Abstract
The use of small animal models for the study of infectious disease is critical for understanding disease progression and for developing prophylactic and therapeutic treatment options. For many diseases, Syrian golden hamsters have emerged as an ideal animal model due to their low cost, small size, ease of handling, and ability to accurately reflect disease progression in humans. Despite the increasing use and popularity of hamsters, there remains a lack of available reagents for studying hamster immune responses. Without suitable reagents for assessing immune responses, researchers are left to examine clinical signs and disease pathology. This becomes an issue for the development of vaccine and treatment options where characterizing the type of immune response generated is critical for understanding protection from disease. Despite the relative lack of reagents for use in hamsters, significant advances have been made recently with several hamster specific immunologic methods being developed. Here we discuss the progress of this development, with focus on classical methods used as well as more recent molecular methods. We outline what methods are currently available for use in hamsters and what is readily used as well as what limitations still exist and future perspectives of reagent and assay development for hamsters. This will provide valuable information to researchers who are deciding whether to use hamsters as an animal model.
Keywords: Syrian hamster, Golden hamster, Syrian golden hamster, Immunological tools, Molecular methods, Mesocricetus auratus, Animal models
Introduction
The need for a reliable and representative animal model for the study of a particular disease is critical for expanding our understanding of the disease and for developing therapeutics and ultimately a cure. The first steps in developing intervention strategies involve establishing a small animal model for study of the disease, typically in rodents where mice are the most common model due to their low cost, ease of handling, availability of reagents, and potential for genetic manipulation. Despite their prominence and widespread use, there are many instances where mice are not suitable candidates for a disease model and alternative model is necessary. The guidelines for selecting an appropriate animal model have been outlined by the Canadian Council for Animal Care (CCAC) (Animal Care 1997) and the United States Food and Drug Administration (FDA) (FDA 2015). A model should be chosen such that the disease is pathophysiologically similar to the human condition in terms of onset, progression, symptoms, pathology, and disease outcome (FDA 2015). Additionally, the challenge agent should reliably cause disease in the model animal consistent with the parameters above and the host should be susceptible to a realistic challenge of the disease agent (FDA 2015).
Due to the ability of Syrian golden hamsters (Mesocricetus auratus; hereafter referred to as hamsters) to satisfy many of the conditions outlined by the CCAC and the FDA, they have been used as an alternative to mice in many disease models. There are many advantages to using hamsters as a disease model. They are outbred animals, allowing for disease modelling with more genetic diversity than inbred mice. Also, the requirements for housing of hamsters is similar to that of mice and rats, and facilities designed for housing rodents can typically accommodate hamsters without the need for additional equipment. Hamsters can be cohoused in small cages, which is a significant advantage over other alternative disease models such as guinea pigs and ferrets. These advantages are why many consider hamsters a superior alternative to other small animals for use in research. For the development of vaccines and therapeutic approaches, some consider hamsters a higher standard as small animal model than mice and as such hamsters have been utilized in a wide range of models, from those examining diabetes, atherosclerosis, neural plasticity, to cancer (Table 1) (Bhathena et al. 2011; Dillard et al. 2010; Jové et al. 2013; Staffend and Meisel 2012; Woods et al. 2015; Vijayalingam et al. 2014). However, the use of hamsters for models of pathogenic human diseases may be the most valuable due to comparable disease progression seen in hamsters to that of humans for many infectious diseases including bacteria, viruses, and parasites (Dondji et al. 2008; da Silva-Couto et al. 2015; Kuehne et al. 2014; Safronetz et al. 2012). Specifically, hamsters are used as a disease model for many high consequence pathogens such as bunyaviruses, arenaviruses, henipaviruses, flaviviruses, alphaviruses, filoviruses, and SARS-corona virus (Table 1) (Safronetz et al. 2009, 2012, 2013; Brown et al. 2011; Schountz et al. 2015; DeBuysscher et al. 2013; Gowen and Holbrook 2008; Steele and Twenhafel 2010; Ebihara et al. 2012; Gowen et al. 2010; Roberts et al. 2010). Additionally, hamsters can be used for the evaluation of vaccines and therapeutic treatments against these viruses. In some cases, as with Andes virus, hamsters are the only lethal model of the disease (Safronetz et al. 2012). The value of hamsters as an animal model is research is only recently being realized. The popularity of hamsters used for infectious disease research has increased significantly the last several years (Fig. 1). This growing use of hamsters highlights the need for the development of hamster-specific reagents for a wide range of applications including immunological assays.
Table 1.
Infectious disease models utilizing Hamsters
| Infectious disease models | References |
|---|---|
| Hantavirus pulmonary syndrome | Safronetz et al. (2012) |
| Eastern equine encephalitis | Steele and Twenhafel (2010) |
| Leishmaniasis | da Silva-Couto et al. (2015) |
| Leptospirosis | Silva et al. (2007) |
| Nipah virus encephalitis | DeBuysscher et al. (2013) |
| Scrapie | Sokolowski et al. (2003) |
| Ebola hemorrhagic fever | Ebihara et al. (2012) |
| Rift Valley virus | Scharton et al. (2014) |
| SARS-corona virus | Roberts et al. (2010) |
| Yellow fever virus | Gowen and Holbrook (2008) |
| Clostridium difficile | Kuehne et al. (2014) |
| Helicobacter spp. | Woods et al. (2015) |
Fig. 1.
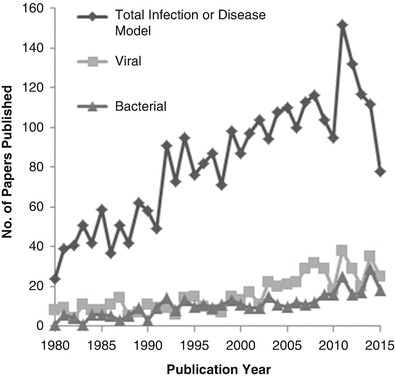
Number of publications using Hamsters as a disease model. The publications using hamsters as an animal model from 1980 through 2015 are shown. For each criterion, the number of publications was determined via a search using the Scopus abstraction and citation database. Searches were performed with the keywords “Syrian-golden-hamster”, “Mesocricetus”, or “Syrian-hamster” and the keyword “model”, as well as either (a) “viral” or “virus”, (b) “bacteria”, (c) “infection” or “disease”
In spite of the growing use of hamsters as disease models, there remains a lack of available immunological reagents developed for assessing immune responses in these animals. Often, researchers are left with examining clinical signs of disease progression and pathology (Zivcec et al. 2011). For the study of disease progression and pathophysiology, this is of little concern, but for the development of vaccines, therapeutic drugs, and determining correlates of immune protection for infectious diseases, evaluating the immune response is critical. Fundamental tools for the study of both innate and adaptive host immune responses commonly used in other models such as mouse and non-human primates have not yet been developed. The result is that researchers who are using or plan to use a hamster model of disease need to use alternative methods for evaluating the immune response.
