Abstract
Infections with flaviviruses are a continuing public health threat. In addition to vaccine development and vector control, the search for antiviral agents that alleviate symptoms in patients are of considerable interest. Among others, the flaviviral protease NS2B-NS3 is a promising drug target to inhibit viral replication. Flaviviral proteases share a high degree of structural similarity and substrate-recognition profile, which may facilitate a strategy towards development of pan-flaviviral protease inhibitors. However, the success of various drug discovery attempts during the last decade has been limited by the nature of the viral enzyme as well as a lack of robust structural templates. Small-molecular, structurally diverse protease inhibitors have been reported to reach affinities in the lower micromolar range. Peptide-based, substrate-derived compounds are often nanomolar inhibitors, however, with highly compromised drug-likeness. With some exceptions, the antiviral cellular activity of most of the reported compounds have been patchy and insufficient for further development. Recent progress has been made in the elucidation of inhibitor binding using different structural methods. This will hopefully lead to more rational attempts for the identification of various lead compounds that may be successful in cellular assays, animal models and ultimately translated to patients.
Keywords: Protease, Inhibitor, Peptides, Small-molecular, Flavivirus, Dengue, West Nile, Zika
The Need for Antivirals Against Flaviviruses
The inhibition of viral enzymes plays an outstanding role in antiviral therapy, especially in cases where vaccines and vector control are not suffieciently robust. Recent progress in flaviviral vaccine development faces crucial challenges with unpredictable outcomes. Four dengue virus (DENV) where there are four serotypes, the serotype-related cross reactivity with human antibodies has been one of the main difficulties. DENV serotypes, are widely spread over the tropical and subtropical countries around the world and have become endemic in more than 100 countries within the last five decades. Antibody dependent enhancement (ADE) during secondary infection after a previous infection with a different serotype can cause severe life-threatening symptoms, such as dengue haemorrhagic fever or shock syndrome. For this reason vaccines that simultaneously create pronounced immunity against all four serotypes are needed. Since late 2015, the first vaccine CYD (Dengvaxia), with significantly varying efficiency among the four dengue serotypes, has been approved in several countries. However, recent studies have demonstrated that the effect of cross reactivity is not limited to the four known dengue serotypes. Immunity to dengue can cause antibody-dependent enhancement of Zika virus infections and potentially increases viremia and severity of the disease [7, 13, 39]. Consequently, it cannot be ruled out that treatment with dengue vaccine may increase the chance of a later enhanced Zika virus infection. Due to such unexpected phenomena, the extensive search for effective antiviral drugs should be promoted in addition to vaccine development. One important approach aims to inhibit viral protease activity, as successfully demonstrated for chronic diseases such as hepatitis C or AIDS, where, among others, inhibitors of HCV and HIV proteases are established in modern combination therapy.
Flaviviral Proteases as Drug Targets
Due to limited druggability, all campaigns towards clinically relevant protease inhibitors for emerging flaviviruses have so far been unsuccessful. However, since the flaviviral proteases share a high degree of similarity in shape, substrate recognition and catalytic function, a successful drug development process may lead to pan-flaviviral protease inhibitors, which could be used against several globally challenging infectious diseases, such as dengue, West Nile or Zika. Therefore, the present chapter aims to briefly summarize the progress that has been made and tries to derive strategies for future attempts focusing on the protease. Within the last decade, a particular focus has been on the proteases of dengue and West Nile viruses. The highlights for these two most prominent examples will be discussed in this chapter. For a more comprehensive and detailed analysis regarding anti-infectives for these two viruses the interested reader is kindly referred to recent reviews [4, 27, 28, 33]. The chapter will also highlight recent campaigns for the identification of suitable inhibitors for Zika virus protease. Outcomes from all these attempts will inform general perspectives for a more efficient and hopefully successful search for drug-like protease inhibitors for flaviviruses.
Function and Structure of Flaviviral Proteases
Flaviviruses consist of a single-stranded positive-sense RNA genome, which is translated into a single polyprotein by the host cell’s ribosomal system. The polyprotein comprises three structural (C, prM, E) and eight non-structural (NS) proteins, which have to be released from the polyprotein after selective protease cleavage [9, 26, 33]. This essential posttranslational procession is executed by host and flaviviral proteases at the membrane of the endoplasmatic reticulum. The viral protease complex comprises a protease unit, located at the N-terminal part of NS3 and requires a hydrophilic core fragment of the membrane-associated protein NS2B as cofactor for catalytic activity. Highly conserved residues S135, His51 and Asp75 assemble the catalytic triad. The flaviviral serine NS2B-NS3 endoproteases show a common tendency to cleave peptidic backbones after two basic residues. However, detailed substrate preferences as well as catalytic efficiency vary among different flaviviruses.
Several crystal structures of dengue and West Nile proteases have been reported during the last decade. The main deviations are related to the role of the cofactor NS2B. Some structures reported NS2B to be disordered (referred as open or inactive form) whereas others resolved the cofactor domain wrapped around the active site of NS3 (referred as closed or active form). NMR studies indicate that, regardless of the presence or absence of ligands, the closed conformation is predominant [12, 47]. The importance of NS2B towards correct folding of the disordered NS3 domain has also been demonstrated [16]. Unfortunately, only a limited number of co-crystal structures with ligands or inhibitors are available (as a reliable basis for rational drug design). None of those comprise a small-molecular drug-like inhibitor. The first X-ray crystal structures of dengue and West Nile virus proteases became available in 2006 [14]. In case of West Nile virus this structure showed the catalytically active closed conformation with a substrate-derived tetrapeptidic aldehyde inhibitor covalently bound to Ser135 (pdb code: 2FP7). In case of dengue serotype 2 only an inactive open protease form without ligand could be crystallized (pdb code: 2FOM). Although inadequate, the latter structure was used as basis for several drug discovery campaigns until a closed and active form of dengue protease serotype 3 with the same tetrapeptidic aldehyde inhibitor became available in 2012 (pdb code: 3U1I) [37].
