets, revealed antibodies to influenza A and B viruses in the majority of samples tested (Leyva-Grado et al. 2012). These results suggest that guinea pigs in close contact with humans naturally acquire influenza virus infection. Nevertheless, among the guinea pigs we have obtained from laboratory animal vendors since 2006, none has been found to be seropositive in hemagglutination inhibition assays (JS, AL and NB, unpublished data); we consider this to be an important advantage of the guinea pig over the ferret model.
Viral Growth in the Guinea Pig Respiratory Tract
Following intranasal inoculation, influenza virus replication in the guinea pig is largely confined to the upper respiratory tract (Lowen et al. 2006; Gabbard et al. 2013; Seibert et al. 2013). While nasal lavage fluid and homogenates of nasal turbinate yield high virus titers, infectious virus is typically present at lower levels or not detected in the lung (Lowen et al. 2006; Gabbard et al. 2013; Seibert et al. 2013). These findings are supported by histological staining for influenza virus antigen: on days 2 and 4 post-infection, viral protein is easily detected in the nasal tissues of guinea pigs infected with the human seasonal A/Panama/2007/1999 (H3N2) [Pan/99] virus, but not found in the lungs of the same animals (Gabbard et al. 2013). In humans, seasonal influenza virus infection is mainly confined to the upper respiratory tract, while growth in the lung is more rare and associated with severe disease (Treanor 2010). Also in ferrets, seasonal influenza viruses target mainly the upper respiratory tract (van der Laan et al. 2008; Zeng et al. 2013). Thus, in broad terms, the tropism of seasonal human influenza viruses in the guinea pig is similar to that typically observed in human hosts, as well as that of ferrets.
Interesting exceptions are seen with influenza A/Anhui/1/2013 (H7N9) virus, an early isolate from the 2013 outbreak in China, and with the highly pathogenic A/duck/Guangxi/35/2001 (H5N1) [DK/35] virus. When inoculated at high dose intranasally (106 PFU or EID50), these viruses initiated productive infection in the lung of guinea pigs (Gao et al. 2009; Gabbard et al. 2013). Under the same conditions, Pan/99 and A/rhea/North Carolina/39482/1993 (H7N1) viruses did not become established in the lung, despite successful delivery of the intranasal inoculum to this tissue (Gabbard et al. 2013). Interestingly, disruption of α2,3 receptor binding in the DK/35 virus background also abrogated the growth of this virus in the guinea pig lung, suggesting that receptor usage is a major determinant of tropism for the guinea pig lower respiratory tract (Gao et al. 2009). Similarly, while human adapted strains remain localized in the upper respiratory tract of macaques, infections with highly pathogenic H5N1 and 2013 H7N9 viruses spread to the lung in these animals (Shinya et al. 2012; Watanabe et al. 2013). In humans, highly pathogenic H5N1 and A/Anhui/1/2013-like H7N9 viruses have caused lower respiratory tract complications such as pneumonia and acute respiratory distress syndrome (ARDS) in a high proportion of the identified cases (Hien et al. 2004; Gao et al. 2013). Thus, tropism of the avian-like H5N1 and H7N9 viruses is broadened not only in guinea pigs, but also in humans and nonhuman primates.
Collection of nasal lavage samples at multiple time points after infection is normally used to track influenza viral replication in guinea pigs. Compared to tissue collection typically used in mice, this method has the advantages of allowing serial sampling from the same animal and reducing animal numbers. Depending on the viral strain and inoculum dose, viral growth in the guinea pig upper respiratory tract peaks between days 2 and 4 after infection, and the infection is usually cleared by day 8 (Fig. 1). At the peak of shedding, titers obtained in guinea pig nasal washings are in the range of 106–107 PFU/ml for human seasonal viruses (Lowen et al. 2006; Mubareka et al. 2009; Steel et al. 2011; Pica et al. 2012) and approximately 105–107 PFU/ml for swine influenza viruses (Steel et al. 2010, 2011; Sun et al. 2010). Avian strains tested to date grew to maximum titers of approximately 104–105 PFU/ml, with the notable exception of A/Anhui/1/2013 (H7N9) virus, which yielded 106–107 TCID50/ml on day 2 post-infection (Steel et al. 2009; Gabbard et al. 2013; Zhang et al. 2013b). These trends in kinetics and peak titers are broadly comparable to those seen in ferrets (Maines et al. 2005, 2006; Belser et al. 2011; Barman et al. 2012) and the growth of human seasonal strains in guinea pigs is similar to that seen in experimentally infected humans (Carrat et al. 2008).
Fig. 1.
Shedding patterns of influenza viruses adapted to human, swine and avian hosts in the guinea pig model. Average viral titers detected in nasal washings collected on days 2, 4, 6, and 8 post-infection are plotted. Groups of four guinea pigs were inoculated with the indicated virus strains at a dose of 103 (human and avian isolates) or 104 (swine isolates) PFU. Error bars indicate standard deviation. Results for A/Panama/2007/1999 (H3N2), A/Netherlands/602/2009 (H1N1) and A/Texas/36/1991 (H1N1) are shown in (a); results for A/swine/Texas/4199-2/1998 (H3N2), A/swine/Spain/53207/2004 (H1N1) and A/swine/Kansas/77778/2007 (H1N1) are shown in (b); and results for A/duck/Alberta/35/1976 (H1N1), A/duck/Ukraine/1963 (H3N8), A/Anhui/1/2013 (H7N9) and A/Viet Nam/1203/2004 (H5N1) are shown in (c). All titers are in units of PFU/ml, except for those of A/Anhui/1/2013, which are in TCID50/ml
Receptor Distribution in the Guinea Pig Respiratory Tract
Influenza viruses attach to their host cells via surface exposed glycoproteins or glycolipids in which sialic acids are linked by an α2,6 or α2,3 bond to galactose. In general, influenza viruses adapted to avian species bind preferentially to α2,3 linked glycans, and strains adapted to humans bind mainly α2,6 linked receptors. The distribution of α2,6 and α2,3 sialylated glycans within host tissues is thought to be a major determinant of host susceptibility and viral tropism, and is therefore an important feature of animal models for influenza. Receptor distribution in the guinea pig respiratory tract has been examined by two methods: (i) staining of fixed tissue sections with lectins specific for α2,6 or α2,3 sialylated glycans (Gao et al. 2009; Sun et al. 2010) and (ii) binding to and detection of whole influenza virus on fixed tissue sections (termed ‘virus histochemistry’) (Gabbard et al. 2013; Siegers et al. In Press).
Staining with the lectins MAA II for α2,3 and SNA for α2,6 sialylated glycans revealed a mixture of both sugar types in the guinea pig nasal respiratory epithelia and trachea, while the lung was decorated mainly with α2,3 sialylated glycans (Gao et al. 2009; Sun et al. 2010). By comparison, similar staining of tissues derived from the human respiratory tract indicated that human upper airway epithelia showed mainly α2,6 linked sialic acid with α2,3 forms present at low levels, while trachea and bronchi showed predominantly α2,6 sialic acids and both α2,6 and α2,3 sialylated glycans were prevalent in the lower lung (Shinya et al. 2006; Nicholls et al. 2007).
By virus histochemistry, the H3N2 subtype human influenza viruses studied (Pan/99 and A/Netherlands/213/03) attached mainly to the guinea pig upper respiratory tract and the trachea, with little to no binding detected on bronchiolar and alveolar epithelia. A 2009 pandemic isolate, A/Netherlands/602/2009 (H1N1), showed high levels of binding to nasal concha as well as low and moderate positivity in the bronchioles and alveoli, respectively. The low pathogenic avian strains examined, including two 2013 H7N9 isolates, bound throughout the respiratory tract of the guinea pig. In contrast, two highly pathogenic avian-like viruses, A/VN/1194/04 (H5N1) and A/NL/219/03 (H7N7), did not show attachment in the upper respiratory tract but bound efficiently to tracheal and lung epithelia. Thus, overall, influenza virus receptors in the guinea pig appear to support attachment of human-adapted viruses mainly to the upper respiratory tract, and avian-adapted viruses either throughout the respiratory tract or to mainly lower respiratory tract tissues (Gabbard et al. 2013; Siegers et al. In Press). There is appreciable overlap between these attachment patterns and those seen in human samples. When human tissues are used as the substrate, avian influenza viruses tend to bind mainly to the lower respiratory tract, while human influenza viruses bind to both upper and lower tracts (van Riel et al. 2006, 2007, 2010, 2013; Siegers et al. In Press).
Pathology in the Guinea Pig Respiratory Tract Following Influenza Virus Infection
Overt signs of disease are not readily apparent in influenza virus infected guinea pigs (Steel et al. 2009; Van Hoeven et al. 2009a). Nevertheless, examination of respiratory tissues from infected guinea pigs has revealed significant histopathological changes. Inspection of the upper airways of infected animals, the main site of viral replication, revealed rhinitis characterized by heavy nasal mucus secretion, mild to severe intraepithelial and lamina proprial inflammation, and mild to severe vacuolation of epithelia (Tang and Chong 2009; Gabbard et al. 2013). When high doses (~106 PFU or TCID50) of virus were used, marked histopathology in the lower respiratory tract of infected guinea pigs is also observed. Mild to severe bronchointerstitial pneumonia characterized by the infiltration of immune cells and stripping of ciliated epithelia have been reported with a number of different influenza strains (Azoulay-Dupuis et al. 1984; Kwon et al. 2009; Tang and Chong 2009; Van Hoeven et al. 2009a). As expected based on results from other animal models, the severity of lung pathology was greater for the 1918 pandemic strain and a highly pathogenic H5N1 subtype virus than either a low pathogenic avian isolate or a seasonal human influenza virus (Van Hoeven et al. 2009a).