The widespread use of laboratory mice over the course of the last century has led to great advances in many fields. The combination of whole genome sequencing for Mus musculus being complete and the nearly universal use of mice for many decades have led to the development of countless mouse-specific reagents used in many disciplines (Chinwalla et al. 2002). The complete sequencing of the hamster genome has been performed at the BROAD Institute (NCBI BioProject 77669) and assembly of the hamster genome only recently completed (http://www.genome.gov/27557963). This recent completion will hopefully lead to a surge in the development of hamster specific research tools. Currently, there are 874 cDNA sequences or expressed sequence tags (ESTs) available from the hamster genome in the NCBI-dbEST database (Boguski et al. 1993). The lack of available sequence data and resulting insufficient tools for molecular biology in hamsters has hamstrung scientists who are looking to use them as a disease model. In addition to the insufficient sequence data for hamsters until very recently, the characterization of many immune-specific markers in the hamster remains to be done. The number of these markers that have been described in the mouse, including cell surface markers, transcription factors, signaling proteins, cytokines, chemokines, and even secreted effectors molecules dwarfs the work that has been done in almost every other species, including hamsters. Additionally, monoclonal antibodies against nearly all of the described immunological markers in mice can be readily found commercially, whereas monoclonal antibodies against hamster specific immune markers are almost completely non-existent. The considerable lag in the time taken to sequence the hamster genome coupled with an almost non-existent commercial collection of monoclonal antibodies developed against hamster specific proteins and immunological markers has limited the advancement of hamster models from an immunological perspective.
Despite the scarcity of available reagents for immunological assays in hamster models, there have been significant advances in the methods used to characterize the immune response in hamsters and there are still many reasons why hamsters are a good choice as an animal model of disease. The development of assays to evaluate immune responses in hamsters has been critical in their use as a model for infectious disease and vaccine development in addition to the aforementioned ability of hamster models to closely mimic the human condition of many diseases and satisfy requirements of a suitable animal model. In this review, we focus on how the progress of immunological assay development in hamsters, from determination of cross-reacting antibodies against hamster markers, hamster specific ELISAs and qRT-PCR, to transcriptome analysis and microarrays. We discuss how the assays that have been developed to this point are being utilized in current hamster models to assess immune responses as well as advantages and disadvantages of these currently available assays in the context of particular models. Finally, we address how recent advances in developing immunological tools for use in hamsters can potentially influence future progress along with what remains to be resolved in this area, providing ideas of what we think would be valuable additions to a growing resource for researchers who plan to use hamsters as an animal model.
Immunological Methods Currently Used in Hamster Models
qRT-PCR
While quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) has been a technique of choice in diagnostics, detection of pathogens, and determination of viral loads, the potential for utilizing qRT-PCR for determination of immune responses in tissues has been realized in many models. For example, in human models qRT-PCR has been employed for the detection of innate cytokines and transcription factors involved in immune responses in cells infected with SARS-coronavirus, the detection of upregulated genes implicated in immune escape in circulating tumor cells, and evaluating immune responses generated in patients given the live attenuated yellow fever vaccine (Zielecki et al. 2013; Steinert et al. 2014; Gaucher et al. 2008). Similarly, in mice, qRT-PCR has been used to describe the mechanisms of immune activation in certain vaccine models, to determine the transcription factors involved in dendritic cell mediated presentation of antigen and the subsequent activation of T cells, to investigate the mechanisms behind macrophage polarization, and to evaluate the role of certain subsets of T cells in infectious disease models (Pollard et al. 2013; Seillet et al. 2013; Davis et al. 2013; Stross et al. 2012). Despite the fact that in many models, qRT-PCR is used to determine the relative gene expression of both innate and adaptive cytokines and chemokines (Zivcec et al. 2011; Safronetz et al. 2011a; Prescott et al. 2013; Overbergh et al. 2003), the examples above illustrate that the ability to detect mRNA of other non-cytokine and non-chemokine genes such as transcription factors and cell surface markers can play a valuable role in evaluating immunity generated in certain instances (Gaucher et al. 2008; Seillet et al. 2013). The fact that qRT-PCR is used in models such as human and mouse models, whereby immunological reagents are readily available and many aspects specific immunity can be analyzed with relatively ease using other methods shows the value and relevance of qRT-PCR among today’s available assays.
In hamsters, qRT-PCR is currently the method of choice for many in evaluating immune responses in infectious disease models. Recently, a panel of TaqMan® prime/probe assays for 51 specifically targeted genes in the hamster that were chosen out of a set of more than 800 reference mRNA sequences in GenBank was described (Zivcec et al. 2011). Each are involved in either pro-inflammatory, anti-inflammatory, innate immune responses, T cell responses, as well as non-immune pathways such as apoptosis, cell junction, or coagulation responses in the hamster (Zivcec et al. 2011). Additionally, the validation of an appropriate housekeeping gene for use in qRT-PCR assays in the hamster was simultaneously performed, with ribosomal protein L18 (RPL18) identified as the most stable of the housekeeping genes tested among β-actin, β-2-microglobulin, and Hypoxanthine phosphoribosyltransferase (Zivcec et al. 2011). Consequently, the use of qRT-PCR in hamster models as an immunological tool has increased greatly. qRT-PCR has been utilized in hamster models studying disease caused by Andes virus (Safronetz et al. 2011a, b; Prescott et al. 2013), ebola virus hemorrhagic fever (Ebihara et al. 2012), Nipah virus (DeBuysscher et al. 2013), and Leishmania spp. (da Silva-Couto et al. 2015) among others.
The development of specific primer/probe sets for use in qRT-PCR in hamsters has improved upon the limited options that are available for researchers who are looking to use hamsters as a small animal model. This advancement allows for detection of a broad range of immune and cellular factors and has been crucial to expanding the use of hamsters as an animal model. That said, while qRT-PCR is in all likelihood the best immunological tool available in hamsters currently, it is still an assay that has its considerable disadvantages. First, the number of immune-related genes that have been sequenced and their mRNA sequences entered into GenBank is still relatively low. The panel of primers that was validated contained only 51 genes that played a role in host immune responses (Table 2) (Zivcec et al. 2011). While many of the genes reported by play an important role in host immunity, the number of immune factors that can be assays pales in comparison to what is available in mouse and human assays. Second, while the study of the immune-related genes involved in disease systems can provide valuable insight into the class of immunity generated or what type of immunity is needed for protection against certain pathogens, the relative amount of mRNA present does not always correlate directly with the amount of expressed protein (Overbergh et al. 2003). In many instances, the presence of expressed protein will be very small, whereby qRT-PCR and relative gene expression must be used instead. In these instances, and particularly in using qRT-PCR for analyzing the expression of immune-related genes in hamsters, a discrepancy between mRNA and protein levels should be considered (Overbergh et al. 2003). In spite of this, there is evidence that there is a good correlation between mRNA and protein levels in some instances (Hein et al. 2001; Blaschke et al. 2000). Finally, the methods used for collection of tissues in hamsters for analysis by qRT-PCR in most models do not allow for the evaluation of specific gene expression in individual cell types. Perhaps the most important flaw to consider when using qRT-PCR for analyzing the expression of immune-related genes is that hamster tissues are often harvested and total RNA is extracted from the tissues for analysis by qRT-PCR (Safronetz et al. 2011a; Prescott et al. 2013; Chattopadhyay et al. 2014). A lack of hamster specific antibodies does not allow for the isolation of individual cell types for analysis of gene expression. Therefore, the detection of antigen-specific immune responses at the individual cell level is nearly impossible, leaving only systemic responses and total cytokine, chemokine, and other markers to be detected. This can cause issues when attempting to detect primary and secondary immune responses, what types of innate cells play a role in protection against pathogens, and determining what the correlates of protection are against certain diseases.
