Abstract
G-quadruplexes (G4s) have become one of the most exciting nucleic acid secondary structures. A noncanonical, four-stranded structure formed in guanine-rich DNA and RNA sequences, G-quadruplexes can readily form under physiologically relevant conditions and are globularly folded structures. DNA is widely recognized as a double-helical structure essential in genetic information storage. However, only ~3% of the human genome is expressed in protein; RNA and DNA may form noncanonical secondary structures that are functionally important. G-quadruplexes are one such example which have gained considerable attention for their formation and regulatory roles in biologically significant regions, such as human telomeres, oncogene-promoter regions, replication initiation sites, and 5′- and 3′-untranslated region (UTR) of mRNA. They are shown to be a regulatory motif in a number of critical cellular processes including gene transcription, translation, replication, and genomic stability. G-quadruplexes are also found in nonhuman genomes, particularly those of human pathogens. Therefore, G-quadruplexes have emerged as a new class of molecular targets for drug development. In addition, there is considerable interest in the use of G-quadruplexes for biomaterials, biosensors, and biocatalysts. The First International Meeting on Quadruplex DNA was held in 2007, and the G-quadruplex field has been growing dramatically over the last decade. The methods used to study G-quadruplexes have been essential to the rapid progress in our understanding of this exciting nucleic acid secondary structure.
Key words: G-quadruplexes, DNA, RNA, Oncogene promoters, Human telomeres, UTR, DNA damage, Transcription, Replication, Translation, Drug target, Cancer, Human diseases
G-Quadruplex Nucleic Acids
G-quadruplexes (G4s) are noncanonical four-stranded nucleic acid structures formed in guanine-rich DNA and RNA sequences (Fig. 1). They have emerged as one of the most exciting nucleic acid secondary structures. DNA is widely recognized as a double-helical structure essential in genetic information storage. Results from the ENCODE project [1] indicate that only ~3% of the human genome is expressed in protein and that RNA and DNA may form noncanonical secondary structures that are functionally important. G-quadruplexes are one such example which have gained considerable attention for their formation and regulatory roles in biologically significant regions. G-quadruplexes are found to be involved in a number of critical cellular processes, including gene transcription, translation, DNA replication, and genomic stability. G-quadruplexes can readily form under physiologically relevant conditions and are globularly folded structures. Many proteins have been identified to interact with G-quadruplex DNA or RNA, including G-quadruplex-stabilizing or destabilizing/unfolding proteins (see reviews: [2–6]). As such, G-quadruplexes have emerged as a new class of molecular targets for drug development. In addition, there is considerable interest in the use of G-quadruplexes for biomaterials [7, 8], biosensors [9, 10], and biocatalysts [11].
Fig. 1.
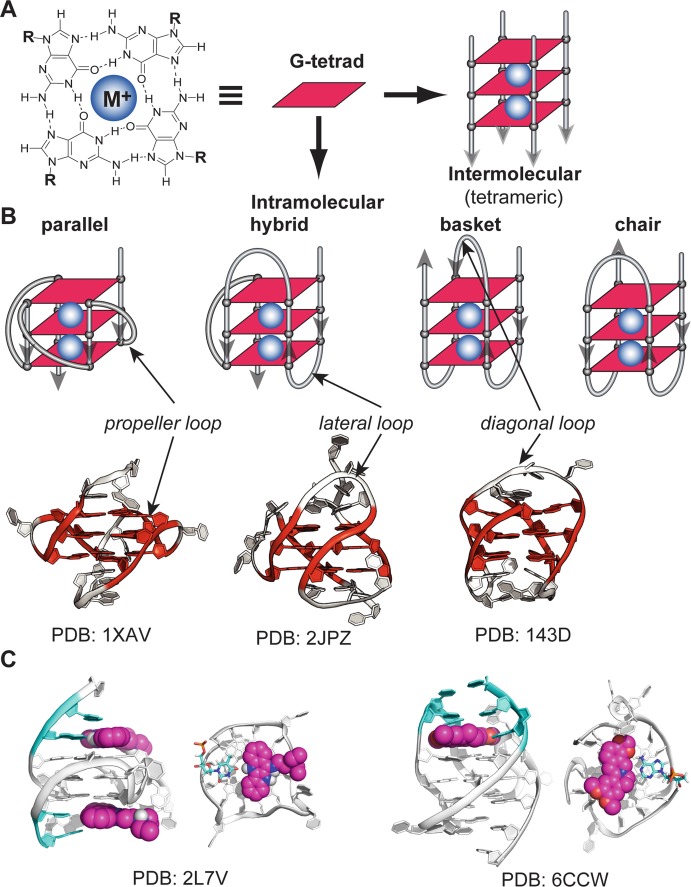
(a) Schematic illustration of a G-tetrad, four guanine bases arranged in a square plane with Hoogsteen hydrogen bonding. Monovalent cations (K+ or Na+, shown as blue spheres) are required to stabilize G-quadruplexes by coordinating with the O6 atoms of the adjacent G-tetrad planes. (b) A schematic intermolecular (tetrameric) G-quadruplex with three G-tetrads. (c) Examples of intramolecular G-quadruplexes with different folding structures and loop conformations. The experimentally determined molecular structures are shown as examples for parallel, hybrid, and basket G-quadruplexes. (d) Example NMR molecular structures of ligand complexes with the c-MYC promoter G-quadruplex and the human telomeric G-quadruplex
First observed in 1910 [12], the G-tetrad structure was not determined until 1962 [13]. The core structure of a G-quadruplex consists of stacked guanine-tetrads (G-tetrads), a square planar platform of four guanine bases that are held together by Hoogsteen hydrogen bonds (Fig. 1a). G-quadruplex structures require cations, particularly K+ or Na+, to stabilize stacked G-tetrads by coordinating with tetrad-guanine O6 atoms [14–16]. The tetrad-guanines can adopt anti or syn glycosidic conformation; tetrad-guanines from G-strands with the same direction, i.e., parallel strands, adopt the same glycosidic conformation, whereas those from G-strands with the opposite direction, i.e., antiparallel strands, adopt different glycosidic conformations [17].
G-quadruplexes can be intramolecular (monomeric) or intermolecular (multimeric), which are formed with one or more than one nucleic acid molecules, respectively. Tetramolecular G-quadruplexes (Fig. 1b) are usually parallel-stranded with tetrad guanines adopting anti glycosidic conformation. Most biologically relevant G-quadruplexes are intramolecular G-quadruplexes, with three-tetrad cores being the most common (Fig. 1c). In contrast to tetramolecular structures, intramolecular G-quadruplexes form quickly and exhibit great conformational diversity, such as in folding topology, loop conformation, and capping structures. Based on G-strand directionality, a G-quadruplex can be parallel with all four G-strands in the same direction, hybrid/mixed with both parallel and antiparallel strands, or antiparallel with all adjacent G-strands antiparallel to each other. G-strands in intramolecular G-quadruplexes are connected by different types of loops, such as propeller for connecting parallel strands, lateral for connecting adjacent antiparallel strands, and diagonal for connecting antiparallel strands across the G-tetrad core. Not only can different sequences adopt distinct topologies, but a given sequence can also fold into different conformations, as in the case of the human telomeric DNA, or form multiple structures, as in the case of human gene promoter sequences [18]. While a number of principles of G-quadruplex folding have been recognized, a G-quadruplex conformation is difficult to predict and requires experimental structure determination.
G-Quadruplex Occurrence and Functions
G-quadruplexes have been found to form in specific human guanine-rich sequences with functional significance, such as telomeres, oncogene-promoter regions, and 5′- and 3′-untranslated region (UTR) of mRNA, as well as in nonhuman genomes.
G-Quadruplexes in Telomeres
The first biologically relevant G-quadruplex was observed in telomeric DNA. Telomeres are specific DNA–protein complexes at the ends of linear chromosomes, providing protection against gene erosion from cell divisions, chromosomal nonhomologous end-joinings, and nuclease attacks [19–21]. Telomeric G-quadruplexes were first reported as novel intramolecular structures containing guanine–guanine base-pairs in single-stranded telomeric sequences of several organisms [22], and as guanine-tetrads between hairpin loops of the Tetrahymena telomeric DNA [23]. The importance of monovalent cations in the stabilization of G-quadruplex structures was revealed by Williamson, Cech in their monovalent cation-induced square-planar G-quartet model using the Oxytricha and Tetrahymena telomeric DNA [14]. Human telomeres consist of tandem repeats of the hexanucleotide d (TTAGGG)n 5–10 kb in length, which terminate in a single-stranded 3′-overhang of 35–600 bases [24]. Telomeres of cancer cells do not shorten upon replication, mainly due to the activation of a reverse transcriptase, telomerase, that extends the telomeric sequence at the chromosome ends [25]. Telomerase is activated in 80–85% of human cancer cells to maintain telomere length and malignant phenotype [25–27]. The G-quadruplex formation can inhibit the activity of telomerase [28], making it an attractive target for cancer therapeutic intervention. In addition to the formation at the telomere end, which most likely involves intramolecular G-quadruplex structures, intermolecular G-quadruplex formation may also be involved in the T-loop invasion complex [29, 30]. Recently, telomeric repeat-containing RNA (TERRA) G-quadruplex was identified and found to inhibit telomerase [31, 32].
Human telomeric DNA is structurally polymorphic and may adopt different intramolecular G-quadruplex conformations, including two equilibrating hybrid-type structures [33–39] and a 2-tetrad structure [40–42] in K+ solution, a parallel structure in the crystalline form in the presence of K+ [43], and a basket-type structure in Na+ solution [44]. The hybrid-type structures can effectively form packed multimers at the telomere ends [33, 45]. Although different human telomeric G-quadruplexes appear to have small energy differences relative to each other, interconversion between them is kinetically slow, indicating a high-energy intermediate (s) [33, 41, 46–48]. The structure polymorphism appears to be an intrinsic property of the highly conserved telomeric sequence in higher eukaryotes, particularly the TTA loop sequence [49]. On the other hand, telomeric repeat-containing RNA was shown to adopt parallel-stranded G-quadruplexes [50–52].
G-Quadruplexes in Gene Promoters
More recently, DNA G-quadruplexes were found to form in the gene promoter regions and function as transcriptional regulators [53, 54], which has been the most active area for G-quadruplex DNA in the past decade. The first experiments that suggested the existence of unusual forms of DNA associated with runs of guanines in gene promoters were reported in 1982 for the chicken β-globulin gene based upon the nuclease hypersensitivity of promoter elements [55–57]. Since then, the occurrence of these elements in the human gene promoters has been reported, including in those of insulin [58], c-MYC [54, 59], VEGF [60, 61], HIF-1α [62], BCL-2 [63–65], MtCK [66], K-RAS [67, 68], c-KIT [69, 70], RET [71], PDGF-A [72], c-MYB [73], hTERT [74], and PDGF-Rβ [75, 76], in addition to mouse α7 integrin [77]. The potential occurrence of DNA G-quadruplexes has been discovered in the promoter regions of human genes involved in growth and proliferation [53, 78, 79]; these genes all contain G-rich/C-rich tracts in the proximal regions of promoters and are mostly TATA-less. In addition, the potential for quadruplex formation is higher within oncogenes as compared to tumor suppressor genes [80]. Computational analyses showed significant enrichment of G-quadruplex-forming sequences in the promoter regions of human genes near transcription start sites (TSS) [81]. The driving force of the formation of promoter G-quadruplexes appears to be the transcription-induced dynamic negative superhelicity [82–85]. The c-MYC gene promoter is the most extensively studied system for the promoter G-quadruplex [54, 86]. A highly conserved G-rich nuclease hypersensitivity element III1 in the proximal region of the c-MYC promoter controls 80–90% of the transcriptional activity regardless of whether the P1 or P2 promoter is used [87–90]. This element in the c-MYC promoter is highly dynamic in its conformation [91], and can form G-quadruplex structures, which function as a transcriptional silencer [54, 59].