Outbred Versus Inbred Guinea Pigs
Almost exclusively, the animals used for influenza virus research since 2006 have been outbred Hartley strain guinea pigs, which are readily available from laboratory animal vendors. Viral growth and signs of disease following infection with seasonal influenza viruses have also been evaluated in inbred strain 2 and strain 13 guinea pigs, but no consistent differences to the Hartley strain were noted (Mubareka et al. 2009 and A. Lowen and P. Palese unpublished data).
Influenza Virus Transmission in the Guinea Pig
To study the transmissibility of influenza viruses in animal models, two different models are used: contact and respiratory droplet. In contact models, an inoculated animal is placed in the same cage with a naïve recipient animal. In this setting transmission can proceed through direct or indirect contact with respiratory secretions, or by the short-range spread of respiratory droplets. In respiratory droplet models, inoculated and naïve animals are placed in separate cages and exposure is achieved by placing the cages in proximity to one another, such that air exchange can occur between them. Thus, in a respiratory droplet model, influenza viruses can transmit only via droplets traveling through the air. Due to the practicalities of performing transmission experiments, most study designs do not distinguish between the spread of small droplet aerosols and larger droplets that settle out of the air more rapidly.
The set up of both contact and respiratory droplet models varies among laboratories. For example, the ratio between inoculated and contact animals used is often 1:1, but may be 1:2 or higher. An important variable for respiratory droplet experiments is the rate and directionality of airflow between cages. While the precise relationship between air exchange rates and transmission levels has not been defined, anecdotal evidence suggests this parameter may account for differing results with a particular strain of influenza virus. Cage size and permeability, ambient humidity, and temperature (discussed further below) and inoculum dose also frequently differ among research groups and may impact transmission outcomes.
Transmission of Human Influenza Viruses in the Guinea Pig
As indicated in Table 1, influenza viruses adapted to humans generally transmit well among guinea pigs. Although there is strain-to-strain variability, the following generalizations can be made. Seasonal H3N2 viruses and 2009 H1N1 pandemic strains show similar and high efficiency of transmission in both contact and respiratory droplet models (Lowen et al. 2006; Steel et al. 2009, 2010, 2011). Influenza B viruses show high levels of transmission in a contact model and intermediate transmissibility in a respiratory droplet model (Pica et al. 2012). Viruses derived from the seasonal H1N1 lineage that circulated prior to 2009 tend to show lower transmissibility in guinea pigs (Mubareka et al. 2009; Bouvier et al. 2012).
Table 1.
Transmission efficiency of influenza A and B viruses in the guinea pig model
| Virus strain | Proportion of contacts infecteda | Reference(s) |
|---|---|---|
| A/Panama/2007/1999 (H3N2) | 29/29 (C), 29/32 (RD) | (Lowen et al. 2006, 2007, 2008, 2009; Bouvier et al. 2008; Mubareka et al. 2009; Steel et al. 2009, 2010) |
| A/Jianxi/262/2005 (H3N2) | 2/3 (C) | (Sun et al. 2010) |
| A/Wisconsin/67/2005 (H3N2) | 5/8 (C) | (Lowen et al. 2009) |
| A/Puerto Rico/8/1934 (H1N1) | 0/13 (C) | (Ince et al. 2013; Seladi-Schulman et al. 2013) |
| A/Texas/36/1991 (H1N1) | 1/4 (RD) | (Mubareka et al. 2009) |
| A/New York/1253/2008 (H1N1) | 12/16 (C), 2/8 (RD) | (Bouvier et al. 2012) |
| A/New York/1326/2008 (H1N1) | 4/4 (C), 3/8 (RD) | (Bouvier et al. 2012) |
| A/Guandong/41/2006 (H1N1) | 1/3 (C) | (Sun et al. 2010) |
| A/Beijing/317/2009 (H1N1) | 3/3 (C) | (Sun et al. 2010) |
| A/California/04/2009 (H1N1) | 11/11 (C), 20/20 (RD) | (Seibert et al. 2010; Steel et al. 2010; Ince et al. 2013) |
| A/Hansa Hamburg/01/2009 (H1N1) | 8/8 (C) | (Seibert et al. 2010; Kaminski et al. 2013) |
| A/Korea/1/2009 (H1N1) | 4/6 (C) | (Kim et al. 2013) |
| A/Netherlands/602/2009 (H1N1) | 4/4 (C), 8/8 (RD) | (Chutinimitkul et al. 2010; Steel et al. 2010; Seladi-Schulman et al. 2013) |
| A/Sichuan/1/2009 (H1N1) | 8/8 (RD) | (Zhang et al. 2012, 2013b) |
| B/Victoria/2/1987 | 4/4 (C), 2/4 (RD) | (Pica et al. 2012) |
| B/Florida/4/2006 | 2/4 (RD) | (Pica et al. 2012) |
| A/Anhui/1/2013 (H7N9) | 4/4 (C) | (Gabbard et al. 2013) |
| A/Shanghai/1/2013 (H7N9) | 1/4 (RD) | (Hai et al. 2013) |
| A/Viet Nam/1203/2004 (H5N1) | 3/4 (C), 0/4 (RD) | (Steel et al. 2009, JS & AL unpublished) |
| A/duck/Fujian/17/2001 (H5N1) | 0/3 (C) | (Gao et al. 2009) |
| A/duck/Guangxi/22/2001 (H5N1) | 0/3 (C) | (Gao et al. 2009) |
| A/duck/Guangxi/35/2001 (H5N1) | 3/3 (C) | (Gao et al. 2009) |
| A/duck/Shanghai/13/2001 (H5N1) | 0/3 (C) | (Gao et al. 2009) |
| A/duck/Guangdong/22/2002 (H5N1) | 0/3 (C) | (Gao et al. 2009) |
| A/bar-headed goose/Qinghai/3/2005 (H5N1) | 3/3 (C) | (Gao et al. 2009) |
| A/quail/Hong Kong/G1/1997 (H9N2) | 0/3 (C) | (Sun et al. 2010) |
| A/chicken/Shandong/ZB/2007 (H9N2) | 0/3 (C) | (Sun et al. 2010) |
| A/chicken/Hebei/LC/2008 (H9N2) | 0/3 (C) | (Sun et al. 2010) |
| A/chicken/Shandong/A/2009 (H9N2) | 1/3 (C), 0/3 (RD) | (Lv et al. 2012) |
| A/chicken/Shandong/M/2009 (H9N2) | 3/3 (C), 0/3 (RD) | (Lv et al. 2012) |
| A/duck/Ukraine/1/1963 (H3N8) | 0/4 (C) | (Gabbard et al. 2013) |
| A/duck/Alberta/35/1976 (H1N1) | 0/4 (C) | (Gabbard et al. 2013) |
| A/rhea/North Carolina/39482/1994 (H7N1) | 0/4 (C) | (Gabbard et al. 2013) |
| A/swine/Texas/4199-2/1998 (H3N2) | 1/4 (RD) | (Steel et al. 2010) |
| A/swine/Guangdong/7/2006 (H3N2) | 0/3 (C) | (Sun et al. 2010) |
| A/swine/Guangdong/211/2006 (H3N2) | 0/3 (C) | (Sun et al. 2010) |
| A/swine/Guangdong/811/2006 (H3N2) | 0/3 (C) | (Sun et al. 2010) |
| A/swine/Guangdong/968/2006 (H3N2) | 0/3 (C) | (Sun et al. 2010) |
| A/swine/Guangdong/1222/2006 (H1N2) | 0/3 (C) | (Sun et al. 2010) |
| A/swine/Guangdong/33/2006 (H1N1) | 0/3 (C) | (Sun et al. 2010) |
| A/swine/Fujian/204/2007 (H1N1) | 0/3 (C) | (Sun et al. 2010) |
Data include those obtained under regulated conditions of 20 °C and 20 % relative humidity, and under standard animal room conditions. RD = respiratory droplet. C = contact
aAs determined by virus detection in nasal washings
For a given strain of influenza virus, transmission in a contact model occurs more rapidly than transmission in a respiratory droplet model (Lowen et al. 2006; Steel et al. 2010; Pica et al. 2012). In addition, the rate and efficiency of transmission among guinea pigs placed in separate cages have been found to decline as the cages are moved further apart (Lowen et al. 2006; Mubareka et al. 2009). These observations most likely reflect the dilution of infectious bio- aerosols with distance from the shedding host.
Transmission of Avian and Swine Influenza Viruses in the Guinea Pig
Most low pathogenic avian influenza viruses tested to date have not transmitted in the guinea pig model (Table 1). Exceptions include the human isolates, A/Anhui/1/2013 and A/Shanghai/1/2013 (H7N9), which are representatives of the 2013 outbreak in China (Gabbard et al. 2013; Hai et al. 2013) and two H9N2 subtype strains isolated from chickens in Shandong (Lv et al. 2012). In addition, certain highly pathogenic avian influenza viruses of the H5N1 subtype have shown efficient transmission among co-caged guinea pigs (Gao et al. 2009; Steel et al. 2009).