Recently, the first crystal structure of Zika virus protease in the active form with a boronate inhibitor, which suits a reasonable model for rational drug discovery campaigns, could be solved (pdb code: 5LC0) [25]. A second crystal structure without inhibitor revealed the open form (apo) of the enzyme with missing resolution for the C-terminal part of NS2B (pdb code: 5GXJ) [11]. A significantly divergent conformation was also obtained for an NS3 loop region between residues 152 and 167, which contributes to the S1 shape. The obvious deviations between these initial crystal structures suggested a conformational activation upon substrate or inhibitor binding. Additional crystallographic and in-solution experiments were necessary to analyse these results in more detail. Since, several new crystal structures and NMR studies in presence and absence of ligands, covering ‘pre-open’ (pdb code: 5T1V), open and closed conformations have been reported [24, 30, 38, 56]. They discovered, in contrast to previous studies with dengue and West Nile protease, a more delicate dependence of open and closed conformations from the construct that was used to fuse NS2B and NS3. The artificial covalently linked construct gZiPro only adopts the closed conformation in presence of substrate-like ligands, but can also bind inhibitors in the open conformation [30]. A construct with an autocleavage site between NS2B and NS3 (eZiPro) showed the closed conformation in the crystal structure (pdb code: 5GJ4), but NMR relaxation data indicated high mobility of NS2B in solution [38]. Apparently, a C-terminal tetrapeptide of NS2B that was found to occupy the active site could not maintain the closed state in solution. A construct without covalent linkage (bZiPro) displayed the closed conformation in solution and in the single crystal (pdb code: 5GPI) [56]. This construct could even be used to capture the structure of a fragment hit in the closed state after being soaked into the crystal (pdb code: 5H4I) [56]. Consequently, bZiPro represents the most suitable construct for compound screening in addition to gZiPro, which may be superior to identify compounds that are able to bind to the open conformation or supposedly perturb the interaction between NS2B and NS3.
Inhibitors of Flaviviral Proteases
In case of dengue protease, which by far has been the most prominent and extensively studied example, several screening campaigns and related inhibitor development approaches were not able to identify promising lead compounds during the last decade [33]. The main reasons for failure have been a lack of structural basis, a relatively flat binding site and a particular focus on often not rationally designed small-molecular compounds. Although viral proteases recognize peptidic substrates, only the minority of studies dealt with peptide-based inhibitors, which have shown to be the only class of compounds that can reach sufficient inhibition in the nanomolar concentration range. However, in contrast to the HIV and HCV success stories, the preference for two permanently charged basic side chains in flaviviral substrates complicates the development of drug-like peptide-derived inhibitors with sufficient bioavailability and antiviral activity in cell culture and animal models (Table 13.1).
Table 13.1.
Biochemical and cellular activities of selected flaviviral protease inhibitors discussed in this chapter
| Compound | Dengue virus [μM]a | West Nile virus [μM] | Zika virus [μM] | |||
|---|---|---|---|---|---|---|
| Biochemicalb | Cellular | Biochemicalb | Cellular | Biochemicalb | Cellular | |
| 1 | IC50 = 1.1 | Inactive | ||||
| 2 | K i = 2.0 | K i = 4.6 | ||||
| 3 | IC50 = 2.0 | EC50 = 59.5 CC50 = 135 | IC50 = 8.7 | EC50 = 42.4 CC50 = 135 | ||
| 4 | IC50 = 2.2 | |||||
| 5 | IC50 = 1.0 | EC50 = 0.8 CC50 > 10 | ||||
| 6 | IC50 = 0.5 | |||||
| 7 | IC50 = 15.4 | EC50 = 0.17 CC50 = 29.3 | ||||
| 8 | IC50 = 1.2 | EC50 = 39.4 CC50 > 100 | ||||
| 9 | IC50 = 8.5 | IC50 = 0.11 | ||||
| 10 | IC50 = 2.8 | EC50 = 40 CC50 = 213 | IC50 = 0.26 | EC50 = 42.3 CC50 = 213 | IC50 = 1.1 | |
| 11 | IC50 > 10 | EC50 = 81.5 CC50 = 236 | IC50 = 0.44 | EC50 = 17 CC50 = 236 | IC50 > 10 | |
| 12 | IC50 = 1.1 | |||||
| 13 | K i = 9.5 | |||||
| 14 | Inactive | EC50 = 0.8 CC50 = 54 | IC50 = 21.6 | EC50 = 13.0 CC50 > 40 | ||
| 15 | IC50 > 10 | EC50 > 100 CC50 = 257 | IC50 = 0.74 | EC50 = 107 CC50 = 257 | IC50 = 0.82 | EC50 ~ 50 CC50 = 257 |
| 16 | K i = 0.051 | EC50 = 30 CC50 > 100 | K i = 0.082 | EC50 = 38 CC50 > 100 | K i = 0.040 | |
| 17 | K i = 0.078 | EC50 = 19 CC50 > 100 | K i = 0.16 | EC50 > 50 CC50 > 100 | IC50 = 2.1 | |
| 18 | K i = 0.012 | EC50 = 20 CC50 > 100 | K i = 0.039 | EC50 = 23 CC50 > 100 | ||
| 19 | IC50 = 0.028 | EC50 = 7.1 CC50 > 100 | IC50 = 0.12 | IC50 = 1.0 | ||
| 20 | IC50 = 0.18 | EC50 = 3.4 CC50 = 100 | IC50 = 0.56 | EC50 = 15.6 CC50 > 100 | ||
aActivities have been reported for various serotypes. The serotype with the best activity results is reported here
bIf reported activities vary by method or report, the lowest (best) value is shown
Small-Molecular Dengue Virus Protease Inhibitors
Approximately 40 approaches towards small-molecular non-peptide-derived inhibitors have been reported in the literature during the last decade. Unfortunately, only a minority of them provided cellular data to confirm that the compounds also achieve proper antiviral activity in cells and are not cytotoxic. A remarkable number of compounds with broad structural variety that are able to inhibit dengue (and West Nile) protease in the concentration range between 25 and 100 μM have been identified. Further ligand derivatizations often only led to limited improvements (maximal affinities in the one-digit micromolar range). Examples are rare where a small structural change causes a pronounced impact on affinity (activity cliff). Consequently, no studies could provide small-molecular non-peptidic compounds that are able to bind to dengue protease in the desirable lower nanomolar range.