In contrast to the repeating tandems in the telomeric sequence, the promoter G-quadruplex-forming sequences are each unique in their number and length of G-tracts and intervening bases. The promoter G-rich sequences often contain more than four G-tracts with unequal numbers of guanines and can form multiple G-quadruplexes through utilizing varying combinations of G-tracts or different loop isomers through utilizing varying guanines on one G-tract [18]. Parallel structures are common to the promoter G-quadruplexes, usually with a three-tetrad core. Structural studies showed that each promoter G-quadruplex adopts unique capping and loop structures determined by its specific sequence, such as c-MYC [92–95], BCL-2 [63, 65, 96, 97], KRAS [98], c-KIT [99–101], VEGF [102], and PDGFR-β [103, 104]. A notable feature in the promoter G-quadruplexes is the prevalence of the G3NG3 motif, a robust parallel-stranded structural motif with a 1-nt propeller loop. This motif was first observed in the major G-quadruplex structure formed in the c-MYC promoter, which showed that the 1-nt propeller loop conformation is highly favored [94]. By having two such motifs, parallel promoter G-quadruplexes can have a long and variable middle loop [65, 97, 105]. In addition, parallel G-quadruplexes exist in variant forms, such as with broken-strand [99, 103], end-insertion [104], or even with an additional hairpin loop conformations [65, 74]. Furthermore, certain promoter sequences can form multiple G-quadruplexes on one overlapping region or on separate regions. For example, the BCL-2 proximal promoter contains two G-quadruplex-forming regions that are separated by 13 nt (Pu39 and P1G4), with two competing G4s, i.e., a hybrid structure [63, 96] and a parallel structure [97], formed in Pu39, and two equilibrating parallel G4s formed in P1G4 [65]. Similar phenomenon was observed in the promoters of KRAS [67, 68, 106–108], c-KIT [69, 70, 99–101], PDGFR-β [75, 76, 103, 104], and hTERT [74, 109–111]. The variations in promoter G-quadruplexes give rise to different overall structure properties that could be specifically recognized by proteins or small-molecule ligands for transcriptional regulation. Moreover, inherent polymorphism and equilibrium between different conformations may provide an additional layer of transcriptional modulation.
G-Quadruplexes in Other Regions of Genome and in RNA
G-quadruplexes have been found in other regions of the human genome, such as immunoglobulin class switch regions [112–114], ribosomal DNA [115], mitochondrial DNA [116–119], replication initiation regions [120], the LINE-1 retrotransposon [121–123], DNA:RNA hybrid-G-quadruplexes in transcription [124], as well as in the extended repeat sequences in neurodegenerative diseases at both DNA and RNA levels, such as the (CGG)n repeat in the 5′-UTR of the FMR1 gene in the Fragile X syndrome (FXS) [125–127] and the hexanucleotide repeat expansion (HRE) (GGGGCC)n in C9orf72 of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) [128]. In addition, G-quadruplexes have been found to form in RNA. G-quadruplexes formed in 5′-UTR have been shown to inhibit translation [129, 130], such as NRAS [131], Zic-1 [132], TRF2 [133], Yin Yang 1 [134], or in internal ribosomal entry sites (IRESs) to initiate cap-independent translation, such as VEGF [135]. In addition, G-quadruplexes are found in 3′-UTRs [136–138] as well as in RNA introns to regulate the alternative splicing, such as TP53 [139] and Bcl-XL [140].
Notably, DNA G-quadruplex structures are recently shown to be involved in genomic instability and DNA damage [141–143].
G-Quadruplexes in Nonhuman Genomes
G-quadruplexes have been identified in nonhuman genomes. As in the human genome, these G-quadruplexes predominantly occur in regions of regulatory importance. Particularly, G-quadruplexes are found in genomes of human pathogens [144]. Many examples of G-quadruplexes were identified in viruses [145, 146], including human immunodeficiency virus (HIV) [147–150], multiple species of herpes virus [151–155], human papillomavirus (HPV) [156], hepatitis C [157], Zika [158], Ebola [159], and a G-quadruplex-binding protein was found in severe acute respiratory syndrome (SARS) coronavirus [160]. In bacteria, G-quadruplexes are found in Escherichia coli [161], Neisseria gonorrhoeae [162], Neisseria meningitidis [163], Mycobacterium tuberculosis [164], and Deinococcus radiodurans [165]. G-quadruplexes were also found in ciliates [14], malaria parasites [166, 167], and yeasts [168, 169]. Notably, helicases that resolve G-quadruplex structures, such as RecQ [170, 171] and Pif1 [142] families, were found in both nonhuman and human systems. Most recently, the presence of G-quadruplexes in plant genomes has also emerged [172].
G-Quadruplex Detection In Vivo
There has been significant progress in the detection of G-quadruplexes structures in vivo [173]. The first direct evidence of the in vivo existence of G-quadruplexes was established by using G-quadruplex-specific single chain variable fragment (scFv) of an antibody to detect G-quadruplexes formed at telomeres in macronuclei of the ciliate Stylonychia lemnae, which was shown to be cell cycle-dependent [174] and controlled by telomere end-binding proteins TEBPα and TEBPβ phosphorylation [175]. More recently, using a G-quadruplex-specific antibodies BG4 (scFv) [176] and 1H6 (monoclonal antibody) [177], G-quadruplex structures were visualized in human cells at both telomeric and non-telomeric sites on chromosomes, and G4-loci number increased after exposure of live cells to G-quadruplex ligands [176] or in the absence of FANCJ, a G-quadruplex DNA-specific helicase [177]. Using BG4 to map endogenous G-quadruplex structures by G4 ChIP-seq in human cells, ~10,000 endogenous G-quadruplex structures were detected in immortalized precancerous HaCaT cells, 10 times higher than in normal human NHEK cells [178]. G-quadruplex structures were found to be enriched in nucleosome-depleted regulatory regions including the promoters, such as c-MYC, and 5′ UTRs, of highly transcribed genes. The detected G-quadruplexes in cells account for less than 1% of the genomic G4-sites identified by G4-seq [179] or predicted by G4 algorithms [81], suggesting the in vivo formation of G-quadruplex is highly context-dependent. In addition, the endogenous potential G4 sites were detected in live human cells by chemical footprinting combined with high-throughput sequencing and were found to enrich in regions involving chromatin reorganization and gene transcription [180]. G-quadruplex formation in vivo was also detected by small molecules probes, such as the radiolabeled G-quadruplex-ligands 360A [181], and fluorescent G-quadruplex-ligands BMVC [182, 183] and DAOTA-M2 [184].
G-Quadruplex-Interactive Small Molecules
Recognition of the biological significance of G-quadruplexes has promoted research and development of G-quadruplex-interactive small molecule ligands (G4-ligands). The identification of genomic G-quadruplex structures in regions of functional importance, such as human telomeres and oncogene promoters, has created the opportunity to selectively target these globular DNA structures for cancer-specific drug development [17, 185–190]. The therapeutic possibilities of targeting telomeric G-quadruplexes to inhibit telomerase were first reported in 1997 [191] and have been actively pursued [185–188]. G-quadruplex-ligands were also shown to inhibit the alternative lengthening of telomeres (ALT) pathway which maintains telomere stability in a telomerase-independent manner in ~15% of cancer cells [192–195]. The discovery of the perylene derivative PIPER to inhibit helicase Sgs1-mediated G-quadruplex unfolding suggested the existence of a broader mechanism for G-quadruplex-ligands [196]. In 2002, a small molecule that stabilizes the G-quadruplex formed in the c-MYC promoter was shown to inhibit c-MYC expression, suggesting therapeutic opportunity of targeting promoter G-quadruplexes for transcriptional modulation [54, 197]. Different groups of compounds, such as quindolines and ellipticines, were reported to suppress c-MYC transcription by stabilization of the c-MYC promoter G-quadruplex [198–201]. Subsequently, transcriptional repression of other oncogenes was shown by compounds stabilizing promoter G-quadruplexes, such as c-KIT [202], BCL-2 [203], KRAS [204–206]. More recently, G-quadruplex-stabilizing compounds were shown to cause DNA damage and genomic instability and exhibit synergistic effect with inhibitors or deficiency of DNA-repair mechanisms [207–211]. Specifically, G-quadruplex-stabilizing compounds were shown to induce selective lethality in BRCA-deficient cancers by targeting the inherent DNA double-strand break (DSB) repair deficiency [212, 213].
A G-quadruplex targeting drug, Quarfloxin (CX-3543) [115], based on the fluoroquinolone compounds developed by Laurence Hurley [214, 215] showed excellent in vivo activity in various solid tumors and had reached Phase II clinical trials. Its second-generation compound, CX-5461, is currently in clinical trials for BRCA1/2 deficient tumors (Canadian trial, NCT02719977) [213]. Diverse families of other small molecule compounds that interact with G-quadruplexes were developed and studied. For example, TMPyP4, a tetra-(N-methyl-4-pyridyl)-porphyrin, is a structure-based designed compound that exhibits significant selectivity for quadruplex DNA over duplex DNA and inhibits telomerase and ALT [216, 217]. Its positional isomer TMPyP2 is a poor G-quadruplex-interactive compound and can be used as a negative control of TMPyP4 [218]. Later studies revealed that TMPyP4 interacts with the c-MYC promoter G-quadruplex and downregulates c-MYC [54, 197]. TMPyP4 and TMPyP2 have been one of the most widely used molecules in G-quadruplex research. Telomestatin is a natural product isolated from Streptomyces anulatus 3533-SV4 to be a highly potent inhibitor of telomerase [219] and active against human cancers by stabilizing telomeric G-quadruplex and inhibit telomere-protein binding [220–223]. BRACO19 is a rationally designed trisubstituted acridine to directly target telomeric G-quadruplex [224] and was shown to inhibit telomerase and induce telomere uncapping in human cancer cells [195] and have high in vivo activity against different cancer xenograft models [225, 226]. 12459 is a triazine G4-ligand that exhibits anti-telomerase activity but also appears to involve BCL-2 and hTERT splicing [192, 227, 228]. Closely related pyridine dicarboxamide derivatives 360A/307A and bisquinolinium compounds Phen-DC(3)/Phen-DC(6) are highly selective G-quadruplex ligands and were shown to be active against both telomeric and c-MYC G-quadruplex and inhibit c-MYC gene transcription in tumor cells, as well as bind to a G-quadruplex formed in the 5′-UTR of TRF2 mRNA to repress translation [133, 229–232]. Phen-DC was also shown to trigger genetic instability in Saccharomyces cerevisiae [207]. PDS is a pyridostatin compound which shows potent G-quadruplex- stabilization and has been widely used to study G-quadruplex functions and G-quadruplex-induced DNA damage [176, 210–212, 233]. In addition, G-quadruplex DNA was also shown to be potential cancer therapeutics. AS1411 (Antisoma, London, UK) is an unmodified 26-nt G-quadruplex forming oligonucleotide that has been in Phase II trials for treatment of renal cancer and acute myeloid leukemia [234].