Results obtained in the guinea pig model with influenza viruses adapted to swine hosts vary among strains tested. A diverse set of Chinese isolates, representing classical, European avian-like, North American triple reassortant, and human-like swine lineages showed no transmission in a contact model (Sun et al. 2010). We have observed inefficient respiratory droplet transmission of the TRIG lineage A/swine/Texas/4199-2/1998 (H3N2) virus (Steel et al. 2010) and limited transmission by a contact route of the TRIG A/swine/Kansas/77778/2007 (H1N1) virus and the European avian-like A/swine/Spain/53207/2004 (H1N1) virus (J. Steel, unpublished data).
Dependence of Influenza Virus Transmission on Environmental Factors
The guinea pig model was used to give much needed experimental insight into the underlying causes of influenza seasonality. Namely, we used the model to test the effects of two environmental conditions that vary with the seasons, relative humidity, and temperature, on influenza virus transmission. Infected and exposed guinea pigs were housed in environmental test chambers for the duration of the exposure period. Transmission efficiency was determined at temperatures of 5, 20, and 30 °C and relative humidities of 20, 35, 50, 65, and 80 %. Using several diverse influenza viruses, including Pan/99, A/Netherlands/602/2009 (H1N1), A/New York/08-1253/2008 (H1N1), B/Victoria/2/1987 and B/Florida/4/2006, transmission was found to be markedly more efficient at 5 °C than at 20 °C (Lowen et al. 2007; Steel et al. 2011; Bouvier et al. 2012; Pica et al. 2012). Continuing this trend, transmission by respiratory droplet was highly inefficient when temperatures were increased to 30 °C (Lowen et al. 2007; Steel et al. 2011). Both high (80 %) and intermediate (50 %) humidities were found to reduce transmission efficiency in a respiratory droplet model (Lowen et al. 2007; Steel et al. 2011), in line with earlier data on the stability of influenza viruses in an aerosol (Schaffer et al. 1976). Although some effect of humidity and temperature have been observed when using a contact transmission model, the effects are dampened compared to those on respiratory droplet transmission, most likely due to the higher overall efficiency of transmission among co-caged animals (Lowen et al. 2008; Pica et al. 2012). Overall, examinations of influenza virus transmission under varying conditions of humidity and temperature strongly suggest that the seasonality of influenza is caused by improved transmission under cold, dry conditions found in the winter-time in temperature regions of the world.
Identification of Viral Determinants of Transmission in the Guinea Pig Model
When coupled with reverse genetics systems that enable the targeted mutagenesis of influenza virus genomes, the guinea pig model allows the identification of viral traits important for transmission. The fact that diverse human-adapted influenza viruses transmit well among guinea pigs, while most avian and swine adapted strains do not, supports the likelihood that viral factors found to be important for guinea pig-to-guinea pig transmission will also be important for human-to-human transmission.
Determinants in the PB2 Protein
Certain amino acids located in the surface-exposed “627 domain” of PB2 have been found to be highly important for optimal replication in mammalian species (Subbarao et al. 1993; Gabriel et al. 2005). In particular, the amino acid changes E627K, D701N and G590S/Q591R, have each been implicated in the adaptation to humans of influenza viruses derived from avian and swine reservoirs (de Jong et al. 2006; Mehle and Doudna 2009; de Wit et al. 2010). The guinea pig model was used to test the roles of 627K and 701N in supporting transmission among mammals (Gao et al. 2009; Steel et al. 2009). K627E and K627E / D701N mutant viruses were generated in the backgrounds of the highly pathogenic A/Viet Nam/1203/2004 (H5N1) virus and the human seasonal Pan/99 (H3N2) virus (Steel et al. 2009). The H5N1 mutants were then compared to the wild-type virus in a contact transmission model, while the H3N2 strains were tested in a respiratory droplet model. In both viral backgrounds, the presence of either human-like adaptation (PB2 627K or 701N) led to greater transmission efficiency over the avian-like amino acid sequence. Similarly, in the context of the highly pathogenic A/duck/Guangxi/35/2001 (H5N1) virus, which is transmissible among co-housed guinea pigs, the presence of the wild-type asparagine at PB2 701 was required for transmission (Gao et al. 2009). This work identified PB2 627K and 701N as determinants of, not only growth and pathogenicity, but also transmission in mammals. Analogous results were also obtained in the ferret model, using the 1918 H1N1 pandemic strain (Van Hoeven et al. 2009b). The observation that these residues contribute to the transmission of H3N2, H1N1, and H5N1 influenza viruses suggests that adaptations in the PB2 627 domain may be a prerequisite for mammalian transmission common to all influenza A subtypes.
Outside of the 627 domain, the position PB2 271 was also shown to affect transmission. Specifically, the mutation A271T in the background of a 2009 pandemic virus abolished transmission by a respiratory droplet route. Both the A and T polymorphisms are prevalent in swine influenza viruses, but 271A is found mainly in the triple reassortant swine lineage and is rare in the classical and Eurasian avian-like swine viruses (Zhang et al. 2012).
Determinants Within the M Segment
Evidence that the influenza virus M segment encodes determinants of transmission was first obtained through the study of the swine-origin 2009 pandemic strain. Although human infection with swine influenza viruses occurs occasionally when individuals are in close contact with pigs, these swine-adapted strains do not typically transmit onward from human-to-human. In contrast, the 2009 pandemic strain originated in the swine reservoir, crossed the species barrier to humans and spread rapidly through the human population, affecting an estimated 1–2 billion people world-wide (Van Kerkhove et al. 2013). These events raised the question, what viral traits differentiating the 2009 pandemic strain from other swine influenza viruses allowed its transmission among humans? One hypothesis related to the highly unusual genotype of this virus: six segments were derived from the North American triple reassortant swine lineage and two segments, NA and M, from the Eurasian swine lineage (Smith et al. 2009).
Studies in the guinea pig model have revealed an important role for the pandemic M segment in supporting the transmissibility of the 2009 virus (Chou et al. 2011). When the M segment from the pandemic isolate, A/California/4/2009 (H1N1) [Cal/09], was included in the background of the laboratory adapted and non-transmissible A/Puerto Rico/8/1934 (H1N1) [PR8] virus, the resultant 7 + 1 reassortant virus transmitted to five of eight respiratory droplet contacts. Similarly, in the background of a North American triple reassortant swine influenza virus, A/swine/Texas/1998 (H3N2), inclusion of the HA, NA, and M segments of Cal/09 resulted in six of eight contacts becoming infected. Inclusion of the Cal/09 HA and NA alone in the A/swine/Texas/1998 background resulted in transmission to only two of eight contact animals. The importance of the Eurasian origin M segment to the highly transmissible phenotype of the 2009 pandemic virus is also supported by data obtained in ferret (Lakdawala et al. 2011) and pig (Ma et al. 2012) models.
A prominent role for the M segment in determining viral fitness and transmissibility in guinea pigs is further supported by two recent efforts to adapt PR8 virus to guinea pigs through serial passage (Ince et al. 2013; Seladi-Schulman et al. 2013). Three independent lineages of guinea pig adapted PR8 virus, generated in two different laboratories, carried coding changes in the M1 open reading frame. All of the viruses passaged in guinea pigs exhibited improved growth and transmission among co-caged animals. M1 mutations identified in the first study, at positions 62 and 166, were confirmed to contribute to these adapted phenotypes (Ince et al. 2013), but individual M1 changes identified in the second study were not sufficient to support improved growth or transmission (Seladi-Schulman et al. 2013).
2009 Pandemic PA and NS Segments Confer Transmissibility to a Highly Pathogenic H5N1 Virus
To gauge the risk of human-transmissible H5 subtype viruses arising through reassortment between a highly pathogenic avian H5N1 influenza virus and strains of the 2009 pandemic lineage, Zhang et al. tested the transmissibility of a broad set of 21 reassortants in the guinea pig model (Zhang et al. 2013b). The viruses were generated by reverse genetics using the isolates A/duck/Guangxi/35/2001 (H5N1) [DK/35] and A/Sichuan/1/2009 (H1N1)[SC/09] and all gene combinations tested included the H5 HA. The wild-type DK/35 H5N1 virus transmitted efficiently among guinea pigs placed in the same cage, but not by a respiratory droplet route. Like other 2009 pandemic strains, SC/09 transmitted with high efficiency by respiratory droplet. The report by Zhang et al. is rich in data, but the most striking results were the highly efficient transmission of DK/35-based reassortant viruses carrying only the PA or NS gene segments of SC/09. It should be noted that the DK/35 virus H5 HA protein shows partial binding to 2,6 linked sialic acids and that this ability to bind human-type receptors was required for its transmission among co-caged guinea pigs (Gao et al. 2009). Thus, when combined with a semi-adapted HA protein in a highly pathogenic H5N1 virus background, the PA or NS gene segments of the 2009 pandemic virus are sufficient to support transmission among guinea pigs in the absence of direct or indirect contact (Zhang et al. 2013b).