Table 2.
Genes validated for qRT-PCR in Hamsters
| Genes validated in Hamsters for qRT-PCR | |
|---|---|
| IL-1β | Complement component 5 |
| IL-2 | Complement component C1qBP |
| IL-2Rα | Chemokine ligand 17 |
| IL-6 | Chemokine ligand 22 |
| IL-6 transducer | Muc1 |
| IL-12p35 | IL-4 |
| IL-12p40 | IL-10 |
| IL-21 | FoxP3 |
| CXCL10 | IRF2 |
| ICAM-1 | TGF-β |
| STAT1 | TGF-β2 |
| STAT1β | TGF-β3 |
| STAT2 | TGFβ type I receptor |
| IFNγ | MHC II α chain |
| Interferon regulatory factor (IRF) 1 | PECAM |
| TNFα | Bcl-2 |
| P75 TNF membrane receptor | Bcl-2 associated protein |
| Myxovirus resistance protein 2 | Ecadherin |
| Protein kinase R | Tight junction protein |
| IFNα inducible protein p27 | Junction adhesion molecule |
| CD83 | Claudulin-1 |
| CCL20/MIP3-α | Occludin |
| NOS2 | Matrix metallinoproteinase-2 |
| Inducible NOS | Tissue inhibitor of matrix metalloproteinase-2 |
| Complement C3d region | Fibrinogen A α chain |
| Vascular endothelial growth factor | |
Reference: Zivcec et al. (2011)
Until more hamster specific monoclonal antibodies become commercially available, qRT-PCR for determining the relative expression levels of immune response genes is one of the best tools at the researcher’s disposal today. Despite certain flaws such as the number of existing analytes to test, and inherent issues with the assay like discrepancies between mRNA and protein levels, and an inability to distinguish between certain cell types, qRT-PCR has become a standard immunological method for use in hamster models. It allows for the simultaneous detection of a large number of immune markers (Table 2), and is a reliable and sensitive assay, with a typical limit of detection between 10−6 and 10−8 ng of gene-specific RNA, corresponding to approximately 9–900 RNA copies (Chattopadhyay et al. 2014). qRT-PCR is one of the most valuable methods we have for immunological analysis in hamster models.
ELISA
Enzyme-linked immunosorbent assays (ELISA) have become standard practice for the detection of cytokines, antibodies, and other proteins since being described in 1971 by Engvall and Perlmann (Engvall and Perlmann 1971). Although originally described as a method for the detection of IgG antibodies, ELISAs have subsequently been developed and optimized for detection of a wide variety of proteins, the most common being cytokines that can be detected in biological samples (Hornbeck 1991). For species like mice, non-human primates, and humans a large number of commercially available kits can be obtained for a reasonable price for the detection of nearly every important cytokine or chemokine. Alternatively, antibodies against cytokines from these species are readily available as well, including antibodies with enzymatic conjugates, allowing for the optimization of individual protocols to each researcher’s liking. In hamsters however, these antibodies for the detection of cytokines are not available. A study examining cross-reactivity of hamster proteins reported that out of 64 antibody-based assays including luminex and ELISA for the detection of cytokines and chemokines in various species, 14 showed significant cross-reactivity with hamster proteins (Zivcec et al. 2011). Out of eight ELISA kits that were tested, only three showed an acceptable level of cross-reactivity with hamster proteins (Zivcec et al. 2011). Due to the high level of sequence homology of the genes for these proteins in the species tested compared with hamsters, this low percentage of cross-reactivity is surprising. The authors concluded from these experiments that ELISA kits and kits for the detection of cytokines and chemokines from other species such as mice and rats were of little value for use in hamsters (Zivcec et al. 2011).
Because of the lack of antibodies for the detection of hamster cytokines, it follows that another valuable and related immune assay, the enzyme-linked immunospot (ELISPOT) assay, is also of little value when using a hamster model. This assay detects individual cytokine producing cells in culture rather than the presence of cytokines in a culture supernatant or serum, but similarly utilizes anti-cytokine antibodies to detect cytokine secreting cells. The lack of antibodies that can detect hamster specific cytokines limits the use of both ELISA and ELISPOT for cytokine detection in hamster models. Development of monoclonal antibodies against various cytokines from hamsters would greatly improve the number of assays that could be utilized for examining immune responses in hamsters.
Despite the inability to use ELISAs for detection of cytokines in hamsters, ELISAs can still be a valuable tool for the detection of antibodies in hamster models. True to its original use, ELISAs can be used to quantify the presence of hamster IgM and IgG. Out of the limited number of commercially available anti-hamster antibodies, anti-hamster IgM and IgG are available and have been used to show the presence of antigen specific antibodies in various hamster models (Safronetz et al. 2009; Prescott et al. 2015; de Wit et al. 2013). In general, the use of a direct ELISA assay is ideal for detection of antibodies in the serum of hamsters. Therefore a recombinant or purified antigen is necessary to coat ELISA plates before the detection of specific antibodies in serum can be achieved. This has been performed in models for hantavirus cardiopulmonary syndrome caused by Andes virus (Safronetz et al. 2009), Ebola hemorrhagic fever (Prescott et al. 2015), and MERS-CoV (de Wit et al. 2013) among others. This has provided a valuable tool for the evaluation of humoral immunity in hamsters in response to infectious agents.
Despite the value of being able to detect IgM and IgG in hamsters, these are the only two isotypes for which there are currently available antibodies against. The detection of different isotypes such as IgG1 and IgG2 would be invaluable to researchers in evaluating the immune response given the other limited options available in hamsters. Additionally, being able to detect IgE, IgA, and IgD, also important players in many immune models, would increase the value of using hamsters as an animal model. Once again, the lack of hamster specific reagents limits the ability to examine the full scope of the immune response. Currently, we are left determining titers of hamster specific IgM and total IgG until further detection antibodies are developed.