G-quadruplex-interactive compounds have contributed immensely to understanding G-quadruplex functions and potential as a therapeutic target. Different G-quadruplex-ligands show various levels of selectivity, between G-quadruplex structures over other forms of DNA and between different G-quadruplexes, and this selectivity is likely to be related to their biological activity. Conventional and in silico screening methods as well as structure-based rational drug design were actively pursued in the development of G-quadruplex-targeting small molecules. A common feature among the G-quadruplex-ligands is the presence of a fused ring system that is capable of stacking with the terminal G-tetrads. In addition, a crescent-shaped asymmetric pharmacophore that can recruit a DNA base and cationic side chain substituents that have the propensity to interact with G-quadruplex grooves can give rise to specific interactions (Fig. 1d) [235, 236]. Structural data of G-quadruplex-ligand complexes has been playing an important role in the understanding of small molecule recognition of G-quadruplexes and the design of G-quadruplex-ligands [18, 237]. This includes a handful NMR solution structures of intramolecular G-quadruplex-ligand complexes, including c-MYC G-quadruplex-ligand complexes [235, 238–240] and telomeric G-quadruplex-ligand complexes [236, 241–243], and X-ray crystallographic structures of intramolecular and intermolecular telomeric G-quadruplex-ligand complexes [43, 237, 244–250].
Methods to Study G-Quadruplexes
A wide variety of experimental tools and methods have been utilized or developed for studying G-quadruplex DNA and RNA. These methods play a pivotal role in enabling researchers to gain an understanding of G-quadruplex structures, properties, and functions. The methods commonly used for studying G-quadruplexes include biophysical, biochemical, molecular biology, and cellular methods, as described in this book.
Biophysical methods are widely used to study physical properties of G-quadruplex such as structures, stability, and binding interactions with ligands and proteins. Circular dichroism (CD) is widely used to study G-quadruplex conformations and stability. Isothermal titration calorimetry (ITC) can directly measure binding enthalpies and provide thermodynamic characterization of G-quadruplex-ligand interactions. Biosensor- surface plasmon resonance (SPR) is a quantitative approach for the study of small molecule and protein ligand-quadruplex nucleic acid interactions in real time. Analytical ultracentrifugation (AUC) method can be used to characterize G-quadruplex formation and to monitor ligand binding. Mass spectroscopy can also be used to characterize G-quadruplex structures and ligand binding. Differential scanning calorimetry (DSC) can be used to obtain thermodynamic and sometimes kinetic parameters of G-quadruplexes. X-ray crystallography and solution NMR spectroscopy provide structural information of G-quadruplexes and ligand complexes, while molecular dynamics simulation can also be used to study G-quadruplex structures and small molecule binding.
Biochemical and molecular biology methods are used to study G-quadruplex formation, functions, and protein interactions. Electrophoretic mobility shift assay (EMSA), dimethyl sulfate (DMS) footprinting, and DNA polymerase stop (Pol-stop) assay are widely used to study G-quadruplex formation, protein complexes, and ligand interactions. Chromatin immunoprecipitation (ChIP) assays are used to probe protein interactions with G-quadruplex-forming DNA sequences. A combination of biochemical and biophysical methods can be used to monitor co-transcriptional formation of G-quadruplexes (transcription assay) and to quantitatively analyze the effects of G-quadruplex formation on DNA replication (replication assay). Single-molecule methods such as optical and magnetic tweezers, atomic-force microscopy (AFM), and single-molecule fluorescence resonance energy transfer (FRET) microscopy can be used to investigate G-quadruplex conformations, ligand interactions, and protein interactions. In addition, methods are used to discover and develop G-quadruplex-targeting molecules, such as FRET-based high-throughput screening of small molecule ligands, and peptide nucleic acid (PNA) oligomers that are designed to bind to G-quadruplexes. G-quadruplexes are also used in nanoparticle-based assays, and as biocatalysts such as G-quadruplex DNAzymes.
More recent and exciting developments include in-cell methods to study the G-quadruplex formation in vivo, such as in vivo chemical footprinting, G-quadruplex detection and visualization, and in-cell NMR. Chemical probing for G-quadruplex formation inside living cells combined with high-throughput sequencing can provide a snapshot of the DNA conformation over the whole genome in vivo. G4-specific antibodies and fluorescence probes are used to detect and visualize G-quadruplexes in cells. NMR spectroscopy is used to study G-quadruplex structures inside living Xenopus laevis oocytes, while 19F NMR can be used to study G-quadruplex conformation in vitro and in living cells.
In conclusion, it is our hope that the protocols described herein will be found both informative and useful.
Acknowledgments
This research was supported by the National Institutes of Health, R01CA122952, 1R01 GM083117, R01CA177585, and P30CA023168 (Purdue Center for Cancer Research).
Contributor Information
Danzhou Yang, Phone: +117654941403, Email: yangdz@purdue.edu.
Clement Lin, Phone: +11765-494-8363, Email: lin835@purdue.edu.
Danzhou Yang, Email: yangdz@purdue.edu.
References
- 1.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fry M. Tetraplex DNA and its interacting proteins. Front Biosci. 2007;12:4336–4351. doi: 10.2741/2391. [DOI] [PubMed] [Google Scholar]
- 3.Oganesian L, Bryan TM. Physiological relevance of telomeric G-quadruplex formation: a potential drug target. BioEssays. 2007;29(2):155–165. doi: 10.1002/bies.20523. [DOI] [PubMed] [Google Scholar]
- 4.Mendoza O, Bourdoncle A, Boule JB, Brosh RM, Jr, Mergny JL. G-quadruplexes and helicases. Nucleic Acids Res. 2016;44(5):1989–2006. doi: 10.1093/nar/gkw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brázda V, Hároníková L, Liao JCC, Fojta M. DNA and RNA Quadruplex-binding proteins. Int J Molecul Sci. 2014;15(10):17493. doi: 10.3390/ijms151017493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McRae EKS, Booy EP, Padilla-Meier GP, McKenna SA. On characterizing the interactions between proteins and guanine quadruplex structures of nucleic acids. J Nucleic Acids. 2017;2017:9675348. doi: 10.1155/2017/9675348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calzolari A, Di Felice R, Molinari E, Garbesi A. G-quartet biomolecular nanowires. Appl Phys Lett. 2002;80(18):3331–3333. doi: 10.1063/1.1476700. [DOI] [Google Scholar]
- 8.Yatsunyk LA, Mendoza O, Mergny JL. “Nano-oddities”: unusual nucleic acid assemblies for DNA-based nanostructures and nanodevices. Acc Chem Res. 2014;47(6):1836–1844. doi: 10.1021/ar500063x. [DOI] [PubMed] [Google Scholar]
- 9.Platella C, Riccardi C, Montesarchio D, Roviello GN, Musumeci D. G-quadruplex-based aptamers against protein targets in therapy and diagnostics. Biochim Biophys Acta Gen Subj. 2017;1861(5):1429–1447. doi: 10.1016/j.bbagen.2016.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma DL, Wu C, Dong ZZ, Tam WS, Wong SW, Yang C, Li G, Leung CH. The development of G-quadruplex-based assays for the detection of small molecules and toxic substances. Chem Asian J. 2017;12(15):1851–1860. doi: 10.1002/asia.201700533. [DOI] [PubMed] [Google Scholar]
- 11.Rioz-Martinez A, Roelfes G. DNA-based hybrid catalysis. Curr Opin Chem Biol. 2015;25:80–87. doi: 10.1016/j.cbpa.2014.12.033. [DOI] [PubMed] [Google Scholar]
- 12.Bang I. Untersuchungen über die Guanylsäure. Biochem Z. 1910;26:293–311. [Google Scholar]
- 13.Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc Natl Acad Sci U S A. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williamson JR, Raghuraman MK, Cech TR. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59(5):871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 15.Sen D, Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344(6265):410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 16.Hud NV, Smith FW, Anet FAL, Feigon J. The selectivity for K+ versus Na+ in DNA quadruplexes is dominated by relative free energies of hydration: a thermodynamic analysis by 1H NMR. Biochemistry. 1996;35(48):15383–15390. doi: 10.1021/bi9620565. [DOI] [PubMed] [Google Scholar]
- 17.Yang D, Okamoto K. Structural insights into G-quadruplexes: towards new anticancer drugs. Future Med Chem. 2010;2(4):619–646. doi: 10.4155/fmc.09.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y, Yang DZ. Sequence, stability, and structure of G-quadruplexes and their interactions with drugs. Curr Protoc Nucl Acid Chem. 2012;50:17.15.11–17.15.17. doi: 10.1002/0471142700.nc1705s50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blackburn EH. Telomere states and cell fates. Nature. 2000;408(6808):53–56. doi: 10.1038/35040500. [DOI] [PubMed] [Google Scholar]
- 20.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92(3):401–413. doi: 10.1016/S0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 21.Hackett JA, Feldser DM, Greider CW. Telomere dysfunction increases mutation rate and genomic instability. Cell. 2001;106(3):275–286. doi: 10.1016/S0092-8674(01)00457-3. [DOI] [PubMed] [Google Scholar]
- 22.Henderson E, Hardin CC, Walk SK, Tinoco I, Blackburn EH. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell. 1987;51(6):899–908. doi: 10.1016/0092-8674(87)90577-0. [DOI] [PubMed] [Google Scholar]