Determinants in the HA Protein
The receptor binding specificity of the HA protein has been identified as an important determinant of transmission efficiency in a number of contexts. In guinea pigs, robust binding to alpha 2,6 linked sialic acids was shown to be required for the respiratory droplet transmission of the pandemic strain A/Sichaun/1/2009 (H1N1) (Zhang et al. 2012) and the contact transmission of A/duck/Guangxi/35/2001 (H5N1) (Gao et al. 2009).
Transmission and overall fitness were also affected by a natural polymorphism found at position 147 of human-adapted, H1 subtype hemagglutinins (Kim et al. 2013). K147 was shown to stabilize the interaction of the 2009 H1 hemagglutinin with sialic acid (Xu et al. 2012). The function of this site within the HA was found to be particularly important to maintaining fitness upon the introduction of glycosylation sites that were acquired by H1N1 viruses circulating in humans from 1918 to 2009, but have not yet been acquired by viruses of the 2009 pandemic lineage. Thus, the presence of K147 may allow the evolution of increased glycosylation as a means of immune escape, without the acquisition of fitness defects (Kim et al. 2013).
Impact of Antiviral Drug Treatment and Resistance on Influenza Virus Transmission
Although timely vaccination is clearly the best way to confer relatively long-lasting protection against influenza-associated morbidity and mortality on an individual or population level, subtype-specific vaccines are generally not immediately available in a pandemic that follows an antigenic shift. In this case, antiviral drugs, used either prophylactically or therapeutically, would be of great value if they could prevent or reduce transmission among humans while a specific vaccine is in preparation.
Very little research has been performed to date on the efficacy of antiviral drugs to prevent influenza virus infection in or transmission among guinea pigs. Because influenza virus-infected guinea pigs do not display obvious clinical signs such as fever or cough, demonstrating a decrease in symptoms or signs is not a practicable endpoint in this model. Antiviral efficacy in this species can, however, be assessed by measuring reductions in nasopharyngeal viral replication in or transmission among susceptible guinea pigs.
Treatment with Oseltamivir
In an unpublished experiment presented in Fig. 2, we investigated the impact of an NA inhibitor, oseltamivir, on the respiratory droplet transmission of influenza virus among guinea pigs. Two groups of guinea pigs were inoculated with Pan/99 virus (103 PFU per animal) and treated with 10 doses of oral oseltamivir in 1.5 % sucrose-water (75 mg/kg/day in two divided doses, administered 12 h apart) or a sucrose-water placebo, starting 4 h before inoculation and continuing for 5 days. The placebo-treated guinea pigs demonstrated normal nasopharyngeal virus shedding kinetics, and 3 of 4 animals transmitted Pan/99 by respiratory droplets to a naive partner animal (Fig. 2). However, among the Pan/99-inoculated, oseltamivir-treated guinea pigs, only one animal displayed a typical virus shedding pattern, while two guinea pigs had no detectable virus in any nasal washes, and five guinea pigs demonstrated protracted viral replication kinetics, with low virus titers (<103 PFU/ml) on day 2 post-inoculation and peak shedding delayed by 2–6 days. Importantly, none of the eight naïve guinea pigs exposed to these Pan/99-inoculated, oseltamivir-treated guinea pigs became infected by respiratory droplet transmission (N.M. Bouvier and M. Michta, unpublished data). These experiments demonstrate that NA enzymatic function is required for maximally efficient influenza virus transmission among guinea pigs and suggest that treatment of influenza patients with NA inhibitors may prevent or reduce transmission among unvaccinated humans, though clinical trials would be required to confirm this hypothesis.
Fig. 2.
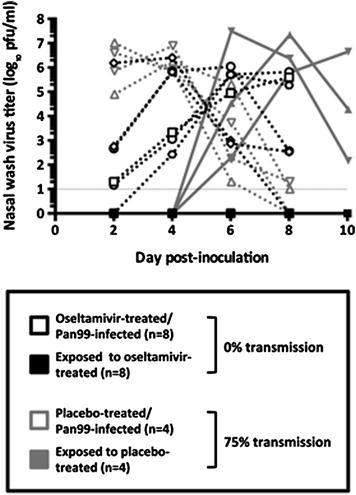
Oseltamivir alters the kinetics of influenza virus shedding and prevents Respiratory droplet transmission in guinea pigs. Lines represent the Pan/99 virus titer in each individual guinea pig nasal wash, plotted as a function of day post-inoculation. Eight Pan/99-inoculated, oseltamivir-treated guinea pigs failed to transmit virus to a naïve partner animal, while 3 of the 4 naïve guinea pigs paired with Pan/99-inoculated, placebo-treated animals became infected by respiratory droplet transmission. Black lines and symbols represent the oseltamivir treatment group, and grey lines and symbols represent the placebo treatment group. Dotted lines and open symbols represent intranasally inoculated guinea pigs, and solid lines and closed symbols represent naïve guinea pigs exposed, starting 24 h post-inoculation, to the respiratory droplets exhaled by inoculated animals. (Unpublished data, N.M. Bouvier and M. Michta)
Treatment with Type I Interferon
Daily intranasal treatment with recombinant human type I interferon (Horisberger and de Staritzky 1987) was also highly effective in limiting the growth of influenza virus in inoculated, treated guinea pigs (Van Hoeven et al. 2009a; Steel et al. 2010) and in preventing transmission from these animals to untreated contacts in the same cage (Steel et al. 2010). Furthermore, treatment of naïve contact guinea pigs was effective in preventing their infection via transmission from untreated, infected cage mates (Steel et al. 2010). Similarly reduced viral titers were observed in interferon-treated ferrets challenged with seasonal influenza viruses (Kugel et al. 2009). Thus, activation of the innate immune response through interferon treatment is an effective means of limiting influenza virus transmission, both from and to interferon treated animals.
Transmission Potential of Oseltamivir Resistant Influenza Viruses
Early in the preclinical development of the NA inhibitor class of antiviral medications, it was observed that mutations in the viral NA could confer relative resistance to the inhibitory action of oseltamivir and, to a lesser degree, zanamivir. Type and subtype-specific point mutations—most commonly, in N2 numbering, H274Y or N294S in the N1 subtype of influenza A viruses; E119V or R292K in the N2 subtype; and R152K or D198N in influenza B viruses (Govorkova 2012)—were observed in cell culture with viruses passaged in the presence of sub-inhibitory NA inhibitor concentrations (Gubareva et al. 1996, 1997; McKimm-Breschkin et al. 1998; Tai et al. 1998; Barnett et al. 1999; Baz et al. 2007), and in the clinic, in influenza patients treated with oseltamivir (Gubareva et al. 1998, 2001; Kiso et al. 2004; de Jong et al. 2005).
Between 2002 and 2005, some research was performed in the ferret contact transmission model to elucidate the impact of oseltamivir resistance mutations on the fitness of several influenza A viruses isolated in the prior decade. The H274Y NA mutation in an A/New Caledonia/20/1999 (H1N1)-like virus was observed to compromise its infectivity and ferret transmissibility, relative to its wild-type parental isolate (Herlocher et al. 2004). Among A/(H3N2) viruses, both A/Wuhan/359/1995-like and A/Sydney/05/1997-like viruses with the NA-R292K mutation transmitted very poorly between donor and recipient ferrets (Herlocher et al. 2002; Yen et al. 2005); however, Wuhan/95-like viruses encoding the NA-E119V substitution transmitted as efficiently between ferrets as the oseltamivir-sensitive parental virus (Herlocher et al. 2004; Yen et al. 2005).
However, the NA-E119V oseltamivir-resistance mutation is rarely seen in human isolates, except those obtained from oseltamivir-treated patients. Thus, it was hypothesized that this mutation may confer a slight fitness deficit sufficient to prevent widespread human-to-human transmission of viruses encoding it, but subtle enough to have been imperceptible in the ferret contact transmission experiments performed previously. Reverse genetics-derived Pan/99 viruses with and without the NA-E119V mutation were found to transmit with equal efficiency in the guinea pig contact transmission model, as had been seen with similar isolates in the ferret model (Herlocher et al. 2004; Yen et al. 2005). However, in the guinea pig respiratory droplet transmission model, wild-type Pan/99 again transmitted efficiently, to 7 of 8 exposed guinea pigs, while Pan/99 encoding the NA-E119V oseltamivir-resistance mutation transmitted to only 2 of 8 exposed guinea pigs (p < 0.05, by Fisher’s exact test) (Bouvier et al. 2008). Thus, the results previously observed in the ferret contact transmission model were successfully replicated in the guinea pig model; however, the guinea pig respiratory droplet transmission experiments suggested that a subtle fitness penalty was indeed associated with acquisition of the E119V oseltamivir-resistance mutation.
Prior to 2008, the vast majority of in vitro and in vivo data suggested that oseltamivir resistance mutations in the influenza virus NA came at some cost to viral fitness (Carr et al. 2002; Herlocher et al. 2002, 2004; Ives et al. 2002; Yen et al. 2005; Zurcher et al. 2006; Bouvier et al. 2008), leading many to conclude that most oseltamivir-resistant viruses were “of limited clinical significance” (Tai et al. 1998) or “unlikely to be of clinical consequence in man” (Ives et al. 2002). However, coincident with the circulation of a new antigenic drift variant characterized by A/Brisbane/59/2007 (H1N1) (Bris/59), in 2008 a sudden increase in the prevalence of oseltamivir resistance among seasonal influenza A(H1N1) (sH1N1) virus isolates was noted. Within 5 months, 25 % of European sH1N1 isolates encoded the NA-H274Y oseltamivir-resistance mutation; by 2009, it was found in 96 % of sH1N1 isolates worldwide (World Health Organization 2009).