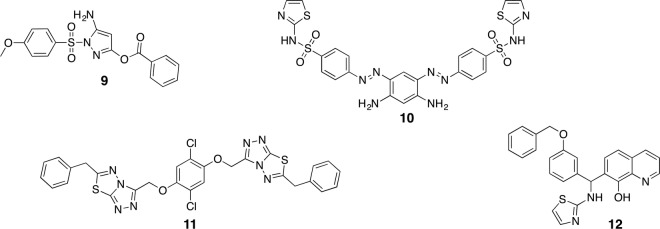
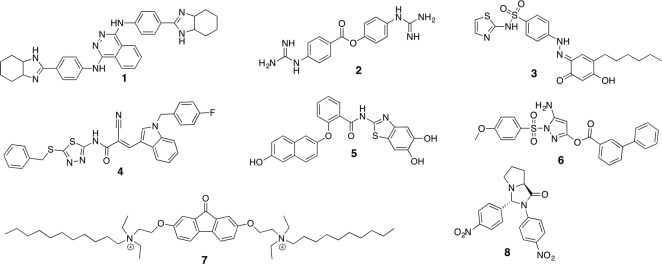
Compound 1 resulted from a campaign including high-throughput screening, scaffold optimization and subsequent derivatization [6]. This compound has an IC50 of 1.1 μM against dengue serotype 2 and decreased potency towards the other serotypes. Specific competitive binding was confirmed by several orthogonal methods; however, despite strong efforts no further optimization of this compound class, which also lacked antiviral activity in cell culture, could be achieved [28]. Compound 2 was identified from a computational screening approach [20] with a K i value of 2.0 μM (serotype 2). Guanidine groups are crucial for sufficient activity, indicating a likely electrostatic interaction with residues in the S1 or S2 pocket of the protease. A covalent interaction of the activated ester bond with Ser135 is possibly, but has not yet been confirmed experimentally. Compound 3 is supposedly an allosteric inhibitor, identified from a virtual screening of compounds, actually aiming at West Nile virus protease, which may inhibit the interactions between NS3 and NS2B [42]. It shows an IC50 value of 2.0 μM, some basic antiviral activity in cells and limited cytotoxicity. A structurally similar but larger derivative 10 (Fig. 13.2) identified from the same campaign showed only slightly lower affinity (IC50 = 2.8 μM), but improved cellular data (EC50 = 40 μM). From a series of thiadiazoloacrylamides, compound 4 showed best activity with an IC50 of 2.2 μM (K d = 2.1 μM) [29]. In correlation with various previously studied compound series the SAR between derivatives remained remarkably flat.
Fig. 13.2.
Selected examples of small-molecular compounds that reached highest inhibition activities against West Nile virus protease
For compound 5, IC50 values of up to 1.0 μM (serotype 3) from a biochemical and 3.2 μM from a cell-based protease assay have been reported [51]. The compound shows cellular antiviral activity in the same range (EC50 = 0.8 μM) but also cytotoxic effects at concentrations above 10 μM. The most active small-molecular non-peptidic inhibitor reported so far for dengue protease is compound 6. It is one of the few examples with affinity significantly lower than 1 μM with an IC50 value of 0.5 μM for serotype 2 [22]. It is also one of the few examples that aim at targeting the catalytically active serine by a covalent interaction. This could be proven by mass spectrometry and may be the key towards small ligands of high affinity. After reaction, the biphenyl-3-carboxylate remains bound to the protease and blocks all further substrate procession until ester hydrolysis may restore the protease activity again.
Another high-throughput screen identified amphiphilic compound 7 with only moderate activity in the biochemical assay (IC50 = 15.4 μM) [52]. However, cellular assays revealed one of the highest reported antiviral activities in cells for any discovered small-molecular dengue protease inhibitor (EC50 = 0.17 μM). Resistance breeding experiments suggested an inhibition of interactions between the NS2B and NS3 domains. The remarkable discrepancy between biochemical and cellular activity may, however, indicate that the protease is not the only target of this compound. An independent screening campaign based on a dengue replicon assay identified a structurally similar compound with related biochemical and virological results [53].
Recently, further derivatizations (including rigidification) of previously published methionine-proline anilides [57] towards non-peptidic analogues revealed compound 8 as small-molecular dengue protease inhibitor with pronounced affinity (IC50 = 1.2 μM) [50]. Selective interaction with the protease is supported by the inactivity of a stereoisomer of 8. In combination, both SARs indicate that the two aromatic nitro substituents are necessary for proper affinity. Although this functional group is highly questionable in terms of drug-likeness, the compound showed no cytotoxic effects at the highest assayed concentration of 100 μM and was proven to inhibit viral replication in cell culture (EC50 = 39.4 μM).
Tolcapone, tannic acid and suramin have been reported as hits from a high-throughput screening with K i values significantly below 1 μM [1]. Tannic acid (K i = 0.34 μM) showed also exceptional activity in a viral plaque assay with an EC50 value of 0.084 μM and only limited cytotoxic effects. However, with a molecular mass of 1700 Da and a polyphenolic structure, this compound would usually not be considered as a suitable lead in drug discovery.
Small-Molecular West Nile Virus Protease Inhibitors
Most of the remarks and conclusions that have been made for the development of dengue virus protease inhibitors can be transferred to the closely related West Nile virus protease. In fact, only a limited number of studies directly aimed at identifying West Nile virus protease inhibitors. However, often West Nile protease activity was additionally assessed within dengue protease inhibitor campaigns. Compound 2 (Fig. 13.1) for example was found to be notably active against West Nile virus protease (K i = 4.6 μM), although the computational screening approach based on a homology model of dengue protease. The highest affinities that could be reached with small-molecular compounds were often up to one order of magnitude better compared to dengue protease.
Fig. 13.1.
Small-molecular inhibitors of dengue virus protease, which either reached highest affinity in biochemical or phenotypic assays
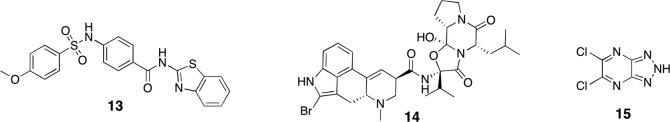
Analogues of compound 6 (Fig. 13.1) have been studied for West Nile virus protease before they were evaluated for dengue protease [18, 45]. For derivate 9 promising IC50 values between 0.11 and 0.16 μM have been reported (only 8.5 μM for dengue protease) [18, 22]. These covalently binding pyrazole esters can be considered as the class of small-molecular compounds with the highest reported affinities and ligand efficiencies for dengue and West Nile virus proteases. However, due to general high reactivity, their chemical stability even in the usual assay buffer is limited [45]. Compounds 10 and 11 were identified from the same virtual screening campaign as compound 3 with IC50 values of 0.26 and 0.44 μM as well as moderate antiviral activity in cells with EC50 values of 42 and 17 μM, respectively [42]. From a high-throughput screening a compound comprising an 8-hydroxyquinoline scaffold was identified as a West Nile virus protease inhibitor with promising activity in cell culture (EC50 = 1.4 μM) [31]. Further derivatizations of this compound class produced 12 with an IC50 value of 1.1 μM [15].