Flowcytometry
Using flowcytometry for the detection of specific cell types has become standard immunological practice. Flowcytometry can identify the presence of specific markers on cells, the activation status of lymphocytes, whether specific subsets of cells are present in tissues, and what cell subsets are producing certain cytokines. In addition, fluorescence activated cell sorting (FACS) can sort cell types upon recognition of fluorescent markers bound to cells via specific antibodies. The specificity and sensitivity of flowcytometry, along with the advent of large panels capable of recognizing over a dozen fluorescent markers on cells make it one of the most valuable tools for immunological analyses. For hamsters, there are currently no monoclonal antibodies against cell surface markers specific to hamster cells for use in flowcytometry. However, many studies have used cross-reactive antibodies against cell surface markers of mice and rats for use in flowcytometry (Prescott et al. 2013; Hammerbeck and Hooper 2012). Hammerbeck and Hooper reported that out of a panel consisting of 52 commercially available antibodies for use in flowcytometry, four were able to identify hamster cells (Hammerbeck and Hooper 2012). The four cross-reactive antibodies included anti-mouse/rat MHC II (I-Ek), anti-mouse CD4, anti-rat CD8β, and anti-mouse Thy1.2 (Hammerbeck and Hooper 2012), confirming some which had been reported to cross-react with hamster cells previously (Dondji et al. 2008; Liu et al. 1990). Antibodies against hamster IgM and IgG can also be used to detect hamster B cells in flowcytometry as well (Hammerbeck and Hooper 2012).
The limitations regarding flowcytometry use with hamster cells is that there are only these very few antibodies available, with many antibodies developed for use with mouse and rat cells non cross-reactive. Another issue is that each specific clone should be tested to confirm cross-reactivity with hamster cells, as the clones generated by different companies may not react with the same specificity with hamster cells in every case. Additionally, the few antibodies that are available that cross-react are limited to cell surface proteins on T and B cells. This results in an inability to determine activation status, cell subset, or identify other non-T and B cells like macrophages, dendritic cells, neutrophils, and NK cells. As with ELISA and ELISPOT assays in hamsters, the lack of antibodies against hamster-specific cytokines prevents using flowcytometry to detect cytokine producing cells by intracellular staining for phenotyping of the immune response. This is a major limitation on the use of flowcytometry in hamster models. Finally, the use of anti-mouse CD4 and anti-rat CD8β antibodies for use in hamsters must be done in conjunction with anti-mouse/rat MHC II to allow for the exclusion of myeloid lineage cells expressing either CD4 or CD8β, which has shown to be the case in mice (Hammerbeck and Hooper 2012). Fortunately the combination of these antibodies has been shown to be effective in identifying CD4 and CD8 T cells in hamsters due to the lack of binding of cells by both anti-mouse CD4 or anti-rat CD8β and anti-mouse/rat MHC II (Hammerbeck and Hooper 2012). Overall, the use of flowcytometry in hamsters in severely limited due to the lack hamster-specific antibodies. The identification and possible sorting of B cells and CD4 and CD8 T cells is possible due to the cross-reactivity of anti-mouse and anti-rat antibodies, but should be done with caution and optimization of protocols by individual researchers to determine cross-reactivity levels of specific clones.
Related to the use of monoclonal antibodies against mouse and rat cell surface markers that are able to cross-react with hamster proteins for use in flowcytometry, these antibodies have been shown to be effective at depletion of CD4 and CD8 T cells in vivo in hamsters (Prescott et al. 2013, 2015; Hammerbeck and Hooper 2012). The ability of these antibodies to recognize hamster T cells and mediate depletion in vivo allows for depletion studies and for determining the importance of CD4 or CD8 T cells in infectious disease models. The ability of cross-reactive antibodies to deplete T cell in vivo is a considerable advantage in examining host immune responses in disease models. With the limited ability to assess certain immune parameters ex vivo in hamsters, this provides a critical tool for immunological studies in hamsters. The same caveats exist for the use of these antibodies for depletion of cells in vivo as for their use in flowcytometry, but this provides an interesting avenue for researchers to pursue when evaluating the immune response against certain pathogens or testing drug and vaccine efficacy.
Immunohistochemistry
Immunohistochemistry has been a powerful method for the detection of specific antigens within formalin-fixed tissues for decades (Schacht and Kern 2015). Since the advent of hybridomas for the production of monoclonal antibodies in 1975, immunohistochemistry has adopted the use of monoclonal antibodies for the detection of specific antigens with great specificity (Schacht and Kern 2015; Köhler and Milstein 1975). The use of specific antibodies allows for the detection within fixed tissues of bacteria, viruses, certain cell types such as lymphocytes, and cellular markers of disease pathology. While immunohistochemistry has important applications in diagnostics, in animal models of disease, it is particularly useful for assessing pathology in affected tissues, and can detect the present of tissue infiltrating immune cells that may be causing immunopathogenesis during the course of disease.
Immunohistochemistry in hamsters for assessing disease pathology has been a popular technique due to the lack of reagents available for classical immunological methods. It has been used to examine the course of disease in many infectious disease models including Andes virus, Sin Nombre virus, Chikungunya virus, Nipah virus, and Ebola virus (Safronetz et al. 2013; DeBuysscher et al. 2013; Ebihara et al. 2012; Safronetz et al. 2011b; Bosco-Lauth et al. 2015). It is also useful in non-infectious disease models in hamsters such as cancer and encephalopathic diseases (Woods et al. 2015; Clouse et al. 2015; Elder et al. 2013). While the use of this technique in hamsters is typically limited to examining preserved tissues that display disease pathology, the advantages are that cross sections of entire tissues can be visualized to give a more representative image of disease tropism and the presence of immune cells or of particular pathogens can easily be detected (Schacht and Kern 2015). As with other immunological methods in hamsters, the detection of most immune cells is limited to few cross-reactive antibodies available, and antibodies against common cell surface markers found in many tissues are not available or their cross-reactivity with antibodies against these markers in other species has not been assessed. Other inherent limitations of immunohistochemistry include the alteration of antigens during the fixation process, the relative insensitivity of the assay as compared to other techniques like ELISA and PCR, and the technical demands involved in the procedure (Schacht and Kern 2015). Particularly in the case of pathogens that require high containment facilities, the procedure for fixation of tissues can be up to several weeks long, increasing the possibility of altered tissues when best results are obtained as soon after euthanization as possible. Despite these issues, the value of using immunohistochemistry in hamster models is the ability to examine multiple tissues that impacted during the course of disease, determining specific tissue tropism of pathogens by visualizing pathogens in infected tissues, and studying possible immune cells infiltration into tissues that could be contributing to immunopathogenesis during certain diseases.