In the ferret contact transmission model, oseltamivir-resistant (NA-H274Y) and -sensitive (NA-H274) viruses were shown to replicate and transmit equally well (Abed et al. 2011). However, enhanced transmission efficiency among humans was one of only a few hypotheses that could account for the unprecedented, exponential increase in prevalence of the NA-H274Y mutation in sH1N1 viruses (Chao et al. 2012). With paired oseltamivir-sensitive and -resistant Bris/59-like clinical isolates from the New York State Department of Health, we demonstrated that the oseltamivir-resistant isolate transmitted more efficiently among guinea pigs than its oseltamivir-sensitive counterpart. Using reverse genetics, we rescued reassortants between the oseltamivir-sensitive (A/New York/08-1253/2008, “NY/1253”) and -resistant (A/New York/08-1326/2008; “NY/1326”) isolates and found that expression of oseltamivir-resistant NY/1326 NA, in a virus backbone comprising the other seven segments from the oseltamivir-sensitive isolate NY/1253, was sufficient to significantly enhance transmission efficiency among guinea pigs (p = 0.009 by the logrank test). Because NY/1253 encodes two non-consensus residues (S336N and M430L) in its NA gene, we also reassorted it with the Bris/59 NA to create a virus that differs only by only two amino acids (NA-H274Y and D354G) from the 7:1 reassortant encoding the NY/1326 NA. This reassortant also transmitted less efficiently than the one encoding the 1326 NA, though it was narrowly nonsignificant (p = 0.078) (Fig. 3) (Bouvier et al. 2012). These data support the hypothesis that the exponential increase in oseltamivir-resistant Bris/59-like sH1N1 influenza viruses resulted from enhanced human-to-human transmissibility conferred primarily by the oseltamivir-resistant NA. These experiments also highlight an advantage of the guinea pig model; namely, lower cost relative to the ferret model, which enables the use of adequately powered experimental groups. A subtle but statistically significant enhancement in transmission efficiency could be demonstrated with experimental groups containing eight guinea pig pairs, whereas prior experiments using groups of four ferret pairs were insufficiently powered to reveal a significant difference in transmissibility between oseltamivir-sensitive and -resistant Bris/59-like viruses (Abed et al. 2011).
Fig. 3.
Guinea pig transmission of A/Brisbane/59/2007-like seasonal influenza A(H1N1) viruses is enhanced by expression of an oseltamivir-resistant NA. A time-to-event (Kaplan-Meier) analysis of data reported previously (Bouvier et al. 2012) demonstrates that a 7:1 reassortant encoding the oseltamivir-resistant NA typical of Brisbane/59-like sH1N1 viruses, expressed in the context of the remaining seven segments from an oseltamivir-sensitive isolate, transmits significantly more rapidly than does the oseltamivir-sensitive isolate itself, similar to the transmission kinetics of the wild-type clinical isolates themselves. Black lines represent reverse genetics-derived viruses, and grey lines represent wild-type clinical isolates. Solid lines represent viruses with an oseltamivir-resistant NA, and dotted lines represent viruses with an oseltamivir-sensitive NA. **, p < 0.01; ***, p < 0.001
The guinea pig model has recently been employed to investigate the mammalian transmissibility of avian-origin influenza A(H7N9) viruses (Gabbard et al. 2013; Hai et al. 2013), which were first isolated from humans in the spring of 2013. These novel viruses have been shown to transmit efficiently by contact in both ferrets and guinea pigs (Belser et al. 2013; Gabbard et al. 2013; Zhu et al. 2013) but generally less efficiently by respiratory droplets in both species (Belser et al. 2013; Gabbard et al. 2013; Hai et al. 2013; Richard et al. 2013; Watanabe et al. 2013; Xu et al. 2013; Zhang et al. 2013a; Zhu et al. 2013). In contrast to previous ferret transmission experiments demonstrating inefficient transmission of H3N2 viruses encoding the NA-R292K oseltamivir-resistance mutation (Herlocher et al. 2002; Yen et al. 2005), a reverse genetics-derived clone of the oseltamivir-resistant clinical isolate A/Shanghai/1/2013 (SH/1), encoding the NA-R292K mutation, transmitted no less efficiently than a 7:1 reassortant that expressed the oseltamivir-sensitive NA-R292 from A/Anhui/1/2013 (AH/1) in a SH/1 backbone. Because PR8 reassortants encoding the NAs from both SH/1 and AH/1 demonstrated roughly equivalent decrements in NA enzyme velocity and substrate affinity and in hemagglutination activity, it was hypothesized that reduced HA expression accompanying the oseltamivir-resistant NA may offset the decrements in NA enzymatic function conferred by the NA-R292K mutation (Hai et al. 2013). These experiments underscore the multigenic nature of fitness in influenza A viruses.
Impact of Vaccination on Influenza Virus Transmission
Potential for Vaccination to Limit Transmission
Blocking transmission through vaccination has the potential to control influenza in all age groups, thereby limiting the burden of disease. Nevertheless, vaccines are not typically evaluated in terms of the efficiency of transmission to or from vaccinated individuals. To demonstrate the value of this approach, we tested immunization through natural infection, intramuscular administration of killed influenza virus, or intranasal infection with a live attenuated influenza virus (Lowen et al. 2009). Immunized guinea pigs were challenged either through intranasal inoculation or through exposure to acutely infected guinea pigs. In addition, naïve animals were housed with vaccinated and challenged animals to assess the reduction in transmission efficiency achieved through vaccination. Immunity acquired through natural infection was found to fully protect from challenge with homologous and heterologous strains and, therefore, block any onward transmission. The live attenuated vaccine tested, encoding a truncated NS1 protein, also fully blocked transmission from vaccinated animals, but allowed partial transmission of the heterologous challenge virus to vaccinated guinea pigs. The killed, whole virus vaccine did not provide sterilizing protection, even against homologous challenge, by either intranasal or contact exposure routes. The killed vaccine, however, reduced but did not block onward transmission from vaccinated guinea pigs to naïve cagemates (Lowen et al. 2009).
A similar study design in the ferret model recently generated comparable results (Houser et al. 2013). Vaccination of ferrets with the 2010/2011 trivalent inactivated vaccine reduced transmission only marginally. The homologous seasonal H3N2 virus and a heterologous H3N2v swine-like virus transmitted from vaccinated and challenged ferrets to the majority of naïve contacts. In contrast, immunization through infection with H3N2 seasonal strains prevented transmission of the heterologous H3N2v challenge virus to all but one of nine contact ferrets (Houser et al. 2013).
Thus, the data obtained in both the guinea pig and ferret models suggest that the public health impact of influenza vaccination could be improved through the optimization of vaccines to more efficiently block transmission.
Role of Secreted IgA in Mediating Protection from Transmission
Expanding upon the work of Lowen and colleagues, (Lowen et al. 2009), Seibert and colleagues (Seibert et al. 2013) investigated the impact of immunoglobulin isotypes on transmission efficiency. With 30D1, a neutralizing mouse monoclonal IgG2b antibody directed at the globular head of the HA of the 2009 pandemic H1N1 (pH1N1) virus A/California/04/2009 (Cal/09), they observed that passive immunization by intramuscular administration of this antibody (at 10 mg/kg) did not protect naïve guinea pigs from infection by transmission of Cal/09 from inoculated partner animals, despite achieving high serum antibody titers after immunization. In contrast, a single intranasal (IN) administration of 30D1 (900 ng per animal) in naïve guinea pigs, which were subsequently exposed to Cal/09-inoculated partners, was sufficient to completely abrogate infection by transmission, suggesting that prevention of infection by respiratory droplets is mediated by the presence of neutralizing antibody at the respiratory mucosa, not in serum. To confirm this finding with a physiologically relevant isotype, the variable region of 30D1 was cloned into the murine IgA heavy chain gene, and expressed with murine κ and J chains. Guinea pigs immunized intramuscularly with this 30D1 IgA construct at 1 mg/kg were not protected from infection with Cal/09 by transmission, but 7 of 8 guinea pigs immunized with 5 mg/kg of 30D1 IgA were protected. The 30D1 IgA antibody was detectible by ELISA in the nasal washes of guinea pigs immunized with 5 mg/kg of 30D1 IgA, but not in those given the lower dose. Collectively, these experiments indicate that mucosal immunity, particularly the expression of sufficient quantities of neutralizing antibodies at the mucosal surfaces of the respiratory tract, is more important than serum IgG in preventing transmission of influenza viruses by respiratory droplets. These results, together with those of Lowen, Steel et al. (Lowen et al. 2009) and Houser et al. (Houser et al. 2013), suggest that inactivated influenza vaccines, which stimulate a primarily IgG antibody response, may not optimally protect against transmission of influenza viruses, and thus the clinical efficacy of influenza vaccines intended to block human-to-human transmission may be more accurately assessed with a correlate of protection other than serum IgG titers.