Small-Molecular Zika Virus Protease Inhibitors
Inspired by previous campaigns, initial progress has been made in the discovery of the first small-molecular inhibitors of Zika virus protease. Several compounds that emerged from dengue or West Nile virus protease screenings, such as 10 and 11, also showed inhibition against Zika virus protease [43]. Compound 13 (K i = 9.5 μM) is an example of a series of lead compounds that have been recycled from an HCV protease high-throughput screening campaign [24]. The dopamine antagonist bromocriptine (14), which has previously been reported to inhibit viral replication for all four dengue serotypes [19], also reduces Zika virus replication in cell culture [8]. In contrast to dengue, the Zika virus protease could be confirmed as potential target of bromocriptine (IC50 = 21.6 μM). Compound 15, which was previously reported to be active against West Nile (IC50 = 0.74 μM) but inactive against dengue protease (at concentrations lower than 10 μM), was found to be also a promising Zika protease inhibitor (IC50 = 0.82 μM), especially from the perspective of ligand efficiency (molecular weight = 190 Da). It inhibited viral replication in cell culture (EC50 ~ 50 μM) and could reduce the level of circulating Zika viruses in mice [43]. Although these data are promising, apart from docking studies, which suggest binding close to the active site and interference with NS2B, structural data that would facilitate a hit-to-lead campaign are missing (Fig. 13.3).
Fig. 13.3.
Small-molecular inhibitors of Zika virus protease that resulted from previous campaigns for related viruses
Peptide-Derived Inhibitors of Dengue, West Nile and Zika Virus Proteases
Substrate-based peptidic inhibitors of dengue and West Nile virus proteases have been studied quite extensively during the earlier attempts. They usually consist of a substrate segment comprising at least two basic side chain residues and may additionally be featured with a C-terminal electrophilic warhead, most often an aldehyde moiety, for covalent binding to Ser135. They often cannot be considered as drug-like leads due to their unsuitable pharmacokinetic and physiochemical properties, such as bioavailability, specificity and plasma stability. Notably, these peptides could reach higher affinity towards West Nile than dengue virus protease. For oligo-d-arginines affinities up to 1 nM have been reported in case of West Nile virus [44]. Peptidomimetics with N-terminal dichloro-substituted phenylacetyl groups and C-terminal arginine mimetics reached IC50 values of up to 0.12 μM [17]. Peptides with an additional possibility for covalent interaction with Ser135 were able to generate affinities of up to 9 nM (K i) for the tripeptidic aldehyde phenacetyl-Lys-Lys-Arg-H [21, 41, 46]. Although this compound showed serum stability, cell permeability and antiviral activity (EC50 = 1.6 μM) no further studies towards more drug-like derivatives have been reported [46].
In case of dengue virus protease, studies with simple substrate-derived peptides reached K i values of only up to 0.3 μM [40]. Recently, cyclic peptides comprising unnatural amino acids were found to be active in cell culture (EC50 = 2.0 μM) [48]. Substrate-like peptides containing C-terminal aldehydes were less active in case of dengue protease with a Ki of 1.5 μM for the most active derivative Bz-Lys-Arg-Arg-H [54]. However, alternatively studied electrophiles such as the trifluoromethylketone in Bz-Nle-Lys-Arg-Arg-CF3 (K i = 0.85 μM) or a boronic acid function in a similar analogue Bz-Nle-Lys-Arg-Arg-B(OH)2 (K i = 0.043 μM) showed increased activity [55].
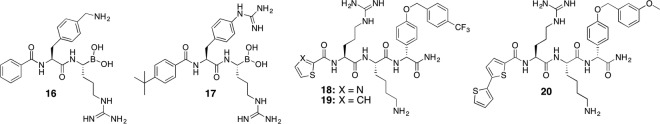
Recent studies have shown that even very small peptide-derived compounds can exhibit extraordinary binding affinities, if they are combined with such a boronic acid moiety, which forms a boronate with the catalytically active residue S135 [36]. These compounds may not only offer a route towards more drug-like small-molecular derivatives, they have also become valuable tools in structural biology to elucidate inhibitor-protease interactions, which will hopefully illuminate the way towards a more structure-based drug design. Although, the drug-likeness of this compound class is limited, a significant reduction of viral titers for West Nile and dengue viruses in cell culture could be observed. Compound 16 showed pronounced pan-flaviviral protease affinity with K i values of 51, 82 and 40 nM for dengue, West Nile and Zika virus proteases, respectively. This derivative was co-crystalized with the proteases of Zika [25] and West Nile [36] viruses. Derivative 17 was used to demonstrate a novel NMR-based approach to identify the binding mode of tightly binding inhibitor molecules towards dengue virus protease from sertotype 2 [10]. The tert-butyl moiety in 17 appears as a sharp and isolated signal in proton NMR spectra. Using the power of paramagnetic NMR spectroscopy [34] combined with NOEs, the positions of the tert-butyl group and aromatic protons in close proximity could be predicted in relation to the 3D structure of the protein. Very recently, compounds 16 and 17 have been used to study the conformational flexibility of the NS2B cofactor of Zika protease in solution [30] (Fig. 13.4).
Fig. 13.4.