Transcriptome/Microarray Analysis
In recent years, transcriptome sequencing and analysis has been utilized to provide sets of mRNA that are expressed in a given species, help provide insights into the expression profiles of these species, and for the development of microarrays for the detection of gene expression (Tchitchek et al. 2014; Ying et al. 2015). The description of the transcriptome of mice, rats, and humans has been critical for the increasing use of microarray for examining expression profiles in tissues in many models (Yu et al. 2009; Okazaki et al. 2002; Yang et al. 2010; Maywood et al. 2009). Recently, hamster transcriptome sequencing and analysis has been performed in several models (Ying et al. 2015; Yu et al. 2009; Yang et al. 2010; Maywood et al. 2009; Hohlweg et al. 2003; Schmucki et al. 2013). In recent years, the use of microarray analysis in hamsters has been limited to cross-reactive hybridization of hamster RNA to cDNA from other species such as rats, mice, and humans (Yu et al. 2009; Wahl-Jensen et al. 2012). cDNAs comprising the hamster transcriptome have been sequence aligned to the transcriptome of species that have been described previously (Tchitchek et al. 2014). This transcriptome sequencing has allowed for the identification of a large set of genes that play a role in a number of biological processes. Subsequently, Ying et al were able to sequence and annotate over 34,000 sequences comprising the hamster transcriptome for the development of a custom hamster microarray (Ying et al. 2015). The microarray that was developed from the hamster transcriptome was validated by comparing gene expression profiles in mice infected with Adenovirus using the custom microarray and qRT-PCR (Ying et al. 2015). This was one of the first descriptions of a hamster specific microarray capable of detecting changes in hamster gene expression. This newly developed microarray following the sequencing and annotation of the hamster transcriptome will hopefully lead to the production of more hamster specific microarrays. While RNA-seq has been recently used to examine the regulation of genes in hamster in a model for Arenavirus infection (Schountz et al. 2015), the use of RNA-seq has not yet become common in hamster models of infectious disease. It is likely that the use of RNA-seq will become more popular in coming years as the genome and transcriptome of hamsters becomes fully characterized and publicly available, the description of the hamster transcriptome and its value in the development of hamster specific microarrays should not be overlooked. This recent work on the hamster transcriptome will hopefully lead to beneficial tools like microarrays for researchers looking to use hamsters as an animal model of disease.
Kinome Analysis
The ability to evaluate host responses to pathogens has historically relied upon examining gene expression or protein synthesis in the form of antibodies or cytokines. Since many of the intracellular pathways involved in immune cell signalling are well known, and many proteins have been characterized, the cell signalling proteins within host cells have become recently become a target for therapeutics. The ability of kinases to phosphorylate proteins is critical in cellular signalling, and allows for rapid responses to environmental stimuli such as stress or infection (Arsenault et al. 2011; Falcinelli et al. 2015). Only recently has the potential of examining the presence of kinases involved in cell signalling, collectively called the kinome, been realized. The study of the kinases involved in immune signalling can give important indicators of the outcome of disease in certain models (Falcinelli et al. 2015). Kinome analysis involves synthesizing peptides representing phosphorylation sites on hundreds of proteins that are immobilized onto an array surface (Arsenault et al. 2011). Samples containing cellular kinases phosphorylate the immobilized peptides, which can then be visualized to determine the level of relative phosphorylation and the activity of the kinases in the sample (Arsenault et al. 2011). The examination of the kinome during the host response to pathogens has been used as a tool to define cellular responses and evaluate host immune responses in different disease models (Kindrachuk et al. 2012, 2014; Arsenault et al. 2013; Kindrachuk and Napper 2013).
In hamsters, a kinome peptide array was recently synthesized by Falcinelli et al. in a model for Arenavirus infection (Falcinelli et al. 2015). This hamster specific kinome was developed with peptides focused primarily on immune pathways, and showed that Arenavirus infection in hamsters is characterized by lung vascular endothelial growth factor and interleukin responses as well as NF-kB and TLR signalling (Falcinelli et al. 2015). This presents a novel assay for assessing hamster-specific immune responses to infection by examining the activity level of host cell kinases.
Genetic Manipulations
The ability to genetically manipulate animals for use in research has been invaluable for decades in countless disease models. Since the first description of genetically manipulated mice in the late 1980s by Martin Evans, Oliver Smithies, and Mario Capecchi, which led to the Nobel Prize in Physiology or Medicine, the use of genetically modified animals has revolutionized biomedical research (Manis 2007; Thomas and Capecchi 1987; Capecchi 2005). For immunologists, the use of knockout and transgenic mice has been essential for determining the roles of cell types, cytokines, and transcription factors as well as providing valuable insights into things like immune memory and regulation (Manis 2007). Genetically modified mice have become so common, that commercially available transgenic and knockout mice for dozens of genes are readily available.
Until recently, gene targeting in hamsters has been limited due to the lack of a completely sequenced genome. However, Fan et al. have reported a CRISPR/Cas9 system for the targeting of hamster specific genes (Fan et al. 2014). They were able to successfully target the STAT2 gene of hamsters with reliable efficiency to produce STAT2 knockout hamsters (Fan et al. 2014). These hamsters have been subsequently used by others in an Adenovirus model to show that type I IFN responses are critical in controlling Adenovirus infection (Toth et al. 2015). These are the first studies to report on and employ genetically modified hamsters. These results show the potential for not only using STAT2 knockout hamsters in studying disease models, but also the potential to knockout other genes that play important roles in immune pathways. These studies will hopefully serve as the first step in developing many more knockout and possible transgenic hamsters for use as models for infectious disease.
Future Directions
The growing use of hamsters in small animal models of disease has brought to light a glaring need for the development of hamster specific reagents for use in immunological assays. The study of the immune response in nearly any disease model is critical for developing therapeutic options and an understanding of the disease course. The number of available reagents for use in hamster models right now is not where it needs to be for sufficient insight into how these animals are protected from or develop disease (Table 3). As of now, the best methods for immunological assays in hamsters are qRT-PCR for detection of expression of immune-related genes and ELISA for the detection of humoral immune responses. Despite being used as an animal model for decades, immunological tools for hamsters remains years behind other animal models such as mice, rats, and non-human primates. For the full potential of hamsters as an appropriate disease model to be realized, developing of many different hamster-specific tools for assays that are specific to hamsters need to be created.
Table 3.
Immunological methods used in Hamster models
| Immunological methods used in Hamsters | References |
|---|---|
| qRT-PCR | Zivcec et al. (2011) and Safronetz et al. (2011b) |
| ELISA | Safronetz et al. (2009) and Brown et al. (2011) |
| Flowcytometry | Prescott et al. (2013) and Hammerbeck and Hooper (2012) |
| Immunohistochemistry | DeBuysscher et al. (2013) and Ebihara et al. (2012) |
| Transcriptome analysis | Tchitchek et al. (2014) and Ying et al. (2015) |
| Microarray | Ying et al. (2015) |
| Kinome analysis | Falcinelli et al. (2015) |
The use of cross-reactive monoclonal antibodies against immune markers and cytokines from other species has given researchers using hamsters a viable option for certain assays, but the development of monoclonal antibodies that are hamster-specific for a variety of immune cell surface markers and cytokines would greatly improve upon the current state of immunological assays done in hamster models. The production of hamster-specific monoclonal antibodies against common immune markers and cytokines should be number one on the wish list of anyone doing immunological assays with hamsters. The ability to perform a plethora of techniques from ELISA, ELISPOT, flowcytometry, and microscopy would increase greatly with the production of only a few dozen hamster-specific monoclonal antibodies.