Conclusions and Perspectives
Research carried out in the guinea pig model over the past eight years has demonstrated the utility of this system for the study of influenza virus transmission. The robust growth and efficient transmission in guinea pigs of influenza viruses adapted to human hosts, compared to the poor growth and lack of transmission of most avian adapted strains, strongly supports the relevance of the model to transmission in the human population. While the absence of measurable signs of disease limits the use of the guinea pig for pathogenesis studies, analysis of tissues using histological methods allows the virulence of an infection to be gauged. Further characterization of the pathology induced by influenza viruses in the guinea pig is warranted. As more research groups adopt the guinea pig as a model system for influenza research, a need has arisen for more data on the differences and similarities between ferret and guinea pig models. A direct comparison, in which influenza virus growth, transmission, and disease are evaluated under standardized conditions in both species, would be highly valuable to the ultimate goal of understanding influenza in humans. Finally, despite significant recent progress, much remains to be learned about the factors driving influenza virus transmission. The reasons for the acute dependence of transmission on humidity and temperature are not yet clear. Similarly, mechanisms underlying the contributions of the M1 and/or M2 proteins and polymerase components to transmission phenotypes remain largely unknown. Ultimately, a comprehensive model of the viral traits required to support influenza virus transmission in avian versus mammalian species is needed. The complex relationships among viral components and between viral and host factors have made, and will continue to make, this aim difficult to attain. If achieved, however, an in-depth understanding of the requirements for sustained influenza virus transmission in both avian and mammalian reservoirs would be invaluable to public health efforts aimed at controlling influenza.
Acknowledgments
Research in the authors’ laboratories is supported by the NIH under R01 AI099000 (to AL) and the Center for Excellence in Influenza Research and Surveillance (CEIRS) contract number HHSN272201400004C (to JS and AL).
Glossary
Abbreviations
- Pan/99
A/Panama/2007/1999 (H3N2)
- PR8
A/Puerto Rico/8/1934 (H1N1)
- SC/09
A/Sichuan/1/2009 (H1N1)
- Bris/59
A/Brisbane/59/2007 (H1N1)
- DK/35
A/duck/Guanxi/35/2001 (H5N1)
- NY/1253
A/New York/08-1253/2008 (H1N1)
- NY/1326
A/New York/08-1326/2008 (H1N1)
- AH/1
A/Anhui/1/2013 (H7N9)
- SH/1
A/Shanghai/1/2013 (H7N9)
- Cal/09
A/California/04/2009 (H1N1pdm09)
- sH1N1
Seasonal influenza A(H1N1)
- pH1N1
2009 pandemic influenza A(H1N1)
- PFU
Plaque-forming unit
- ID50
50 % infectious dose
- EID50
50 % egg infectious dose
- TCID50
50 % tissue culture infectious dose
- MUNANA
2’-(4-methylumbelliferyl)-α-D-N-acetylneuraminic acid
- MAA II
Maakia amurensis agglutinin II
- SNA
Sambucus nigra agglutinin
Contributor Information
Richard W. Compans, Phone: +1(404)727-2015, Email: rcompan@emory.edu
Michael B. A. Oldstone, Phone: +1858-784-8054, Email: mbaobo@scripps.edu
John Steel, Email: john.steel@emory.edu.
References
- Abed Y, Pizzorno A, Bouhy X, Boivin G. Role of permissive neuraminidase mutations in influenza A/Brisbane/59/2007-like (H1N1) viruses. PLoS Pathog. 2011;7:e1002431. doi: 10.1371/journal.ppat.1002431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alford RH, Kasel JA, Gerone PJ, Knight V. Human influenza resulting from aerosol inhalation. Proc Soc Exp Biol Med. 1966;122:800–804. doi: 10.3181/00379727-122-31255. [DOI] [PubMed] [Google Scholar]
- Azoulay-Dupuis E, Lambre CR, Soler P, Moreau J, Thibon M. Lung alterations in guinea-pigs infected with influenza virus. J Comp Pathol. 1984;94:273–283. doi: 10.1016/0021-9975(84)90046-X. [DOI] [PubMed] [Google Scholar]
- Barman S, Krylov PS, Fabrizio TP et al (2012) Pathogenicity and transmissibility of North American triple reassortant swine influenza A viruses in ferrets. PLoS Pathog 8:e1002791. doi:10.1371/journal.ppat.1002791, PPATHOGENS-D-12-00034 [DOI] [PMC free article] [PubMed]
- Barnett JM, Cadman A, Burrell FM, Madar SH, Lewis AP, Tisdale M, Bethell R. In vitro selection and characterisation of influenza B/Beijing/1/87 isolates with altered susceptibility to zanamivir. Virology. 1999;265:286–295. doi: 10.1006/viro.1999.0058. [DOI] [PubMed] [Google Scholar]
- Baz M, Abed Y, Boivin G. Characterization of drug-resistant recombinant influenza A/H1N1 viruses selected in vitro with peramivir and zanamivir. Antiviral Res. 2007;74:159–162. doi: 10.1016/j.antiviral.2006.10.012. [DOI] [PubMed] [Google Scholar]
- Belser JA, Gustin KM, Maines TR, Blau DM, Zaki SR, Katz JM, Tumpey TM. Pathogenesis and transmission of triple-reassortant swine H1N1 influenza viruses isolated before the 2009 H1N1 pandemic. J Virol. 2011;85:1563–1572. doi: 10.1128/JVI.02231-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser JA, Gustin KM, Pearce MB, et al. Pathogenesis and transmission of avian influenza A (H7N9) virus in ferrets and mice. Nature. 2013;501:556–559. doi: 10.1038/nature12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier NM, Lowen AC, Palese P. Oseltamivir-resistant influenza A viruses are transmitted efficiently among guinea pigs by direct contact but not by aerosol. J Virol. 2008;82:10052–10058. doi: 10.1128/JVI.01226-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier NM, Rahmat S, Pica N. Enhanced mammalian transmissibility of seasonal influenza A/H1N1 viruses encoding an oseltamivir-resistant neuraminidase. J Virol. 2012;86:7268–7279. doi: 10.1128/JVI.07242-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr J, Ives J, Kelly L, et al. Influenza virus carrying neuraminidase with reduced sensitivity to oseltamivir carboxylate has altered properties in vitro and is compromised for infectivity and replicative ability in vivo. Antiviral Res. 2002;54:79–88. doi: 10.1016/S0166-3542(01)00215-7. [DOI] [PubMed] [Google Scholar]
- Carrat F, Vergu E, Ferguson NM, Lemaitre M, Cauchemez S, Leach S, Valleron AJ. Time lines of infection and disease in human influenza: a review of volunteer challenge studies. Am J Epidemiol. 2008;167:775–785. doi: 10.1093/aje/kwm375. [DOI] [PubMed] [Google Scholar]
- Chao DL, Bloom JD, Kochin BF, Antia R, Longini IM., Jr The global spread of drug-resistant influenza. J R Soc Interface R Soc. 2012;9:648–656. doi: 10.1098/rsif.2011.0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou YY, Albrecht RA, Pica N, et al. The M segment of the 2009 new pandemic H1N1 influenza virus is critical for its high transmission efficiency in the guinea pig model. J Virol. 2011;85:11235–11241. doi: 10.1128/JVI.05794-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chutinimitkul S, Herfst S, Steel J, et al. Virulence-associated substitution D222G in the hemagglutinin of 2009 pandemic influenza A(H1N1) virus affects receptor binding. J Virol. 2010;84:11802–11813. doi: 10.1128/JVI.01136-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong MD, Simmons CP, Thanh TT, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12:1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong MD, Tran TT, Truong HK, et al. Oseltamivir resistance during treatment of influenza A (H5N1) infection. N Engl j Med. 2005;353:2667–2672. doi: 10.1056/NEJMoa054512. [DOI] [PubMed] [Google Scholar]
- de Wit E, Munster VJ, van Riel D, et al. Molecular determinants of adaptation of highly pathogenic avian influenza H7N7 viruses to efficient replication in the human host. J Virol. 2010;84:1597–1606. doi: 10.1128/JVI.01783-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabbard JD, Dlugolenski D, Van Riel D, et al. Novel H7N9 influenza virus shows low infectious dose, high growth and efficient contact transmission in the guinea pig model. J Virol. 2013 doi: 10.1128/JVI.02959-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel G, Dauber B, Wolff T, Planz O, Klenk HD, Stech J. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc Natl Acad Sci U S A. 2005;102:18590–18595. doi: 10.1073/pnas.0507415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao HN, Lu HZ, Cao B, et al. Clinical findings in 111 cases of influenza A (H7N9) virus infection. N Engl J Med. 2013;368:2277–2285. doi: 10.1056/NEJMoa1305584. [DOI] [PubMed] [Google Scholar]