Recently published high-affinity peptide-derived inhibitors of flaviviral proteases
Finally, over the last 5 years the stepwise elaboration of non-covalently binding tripeptidic inhibitors, comprising two basic side chains, has been reported regularly [2, 3, 5, 32, 35, 49]. The optimization focused so far only on enzymatic inhibition in biochemical assays, limiting the drug-likeness and pharmacokinetic properties of this compound class. However, with a K i value of 12 nM, 18 is the compound of highest affinity towards dengue serotype 2 protease reported so far [3]. It also shows remarkable affinity against West Nile virus protease with a K i of 39 nM. Due to limited permeability, the antiviral cellular activity is only moderate with EC50 values of 20 and 23 μM for dengue and West Nile viruses, respectively. Recently, the highly related derivative 19 was analysed against Zika protease [23]. It showed reduced inhibition potential for Zika (IC50 = 1.0 μM) compared to previous reported results for dengue (IC50 = 0.028 μM) and West Nile proteases (IC50 = 0.12 μM) [3, 23]. Increased cellular activity was found for analogue 20 with slightly improved permeability (dengue: EC50 = 3.4 μM; West Nile: EC50 = 15.6 μM) [3]. However, the affinity of 20 towards the proteases dropped significantly compared to 18 or 19 with IC50 values of 176 and 557 nM for dengue and West Nile proteases, respectively. Structural evidences, such as NMR or crystallographic data, for the binding mode of this compound class are unfortunately missing. This information would be very valuable to rationalize the pronounced SAR.
Perspectives Towards Zika Virus Protease Inhibitors
All compounds that have recently been described to inhibit Zika protease, either small molecules or peptides, originated from previous drug discovery campaigns for related viruses. The co-crystal structures of compound 16 [25] and the fragment benzimidazol-1-ylmethanol [56] in the active form present a unique chance for rationality in the upcoming drug discovery attempts. This opportunity was not available during a long period of early dengue protease inhibitor investigations. The main lessons for Zika learned from a decade of challenging dengue protease research are: to balance the focus between small-molecular and substrate-derived inhibitors; to take advantage of the virtue of covalence in enzyme inhibition; and to generate as much structural information as possible.
The latter aspect becomes obvious by taking a closer look into the co-crystal structure of Zika virus protease with compound 16. In contrast to other flaviviral proteases, such as those from dengue or West Nile viruses, Zika comprises a negatively charged aspartate residue in position 83 of the cofactor domain NS2B. The West Nile protease contains a structurally very similar, although uncharged, asparagine residue in that position. Dengue protease serotypes have either a serine or threonine in that position. Without any structural information this situation may not have led to any special attention for drug development purposes with Zika virus protease. However, from the crystal structure it turned out that this NS2B aspartate residue is responsible for a salt-bridge formation with the aminomethyl-phenyl moiety (P2) of compound 16 leading to an extra tight binding of inhibitors and substrates. As the possibilities for tight interactions of inhibitors with flaviviral proteases are usually limited, this information is highly relevant for further drug design campaigns.
Conclusion
Although alternative flaviviral proteins (e.g. NS1 and NS5) as well as virus-host interactions have become attractive drug targets, the NS2B-NS3 protease is still of considerable interest. However, the progress towards drug-like compounds with promising intracellular activity was limited. It may take another decade until the first selective flaviviral protease inhibitor that convincingly works in an animal model will become available. The main challenges are the rather flat binding sites, the absence of product inhibition (in contrast to HCV) and the strong recognition preference for basic moieties, which highly complicates the development of compounds with high affinity and a desirable ADME profile.
As new methods and technologies have also become available during the last decade of struggling inhibitor development, they may open the way for alternative strategies. Many of the earlier attempts (especially with small-molecular compounds) were pursued without any reasonable structural basis. Co-crystal structures of flaviviral proteases with small-molecular ligands are still remarkably rare compared to other drug discovery campaigns. Technologies that do not rely on protease crystals, such as modern NMR, can help to elucidate the binding mode of inhibitor candidates. In this context, recently emerging fragment-based screening approaches in combination with NMR, which have so far been neglected for flaviviral proteases, may also revitalise the drug discovery process. The identification of new small scaffolds with only weak affinity and their consequent rational elaboration into drug-like inhibitors may be superior compared to previous approaches.
In addition to new strategies regarding the identification and optimization of lead compounds, new approaches regarding cellular assays will be required to address the present delay or total lack of cellular data. Some initial progress on intracellular protease assays has been made, but further advance in this area, also concerning imaging techniques to track potential drug candidates within the cell, is necessary.
Note
Since the ‘Tofo Advanced Study Week on Arboviruses’ significant progress has been achieved particularly in the field of Zika virus protease. These recent results are reflected in this chapter, although they were not content of the conference presentation and discussion.
Acknowledgements
Funding by the Alexander von Humboldt Foundation is gratefully acknowledged.
Contributor Information
Rolf Hilgenfeld, Email: hilgenfeld@biochem.uni-luebeck.de.
Subhash G. Vasudevan, Phone: +65656565166718, FAX: +6565+65-6221-2529, Email: subhash.vasudevan@duke-nus.edu.sg
Christoph Nitsche, Email: christoph.nitsche@anu.edu.au.