- 23.Sundquist WI, Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342(6251):825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- 24.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci U S A. 1988;85(18):6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43(2):405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 26.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266(5193):2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 27.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 28.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350(6320):718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 29.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97(4):503–514. doi: 10.1016/S0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 30.Stansel RM, de Lange T, Griffith JD. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 2001;20(19):5532–5540. doi: 10.1093/emboj/20.19.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat–containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318(5851):798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 32.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol. 2008;10(2):228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 33.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34(9):2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai J, Punchihewa C, Ambrus A, Chen D, Jones RA, Yang DZ. Structure of the intramolecular human telomeric G-quadruplex in potassium solution: a novel adenine triple formation. Nucleic Acids Res. 2007;35(7):2440–2450. doi: 10.1093/nar/gkm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dai J, Carver M, Punchihewa C, Jones RA, Yang DZ. Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007;35(15):4927–4940. doi: 10.1093/nar/gkm522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Y, Noguchi Y, Sugiyama H. The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorg Med Chem. 2006;14(16):5584–5591. doi: 10.1016/j.bmc.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 37.Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the human telomere in K+ solution: an intramolecular (3+1) G-quadruplex scaffold. J Am Chem Soc. 2006;128(30):9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phan AT, Luu KN, Patel DJ. Different loop arrangements of intramolecular human telomeric (3+1) G-quadruplexes in K+ solution. Nucleic Acids Res. 2006;34(19):5715–5719. doi: 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phan AT, Kuryavyi V, Luu KN, Patel DJ. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007;35(19):6517–6525. doi: 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim KW, Amrane S, Bouaziz S, Xu WX, Mu YG, Patel DJ, Luu KN, Phan AT. Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers. J Am Chem Soc. 2009;131(12):4301–4309. doi: 10.1021/ja807503g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z, Dai J, Veliath E, Jones RA, Yang DZ. Structure of a two-G-tetrad intramolecular G-quadruplex formed by a variant human telomeric sequence in K+ solution: insights into the interconversion of human telomeric G-quadruplex structures. Nucleic Acids Res. 2010;38(3):1009–1021. doi: 10.1093/nar/gkp1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansel R, Loehr F, Trantirek L, Doetsch V. High-resolution insights into G-overhang architecture. J Am Chem Soc. 2013;135(7):2816–2824. doi: 10.1021/ja312403b. [DOI] [PubMed] [Google Scholar]
- 43.Parkinson GN, Lee MPH, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417(6891):876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1(4):263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 45.Petraccone L, Spink C, Trent JO, Garbett NC, Mekmaysy CS, Giancola C, Chaires JB. Structure and stability of higher-order human telomeric quadruplexes. J Am Chem Soc. 2011;133(51):20951–20961. doi: 10.1021/ja209192a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gray RD, Chaires JB. Kinetics and mechanism of K+- and Na+−induced folding of models of human telomeric DNA into G-quadruplex structures. Nucleic Acids Res. 2008;36(12):4191–4203. doi: 10.1093/nar/gkn379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rajendran A, Endo M, Hidaka K, Sugiyama H. Direct and single-molecule visualization of the solution-state structures of G-hairpin and G-triplex intermediates. Angew Chem Int Ed Engl. 2014;53(16):4107–4112. doi: 10.1002/anie.201308903. [DOI] [PubMed] [Google Scholar]
- 48.Mashimo T, Yagi H, Sannohe Y, Rajendran A, Sugiyama H. Folding pathways of human telomeric type-1 and type-2 G-quadruplex structures. J Am Chem Soc. 2010;132(42):14910–14918. doi: 10.1021/ja105806u. [DOI] [PubMed] [Google Scholar]
- 49.Dai J, Carver M, Yang D. Polymorphism of human telomeric quadruplex structures. Biochimie. 2008;90(8):1172–1183. doi: 10.1016/j.biochi.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu Y, Kaminaga K, Komiyama M. G-quadruplex formation by human telomeric repeats-containing RNA in Na+ solution. J Am Chem Soc. 2008;130(33):11179–11184. doi: 10.1021/ja8031532. [DOI] [PubMed] [Google Scholar]
- 51.Collie GW, Haider SM, Neidle S, Parkinson GN. A crystallographic and modelling study of a human telomeric RNA (TERRA) quadruplex. Nucleic Acids Res. 2010;38(16):5569–5580. doi: 10.1093/nar/gkq259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martadinata H, Phan AT. Structure of human telomeric RNA (TERRA): stacking of two G-quadruplex blocks in K(+) solution. Biochemistry. 2013;52(13):2176–2183. doi: 10.1021/bi301606u. [DOI] [PubMed] [Google Scholar]
- 53.Qin Y, Hurley LH. Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie. 2008;90(8):1149–1171. doi: 10.1016/j.biochi.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci U S A. 2002;99(18):11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Larsen A, Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982;29(2):609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- 56.Wood WI, Felsenfeld G. Chromatin structure of the chicken beta-globin gene region - sensitivity to Dnase-I, micrococcal nuclease, and Dnase-II. J Biol Chem. 1982;257(13):7730–7736. [PubMed] [Google Scholar]
- 57.Woodford KJ, Howell RM, Usdin K. A novel K+-dependent DNA-synthesis arrest site in a commonly occurring sequence motif in eukaryotes. J Biol Chem. 1994;269(43):27029–27035. [PubMed] [Google Scholar]
- 58.Hammond-Kosack MC, Dobrinski B, Lurz R, Docherty K, Kilpatrick MW. The human insulin gene linked polymorphic region exhibits an altered DNA structure. Nucleic Acids Res. 1992;20(2):231–236. doi: 10.1093/nar/20.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simonsson T, Pecinka P, Kubista M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26(5):1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun D, Guo K, Rusche JJ, Hurley LH. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33(18):6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo K, Gokhale V, Hurley LH, Sun D. Intramolecularly folded G-quadruplex and i-motif structures in the proximal promoter of the vascular endothelial growth factor gene. Nucleic Acids Res. 2008;36(14):4598–4608. doi: 10.1093/nar/gkn380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De Armond R, Wood S, Sun D, Hurley LH, Ebbinghaus SW. Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1alpha promoter. Biochemistry. 2005;44(49):16341–16350. doi: 10.1021/bi051618u. [DOI] [PubMed] [Google Scholar]
- 63.Dai J, Dexheimer TS, Chen D, Carver M, Ambrus A, Jones RA, Yang DZ. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J Am Chem Soc. 2006;128(4):1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dexheimer TS, Sun D, Hurley LH. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J Am Chem Soc. 2006;128(16):5404–5415. doi: 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Onel B, Carver M, Wu G, Timonina D, Kalarn S, Larriva M, Yang D. A new G-quadruplex with hairpin loop immediately upstream of the human BCL2 P1 promoter modulates transcription. J Am Chem Soc. 2016;138(8):2563–2570. doi: 10.1021/jacs.5b08596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yafe A, Etzioni S, Weisman-Shomer P, Fry M. Formation and properties of hairpin and tetraplex structures of guanine-rich regulatory sequences of muscle-specific genes. Nucleic Acids Res. 2005;33(9):2887–2900. doi: 10.1093/nar/gki606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cogoi S, Paramasivam M, Spolaore B, Xodo LE. Structural polymorphism within a regulatory element of the human KRAS promoter: formation of G4-DNA recognized by nuclear proteins. Nucleic Acids Res. 2008;36(11):3765–3780. doi: 10.1093/nar/gkn120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34(9):2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rankin S, Reszka AP, Huppert J, Zloh M, Parkinson GN, Todd AK, Ladame S, Balasubramanian S, Neidle S. Putative DNA quadruplex formation within the human c-kit oncogene. J Am Chem Soc. 2005;127(30):10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fernando H, Reszka AP, Huppert J, Ladame S, Rankin S, Venkitaraman AR, Neidle S, Balasubramanian S. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry. 2006;45(25):7854–7860. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guo K, Pourpak A, Beetz-Rogers K, Gokhale V, Sun D, Hurley LH. Formation of pseudosymmetrical G-quadruplex and i-motif structures in the proximal promoter region of the RET oncogene. J Am Chem Soc. 2007;129(33):10220–10228. doi: 10.1021/ja072185g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qin Y, Rezler EM, Gokhale V, Sun D, Hurley LH. Characterization of the G-quadruplexes in the duplex nuclease hypersensitive element of the PDGF-A promoter and modulation of PDGF-A promoter activity by TMPyP4. Nucleic Acids Res. 2007;35(22):7698–7713. doi: 10.1093/nar/gkm538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Palumbo SL, Memmott RM, Uribe DJ, Krotova-Khan Y, Hurley LH, Ebbinghaus SW. A novel G-quadruplex-forming GGA repeat region in the c-myb promoter is a critical regulator of promoter activity. Nucleic Acids Res. 2008;36(6):1755–1769. doi: 10.1093/nar/gkm1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Palumbo SL, Ebbinghaus SW, Hurley LH. Formation of a unique end-to-end stacked pair of G-quadruplexes in the hTERT core promoter with implications for inhibition of telomerase by G-quadruplex-interactive ligands. J Am Chem Soc. 2009;131(31):10878–10891. doi: 10.1021/ja902281d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qin Y, Fortin JS, Tye D, Gleason-Guzman M, Brooks TA, Hurley LH. Molecular cloning of the human platelet-derived growth factor receptor beta (PDGFR-beta) promoter and drug targeting of the G-quadruplex-forming region to repress PDGFR-beta expression. Biochemistry. 2010;49(19):4208–4219. doi: 10.1021/bi100330w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brown RV, Wang T, Chappeta VR, Wu G, Onel B, Chawla R, Quijada H, Camp SM, Chiang ET, Lassiter QR. The consequences of overlapping G-quadruplexes and i-motifs in the platelet-derived growth factor receptor β core promoter nuclease hypersensitive element can explain the unexpected effects of mutations and provide opportunities for selective targeting of both structures by small molecules to downregulate gene expression. J Am Chem Soc. 2017;139(22):7456–7475. doi: 10.1021/jacs.6b10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Etzioni S, Yafe A, Khateb S, Weisman-Shomer P, Bengal E, Fry M. Homodimeric MyoD preferentially binds tetraplex structures of regulatory sequences of muscle-specific genes. J Biol Chem. 2005;280(29):26805–26812. doi: 10.1074/jbc.M500820200. [DOI] [PubMed] [Google Scholar]
- 78.Rustighi A, Tessari MA, Vascotto F, Sgarra R, Giancotti V, Manfioletti G. A polypyrimidine/polypurine tract within the Hmga2 minimal promoter: a common feature of many growth-related genes. Biochemistry. 2002;41(4):1229–1240. doi: 10.1021/bi011666o. [DOI] [PubMed] [Google Scholar]
- 79.Hurley LH, Siddiqui-Jain A. Developing therapeutics to target oncogenes. Genet Eng News. 2005;25(1):26. [Google Scholar]
- 80.Eddy J, Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34(14):3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35(2):406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat Struct Mol Biol. 2008;15(2):146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 83.Kouzine F, Levens D. Supercoil-driven DNA structures regulate genetic transactions. Front Biosci. 2007;12:4409–4423. doi: 10.2741/2398. [DOI] [PubMed] [Google Scholar]