In the coming years, with the ability to produce monoclonal antibodies with increased efficiency and the increase in the use of hamsters as an animal model of diseases where understanding the immune response is critical, antibodies for use in hamsters will hopefully see an increase in demand and production. With the sequencing of the hamster genome and transcriptome, we have seen the development of novel assays for use in hamster models like microarrays and kinome analysis via peptide arrays. The first use of gene knockout hamsters has been reported. These advances will hopefully open the flood gates in terms of what becomes available for researchers in the near future. The recent sequencing projects that have gone on have given us the ability to uncover the gene sequences of many immune-related genes and the proteins that are encoded by them. The demand for the reagents available for use in other species like mice and rats should increase by a large amount as hamsters become more and more popular as animal models of disease. We now have the ability to develop the repertoire of reagents that is available in commonly used species. The recent advances in hamster-specific immunological tools gives us reason to hope that soon researchers will have a number of assays at their disposal when conducting experiments in hamsters.
Concluding Remarks
The use of hamsters as an animal model has increased greatly in recent years due to their ability to recapitulate human disease in models for diseases like Hantavirus, Ebola virus, Nipah virus, C difficile, Leishmania spp., as well as cancers, and atherosclerosis (Dillard et al. 2010; Jové et al. 2013; Woods et al. 2015; da Silva-Couto et al. 2015; Kuehne et al. 2014; Safronetz et al. 2009, 2012; DeBuysscher et al. 2013; Ebihara et al. 2012). The realization of hamsters as valuable animal models has led to their use in studying disease course for many pathogens. However, the lack of immunological reagents available for use in hamsters limits their value when it comes to developing therapeutic options, vaccines, and determining correlates of protection or immunopathogenesis of disease. The methods available currently pale in comparison to those available for use in other species like mice, rats, and non-human primates and the ones that are available have many inherent disadvantages. For hamsters to continue to grow into a common animal model of disease, and one that can hopefully lead to important therapeutic and vaccine developments in the coming years as the number of available immunological reagents in this species needs to improve greatly.
Contributor Information
Giovanni Rezza, Email: giovanni.rezza@iss.it.
Giuseppe Ippolito, Email: giuseppe.ippolito@inmi.it.
David Safronetz, Email: david.safronetz@phac-aspc.gc.ca.
Gary P. Kobinger, Email: Gary.Kobinger@crchudequebec.ulaval.ca
References
- Arsenault R, Griebel P, Napper S. Peptide arrays for kinome analysis: new opportunities and remaining challenges. Proteomics. 2011;11(24):4595–4609. doi: 10.1002/pmic.201100296. [DOI] [PubMed] [Google Scholar]
- Arsenault RJ, Li Y, Maattanen P, et al. Altered Toll-like receptor 9 signaling in Mycobacterium avium subsp. paratuberculosis-infected bovine monocytes reveals potential therapeutic targets. Infect Immun. 2013;81(1):226–237. doi: 10.1128/IAI.00785-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhathena J, Kulamarva A, Martoni C, et al. Diet-induced metabolic hamster model of nonalcoholic fatty liver disease. Diabetes Metab Syndr Obes. 2011;4:195–203. doi: 10.2147/DMSO.S18435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke V, Reich K, Blaschke S, et al. Rapid quantitation of proinflammatory and chemoattractant cytokine expression in small tissue samples and monocyte-derived dendritic cells: validation of a new real-time RT-PCR technology. J Immunol Methods. 2000;246(1):79–90. doi: 10.1016/S0022-1759(00)00304-5. [DOI] [PubMed] [Google Scholar]
- Boguski MS, Lowe TM, Tolstoshev CM. dbEST—database for “expressed sequence tags”. Nat Genet. 1993;4(4):332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- Bosco-Lauth AM, Han S, Hartwig A, et al. Development of a hamster model for Chikungunya virus infection and pathogenesis. PLoS One. 2015;10(6):e0130150. doi: 10.1371/journal.pone.0130150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KS, Safronetz D, Marzi A, et al. Vesicular stomatitis virus-based vaccine protects hamsters against lethal challenge with Andes virus. J Virol. 2011;85(23):12781–12791. doi: 10.1128/JVI.00794-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canadian Council on Animal Care (1997) CCAC guidelines on: animal use protocol review
- Capecchi MR. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet. 2005;6(6):507–512. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay PK, Gierahn TM, Roederer M, et al. Single-cell technologies for monitoring immune systems. Nat Immunol. 2014;15(2):128–135. doi: 10.1038/ni.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinwalla AT, Cook LL, Delehaunty KD, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420(6915):520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Clouse MD, Shikiya RA, Bartz JC, et al. Nasal associated lymphoid tissue of the Syrian golden hamster expresses high levels of PrP C. PLoS One. 2015;10(2):e0117935. doi: 10.1371/journal.pone.0117935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva-Couto L, Ribeiro-Romão RP, Saavedra AF, et al. Intranasal vaccination with Leishmanial antigens protects golden hamsters (Mesocricetus auratus) against Leishmania (Viannia) braziliensis infection. PLoS Negl Trop Dis. 2015;9(1):e3439. doi: 10.1371/journal.pntd.0003439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MJ, Tsang TM, Qiu Y, et al. Macrophage M1/M2 polarization dynamically adapts to changes in cytokine microenvironments in Cryptococcus neoformans infection. MBio. 2013;4(3):e00264–13. doi: 10.1128/mBio.00264-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Prescott J, Baseler L, et al. The Middle East Respiratory Syndrome Coronavirus (MERS-CoV) does not replicate in Syrian hamsters. PLoS One. 2013;8(7):e69127. doi: 10.1371/journal.pone.0069127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBuysscher BL, de Wit E, Munster VJ, et al. Comparison of the pathogenicity of Nipah virus isolates from Bangladesh and Malaysia in the Syrian hamster. PLoS Negl Trop Dis. 2013;7(1):e2024. doi: 10.1371/journal.pntd.0002024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillard A, Matthan NR, Lichtenstein AH. Use of hamster as a model to study diet-induced atherosclerosis. Nutr Metab. 2010;7(1):1. doi: 10.1186/1743-7075-7-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondji B, Bungiro RD, Harrison LM, et al. Role for nitric oxide in hookworm-associated immune suppression. Infect Immun. 2008;76(6):2560–2567. doi: 10.1128/IAI.00094-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebihara H, Zivcec M, Gardner D et al (2012) A Syrian golden hamster model recapitulating ebola hemorrhagic fever. J Infect Dis: jis626 [DOI] [PMC free article] [PubMed]