References
- 1.Balasubramanian A, Manzano M, Teramoto T, Pilankatta R, Padmanabhan R. High-throughput screening for the identification of small-molecule inhibitors of the flaviviral protease. Antivir Res. 2016;134:6–16. doi: 10.1016/j.antiviral.2016.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bastos Lima A, Behnam MA, El Sherif Y, Nitsche C, Vechi SM, Klein CD. Dual inhibitors of the dengue and West Nile virus NS2B-NS3 proteases: synthesis, biological evaluation and docking studies of novel peptide-hybrids. Bioorg Med Chem. 2015;23(17):5748–5755. doi: 10.1016/j.bmc.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 3.Behnam MA, Graf D, Bartenschlager R, Zlotos DP, Klein CD. Discovery of nanomolar dengue and West Nile virus protease inhibitors containing a 4-benzyloxyphenylglycine residue. J Med Chem. 2015;58(23):9354–9370. doi: 10.1021/acs.jmedchem.5b01441. [DOI] [PubMed] [Google Scholar]
- 4.Behnam MA, Nitsche C, Boldescu V, Klein CD. The medicinal chemistry of dengue virus. J Med Chem. 2016;59(12):5622–5649. doi: 10.1021/acs.jmedchem.5b01653. [DOI] [PubMed] [Google Scholar]
- 5.Behnam MA, Nitsche C, Vechi SM, Klein CD. C-terminal residue optimization and fragment merging: discovery of a potent peptide-hybrid inhibitor of dengue protease. ACS Med Chem Lett. 2014;5(9):1037–1042. doi: 10.1021/ml500245v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodenreider C, Beer D, Keller TH, Sonntag S, Wen D, Yap L, Yau YH, Shochat SG, Huang D, Zhou T, Caflisch A, Su X-C, Ozawa K, Otting G, Vasudevan SG, Lescar J, Lim SP. A fluorescence quenching assay to discriminate between specific and nonspecific inhibitors of dengue virus protease. Anal Biochem. 2009;395(2):195–204. doi: 10.1016/j.ab.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 7.Castanha PMS, Nascimento EJM, Braga C, Cordeiro MT, de Carvalho OV, de Mendonça LR, Azevedo EAN, França RFO, Dhalia R, Marques ETA. Dengue virus–specific antibodies enhance Brazilian Zika virus infection. J Infect Dis. 2017;215(5):781–785. doi: 10.1093/infdis/jiw638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan JF-W, Chik KK-H, Yuan S, Yip CC-Y, Zhu Z, Tee K-M, Tsang JO-L, Chan CC-S, Poon VK-M, Lu G, Zhang AJ, Lai K-K, Chan K-H, Kao RY-T, Yuen K-Y. Novel antiviral activity and mechanism of bromocriptine as a Zika virus NS2B-NS3 protease inhibitor. Antivir Res. 2017;141:29–37. doi: 10.1016/j.antiviral.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Chappell KJ, Stoermer MJ, Fairlie DP, Young PR. West Nile virus NS2B/NS3 protease as an antiviral target. Curr Med Chem. 2008;15(27):2771–2784. doi: 10.2174/092986708786242804. [DOI] [PubMed] [Google Scholar]
- 10.Chen WN, Nitsche C, Pilla KB, Graham B, Huber T, Klein CD, Otting G. Sensitive NMR approach for determining the binding mode of tightly binding ligand molecules to protein targets. J Am Chem Soc. 2016;138(13):4539–4546. doi: 10.1021/jacs.6b00416. [DOI] [PubMed] [Google Scholar]
- 11.Chen X, Yang K, Wu C, Chen C, Hu C, Buzovetsky O, Wang Z, Ji X, Xiong Y, Yang H. Mechanisms of activation and inhibition of Zika virus NS2B-NS3 protease. Cell Res. 2016;26(11):1260–1263. doi: 10.1038/cr.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de la Cruz L, Chen WN, Graham B, Otting G. Binding mode of the activity-modulating C-terminal segment of NS2B to NS3 in the dengue virus NS2B-NS3 protease. FEBS J. 2014;281(6):1517–1533. doi: 10.1111/febs.12729. [DOI] [PubMed] [Google Scholar]
- 13.Dejnirattisai W, Supasa P, Wongwiwat W, Rouvinski A, Barba-Spaeth G, Duangchinda T, Sakuntabhai A, Cao-Lormeau VM, Malasit P, Rey FA, Mongkolsapaya J, Screaton GR. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with Zika virus. Nat Immunol. 2016;17(9):1102–1108. doi: 10.1038/ni.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erbel P, Schiering N, D’Arcy A, Renatus M, Kroemer M, Lim SP, Yin Z, Keller TH, Vasudevan SG, Hommel U. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat Struct Mol Biol. 2006;13(4):372–373. doi: 10.1038/nsmb1073. [DOI] [PubMed] [Google Scholar]
- 15.Ezgimen M, Lai H, Mueller NH, Lee K, Cuny G, Ostrov DA, Padmanabhan R. Characterization of the 8-hydroxyquinoline scaffold for inhibitors of West Nile virus serine protease. Antivir Res. 2012;94(1):18–24. doi: 10.1016/j.antiviral.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta G, Lim L, Song J. NMR and MD studies reveal that the isolated dengue NS3 protease is an intrinsically disordered chymotrypsin fold which absolutely requests NS2B for correct folding and functional dynamics. PLoS One. 2015;10(8):e0134823. doi: 10.1371/journal.pone.0134823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammamy MZ, Haase C, Hammami M, Hilgenfeld R, Steinmetzer T. Development and characterization of new peptidomimetic inhibitors of the West Nile virus NS2B-NS3 protease. ChemMedChem. 2013;8(2):231–241. doi: 10.1002/cmdc.201200497. [DOI] [PubMed] [Google Scholar]
- 18.Johnston PA, Phillips J, Shun TY, Shinde S, Lazo JS, Huryn DM, Myers MC, Ratnikov BI, Smith JW, Su Y, Dahl R, Cosford ND, Shiryaev SA, Strongin AY. HTS identifies novel and specific uncompetitive inhibitors of the two-component NS2B-NS3 proteinase of West Nile virus. Assay Drug Dev Technol. 2007;5(6):737–750. doi: 10.1089/adt.2007.101. [DOI] [PubMed] [Google Scholar]
- 19.Kato F, Ishida Y, Oishi S, Fujii N, Watanabe S, Vasudevan SG, Tajima S, Takasaki T, Suzuki Y, Ichiyama K, Yamamoto N, Yoshii K, Takashima I, Kobayashi T, Miura T, Igarashi T, Hishiki T. Novel antiviral activity of bromocriptine against dengue virus replication. Antivir Res. 2016;131:141–147. doi: 10.1016/j.antiviral.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 20.Knehans T, Schuller A, Doan DN, Nacro K, Hill J, Guntert P, Madhusudhan MS, Weil T, Vasudevan SG. Structure-guided fragment-based in silico drug design of dengue protease inhibitors. J Comput Aided Mol Des. 2011;25(3):263–274. doi: 10.1007/s10822-011-9418-0. [DOI] [PubMed] [Google Scholar]