- 84.Sun D, Hurley LH. The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: implications for drug targeting and control of gene expression. J Medicin Chem. 2009;52(9):2863–2874. doi: 10.1021/jm900055s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zheng KW, He YD, Liu HH, Li XM, Hao YH, Tan Z. Superhelicity constrains a localized and R-loop-dependent formation of G-quadruplexes at the upstream region of transcription. ACS Chem Biol. 2017;12(10):2609–2618. doi: 10.1021/acschembio.7b00435. [DOI] [PubMed] [Google Scholar]
- 86.Brooks TA, Hurley LH. The role of supercoiling in transcriptional control of MYC and its importance in molecular therapeutics. Nat Rev Cancer. 2009;9(12):849–861. doi: 10.1038/nrc2733. [DOI] [PubMed] [Google Scholar]
- 87.Michelotti EF, Tomonaga T, Krutzsch H, Levens D. Cellular nucleic acid binding protein regulates the CT element of the human c- myc protooncogene. J Biol Chem. 1995;270(16):9494–9499. doi: 10.1074/jbc.270.16.9494. [DOI] [PubMed] [Google Scholar]
- 88.Sakatsume O, Tsutsui H, Wang Y, Gao H, Tang X, Yamauchi T, Murata T, Itakura K, Yokoyama KK. Binding of THZif-1, a MAZ-like zinc finger protein to the nuclease-hypersensitive element in the promoter region of the c-MYC protooncogene. J Biol Chem. 1996;271(49):31322–31333. doi: 10.1074/jbc.271.49.31322. [DOI] [PubMed] [Google Scholar]
- 89.Tomonaga T, Levens D. Activating transcription from single stranded DNA. Proc Natl Acad Sci U S A. 1996;93(12):5830–5835. doi: 10.1073/pnas.93.12.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Berberich SJ, Postel EH. PuF/NM23-H2/NDPK-B transactivates a human c-myc promoter-CAT gene via a functional nuclease hypersensitive element. Oncogene. 1995;10(12):2343–2347. [PubMed] [Google Scholar]
- 91.Michelotti GA, Michelotti EF, Pullner A, Duncan RC, Eick D, Levens D. Multiple single-stranded cis elements are associated with activated chromatin of the human c-myc gene in vivo. Molecul Cell Biol. 1996;16(6):2656–2669. doi: 10.1128/MCB.16.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Seenisamy J, Rezler EM, Powell TJ, Tye D, Gokhale V, Joshi CS, Siddiqui-Jain A, Hurley LH. The dynamic character of the G-quadruplex element in the c-MYC promoter and modification by TMPyP4. J Am Chem Soc. 2004;126(28):8702–8709. doi: 10.1021/ja040022b. [DOI] [PubMed] [Google Scholar]
- 93.Phan AT, Modi YS, Patel DJ. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J Am Chem Soc. 2004;126(28):8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44(6):2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 95.Mathad RI, Hatzakis E, Dai J, Yang DZ. C-MYC promoter G-quadruplex formed at the 5′-end of NHE III1 element: insights into biological relevance and parallel-stranded G-quadruplex stability. Nucleic Acids Res. 2011;39(20):9023–9033. doi: 10.1093/nar/gkr612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dai J, Chen D, Jones RA, Hurley LH, Yang DZ. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res. 2006;34(18):5133–5144. doi: 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Agrawal P, Lin C, Mathad RI, Carver M, Yang D. The major G-quadruplex formed in the human BCL-2 proximal promoter adopts a parallel structure with a 13-nt loop in K+ solution. J Am Chem Soc. 2014;136(5):1750–1753. doi: 10.1021/ja4118945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kerkour A, Marquevielle J, Ivashchenko S, Yatsunyk LA, Mergny JL, Salgado GF. High-resolution three-dimensional NMR structure of the KRAS proto-oncogene promoter reveals key features of a G-quadruplex involved in transcriptional regulation. J Biol Chem. 2017;292(19):8082–8091. doi: 10.1074/jbc.M117.781906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Phan AT, Kuryavyi V, Burge S, Neidle S, Patel DJ. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J Am Chem Soc. 2007;129(14):4386–4392. doi: 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hsu ST, Varnai P, Bugaut A, Reszka AP, Neidle S, Balasubramanian S. A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics. J Am Chem Soc. 2009;131(37):13399–13409. doi: 10.11021/ja904007p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kuryavyi V, Phan AT, Patel DJ. Solution structures of all parallel-stranded monomeric and dimeric G-quadruplex scaffolds of the human c-kit2 promoter. Nucleic Acids Res. 2010;38(19):6757–6773. doi: 10.1093/nar/gkq558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Agrawal P, Hatzakis E, Guo K, Carver M, Yang D. Solution structure of the major G-quadruplex formed in the human VEGF promoter in K+: insights into loop interactions of the parallel G-quadruplexes. Nucleic Acids Res. 2013;41(22):10584–10592. doi: 10.1093/nar/gkt784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen Y, Agrawal P, Brown RV, Hatzakis E, Hurley L, Yang D. The major G-quadruplex formed in the human platelet-derived growth factor receptor beta promoter adopts a novel broken-strand structure in K+ solution. J Am Chem Soc. 2012;134(32):13220–13223. doi: 10.1021/ja305764d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Onel B, Carver M, Agrawal P, Hurley LH, Yang D. The 3′-end region of the human PDGFR-beta core promoter nuclease hypersensitive element forms a mixture of two unique end-insertion G-quadruplexes. Biochim Biophys Acta Gen Subj. 2018;1862(4):846–854. doi: 10.1016/j.bbagen.2017.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Guédin A, Gros J, Alberti P, Mergny J-L. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010;38(21):7858–7868. doi: 10.1093/nar/gkq639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cogoi S, Paramasivam M, Filichev V, Geci I, Pedersen EB, Xodo LE. Identification of a new G-quadruplex motif in the KRAS promoter and design of pyrene-modified G4-decoys with antiproliferative activity in pancreatic cancer cells. J Medicin Chem. 2009;52(2):564–568. doi: 10.1021/jm800874t. [DOI] [PubMed] [Google Scholar]
- 107.Paramasivam M, Cogoi S, Xodo LE. Primer extension reactions as a tool to uncover folding motifs within complex G-rich sequences: analysis of the human KRAS NHE. Chem Commun. 2011;47(17):4965–4967. doi: 10.1039/c1cc10269a. [DOI] [PubMed] [Google Scholar]
- 108.Kaiser CE, Van Ert NA, Agrawal P, Chawla R, Yang D, Hurley LH. Insight into the complexity of the i-motif and G-quadruplex DNA structures formed in the KRAS promoter and subsequent drug-induced gene repression. J Am Chem Soc. 2017;139(25):8522–8536. doi: 10.1021/jacs.7b02046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lim KW, Lacroix L, Yue DJ, Lim JK, Lim JM, Phan AT. Coexistence of two distinct G-quadruplex conformations in the hTERT promoter. J Am Chem Soc. 2010;132(35):12331–12342. doi: 10.1021/ja101252n. [DOI] [PubMed] [Google Scholar]
- 110.Chaires JB, Trent JO, Gray RD, Dean WL, Buscaglia R, Thomas SD, Miller DM. An improved model for the hTERT promoter quadruplex. PLoS One. 2014;9(12):e115580. doi: 10.1371/journal.pone.0115580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kang HJ, Cui Y, Yin H, Scheid A, Hendricks WP, Schmidt J, Sekulic A, Kong D, Trent JM, Gokhale V, Mao H, Hurley LH. A pharmacological chaperone molecule induces cancer cell death by restoring tertiary DNA structures in mutant hTERT promoters. J Am Chem Soc. 2016;138(41):13673–13692. doi: 10.1021/jacs.6b07598. [DOI] [PubMed] [Google Scholar]
- 112.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334(6180):364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 113.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18(13):1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Larson ED, Duquette ML, Cummings WJ, Streiff RJ, Maizels N. MutS[alpha] binds to and promotes synapsis of transcriptionally activated immunoglobulin switch regions. Curr Biol. 2005;15(5):470–474. doi: 10.1016/j.cub.2004.12.077. [DOI] [PubMed] [Google Scholar]
- 115.Drygin D, Siddiqui-Jain A, O’Brien S, Schwaebe M, Lin A, Bliesath J, Ho CB, Proffitt C, Trent K, Whitten JP, Lim JK, Von Hoff D, Anderes K, Rice WG. Anticancer activity of CX-3543: a direct inhibitor of rRNA biogenesis. Cancer Res. 2009;69(19):7653–7661. doi: 10.1158/0008-5472.CAN-09-1304. [DOI] [PubMed] [Google Scholar]
- 116.Wanrooij PH, Uhler JP, Simonsson T, Falkenberg M, Gustafsson CM. G-quadruplex structures in RNA stimulate mitochondrial transcription termination and primer formation. Proc Natl Acad Sci U S A. 2010;107(37):16072–16077. doi: 10.1073/pnas.1006026107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wanrooij PH, Uhler JP, Shi Y, Westerlund F, Falkenberg M, Gustafsson CM. A hybrid G-quadruplex structure formed between RNA and DNA explains the extraordinary stability of the mitochondrial R-loop. Nucleic Acids Res. 2012;40(20):10334–10344. doi: 10.1093/nar/gks802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bharti SK, Sommers JA, Zhou J, Kaplan DL, Spelbrink JN, Mergny JL, Brosh RM., Jr DNA sequences proximal to human mitochondrial DNA deletion breakpoints prevalent in human disease form G-quadruplexes, a class of DNA structures inefficiently unwound by the mitochondrial replicative twinkle helicase. J Biol Chem. 2014;289(43):29975–29993. doi: 10.1074/jbc.M114.567073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Huang WC, Tseng TY, Chen YT, Chang CC, Wang ZF, Wang CL, Hsu TN, Li PT, Chen CT, Lin JJ, Lou PJ, Chang TC. Direct evidence of mitochondrial G-quadruplex DNA by using fluorescent anti-cancer agents. Nucleic Acids Res. 2015;43(21):10102–10113. doi: 10.1093/nar/gkv1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Besnard E, Babled A, Lapasset L, Milhavet O, Parrinello H, Dantec C, Marin JM, Lemaitre JM. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat Struct Mol Biol. 2012;19(8):837–844. doi: 10.1038/nsmb.2339. [DOI] [PubMed] [Google Scholar]
- 121.Howell R, Usdin K. The ability to form intrastrand tetraplexes is an evolutionarily conserved feature of the 3′ end of L1 retrotransposons. Molecul Biol Evol. 1997;14(2):144–155. doi: 10.1093/oxfordjournals.molbev.a025747. [DOI] [PubMed] [Google Scholar]
- 122.Lexa M, Steflova P, Martinek T, Vorlickova M, Vyskot B, Kejnovsky E. Guanine quadruplexes are formed by specific regions of human transposable elements. BMC Genomics. 2014;15:1032. doi: 10.1186/1471-2164-15-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sahakyan AB, Murat P, Mayer C, Balasubramanian S. G-quadruplex structures within the 3′ UTR of LINE-1 elements stimulate retrotransposition. Nat Struct Mol Biol. 2017;24(3):243–247. doi: 10.1038/nsmb.3367. [DOI] [PubMed] [Google Scholar]
- 124.Zhang JY, Zheng KW, Xiao S, Hao YH, Tan Z. Mechanism and manipulation of DNA:RNA hybrid G-quadruplex formation in transcription of G-rich DNA. J Am Chem Soc. 2014;136(4):1381–1390. doi: 10.1021/ja4085572. [DOI] [PubMed] [Google Scholar]