- Gao Y, Zhang Y, Shinya K, et al. Identification of amino acids in HA and PB2 critical for the transmission of H5N1 avian influenza viruses in a mammalian host. PLoS Pathog. 2009;5:e1000709. doi: 10.1371/journal.ppat.1000709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorkova EA. Consequences of resistance: in vitro fitness, in vivo infectivity, and transmissibility of oseltamivir-resistant influenza A viruses. Influenza Other Respir Viruses. 2012;7:50–57. doi: 10.1111/irv.12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubareva LV, Bethell R, Hart GJ, Murti KG, Penn CR, Webster RG. Characterization of mutants of influenza A virus selected with the neuraminidase inhibitor 4-guanidino-Neu5Ac2en. J Virol. 1996;70:1818–1827. doi: 10.1128/jvi.70.3.1818-1827.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubareva LV, Kaiser L, Matrosovich MN, Soo-Hoo Y, Hayden FG. Selection of influenza virus mutants in experimentally infected volunteers treated with oseltamivir. J Infect Dis. 2001;183:523–531. doi: 10.1086/318537. [DOI] [PubMed] [Google Scholar]
- Gubareva LV, Matrosovich MN, Brenner MK, Bethell RC, Webster RG. Evidence for zanamivir resistance in an immunocompromised child infected with influenza B virus. J Infect Dis. 1998;178:1257–1262. doi: 10.1086/314440. [DOI] [PubMed] [Google Scholar]
- Gubareva LV, Robinson MJ, Bethell RC, Webster RG. Catalytic and framework mutations in the neuraminidase active site of influenza viruses that are resistant to 4-guanidino-Neu5Ac2en. J Virol. 1997;71:3385–3390. doi: 10.1128/jvi.71.5.3385-3390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin KM, Belser JA, Wadford DA, Pearce MB, Katz JM, Tumpey TM, Maines TR. Influenza virus aerosol exposure and analytical system for ferrets. Proc Natl Acad Sci U S A. 2011;108:8432–8437. doi: 10.1073/pnas.1100768108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai R, Schmolke M, Leyva-Grado VH, et al. Influenza A(H7N9) virus gains neuraminidase inhibitor resistance without loss of in vivo virulence or transmissibility. Nature Commun. 2013;4:2854. doi: 10.1038/ncomms3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlocher ML, Carr J, Ives J, Elias S, Truscon R, Roberts N, Monto AS. Influenza virus carrying an R292K mutation in the neuraminidase gene is not transmitted in ferrets. Antiviral Res. 2002;54:99–111. doi: 10.1016/S0166-3542(01)00214-5. [DOI] [PubMed] [Google Scholar]
- Herlocher ML, Truscon R, Elias S, Yen HL, Roberts NA, Ohmit SE, Monto AS. Influenza viruses resistant to the antiviral drug oseltamivir: transmission studies in ferrets. J Infect Dis. 2004;190:1627–1630. doi: 10.1086/424572. [DOI] [PubMed] [Google Scholar]
- Hien TT, de Jong M, Farrar J. Avian influenza–a challenge to global health care structures. N Engl J Med. 2004;351:2363–2365. doi: 10.1056/NEJMp048267. [DOI] [PubMed] [Google Scholar]
- Horisberger MA, de Staritzky K. A recombinant human interferon-alpha B/D hybrid with a broad host-range. J Gen Virol. 1987;68(Pt 3):945–948. doi: 10.1099/0022-1317-68-3-945. [DOI] [PubMed] [Google Scholar]
- Houser KV, Pearce MB, Katz JM, Tumpey TM. Impact of prior seasonal H3N2 influenza vaccination or infection on protection and transmission of emerging variants of influenza A(H3N2)v virus in ferrets. J Virol. 2013;87:13480–13489. doi: 10.1128/JVI.02434-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince WL, Gueye-Mbaye A, Bennink JR, Yewdell JW. Reassortment complements spontaneous mutation in influenza A virus NP and M1 genes to accelerate adaptation to a new host. J Virol. 2013;87:4330–4338. doi: 10.1128/JVI.02749-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ives JA, Carr JA, Mendel DB, et al. The H274Y mutation in the influenza A/H1N1 neuraminidase active site following oseltamivir phosphate treatment leave virus severely compromised both in vitro and in vivo. Antiviral Res. 2002;55:307–317. doi: 10.1016/S0166-3542(02)00053-0. [DOI] [PubMed] [Google Scholar]
- Kaminski MM, Ohnemus A, Staeheli P, Rubbenstroth D. Pandemic 2009 H1N1 influenza A virus carrying a Q136K mutation in the neuraminidase gene is resistant to zanamivir but exhibits reduced fitness in the guinea pig transmission model. J Virol. 2013;87:1912–1915. doi: 10.1128/JVI.02507-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, Lee I, Park S, et al. Genetic requirement for hemagglutinin glycosylation and its implications for influenza A H1N1 virus evolution. J Virol. 2013;87:7539–7549. doi: 10.1128/JVI.00373-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiso M, Mitamura K, Sakai-Tagawa Y, et al. Resistant influenza A viruses in children treated with oseltamivir: descriptive study. Lancet. 2004;364:759–765. doi: 10.1016/S0140-6736(04)16934-1. [DOI] [PubMed] [Google Scholar]
- Kugel D, Kochs G, Obojes K, et al. Intranasal administration of alpha interferon reduces seasonal influenza A virus morbidity in ferrets. J Virol. 2009;83:3843–3851. doi: 10.1128/JVI.02453-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YK, Lipatov AS, Swayne DE. Bronchointerstitial pneumonia in guinea pigs following inoculation with H5N1 high pathogenicity avian influenza virus. Vet Pathol. 2009;46:138–141. doi: 10.1354/vp.46-1-138. [DOI] [PubMed] [Google Scholar]
- Lakdawala SS, Lamirande EW, Jr Suguitan AL et al (2011) Eurasian-origin gene segments contribute to the transmissibility, aerosol release, and morphology of the 2009 pandemic H1N1 influenza virus. PLoS Pathog 7:e1002443. doi: 10.1371/journal.ppat.1002443, PPATHOGENS-D-11-01769 [DOI] [PMC free article] [PubMed]
- Leyva-Grado VH, Mubareka S, Krammer F, Cardenas WB, Palese P. Influenza virus infection in guinea pigs raised as livestock, Ecuador. Emerg Infect Dis. 2012;18:1135–1138. doi: 10.3201/eid1807.111930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowen AC, Mubareka S, Steel J, Palese P (2007) Influenza virus transmission is dependent on relative humidity and temperature. PLoS Pathog 3:1470–1476. doi:07-PLPA-RA-0426, doi:10.1371/journal.ppat.0030151 [DOI] [PMC free article] [PubMed]
- Lowen AC, Mubareka S, Tumpey TM, Garcia-Sastre A, Palese P. The guinea pig as a transmission model for human influenza viruses. Proc Natl Acad Sci U S A. 2006;103:9988–9992. doi: 10.1073/pnas.0604157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowen AC, Steel J, Mubareka S, Carnero E, Garcia-Sastre A, Palese P. Blocking interhost transmission of influenza virus by vaccination in the guinea pig model. J Virol. 2009;83:2803–2818. doi: 10.1128/JVI.02424-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowen AC, Steel J, Mubareka S, Palese P. High temperature (30 degrees C) blocks aerosol but not contact transmission of influenza virus. J Virol. 2008;82:5650–5652. doi: 10.1128/JVI.00325-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J, Wei B, Yang Y et al (2012) Experimental transmission in guinea pigs of H9N2 avian influenza viruses from indoor air of chicken houses. Virus Res 170:102–108. doi:10.1016/j.virusres.2012.09.003, S0168-1702(12)00321-8 [DOI] [PubMed]
- Ma W, Liu Q, Bawa B, et al. The neuraminidase and matrix genes of the 2009 pandemic influenza H1N1 virus cooperate functionally to facilitate efficient replication and transmissibility in pigs. J Gen Virol. 2012;93:1261–1268. doi: 10.1099/vir.0.040535-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines TR, Chen LM, Matsuoka Y, et al. Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. Proc Natl Acad Sci U S A. 2006;103:12121–12126. doi: 10.1073/pnas.0605134103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines TR, Lu XH, Erb SM, et al. Avian influenza (H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals. J Virol. 2005;79:11788–11800. doi: 10.1128/JVI.79.18.11788-11800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKimm-Breschkin JL, Sahasrabudhe A, Blick TJ, et al. Mutations in a conserved residue in the influenza virus neuraminidase active site decreases sensitivity to Neu5Ac2en-derived inhibitors. J Virol. 1998;72:2456–2462. doi: 10.1128/jvi.72.3.2456-2462.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle A, Doudna JA. Adaptive strategies of the influenza virus polymerase for replication in humans. Proc Natl Acad Sci U S A. 2009;106:21312–21316. doi: 10.1073/pnas.0911915106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mubareka S, Lowen AC, Steel J, Coates AL, Garcia-Sastre A, Palese P. Transmission of influenza virus via aerosols and fomites in the guinea pig model. J Infect Dis. 2009;199:858–865. doi: 10.1086/597073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls JM, Bourne AJ, Chen H, Guan Y, Peiris JS. Sialic acid receptor detection in the human respiratory tract: evidence for widespread distribution of potential binding sites for human and avian influenza viruses. Respir Res. 2007;8:73. doi: 10.1186/1465-9921-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair JP, Kauffman CA, Jennings R, Potter CW. Influenza virus infection of the guinea pig: immune response and resistance. Med Microbiol Immunol. 1979;165:241–254. doi: 10.1007/BF02152923. [DOI] [PubMed] [Google Scholar]