- 21.Knox JE, Ma NL, Yin Z, Patel SJ, Wang WL, Chan WL, Ranga Rao KR, Wang G, Ngew X, Patel V, Beer D, Lim SP, Vasudevan SG, Keller TH. Peptide inhibitors of West Nile NS3 protease: SAR study of tetrapeptide aldehyde inhibitors. J Med Chem. 2006;49(22):6585–6590. doi: 10.1021/jm0607606. [DOI] [PubMed] [Google Scholar]
- 22.Koh-Stenta X, Joy J, Wang SF, Kwek PZ, Wee JL, Wan KF, Gayen S, Chen AS, Kang C, Lee MA, Poulsen A, Vasudevan SG, Hill J, Nacro K. Identification of covalent active site inhibitors of dengue virus protease. Drug Des Devel Ther. 2015;9:6389–6399. doi: 10.2147/DDDT.S94207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuiper BD, Slater K, Spellmon N, Holcomb J, Medapureddy P, Muzzarelli KM, Yang Z, Ovadia R, Amblard F, Kovari IA, Schinazi RF, Kovari LC. Increased activity of unlinked Zika virus NS2B/NS3 protease compared to linked Zika virus protease. Biochem Biophys Res Commun. 2017;492:668–673. doi: 10.1016/j.bbrc.2017.03.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee H, Ren J, Nocadello S, Rice AJ, Ojeda I, Light S, Minasov G, Vargas J, Nagarathnam D, Anderson WF, Johnson ME. Identification of novel small molecule inhibitors against NS2B/NS3 serine protease from Zika virus. Antivir Res. 2017;139:49–58. doi: 10.1016/j.antiviral.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lei J, Hansen G, Nitsche C, Klein CD, Zhang L, Hilgenfeld R. Crystal structure of Zika virus NS2B-NS3 protease in complex with a boronate inhibitor. Science. 2016;353(6298):503–505. doi: 10.1126/science.aag2419. [DOI] [PubMed] [Google Scholar]
- 26.Lescar J, Luo D, Xu T, Sampath A, Lim SP, Canard B, Vasudevan SG. Towards the design of antiviral inhibitors against flaviviruses: the case for the multifunctional NS3 protein from dengue virus as a target. Antivir Res. 2008;80(2):94–101. doi: 10.1016/j.antiviral.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 27.Lim SP, Shi PY. West Nile virus drug discovery. Virus. 2013;5(12):2977–3006. doi: 10.3390/v5122977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim SP, Wang QY, Noble CG, Chen YL, Dong H, Zou B, Yokokawa F, Nilar S, Smith P, Beer D, Lescar J, Shi PY. Ten years of dengue drug discovery: progress and prospects. Antivir Res. 2013;100(2):500–519. doi: 10.1016/j.antiviral.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 29.Liu H, Wu R, Sun Y, Ye Y, Chen J, Luo X, Shen X, Liu H. Identification of novel thiadiazoloacrylamide analogues as inhibitors of dengue-2 virus NS2B/NS3 protease. Bioorg Med Chem. 2014;22(22):6344–6352. doi: 10.1016/j.bmc.2014.09.057. [DOI] [PubMed] [Google Scholar]
- 30.Mahawaththa MC, Pearce BJG, Szabo M, Graham B, Klein CD, Nitsche C, Otting G. Solution conformations of a linked construct of the Zika virus NS2B-NS3 protease. Antivir Res. 2017;142:141–147. doi: 10.1016/j.antiviral.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 31.Mueller NH, Pattabiraman N, Ansarah-Sobrinho C, Viswanathan P, Pierson TC, Padmanabhan R. Identification and biochemical characterization of small-molecule inhibitors of West Nile virus serine protease by a high-throughput screen. Antimicrob Agents Chemother. 2008;52(9):3385–3393. doi: 10.1128/AAC.01508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nitsche C, Behnam MA, Steuer C, Klein CD. Retro peptide-hybrids as selective inhibitors of the dengue virus NS2B-NS3 protease. Antivir Res. 2012;94(1):72–79. doi: 10.1016/j.antiviral.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 33.Nitsche C, Holloway S, Schirmeister T, Klein CD. Biochemistry and medicinal chemistry of the dengue virus protease. Chem Rev. 2014;114(22):11348–11381. doi: 10.1021/cr500233q. [DOI] [PubMed] [Google Scholar]
- 34.Nitsche C, Otting G. Pseudocontact shifts in biomolecular NMR using paramagnetic metal tags. Prog Nucl Magn Reson Spectrosc. 2017;98–99:20–49. doi: 10.1016/j.pnmrs.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Nitsche C, Schreier VN, Behnam MA, Kumar A, Bartenschlager R, Klein CD. Thiazolidinone-peptide hybrids as dengue virus protease inhibitors with antiviral activity in cell culture. J Med Chem. 2013;56(21):8389–8403. doi: 10.1021/jm400828u. [DOI] [PubMed] [Google Scholar]
- 36.Nitsche C, Zhang L, Weigel LF, Schilz J, Graf D, Bartenschlager R, Hilgenfeld R, Klein CD. Peptide–boronic acid inhibitors of flaviviral proteases: medicinal chemistry and structural biology. J Med Chem. 2017;60(1):511–516. doi: 10.1021/acs.jmedchem.6b01021. [DOI] [PubMed] [Google Scholar]
- 37.Noble CG, Seh CC, Chao AT, Shi PY. Ligand-bound structures of the dengue virus protease reveal the active conformation. J Virol. 2012;86(1):438–446. doi: 10.1128/JVI.06225-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phoo WW, Li Y, Zhang Z, Lee MY, Loh YR, Tan YB, Ng EY, Lescar J, Kang C, Luo D. Structure of the NS2B-NS3 protease from Zika virus after self-cleavage. Nat Commun. 2016;7:13410. doi: 10.1038/ncomms13410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Priyamvada L, Quicke KM, Hudson WH, Onlamoon N, Sewatanon J, Edupuganti S, Pattanapanyasat K, Chokephaibulkit K, Mulligan MJ, Wilson PC, Ahmed R, Suthar MS, Wrammert J. Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proc Natl Acad Sci U S A. 2016;113(28):7852–7857. doi: 10.1073/pnas.1607931113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prusis P, Junaid M, Petrovska R, Yahorava S, Yahorau A, Katzenmeier G, Lapins M, Wikberg JES. Design and evaluation of substrate-based octapeptide and non substrate-based tetrapeptide inhibitors of dengue virus NS2B–NS3 proteases. Biochem Biophys Res Commun. 2013;434(4):767–772. doi: 10.1016/j.bbrc.2013.03.139. [DOI] [PubMed] [Google Scholar]