- Elder AM, Henderson DM, Nalls AV, et al. In vitro detection of prionemia in TSE-infected cervids and hamsters. PLoS One. 2013;8(11):e80203. doi: 10.1371/journal.pone.0080203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engvall E, Perlmann P. Enzyme-linked immunosorbent assay (ELISA) quantitative assay of immunoglobulin G. Immunochemistry. 1971;8(9):871–874. doi: 10.1016/0019-2791(71)90454-X. [DOI] [PubMed] [Google Scholar]
- Falcinelli S, Gowen BB, Trost B. Characterization of the host response to Pichinde virus infection in the Syrian golden hamster by species-specific kinome analysis. Mol Cell Proteomics. 2015;14(3):646–657. doi: 10.1074/mcp.M114.045443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z, Li W, Lee SR, et al. Efficient gene targeting in golden Syrian hamsters by the CRISPR/Cas9 system. PLoS One. 2014;9(10):e109755. doi: 10.1371/journal.pone.0109755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA (2015) Product development under the animal rule guidelines for industry
- Gaucher D, Therrien R, Kettaf N, et al. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med. 2008;205(13):3119–3131. doi: 10.1084/jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowen BB, Holbrook MR. Animal models of highly pathogenic RNA viral infections: hemorrhagic fever viruses. Antivir Res. 2008;78(1):79–90. doi: 10.1016/j.antiviral.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Gowen BB, Julander JG, London NR, et al. Assessing changes in vascular permeability in a hamster model of viral hemorrhagic fever. Virol J. 2010;7(1):1. doi: 10.1186/1743-422X-7-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerbeck CD, Hooper JW. T cells are not required for pathogenesis in the Syrian hamster model of hantavirus pulmonary syndrome. J Virol. 2012;86(7):4043. doi: 10.1128/JVI.00203-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein J, Schellenberg U, Bein G, et al. Quantification of Murine IFN‐γ mRNA and protein expression: impact of real‐time kinetic RT–PCR using SYBR green I Dye. Scand J Immunol. 2001;54(3):285–291. doi: 10.1046/j.1365-3083.2001.00928.x. [DOI] [PubMed] [Google Scholar]
- Hohlweg U, Hösel M, Dorn A, et al. Intraperitoneal dissemination of Ad12-induced undifferentiated neuroectodermal hamster tumors: de novo methylation and transcription patterns of integrated viral and of cellular genes. Virus Res. 2003;98(1):45–56. doi: 10.1016/j.virusres.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Hornbeck PV (1991) Enzyme‐linked immunosorbent assays. Curr Protoc Immunol:2–1 [DOI] [PubMed]
- Jové M, Ayala V, Ramírez-Núñez O, et al. Lipidomic and metabolomic analyses reveal potential plasma biomarkers of early atheromatous plaque formation in hamsters. Cardiovasc Res. 2013;97(4):642–652. doi: 10.1093/cvr/cvs368. [DOI] [PubMed] [Google Scholar]
- Kindrachuk J, Napper S (2013) Probing the kinome for biomakers and therapeutic targets: peptide arrays for global phosphorylation-mediated signal transduction. Comprehensive biomarker discover and validation for clinical applications
- Kindrachuk J, Arsenault R, Kusalik A, et al. Systems kinomics demonstrates Congo Basin monkeypox virus infection selectively modulates host cell signaling responses as compared to West African monkeypox virus. Mol Cell Proteomics. 2012;11(6):M111–M015701. doi: 10.1074/mcp.M111.015701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindrachuk J, Wahl-Jensen V, Safronetz D, et al. Ebola virus modulates transforming growth factor β signaling and cellular markers of mesenchyme-like transition in hepatocytes. J Virol. 2014;88(17):9877–9892. doi: 10.1128/JVI.01410-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- Kuehne SA, Collery MM, Kelly ML, et al. Importance of toxin A, toxin B, and CDT in virulence of an epidemic Clostridium difficile strain. J Infect Dis. 2014;209(1):83–86. doi: 10.1093/infdis/jit426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Steiner BM, Alder JD, et al. Immune T cells sorted by flow cytometry confer protection against infection with Treponema pallidum subsp. pertenue in hamsters. Infect Immun. 1990;58(6):1685–1690. doi: 10.1128/iai.58.6.1685-1690.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manis JP. Knock out, knock in, knock down—genetically manipulated mice and the Nobel Prize. N Engl J Med. 2007;357(24):2426–2429. doi: 10.1056/NEJMp0707712. [DOI] [PubMed] [Google Scholar]
- Maywood ES, Chahad-Ehlers S, Garabette ML, et al. Differential testicular gene expression in seasonal fertility. J Biol Rhythm. 2009;24(2):114–125. doi: 10.1177/0748730409332029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y, Furuno M, Kasukawa T, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420(6915):563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- Overbergh L, Giulietti A, Valckx D, et al. The use of real-time reverse transcriptase PCR for the quantification of cytokine gene expression. J Biomol Tech. 2003;14(1):33. [PMC free article] [PubMed] [Google Scholar]
- Pollard C, Rejman J, De Haes W, et al. Type I IFN counteracts the induction of antigen-specific immune responses by lipid-based delivery of mRNA vaccines. Mol Ther. 2013;21(1):251–259. doi: 10.1038/mt.2012.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott J, Safronetz D, Haddock E, et al. The adaptive immune response does not influence hantavirus disease or persistence in the Syrian hamster. Immunology. 2013;140(2):168–178. doi: 10.1111/imm.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott J, Falzarano D, Feldmann H (2015) Natural immunity to ebola virus in the Syrian hamster requires antibody responses. J Infect Dis: jiv203 [DOI] [PMC free article] [PubMed]