- 125.Fry M, Loeb LA. The fragile-X syndrome D(Cgg)(N) nucleotide repeats form a stable tetrahelical structure. Proc Natl Acad Sci U S A. 1994;91(11):4950–4954. doi: 10.1073/pnas.91.11.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Usdin K, Woodford KJ. Cgg repeats associated with DNA instability and chromosome fragility form structures that block DNA-synthesis in-vitro. Nucleic Acids Res. 1995;23(20):4202–4209. doi: 10.1093/nar/23.20.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Brown V, Jin P, Ceman S, Darnell JC, O’Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, Darnell RB, Warren ST. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107(4):477–487. doi: 10.1016/S0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 128.Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS, Maragakis NJ, Troncoso JC, Pandey A, Sattler R, Rothstein JD, Wang J. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507(7491):195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bugaut A, Balasubramanian S. 5′-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012;40(11):4727–4741. doi: 10.1093/nar/gks068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Beaudoin J-D, Perreault J-P. 5′-UTR G-quadruplex structures acting as translational repressors. Nucleic Acids Res. 2010;38(20):7022–7036. doi: 10.1093/nar/gkq557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat Chem Biol. 2007;3(4):218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Arora A, Dutkiewicz M, Scaria V, Hariharan M, Maiti S, Kurreck J. Inhibition of translation in living eukaryotic cells by an RNA G-quadruplex motif. RNA. 2008;14(7):1290–1296. doi: 10.1261/rna.1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gomez D, Guédin A, Mergny JL, Salles B, Riou JF, Teulade-Fichou MP, Calsou P. A G-quadruplex structure within the 5′-UTR of TRF2 mRNA represses translation in human cells. Nucleic Acids Res. 2010;38(20):7187–7198. doi: 10.1093/nar/gkq563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Huang W, Smaldino PJ, Zhang Q, Miller LD, Cao P, Stadelman K, Wan M, Giri B, Lei M, Nagamine Y. Yin Yang 1 contains G-quadruplex structures in its promoter and 5′-UTR and its expression is modulated by G4 resolvase 1. Nucleic Acids Res. 2011;40(3):1033–1049. doi: 10.1093/nar/gkr849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Morris MJ, Negishi Y, Pazsint C, Schonhoft JD, Basu S. An RNA G-quadruplex is essential for cap-independent translation initiation in human VEGF IRES. J Am Chem Soc. 2010;132(50):17831–17839. doi: 10.1021/ja106287x. [DOI] [PubMed] [Google Scholar]
- 136.Christiansen J, Kofod M, Nielsen FC. A guanosine quadruplex and two stable hairpins flank a major cleavage site in insulin-like growth factor II mRNA. Nucleic Acids Res. 1994;22(25):5709–5716. doi: 10.1093/nar/22.25.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wieland M, Hartig JS. RNA quadruplex-based modulation of gene expression. Chem Biol. 2007;14(7):757–763. doi: 10.1016/j.chembiol.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 138.Beaudoin JD, Perreault JP. Exploring mRNA 3′-UTR G-quadruplexes: evidence of roles in both alternative polyadenylation and mRNA shortening. Nucleic Acids Res. 2013;41(11):5898–5911. doi: 10.1093/nar/gkt265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Marcel V, Tran PL, Sagne C, Martel-Planche G, Vaslin L, Teulade-Fichou MP, Hall J, Mergny JL, Hainaut P, Van Dyck E. G-quadruplex structures in TP53 intron 3: role in alternative splicing and in production of p53 mRNA isoforms. Carcinogenesis. 2011;32(3):271–278. doi: 10.1093/carcin/bgq253. [DOI] [PubMed] [Google Scholar]
- 140.Weldon C, Dacanay JG, Gokhale V, Boddupally PVL, Behm-Ansmant I, Burley GA, Branlant C, Hurley LH, Dominguez C, Eperon IC. Specific G-quadruplex ligands modulate the alternative splicing of Bcl-X. Nucleic Acids Res. 2018;46(2):886–896. doi: 10.1093/nar/gkx1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ribeyre C, Lopes J, Boule JB, Piazza A, Guedin A, Zakian VA, Mergny JL, Nicolas A. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5(5):e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Paeschke K, Bochman ML, Garcia PD, Cejka P, Friedman KL, Kowalczykowski SC, Zakian VA. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497(7450):458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhao J, Bacolla A, Wang G, Vasquez KM. Non-B DNA structure-induced genetic instability and evolution. Cell Mol Life Sci. 2010;67(1):43–62. doi: 10.1007/s00018-009-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Harris LM, Merrick CJ. G-quadruplexes in pathogens: a common route to virulence control? PLoS Pathog. 2015;11(2):e1004562. doi: 10.1371/journal.ppat.1004562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ruggiero E, Richter SN. G-quadruplexes and G-quadruplex ligands: targets and tools in antiviral therapy. Nucleic Acids Res. 2018;46(7):3270–3283. doi: 10.1093/nar/gky187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Metifiot M, Amrane S, Litvak S, Andreola ML. G-quadruplexes in viruses: function and potential therapeutic applications. Nucleic Acids Res. 2014;42(20):12352–12366. doi: 10.1093/nar/gku999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sundquist WI, Heaphy S. Evidence for interstrand quadruplex formation in the dimerization of human immunodeficiency virus 1 genomic RNA. Proc Natl Acad Sci U S A. 1993;90(8):3393–3397. doi: 10.1073/pnas.90.8.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Perrone R, Nadai M, Frasson I, Poe JA, Butovskaya E, Smithgall TE, Palumbo M, Palu G, Richter SN. A dynamic G-quadruplex region regulates the HIV-1 long terminal repeat promoter. J Med Chem. 2013;56(16):6521–6530. doi: 10.1021/jm400914r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Perrone R, Nadai M, Poe JA, Frasson I, Palumbo M, Palu G, Smithgall TE, Richter SN. Formation of a unique cluster of G-quadruplex structures in the HIV-1 Nef coding region: implications for antiviral activity. PLoS One. 2013;8(8):e73121. doi: 10.1371/journal.pone.0073121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Piekna-Przybylska D, Sullivan MA, Sharma G, Bambara RA. U3 region in the HIV-1 genome adopts a G-quadruplex structure in its RNA and DNA sequence. Biochemistry. 2014;53(16):2581–2593. doi: 10.1021/bi4016692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Norseen J, Johnson FB, Lieberman PM. Role for G-quadruplex RNA binding by Epstein-Barr virus nuclear antigen 1 in DNA replication and metaphase chromosome attachment. J Virol. 2009;83(20):10336–10346. doi: 10.1128/JVI.00747-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Artusi S, Nadai M, Perrone R, Biasolo MA, Palu G, Flamand L, Calistri A, Richter SN. The herpes simplex virus-1 genome contains multiple clusters of repeated G-quadruplex: implications for the antiviral activity of a G-quadruplex ligand. Antivir Res. 2015;118:123–131. doi: 10.1016/j.antiviral.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Biswas B, Kandpal M, Jauhari UK, Vivekanandan P. Genome-wide analysis of G-quadruplexes in herpesvirus genomes. BMC Genomics. 2016;17(1):949. doi: 10.1186/s12864-016-3282-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Madireddy A, Purushothaman P, Loosbroock CP, Robertson ES, Schildkraut CL, Verma SC. G-quadruplex-interacting compounds alter latent DNA replication and episomal persistence of KSHV. Nucleic Acids Res. 2016;44(8):3675–3694. doi: 10.1093/nar/gkw038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gilbert-Girard S, Gravel A, Artusi S, Richter SN, Wallaschek N, Kaufer BB, Flamand L. Stabilization of telomere G-quadruplexes interferes with human herpesvirus 6A chromosomal integration. J Virol. 2017;91(14):00402–00417. doi: 10.1128/JVI.00402-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tluckova K, Marusic M, Tothova P, Bauer L, Sket P, Plavec J, Viglasky V. Human papillomavirus G-quadruplexes. Biochemistry. 2013;52(41):7207–7216. doi: 10.1021/bi400897g. [DOI] [PubMed] [Google Scholar]
- 157.Wang SR, Min YQ, Wang JQ, Liu CX, Fu BS, Wu F, Wu LY, Qiao ZX, Song YY, Xu GH, Wu ZG, Huang G, Peng NF, Huang R, Mao WX, Peng S, Chen YQ, Zhu Y, Tian T, Zhang XL, Zhou X. A highly conserved G-rich consensus sequence in hepatitis C virus core gene represents a new anti-hepatitis C target. Sci Adv. 2016;2(4):e1501535. doi: 10.1126/sciadv.1501535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fleming AM, Ding Y, Alenko A, Burrows CJ. Zika virus genomic RNA possesses conserved G-quadruplexes characteristic of the flaviviridae family. ACS Infect Dis. 2016;2(10):674–681. doi: 10.1021/acsinfecdis.6b00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Krafcikova P, Demkovicova E, Viglasky V. Ebola virus derived G-quadruplexes: thiazole orange interaction. BBA-Gen Subjects. 2017;1861 ((5):1321–1328. doi: 10.1016/j.bbagen.2016.12.009. [DOI] [PubMed] [Google Scholar]
- 160.Kusov Y, Tan J, Alvarez E, Enjuanes L, Hilgenfeld R. A G-quadruplex-binding macrodomain within the "SARS-unique domain" is essential for the activity of the SARS-coronavirus replication-transcription complex. Virology. 2015;484:313–322. doi: 10.1016/j.virol.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rawal P, Kummarasetti VBR, Ravindran J, Kumar N, Halder K, Sharma R, Mukerji M, Das SK, Chowdhury S. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 2006;16(5):644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cahoon LA, Seifert HS. An alternative DNA structure is necessary for pilin antigenic variation in neisseria gonorrhoeae. Science. 2009;325(5941):764–767. doi: 10.1126/science.1175653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wormann ME, Horien CL, Bennett JS, Jolley KA, Maiden MCJ, Tang CM, Aho EL, Exley RM. Sequence, distribution and chromosomal context of class I and class II pilin genes of Neisseria meningitidis identified in whole genome sequences. BMC Genomics. 2014;15(1):253. doi: 10.1186/1471-2164-15-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Thakur RS, Desingu A, Basavaraju S, Subramanya S, Rao DN, Nagaraju G. Mycobacterium tuberculosis DinG is a structure-specific helicase that unwinds G4 DNA implications for targeting g4 dna as a novel therapeutic approach. J Biol Chem. 2014;289(36):25112–25136. doi: 10.1074/jbc.M114.563569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Beaume N, Pathak R, Yadav VK, Kota S, Misra HS, Gautam HK, Chowdhury S. Genome-wide study predicts promoter-G4 DNA motifs regulate selective functions in bacteria: radioresistance of D. radiodurans involves G4 DNA-mediated regulation. Nucleic Acids Res. 2013;41(1):76–89. doi: 10.1093/nar/gks1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Smargiasso N, Gabelica V, Damblon C, Rosu F, De Pauw E, Teulade-Fichou MP, Rowe JA, Claessens A. Putative DNA G-quadruplex formation within the promoters of Plasmodium falciparum var genes. BMC Genomics. 2009;10(1):362. doi: 10.1186/1471-2164-10-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Stanton A, Harris LM, Graham G, Merrick CJ. Recombination events among virulence genes in malaria parasites are associated with G-quadruplex-forming DNA motifs. BMC Genomics. 2016;17(1):859. doi: 10.1186/s12864-016-3183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hershman SG, Chen Q, Lee JY, Kozak ML, Yue P, Wang LS, Johnson FB. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36(1):144–156. doi: 10.1093/nar/gkm986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Smith JS, Chen Q, Yatsunyk LA, Nicoludis JM, Garcia MS, Kranaster R, Balasubramanian S, Monchaud D, Teulade-Fichou M-P, Abramowitz L. Rudimentary G-quadruplex–based telomere capping in Saccharomyces cerevisiae. Nat Struct Mol Biol. 2011;18(4):478. doi: 10.1038/nsmb.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hickson ID. RecQ helicases: caretakers of the genome. Nat Rev Cancer. 2003;3(3):169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 171.Lee JY, Kozak M, Martin JD, Pennock E, Johnson FB. Evidence that a RecQ helicase slows senescence by resolving recombining telomeres. PLoS Biol. 2007;5(6):e160. doi: 10.1371/journal.pbio.0050160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yadav V, Hemansi KN, Tuteja N, Yadav P. G quadruplex in plants: a ubiquitous regulatory element and its biological relevance. Front Plant Sci. 2017;8(1163):1163. doi: 10.3389/fpls.2017.01163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hansel-Hertsch R, Di Antonio M, Balasubramanian S. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat Rev Mol Cell Biol. 2017;18(5):279–284. doi: 10.1038/nrm.2017.3. [DOI] [PubMed] [Google Scholar]