- 41.Schüller A, Yin Z, Brian Chia CS, Doan DN, Kim HK, Shang L, Loh TP, Hill J, Vasudevan SG. Tripeptide inhibitors of dengue and West Nile virus NS2B-NS3 protease. Antivir Res. 2011;92(1):96–101. doi: 10.1016/j.antiviral.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 42.Shiryaev SA, Cheltsov AV, Gawlik K, Ratnikov BI, Strongin AY. Virtual ligand screening of the National Cancer Institute (NCI) compound library leads to the allosteric inhibitory scaffolds of the West Nile virus NS3 proteinase. Assay Drug Dev Technol. 2011;9(1):69–78. doi: 10.1089/adt.2010.0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shiryaev SA, Farhy C, Pinto A, Huang CT, Simonetti N, Ngono AE, Dewing A, Shresta S, Pinkerton AB, Cieplak P, Strongin AY, Terskikh AV. Characterization of the Zika virus two-component NS2B-NS3 protease and structure-assisted identification of allosteric small-molecule antagonists. Antivir Res. 2017;143:218–229. doi: 10.1016/j.antiviral.2017.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shiryaev SA, Ratnikov BI, Chekanov AV, Sikora S, Rozanov DV, Godzik A, Wang J, Smith JW, Huang Z, Lindberg I, Samuel MA, Diamond MS, Strongin AY. Cleavage targets and the D-arginine-based inhibitors of the West Nile virus NS3 processing proteinase. Biochem J. 2006;393(Pt 2):503–511. doi: 10.1042/BJ20051374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sidique S, Shiryaev SA, Ratnikov BI, Herath A, Su Y, Strongin AY, Cosford ND. Structure-activity relationship and improved hydrolytic stability of pyrazole derivatives that are allosteric inhibitors of West Nile virus NS2B-NS3 proteinase. Bioorg Med Chem Lett. 2009;19(19):5773–5777. doi: 10.1016/j.bmcl.2009.07.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stoermer MJ, Chappell KJ, Liebscher S, Jensen CM, Gan CH, Gupta PK, Xu WJ, Young PR, Fairlie DP. Potent cationic inhibitors of West Nile virus NS2B/NS3 protease with serum stability, cell permeability and antiviral activity. J Med Chem. 2008;51(18):5714–5721. doi: 10.1021/jm800503y. [DOI] [PubMed] [Google Scholar]
- 47.Su XC, Ozawa K, Qi R, Vasudevan SG, Lim SP, Otting G. NMR analysis of the dynamic exchange of the NS2B cofactor between open and closed conformations of the West Nile virus NS2B-NS3 protease. PLoS Negl Trop Dis. 2009;3(12):e561. doi: 10.1371/journal.pntd.0000561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takagi Y, Matsui K, Nobori H, Maeda H, Sato A, Kurosu T, Orba Y, Sawa H, Hattori K, Higashino K, Numata Y, Yoshida Y. Discovery of novel cyclic peptide inhibitors of dengue virus NS2B-NS3 protease with antiviral activity. Bioorg Med Chem Lett. 2017;27:3586–3590. doi: 10.1016/j.bmcl.2017.05.027. [DOI] [PubMed] [Google Scholar]
- 49.Weigel LF, Nitsche C, Graf D, Bartenschlager R, Klein CD. Phenylalanine and phenylglycine analogues as arginine mimetics in dengue protease inhibitors. J Med Chem. 2015;58(19):7719–7733. doi: 10.1021/acs.jmedchem.5b00612. [DOI] [PubMed] [Google Scholar]
- 50.Weng Z, Shao X, Graf D, Wang C, Klein CD, Wang J, Zhou GC. Identification of fused bicyclic derivatives of pyrrolidine and imidazolidinone as dengue virus-2 NS2B-NS3 protease inhibitors. Eur J Med Chem. 2016;125:751–759. doi: 10.1016/j.ejmech.2016.09.063. [DOI] [PubMed] [Google Scholar]
- 51.Wu H, Bock S, Snitko M, Berger T, Weidner T, Holloway S, Kanitz M, Diederich WE, Steuber H, Walter C, Hofmann D, Weissbrich B, Spannaus R, Acosta EG, Bartenschlager R, Engels B, Schirmeister T, Bodem J. Novel dengue virus NS2B/NS3 protease inhibitors. Antimicrob Agents Chemother. 2015;59(2):1100–1109. doi: 10.1128/AAC.03543-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang CC, Hsieh YC, Lee SJ, Wu SH, Liao CL, Tsao CH, Chao YS, Chern JH, Wu CP, Yueh A. Novel dengue virus-specific NS2B/NS3 protease inhibitor, BP2109, discovered by a high-throughput screening assay. Antimicrob Agents Chemother. 2011;55(1):229–238. doi: 10.1128/AAC.00855-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang CC, Hu HS, Wu RH, Wu SH, Lee SJ, Jiaang WT, Chern JH, Huang ZS, Wu HN, Chang CM, Yueh A. A novel dengue virus inhibitor, BP13944, discovered by high-throughput screening with dengue virus replicon cells selects for resistance in the viral NS2B/NS3 protease. Antimicrob Agents Chemother. 2014;58(1):110–119. doi: 10.1128/AAC.01281-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yin Z, Patel SJ, Wang W-L, Chan W-L, Ranga Rao KR, Wang G, Ngew X, Patel V, Beer D, Knox JE, Ma NL, Ehrhardt C, Lim SP, Vasudevan SG, Keller TH. Peptide inhibitors of dengue virus NS3 protease. Part 2: SAR study of tetrapeptide aldehyde inhibitors. Bioorg Med Chem Lett. 2006;16(1):40–43. doi: 10.1016/j.bmcl.2005.09.049. [DOI] [PubMed] [Google Scholar]
- 55.Yin Z, Patel SJ, Wang W-L, Wang G, Chan W-L, Rao KRR, Alam J, Jeyaraj DA, Ngew X, Patel V, Beer D, Lim SP, Vasudevan SG, Keller TH. Peptide inhibitors of dengue virus NS3 protease. Part 1: warhead. Bioorg Med Chem Lett. 2006;16(1):36–39. doi: 10.1016/j.bmcl.2005.09.062. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Z, Li Y, Loh YR, Phoo WW, Hung AW, Kang C, Luo D. Crystal structure of unlinked NS2B-NS3 protease from Zika virus. Science. 2016;354(6319):1597–1600. doi: 10.1126/science.aai9309. [DOI] [PubMed] [Google Scholar]
- 57.Zhou GC, Weng Z, Shao X, Liu F, Nie X, Liu J, Wang D, Wang C, Guo K. Discovery and SAR studies of methionine-proline anilides as dengue virus NS2B-NS3 protease inhibitors. Bioorg Med Chem Lett. 2013;23(24):6549–6554. doi: 10.1016/j.bmcl.2013.10.071. [DOI] [PubMed] [Google Scholar]