- Pica N, Chou YY, Bouvier NM, Palese P. Transmission of influenza B viruses in the guinea pig. J Virol. 2012;86:4279–4287. doi: 10.1128/JVI.06645-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard M, Schrauwen EJ, de Graaf M, et al. Limited airborne transmission of H7N9 influenza A virus between ferrets. Nature. 2013;501:560–563. doi: 10.1038/nature12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer FL, Soergel ME, Straube DC. Survival of airborne influenza virus: effects of propagating host, relative humidity, and composition of spray fluids. Arch Virol. 1976;51:263–273. doi: 10.1007/BF01317930. [DOI] [PubMed] [Google Scholar]
- Seibert CW, Kaminski M, Philipp J, et al. Oseltamivir-resistant variants of the 2009 pandemic H1N1 influenza A virus are not attenuated in the guinea pig and ferret transmission models. J Virol. 2010;84:11219–11226. doi: 10.1128/JVI.01424-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibert CW, Rahmat S, Krause JC, et al. Recombinant IgA is sufficient to prevent influenza virus transmission in guinea pigs. J Virol. 2013;87:7793–7804. doi: 10.1128/JVI.00979-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seladi-Schulman J, Steel J, Lowen AC. Spherical influenza viruses have a fitness advantage in embryonated eggs, while filament-producing strains are selected in vivo. J Virol. 2013;87:13343–13353. doi: 10.1128/JVI.02004-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Avian flu: influenza virus receptors in the human airway. Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- Shinya K, Gao Y, Cilloniz C, et al. Integrated clinical, pathologic, virologic, and transcriptomic analysis of H5N1 influenza virus-induced viral pneumonia in the rhesus macaque. J Virol. 2012;86:6055–6066. doi: 10.1128/JVI.00365-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegers JY, Short KR, Leijten LME et al (In Press) Novel avian-origin influenza A (H7N9) virus attachment to the mammalian respiratory tract. J Virol [DOI] [PMC free article] [PubMed]
- Smith GJ, Vijaykrishna D, Bahl J, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459:1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- Steel J, Lowen AC, Mubareka S, Palese P. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog. 2009;5:e1000252. doi: 10.1371/journal.ppat.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel J, Palese P, Lowen AC. Transmission of a 2009 pandemic influenza virus shows a sensitivity to temperature and humidity similar to that of an H3N2 seasonal strain. J Virol. 2011;85:1400–1402. doi: 10.1128/JVI.02186-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel J, Staeheli P, Mubareka S, Garcia-Sastre A, Palese P, Lowen AC. Transmission of pandemic H1N1 influenza virus and impact of prior exposure to seasonal strains or interferon treatment. J Virol. 2010;84:21–26. doi: 10.1128/JVI.01732-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao EK, London W, Murphy BR. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J Virol. 1993;67:1761–1764. doi: 10.1128/jvi.67.4.1761-1764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Bi Y, Pu J, et al. Guinea pig model for evaluating the potential public health risk of swine and avian influenza viruses. PLoS One. 2010;5:e15537. doi: 10.1371/journal.pone.0015537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai CY, Escarpe PA, Sidwell RW, et al. Characterization of human influenza virus variants selected in vitro in the presence of the neuraminidase inhibitor GS 4071. Antimicrobial agents and chemotherapy. 1998;42:3234–3241. doi: 10.1128/aac.42.12.3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Chong KT. Histopathology and growth kinetics of influenza viruses (H1N1 and H3N2) in the upper and lower airways of guinea pigs. J Gen Virol. 2009;90:386–391. doi: 10.1099/vir.0.007054-0. [DOI] [PubMed] [Google Scholar]
- Treanor JJ. Influenza viruses, including avian influenza and swine influenza. In: Mandell GL, Bennett JE, Dolin R, editors. Mandell, Douglas, and Bennett’s principles and practices of infectious diseases. 7. Philadelphia: Elsevier; 2010. [Google Scholar]
- van der Laan JW, Herberts C, Lambkin-Williams R, Boyers A, Mann AJ, Oxford J. Animal models in influenza vaccine testing. Expert Rev Vaccines. 2008;7:783–793. doi: 10.1586/14760584.7.6.783. [DOI] [PubMed] [Google Scholar]
- Van Hoeven N, Belser JA, Szretter KJ, et al. Pathogenesis of 1918 pandemic and H5N1 influenza virus infections in a guinea pig model: antiviral potential of exogenous alpha interferon to reduce virus shedding. J Virol. 2009;83:2851–2861. doi: 10.1128/JVI.02174-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoeven N, Pappas C, Belser JA, et al. Human HA and polymerase subunit PB2 proteins confer transmission of an avian influenza virus through the air. Proc Natl Acad Sci U S A. 2009;106:3366–3371. doi: 10.1073/pnas.0813172106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kerkhove MD, Hirve S, Koukounari A, Mounts AW. Estimating age-specific cumulative incidence for the 2009 influenza pandemic: a meta-analysis of A(H1N1)pdm09 serological studies from 19 countries. Influenza Other Respir Viruses. 2013;7:872–886. doi: 10.1111/irv.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Riel D, den Bakker MA, Leijten LM et al (2010) Seasonal and pandemic human influenza viruses attach better to human upper respiratory tract epithelium than avian influenza viruses. Am J Pathol 176:1614–1618. doi: S0002-9440(10)60476-2, doi:10.2353/ajpath.2010.090949 [DOI] [PMC free article] [PubMed]
- van Riel D, Leijten LM, de Graaf M et al (2013) Novel avian-origin influenza A (H7N9) virus attaches to epithelium in both upper and lower respiratory tract of humans. Am J Pathol 183:1137–1143. doi: S0002-9440(13)00457-4, doi:10.1016/j.ajpath.2013.06.011 [DOI] [PMC free article] [PubMed]
- van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, Kuiken T. H5N1 virus attachment to lower respiratory tract. Science. 2006;312:399. doi: 10.1126/science.1125548. [DOI] [PubMed] [Google Scholar]
- van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, Kuiken T (2007) Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am J Pathol 171:1215–1223. doi: S0002-9440(10)62385-1, doi:10.2353/ajpath.2007.070248 [DOI] [PMC free article] [PubMed]
- Watanabe T, Kiso M, Fukuyama S, et al. Characterization of H7N9 influenza A viruses isolated from humans. Nature. 2013;501:551–555. doi: 10.1038/nature12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetherbee RE. Induction of systemic delayed hypersensitivity during experimental viral infection of the respiratory tract with a myxovirus or paramyxovirus. J Immunol. 1973;111:157–163. [PubMed] [Google Scholar]
- World Health Organization (2009) Table 2: seasonal influenza A(H1N1) virus resistant to oseltamivir, last quarter 2008 to 31 March 2009. World Health Organization, Geneva, Switzerland
- Xu L, Bao L, Deng W, et al. Novel avian-origin human influenza A(H7N9) can be transmitted between ferrets via respiratory droplets. J Infect Dis. 2013 doi: 10.1093/infdis/jit474. [DOI] [PubMed] [Google Scholar]
- Xu R, McBride R, Nycholat CM, Paulson JC, Wilson IA. Structural characterization of the hemagglutinin receptor specificity from the 2009 H1N1 influenza pandemic. J Virol. 2012;86:982–990. doi: 10.1128/JVI.06322-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen HL, Herlocher LM, Hoffmann E, Matrosovich MN, Monto AS, Webster RG, Govorkova EA. Neuraminidase inhibitor-resistant influenza viruses may differ substantially in fitness and transmissibility. Antimicrob Agents Chemother. 2005;49:4075–4084. doi: 10.1128/AAC.49.10.4075-4084.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Goldsmith CS, Maines TR, et al. Tropism and infectivity of influenza virus, including highly pathogenic avian H5N1 virus, in ferret tracheal differentiated primary epithelial cell cultures. J Virol. 2013;87:2597–2607. doi: 10.1128/JVI.02885-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Shi J, Deng G, et al. H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science. 2013;341:410–414. doi: 10.1126/science.1240532. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang Q, Gao Y, et al. Key molecular factors in hemagglutinin and PB2 contribute to efficient transmission of the 2009 H1N1 pandemic influenza virus. J Virol. 2012;86:9666–9674. doi: 10.1128/JVI.00958-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang Q, Kong H, et al. H5N1 hybrid viruses bearing 2009/H1N1 virus genes transmit in guinea pigs by respiratory droplet. Science. 2013;340:1459–1463. doi: 10.1126/science.1229455. [DOI] [PubMed] [Google Scholar]
- Zhu H, Wang D, Kelvin DJ, et al. Infectivity, transmission, and pathology of human-isolated H7N9 influenza virus in ferrets and pigs. Science. 2013;341:183–186. doi: 10.1126/science.1239844. [DOI] [PubMed] [Google Scholar]
- Zurcher T, Yates PJ, Daly J, et al. Mutations conferring zanamivir resistance in human influenza virus N2 neuraminidases compromise virus fitness and are not stably maintained in vitro. J Antimicrob Chemother. 2006;58:723–732. doi: 10.1093/jac/dkl321. [DOI] [PubMed] [Google Scholar]