- Roberts A, Lamirande EW, Vogel L, et al. Immunogenicity and protective efficacy in mice and hamsters of a β-propiolactone inactivated whole virus SARS-CoV vaccine. Viral Immunol. 2010;23(5):509–519. doi: 10.1089/vim.2010.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safronetz D, Hegde NR, Ebihara H, et al. Adenovirus vectors expressing hantavirus proteins protect hamsters against lethal challenge with Andes virus. J Virol. 2009;83(14):7285–7295. doi: 10.1128/JVI.00373-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safronetz D, Haddock E, Feldmann F, et al. In vitro and in vivo activity of ribavirin against Andes virus infection. PLoS One. 2011;6(8):e23560. doi: 10.1371/journal.pone.0023560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safronetz D, Zivcec M, LaCasse R, et al. Pathogenesis and host response in Syrian hamsters following intranasal infection with Andes virus. PLoS Pathog. 2011;7(12):e1002426. doi: 10.1371/journal.ppat.1002426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safronetz D, Ebihara H, Feldmann H, et al. The Syrian hamster model of hantavirus pulmonary syndrome. Antivir Res. 2012;95(3):282–292. doi: 10.1016/j.antiviral.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safronetz D, Prescott J, Haddock E, et al. Hamster-adapted Sin Nombre virus causes disseminated infection and efficiently replicates in pulmonary endothelial cells without signs of disease. J Virol. 2013;87(8):4778–4782. doi: 10.1128/JVI.03291-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacht V, Kern JS. Basics of immunohistochemistry. J Investig Dermatol. 2015;135(3):1–4. doi: 10.1038/jid.2014.541. [DOI] [PubMed] [Google Scholar]
- Scharton Dionna, Bailey Kevin W., Vest Zachary, Westover Jonna B., Kumaki Yohichi, Van Wettere Arnaud, Furuta Yousuke, Gowen Brian B. Favipiravir (T-705) protects against peracute Rift Valley fever virus infection and reduces delayed-onset neurologic disease observed with ribavirin treatment. Antiviral Research. 2014;104:84–92. doi: 10.1016/j.antiviral.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucki R, Berrera M, Küng E, et al. High throughput transcriptome analysis of lipid metabolism in Syrian hamster liver in absence of an annotated genome. BMC Genomics. 2013;14(1):237. doi: 10.1186/1471-2164-14-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schountz T, Phillips A, Rico A, et al. Interferon response in a hamster model of arenavirus hemorrhagic disease. New Horiz Transl Med. 2015;2(4):135. doi: 10.1016/j.nhtm.2015.07.072. [DOI] [Google Scholar]
- Seillet C, Jackson JT, Markey KA, et al. CD8α + DCs can be induced in the absence of transcription factors Id2, Nfil3, and Batf3. Blood. 2013;121(9):1574–1583. doi: 10.1182/blood-2012-07-445650. [DOI] [PubMed] [Google Scholar]
- Silva Éverton F., Medeiros Marco A., McBride Alan J.A., Matsunaga Jim, Esteves Gabriela S., Ramos João G.R., Santos Cleiton S., Croda Júlio, Homma Akira, Dellagostin Odir A., Haake David A., Reis Mitermayer G., Ko Albert I. The terminal portion of leptospiral immunoglobulin-like protein LigA confers protective immunity against lethal infection in the hamster model of leptospirosis. Vaccine. 2007;25(33):6277–6286. doi: 10.1016/j.vaccine.2007.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolowski Fabian, Modler Andreas Johannes, Masuch Ralf, Zirwer Dietrich, Baier Michael, Lutsch Gudrun, Moss David Alan, Gast Klaus, Naumann Dieter. Formation of Critical Oligomers Is a Key Event during Conformational Transition of Recombinant Syrian Hamster Prion Protein. Journal of Biological Chemistry. 2003;278(42):40481–40492. doi: 10.1074/jbc.M304391200. [DOI] [PubMed] [Google Scholar]
- Staffend NA, Meisel RL. Aggressive experience increases dendritic spine density within the nucleus accumbens core in female syrian hamsters. Neuroscience. 2012;227:163–169. doi: 10.1016/j.neuroscience.2012.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele KE, Twenhafel NA. REVIEW PAPER pathology of animal models of alphavirus encephalitis. Vet Pathol Online. 2010;47(5):790–805. doi: 10.1177/0300985810372508. [DOI] [PubMed] [Google Scholar]
- Steinert G, Schölch S, Niemietz T, et al. Immune escape and survival mechanisms in circulating tumor cells of colorectal cancer. Cancer Res. 2014;74(6):1694–1704. doi: 10.1158/0008-5472.CAN-13-1885. [DOI] [PubMed] [Google Scholar]
- Stross L, Günther J, Gasteiger G, et al. Foxp3+ regulatory T cells protect the liver from immune damage and compromise virus control during acute experimental hepatitis B virus infection in mice. Hepatology. 2012;56(3):873–883. doi: 10.1002/hep.25765. [DOI] [PubMed] [Google Scholar]
- Tchitchek N, Safronetz D, Rasmussen AL, et al. Sequencing, annotation and analysis of the Syrian hamster (Mesocricetus auratus) transcriptome. PLoS One. 2014;9(11):e112617. doi: 10.1371/journal.pone.0112617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51(3):503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- Toth K, Lee SR, Ying B, et al. STAT2 knockout Syrian hamsters support enhanced replication and pathogenicity of human adenovirus, revealing an important role of type I interferon response in viral control. PLoS Pathog. 2015;11(8):e100508. doi: 10.1371/journal.ppat.1005084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayalingam S, Kuppuswamy M, Subramanian T, et al. Evaluation of apoptogenic adenovirus type 5 oncolytic vectors in a Syrian hamster head and neck cancer model. Cancer Gene Ther. 2014;21(6):228–237. doi: 10.1038/cgt.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl-Jensen V, Bollinger L, Safronetz D, et al. Use of the Syrian hamster as a new model of ebola virus disease and other viral hemorrhagic fevers. Viruses. 2012;4(12):3754–3784. doi: 10.3390/v4123754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods SE, Ek C, Shen Z et al (2015) Male Syrian hamsters experimentally infected with helicobacter spp. of the H. bilis cluster develop MALT‐associated gastrointestinal lymphomas. Helicobacter [DOI] [PMC free article] [PubMed]
- Yang K, Zhang G, Mei J, et al. Screening and analysis of pathogenic genes during DMBA-induced buccal mucosa carcinogenesis in golden hamsters. Oncol Rep. 2010;23(6):1619–1624. doi: 10.3892/or_00000803. [DOI] [PubMed] [Google Scholar]
- Ying B, Toth K, Spencer JF, et al. Transcriptome sequencing and development of an expression microarray platform for liver infection in adenovirus type 5-infected Syrian golden hamsters. Virology. 2015;485:305–312. doi: 10.1016/j.virol.2015.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Wang XY, Gong RG, et al. The expression profile of microRNAs in a model of 7, 12-dimethyl-benz [a] anthrance-induced oral carcinogenesis in Syrian hamster. J Exp Clin Cancer Res. 2009;28(1):1. doi: 10.1186/1756-9966-28-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielecki F, Weber M, Eickmann M, et al. Human cell tropism and innate immune system interactions of human respiratory coronavirus EMC compared to those of severe acute respiratory syndrome coronavirus. J Virol. 2013;87(9):5300–5304. doi: 10.1128/JVI.03496-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivcec M, Safronetz D, Haddock E, et al. Validation of assays to monitor immune responses in the Syrian golden hamster (Mesocricetus auratus) J Immunol Methods. 2011;368(1):24–35. doi: 10.1016/j.jim.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]