- 174.Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc Natl Acad Sci U S A. 2001;98(15):8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat Struct Mol Biol. 2005;12(10):847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 176.Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat Chem. 2013;5(3):182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Henderson A, Wu Y, Huang YC, Chavez EA, Platt J, Johnson FB, Brosh RM, Jr, Sen D, Lansdorp PM. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014;42(2):860–869. doi: 10.1093/nar/gkt957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hansel-Hertsch R, Beraldi D, Lensing SV, Marsico G, Zyner K, Parry A, Di Antonio M, Pike J, Kimura H, Narita M, Tannahill D, Balasubramanian S. G-quadruplex structures mark human regulatory chromatin. Nat Genet. 2016;48(10):1267–1272. doi: 10.1038/ng.3662. [DOI] [PubMed] [Google Scholar]
- 179.Chambers VS, Marsico G, Boutell JM, Di Antonio M, Smith GP, Balasubramanian S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat Biotechnol. 2015;33(8):877–881. doi: 10.1038/nbt.3295. [DOI] [PubMed] [Google Scholar]
- 180.Kouzine F, Wojtowicz D, Baranello L, Yamane A, Nelson S, Resch W, Kieffer-Kwon KR, Benham CJ, Casellas R, Przytycka TM, Levens D. Permanganate/S1 nuclease footprinting reveals non-B DNA structures with regulatory potential across a mammalian genome. Cell Syst. 2017;4(3):344–356 e347. doi: 10.1016/j.cels.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Granotier C, Pennarun G, Riou L, Hoffschir F, Gauthier LR, De Cian A, Gomez D, Mandine E, Riou JF, Mergny JL, Mailliet P, Dutrillaux B, Boussin FD. Preferential binding of a G-quadruplex ligand to human chromosome ends. Nucleic Acids Res. 2005;33(13):4182–4190. doi: 10.1093/nar/gki722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Chang CC, Kuo IC, Lin JJ, Lu YC, Chen CT, Back HT, Lou PJ, Chang TC. A novel carbazole derivative, BMVC: a potential antitumor agent and fluorescence marker of cancer cells. Chem Biodivers. 2004;1(9):1377–1384. doi: 10.1002/cbdv.200490100. [DOI] [PubMed] [Google Scholar]
- 183.Kang CC, Chang CC, Chang TC, Liao LJ, Lou PJ, Xie W, Yeung ES. A handheld device for potential point-of-care screening of cancer. Analyst. 2007;132(8):745–749. doi: 10.1039/b617733f. [DOI] [PubMed] [Google Scholar]
- 184.Shivalingam A, Izquierdo MA, Le Marois A, Vysniauskas A, Suhling K, Kuimova MK, Vilar R. The interactions between a small molecule and G-quadruplexes are visualized by fluorescence lifetime imaging microscopy. Nat Commun. 2015;6:8178. doi: 10.1038/ncomms9178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hurley LH, Wheelhouse RT, Sun D, Kerwin SM, Salazar M, Fedoroff OY, Han FX, Han H, Izbicka E, Von Hoff DD. G-quadruplexes as targets for drug design. Pharmacol Therapeut. 2000;85(3):141–158. doi: 10.1016/S0163-7258(99)00068-6. [DOI] [PubMed] [Google Scholar]
- 186.Hurley LH. DNA and its associated processes as targets for cancer therapy. Nat Rev Cancer. 2002;2(3):188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 187.Mergny JL, Helene C. G-quadruplex DNA: a target for drug design. Nat Med. 1998;4(12):1366–1367. doi: 10.1038/3949. [DOI] [PubMed] [Google Scholar]
- 188.Neidle S, Parkinson G. Telomere maintenance as a target for anticancer drug discovery. Nat Rev Drug Discov. 2002;1(5):383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- 189.Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat Rev Drug Discov. 2011;10(4):261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Neidle S. Quadruplex nucleic acids as targets for anticancer therapeutics. Nature Reviews Chemistry. 2017;1:0041. doi: 10.1038/s41570-017-0041. [DOI] [Google Scholar]
- 191.Sun D, Thompson B, Cathers BE, Salazar M, Kerwin SM, Trent JO, Jenkins TC, Neidle S, Hurley LH. Inhibition of human telomerase by a G-quadruplex-interactive compound. J Med Chem. 1997;40(14):2113–2116. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 192.Riou JF, Guittat L, Mailliet P, Laoui A, Renou E, Petitgenet O, Megnin-Chanet F, Helene C, Mergny JL. Cell senescence and telomere shortening induced by a new series of specific G-quadruplex DNA ligands. Proc Natl Acad Sci U S A. 2002;99(5):2672–2677. doi: 10.1073/pnas.052698099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Gowan SM, Heald R, Stevens MF, Kelland LR. Potent inhibition of telomerase by small-molecule pentacyclic acridines capable of interacting with G-quadruplexes. Mol Pharmacol. 2001;60(5):981–988. doi: 10.1124/mol.60.5.981. [DOI] [PubMed] [Google Scholar]
- 194.Harrison RJ, Reszka AP, Haider SM, Romagnoli B, Morrell J, Read MA, Gowan SM, Incles CM, Kelland LR, Neidle S. Evaluation of by disubstituted acridone derivatives as telomerase inhibitors: the importance of G-quadruplex binding. Bioorg Med Chem Lett. 2004;14(23):5845–5849. doi: 10.1016/j.bmcl.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 195.Incles CM, Schultes CM, Kempski H, Koehler H, Kelland LR, Neidle S. A G-quadruplex telomere targeting agent produces p16-associated senescence and chromosomal fusions in human prostate cancer cells. Molecul Cancer Therapeut. 2004;3(10):1201–1206. [PubMed] [Google Scholar]
- 196.Han HY, Bennett RJ, Hurley LH. Inhibition of unwinding of G-quadruplex structures by Sgs1 helicase in the presence of N,N′-bis[2-(1-piperidino)ethyl]-3,4,9,10-perylenetetracarboxylic diimide, a G-quadruplex-interactive ligand. Biochemistry. 2000;39(31):9311–9316. doi: 10.1021/bi000482r. [DOI] [PubMed] [Google Scholar]
- 197.Grand CL, Han H, Munoz RM, Weitman S, Von Hoff DD, Hurley LH, Bearss DJ. The cationic porphyrin TMPyP4 down-regulates c-MYC and human telomerase reverse transcriptase expression and inhibits tumor growth in vivo. Molecul Cancer Therapeut. 2002;1(8):565–573. [PubMed] [Google Scholar]
- 198.Ou TM, Lu YJ, Zhang C, Huang ZS, Wang XD, Tan JH, Chen Y, Ma DL, Wong KY, Tang JC, Chan AS, Gu LQ. Stabilization of G-quadruplex DNA and down-regulation of oncogene c-myc by quindoline derivatives. J Med Chem. 2007;50(7):1465–1474. doi: 10.1021/jm0610088. [DOI] [PubMed] [Google Scholar]
- 199.Liu J-N, Deng R, Guo J-F, Zhou J-M, Feng G-K, Huang Z-S, Gu L-Q, Zeng Y-X, Zhu X-F. Inhibition of myc promoter and telomerase activity and induction of delayed apoptosis by SYUIQ-5, a novel G-quadruplex interactive agent in leukemia cells. Leukemia. 2007;21(6):1300–1302. doi: 10.1038/sj.leu.2404652. [DOI] [PubMed] [Google Scholar]
- 200.Kang HJ, Park HJ. Novel molecular mechanism for actinomycin D activity as an oncogenic promoter G-quadruplex binder. Biochemistry. 2009;48(31):7392–7398. doi: 10.1021/bi9006836. [DOI] [PubMed] [Google Scholar]
- 201.Brown RV, Danford FL, Gokhale V, Hurley LH, Brooks TA. Demonstration that drug-targeted downregulation of MYC in non-hodgkins lymphoma is directly mediated through the promoter G-quadruplex. J Biol Chem. 2011;286(47):41018–41027. doi: 10.1074/jbc.M111.274720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.McLuckie KI, Waller ZA, Sanders DA, Alves D, Rodriguez R, Dash J, McKenzie GJ, Venkitaraman AR, Balasubramanian S. G-quadruplex-binding benzo[a]phenoxazines down-regulate c-KIT expression in human gastric carcinoma cells. J Am Chem Soc. 2011;133(8):2658–2663. doi: 10.1021/ja109474c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wang XD, Ou TM, Lu YJ, Li Z, Xu Z, Xi C, Tan JH, Huang SL, An LK, Li D, Gu LQ, Huang ZS. Turning off transcription of the bcl-2 gene by stabilizing the bcl-2 promoter quadruplex with quindoline derivatives. J Med Chem. 2010;53(11):4390–4398. doi: 10.1021/jm100445e. [DOI] [PubMed] [Google Scholar]
- 204.Lavrado J, Borralho PM, Ohnmacht SA, Castro RE, Rodrigues CM, Moreira R, dos Santos DJ, Neidle S, Paulo A. Synthesis, G-quadruplex stabilisation, docking studies, and effect on cancer cells of indolo[3,2-b]quinolines with one, two, or three basic side chains. ChemMedChem. 2013;8(10):1648–1661. doi: 10.1002/cmdc.201300288. [DOI] [PubMed] [Google Scholar]
- 205.Lavrado J, Brito H, Borralho PM, Ohnmacht SA, Kim NS, Leitao C, Pisco S, Gunaratnam M, Rodrigues CM, Moreira R, Neidle S, Paulo A. KRAS oncogene repression in colon cancer cell lines by G-quadruplex binding indolo[3,2-c]quinolines. Sci Rep. 2015;5:9696. doi: 10.1038/srep09696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Ohnmacht SA, Marchetti C, Gunaratnam M, Besser RJ, Haider SM, Di Vita G, Lowe HL, Mellinas-Gomez M, Diocou S, Robson M, Sponer J, Islam B, Pedley RB, Hartley JA, Neidle S. A G-quadruplex-binding compound showing anti-tumour activity in an in vivo model for pancreatic cancer. Sci Rep. 2015;5:11385. doi: 10.1038/srep11385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Piazza A, Boule JB, Lopes J, Mingo K, Largy E, Teulade-Fichou MP, Nicolas A. Genetic instability triggered by G-quadruplex interacting Phen-DC compounds in Saccharomyces cerevisiae. Nucleic Acids Res. 2010;38(13):4337–4348. doi: 10.1093/nar/gkq136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Salvati E, Scarsella M, Porru M, Rizzo A, Iachettini S, Tentori L, Graziani G, D’Incalci M, Stevens MFG, Orlandi A, Passeri D, Gilson E, Zupi G, Leonetti C, Biroccio A. PARP1 is activated at telomeres upon G4 stabilization: possible target for telomere-based therapy. Oncogene. 2010;29(47):6280–6293. doi: 10.1038/onc.2010.344. [DOI] [PubMed] [Google Scholar]
- 209.Aggarwal M, Sommers JA, Shoemaker RH, Brosh RM., Jr Inhibition of helicase activity by a small molecule impairs Werner syndrome helicase (WRN) function in the cellular response to DNA damage or replication stress. Proc Natl Acad Sci U S A. 2011;108(4):1525–1530. doi: 10.1073/pnas.1006423108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Rodriguez R, Miller KM, Forment JV, Bradshaw CR, Nikan M, Britton S, Oelschlaegel T, Xhemalce B, Balasubramanian S, Jackson SP. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat Chem Biol. 2012;8(3):301–310. doi: 10.1038/nchembio.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.McLuckie KIE, Di Antonio M, Zecchini H, Xian J, Caldas C, Krippendorff BF, Tannahill D, Lowe C, Balasubramanian S. G-quadruplex DNA as a molecular target for induced synthetic lethality in cancer cells. J Am Chem Soc. 2013;135(26):9640–9643. doi: 10.1021/ja404868t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Zimmer J, Tacconi EMC, Folio C, Badie S, Porru M, Klare K, Tumiati M, Markkanen E, Halder S, Ryan A, Jackson SP, Ramadan K, Kuznetsov SG, Biroccio A, Sale JE, Tarsounas M. Targeting BRCA1 and BRCA2 deficiencies with G-quadruplex-interacting compounds. Mol Cell. 2016;61(3):449–460. doi: 10.1016/j.molcel.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Xu H, Di Antonio M, McKinney S, Mathew V, Ho B, O’Neil NJ, Dos Santos N, Silvester J, Wei V, Garcia J. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat Commun. 2017;8:14432. doi: 10.1038/ncomms14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Duan W, Rangan A, Vankayalapati H, Kim MY, Zeng Q, Sun D, Han H, Fedoroff OY, Nishioka D, Rha SY, Izbicka E, Von Hoff DD, Hurley LH. Design and synthesis of fluoroquinophenoxazines that interact with human telomeric G-quadruplexes and their biological effects. Mol Cancer Ther. 2001;1(2):103–120. [PubMed] [Google Scholar]
- 215.Kim M-Y, Duan W, Gleason-Guzman Mary, Hurley LH. Design, synthesis, and biological evaluation of a series of fluoroquinoanthroxazines with contrasting dual mechanisms of action against topoisomerase II and G-Quadruplexes. J Med Chem. 2003;46(4):571–583. doi: 10.1021/jm0203377. [DOI] [PubMed] [Google Scholar]
- 216.Wheelhouse RT, Sun D, Han H, Han FX, Hurley LH. Cationic porphyrins as telomerase inhibitors: the interaction of tetra-(N-methyl-4-pyridyl)porphine with quadruplex DNA. J Am Chem Soc. 1998;120(13):3261–3262. doi: 10.1021/ja973792e. [DOI] [Google Scholar]
- 217.Kim MY, Gleason-Guzman M, Izbicka E, Nishioka D, Hurley LH. The different biological effects of telomestatin and TMPyP4 can be attributed to their selectivity for interaction with intramolecular or intermolecular G-quadruplex structures. Cancer Res. 2003;63(12):3247–3256. [PubMed] [Google Scholar]
- 218.Han FXG, Wheelhouse RT, Hurley LH. Interactions of TMPyP4 and TMPyP2 with quadruplex DNA. Structural basis for the differential effects on telomerase inhibition. J Am Chem Soc. 1999;121(15):3561–3570. doi: 10.1021/ja984153m. [DOI] [Google Scholar]
- 219.Shin-ya K, Wierzba K, Matsuo K, Ohtani T, Yamada Y, Furihata K, Hayakawa Y, Seto H. Telomestatin, a novel telomerase inhibitor from Streptomyces anulatus. J Am Chem Soc. 2001;123(6):1262–1263. doi: 10.1021/ja005780q. [DOI] [PubMed] [Google Scholar]
- 220.Kim MY, Vankayalapati H, Shin-Ya K, Wierzba K, Hurley LH. Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular G-quadruplex. J Am Chem Soc. 2002;124(10):2098–2099. doi: 10.1021/ja017308q. [DOI] [PubMed] [Google Scholar]
- 221.Tauchi T, Shin-ya K, Sashida G, Sumi M, Okabe S, Ohyashiki JH, Ohyashiki K. Telomerase inhibition with a novel G-quadruplex-interactive agent, telomestatin: in vitro and in vivo studies in acute leukemia. Oncogene. 2006;25(42):5719–5725. doi: 10.1038/sj.onc.1209577. [DOI] [PubMed] [Google Scholar]
- 222.Gomez D, O’Donohue MF, Wenner T, Douarre C, Macadre J, Koebel P, Giraud-Panis MJ, Kaplan H, Kolkes A, Shin-ya K, Riou JF. The G-quadruplex ligand telomestatin inhibits POT1 binding to telomeric sequences in vitro and induces GFP-POT1 dissociation from telomeres in human cells. Cancer Res. 2006;66(14):6908–6912. doi: 10.1158/0008-5472.CAN-06-1581. [DOI] [PubMed] [Google Scholar]
- 223.Tahara H, Shin-Ya K, Seimiya H, Yamada H, Tsuruo T, Ide T. G-Quadruplex stabilization by telomestatin induces TRF2 protein dissociation from telomeres and anaphase bridge formation accompanied by loss of the 3′ telomeric overhang in cancer cells. Oncogene. 2006;25(13):1955–1966. doi: 10.1038/sj.onc.1209217. [DOI] [PubMed] [Google Scholar]
- 224.Read M, Harrison RJ, Romagnoli B, Tanious FA, Gowan SH, Reszka AP, Wilson WD, Kelland LR, Neidle S. Structure-based design of selective and potent G-quadruplex-mediated telomerase inhibitors. Proc Natl Acad Sci U S A. 2001;98(9):4844–4849. doi: 10.1073/pnas.081560598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Kelland LR. Overcoming the immortality of tumour cells by telomere and telomerase based cancer therapeutics - current status and future prospects. Eur J Cancer. 2005;41(7):971–979. doi: 10.1016/j.ejca.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 226.Burger AM, Dai FP, Schultes CM, Reszka AP, Moore MJ, Double JA, Neidle S. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005;65(4):1489–1496. doi: 10.1158/0008-5472.CAN-04-2910. [DOI] [PubMed] [Google Scholar]
- 227.Douarre C, Gomez D, Morjani H, Zahm JM, O’Donohue MF, Eddabra L, Mailliet P, Riou JF, Trentesaux C. Overexpression of Bcl-2 is associated with apoptotic resistance to the G-quadruplex ligand 12459 but is not sufficient to confer resistance to long-term senescence. Nucleic Acids Res. 2005;33(7):2192–2203. doi: 10.1093/nar/gki514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Gomez D, Lemarteleur T, Lacroix L, Mailliet P, Mergny J-L, Riou J-F. Telomerase downregulation induced by the G-quadruplex ligand 12459 in A549 cells is mediated by hTERT RNA alternative splicing. Nucleic Acids Res. 2004;32(1):371–379. doi: 10.1093/nar/gkh181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Lemarteleur T, Gomez D, Paterski R, Mandine E, Mailliet P, Riou J-F. Stabilization of the c-myc gene promoter quadruplex by specific ligands’ inhibitors of telomerase. Biochem Biophys Res Commun. 2004;323(3):802–808. doi: 10.1016/j.bbrc.2004.08.150. [DOI] [PubMed] [Google Scholar]
- 230.Pennarun G, Granotier C, Gauthier LR, Gomez D, Boussin FD. Apoptosis related to telomere instability and cell cycle alterations in human glioma cells treated by new highly selective G-quadruplex ligands. Oncogene. 2005;24(18):2917–2928. doi: 10.1038/sj.onc.1208468. [DOI] [PubMed] [Google Scholar]
- 231.De Cian A, Mergny JL. Quadruplex ligands may act as molecular chaperones for tetramolecular quadruplex formation. Nucleic Acids Res. 2007;35(8):2483–2493. doi: 10.1093/nar/gkm098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.De Cian A, Delemos E, Mergny JL, Teulade-Fichou MP, Monchaud D. Highly efficient G-quadruplex recognition by bisquinolinium compounds. J Am Chem Soc. 2007;129(7):1856–1857. doi: 10.1021/ja067352b. [DOI] [PubMed] [Google Scholar]
- 233.Rodriguez MC, Sonyang Z. BRCT domains: phosphopeptide binding and signaling modules. Front Biosci. 2008;13:5905–5915. doi: 10.2741/3125. [DOI] [PubMed] [Google Scholar]
- 234.Bates PJ, Laber DA, Miller DM, Thomas SD, Trent JO. Discovery and development of the G-rich oligonucleotide AS1411 AS a novel treatment for cancer. Exp Mol Pathol. 2009;86(3):151–164. doi: 10.1016/j.yexmp.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Dai J, Carver M, Hurley LH, Yang D. Solution structure of a 2:1 quindoline-c-MYC G-quadruplex: insights into G-quadruplex-interactive small molecule drug design. J Am Chem Soc. 2011;133(44):17673–17680. doi: 10.1021/ja205646q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Lin C, Wu G, Wang K, Onel B, Sakai S, Shao Y, Yang D. Molecular recognition of the hybrid-2 human telomeric G-quadruplex by epiberberine: insights into conversion of telomeric g-quadruplex structures. Angew Chem Int Ed. 2018;57(34):10888–10893. doi: 10.1002/anie.201804667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Neidle S. The structures of quadruplex nucleic acids and their drug complexes. Curr Opin Struct Biol. 2009;19(3):239–250. doi: 10.1016/j.sbi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 238.Phan AT, Kuryavyi V, Gaw HY, Patel DJ. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat Chem Biol. 2005;1(3):167–173. doi: 10.1038/nchembio723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Chung WJ, Heddi B, Hamon F, Teulade-Fichou MP, Phan AT. Solution structure of a G-quadruplex bound to the bisquinolinium compound Phen-DC(3) Angew Chem Int Ed Engl. 2014;53(4):999–1002. doi: 10.1002/anie.201308063. [DOI] [PubMed] [Google Scholar]
- 240.Kotar A, Wang B, Shivalingam A, Gonzalez-Garcia J, Vilar R, Plavec J. NMR structure of a triangulenium-based long-lived fluorescence probe bound to a G-quadruplex. Angew Chem Int Ed Engl. 2016;55(40):12508–12511. doi: 10.1002/anie.201606877. [DOI] [PubMed] [Google Scholar]
- 241.Chung WJ, Heddi B, Tera M, Iida K, Nagasawa K, Phan AT. Solution structure of an intramolecular (3 + 1) human telomeric G-quadruplex bound to a telomestatin derivative. J Am Chem Soc. 2013;135(36):13495–13501. doi: 10.1021/ja405843r. [DOI] [PubMed] [Google Scholar]
- 242.Wirmer-Bartoschek J, Bendel LE, Jonker HR, Grün JT, Papi F, Bazzicalupi C, Messori L, Gratteri P, Schwalbe H. Solution NMR structure of a ligand/hybrid-2-G-quadruplex complex reveals rearrangements that affect ligand binding. Angew Chem. 2017;129(25):7208–7212. doi: 10.1002/ange.201702135. [DOI] [PubMed] [Google Scholar]
- 243.Liu W, Zhong Y-F, Liu L-Y, Shen C-T, Zeng W, Wang F, Yang D, Mao Z-W. Solution structures of multiple G-quadruplex complexes induced by a platinum (II)-based tripod reveal dynamic binding. Nat Commun. 2018;9(1):3496. doi: 10.1038/s41467-018-05810-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Clark GR, Pytel PD, Squire CJ, Neidle S. Structure of the first parallel DNA quadruplex-drug complex. J Am Chem Soc. 2003;125(14):4066–4067. doi: 10.1021/ja0297988. [DOI] [PubMed] [Google Scholar]
- 245.Haider SM, Parkinson GN, Neidle S. Structure of a G-quadruplex-ligand complex. J Molecul Biol. 2003;326(1):117–125. doi: 10.1016/S0022-2836(02)01354-2. [DOI] [PubMed] [Google Scholar]
- 246.Parkinson GN, Ghosh R, Neidle S. Structural basis for binding of porphyrin to human telomeres. Biochemistry. 2007;46(9):2390–2397. doi: 10.1021/bi062244n. [DOI] [PubMed] [Google Scholar]
- 247.Parkinson GN, Cuenca F, Neidle S. Topology conservation and loop flexibility in quadruplex-drug recognition: crystal structures of inter- and intramolecular telomeric DNA quadruplex-drug complexes. J Molecul Biol. 2008;381(5):1145–1156. doi: 10.1016/j.jmb.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 248.Campbell NH, Parkinson GN, Reszka AP, Neidle S. Structural basis of DNA quadruplex recognition by an acridine drug. J Am Chem Soc. 2008;130(21):6722–6724. doi: 10.1021/ja8016973. [DOI] [PubMed] [Google Scholar]
- 249.Collie GW, Promontorio R, Hampel SM, Micco M, Neidle S, Parkinson GN. Structural basis for telomeric G-quadruplex targeting by naphthalene diimide ligands. J Am Chem Soc. 2012;134(5):2723–2731. doi: 10.1021/ja2102423. [DOI] [PubMed] [Google Scholar]
- 250.Bazzicalupi C, Ferraroni M, Bilia AR, Scheggi F, Gratteri P. The crystal structure of human telomeric DNA complexed with berberine: an interesting case of stacked ligand to G-tetrad ratio higher than 1:1. Nucleic Acids Res. 2013;41(1):632–638. doi: 10.1093/nar/gks1001. [DOI] [PMC free article] [PubMed] [Google Scholar]