Abstract
The emergence of severe acute respiratory syndrome (SARS) and its subsequent worldwide spread challenged the global public health community to confront a novel infectious disease. The infection is caused by a coronavirus of animal origin. In this epidemic, molecular detections of SARS coronavirus RNA were shown to be useful for the early diagnosis of SARS. Although this pathogen was eradicated in humans, SARS or SARS-like viruses might reemerge from animals or from laboratory incidents. In this chapter, we describe several polymerase chain reaction (PCR) protocols for detecting SARS coronaviruses. These assays were routinely used for clinical diagnosis during the SARS outbreak.
Key words: SARS coronavirus, RT-PCR, Molecular detection, Clinical diagnosis
Introduction
Coronavirus is a genus of viruses in the family Coronaviridae under the order Nidovirales (1). The virus has a corona-like morphology and its genome contains a single segment of single-stranded RNA of positive polarity. Viruses under this genus have genome sizes ranging from 28 to 32 kb, which are the biggest among the RNA viruses. The genomic RNA of these viruses contains five major open reading frames (Orfs) that encode the replicase polyproteins (Orf 1a and Orf 1ab), spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins, in that order. Based on antigenic and genetic analysis, coronaviruses are traditionally classified into three groups. Coronaviruses from groups 1 and 2 have been found to infect mammals. By contrast, group 3 viruses are primarily avian viruses. Many coronaviruses are also pathogenic to their hosts (2). Animal coronaviruses can cause severe respiratory, enteric, neurological, or hepatic disease in their hosts. With the exception of severe acute respiratory syndrome (SARS) coronavirus, all human coronaviruses are often only associated with mild respiratory and gastrointestinal diseases. Interestingly, virus surveillance studies over the last few years have identified many novel coronaviruses from different animals. These suggest that there might be a wealth of “unknown” coronaviruses which are yet to be identified.
SARS is the first novel infectious respiratory disease in this century. The disease is caused by a coronavirus originating from animals and the clinical presentations of the disease have been extensively reviewed (3). Further studies also indicated that the virus is a distant relative of bat coronaviruses (4), suggesting bats might be natural carriers of the precursor of SARS coronavirus. However, the nature reservoir of SARS coronavirus is still not confirmed. As the majority of SARS patients seroconverted in the second week of disease onset, serological tests might not be a practical approach for early SARS diagnosis (3). Because of these reasons, the focus of early diagnosis was mainly concentrated on the development of conventional and quantitative reverse transcriptase (RT) polymerase chain reaction (PCR) assays (3). Besides, several molecular tests which employ non-PCR-based methods, such as loop mediated isothermal amplification (5), rolling circle amplification (6) and nucleic acid sequence-based amplification (7), were also developed for the detection of SARS coronavirus RNA.
Here, we share our experiences on the molecular diagnosis of SARS and other coronaviruses. The protocols described in this chapter are directly adopted from our previous publications (8, 9). The first and second assays are manual RT-PCR (Subheadings 3.2, 3.3, and 3.4) and real-time quantitative RT-PCR (Subheading 3.5) assays, respectively, for SARS coronavirus detections (see Notes 1–3). As there is a possibility that other SARS-like coronaviruses found in bats or other mammals might have zoonotic potential, we also present another PCR assay which is able to detect groups 1 and 2 viruses (Subheading 3.6). This assay might be useful to screen the SARS-like patients whom are negative in the first two assays. It should be noted that the primer set used in the third assay can cross react with a wide range of coronaviruses. Therefore, the identities of all the positive PCR products from the third assay should be formally confirmed by DNA sequencing. In our experiences, this assay is able to detect other common human coronaviruses (e.g. HKU1, NL63, OC43, and 229E).
Materials
RNA Extraction
QIAamp virus RNA mini kit (Qiagen).
Ethanol, 96–100%.
Autoclaved RNase-free water or its equivalents.
Clinical samples stored in 1–3 mL of viral transport medium. For 1 L of viral transport medium, dissolve 2 g of sodium bicarbonate (Merck), 5 g of bovine serum albumin (Sigma-Aldrich), 200 μg of vancomycin (Sigma-Aldrich), 18 μg of amikacin (Sigma-Aldrich), and 160 U of nystatin (Sigma-Aldrich) in 1 L of Earle’s balanced salt solution (Sigma-Aldrich) and filter the solution using a 0.22 μm pore size filter (see Note 4).
Reverse Transcription
SuperScript II reverse transcriptase, 200 U/μL (Invitrogen).
5× First strand buffer [250 mM Tris–HCl (pH 8.3), 375 mM KCl, 15 mM MgCl2] (Invitrogen).
0.1 mM dithiothreitol (Invitrogen).
Random hexamers, 150 ng/μL (Invitrogen).
RNaseOUT recombinant ribonuclease inhibitor, 40 U/μL (Invitrogen).
Deoxynucleotide triphosphates (dNTP) mix, 10 mM each.
Autoclaved RNase-free water or equivalents.
Heating block or equivalents.
PCR for SARS Coronavirus
AmpliTaq Gold DNA polymerase, 5 U/μL (Applied Biosystems).
10× Gold PCR buffer (Applied Biosystems).
dNTP mix, 10 mM each.
25 mM MgCl2 solution (Applied Biosystems).
10 μM PCR forward primer, 5′-TACACACCT CAGCGTTG-3′.
10 μM PCR reverse primer, 5′-CACGAACGTGACGAAT-3′.
Themocycler (GeneAmp 9700, Applied Biosystems) (see Note 5).
Gel Electrophoresis
50× TAE buffer (Bio-Rad).
Seakam LE agarose powder (Cambrex).
6× Gel loading buffer [10 mM Tris–HCl (pH 7.6), 0.03% bromophenol blue, 0.03% xylene cyanol, 60% glycerol, and 60 mM EDTA].
1 kb plus DNA ladder markers (Invitrogen).
Ethidium bromide, 10 mg/mL.
Agarose gel electrophoresis apparatus.
Power supply (PowerPac Basic, Bio-Rad).
Gel documentary machine or equivalents.
Quantitative RT-PCR
TaqMan EZ RT-PCR Core Reagents kits (Applied Biosystems).
50 μM PCR forward primer, 5′-CAGAACGCTGTAGCTTCAAAAATCT-3′.
50 μM PCR reverse primer, 5′-TCAGAACCCTGTGATGAATCAACAG-3′.
10 μM probe, 5′-(FAM)TCTGCGTAGGCAATCC(NFQ)-3′ (FAM, 6-carboxyfluorescein; NFQ, nonfluorescent quencher; Applied Biosystems).
Quantitative PCR machine (ABI Prism 7000 Sequence Detection System, Applied Biosystems).
PCR reaction plates (MicroAmp optical 96-well reaction plate, Applied Biosystems).
Optical adhesive covers (Applied Biosystems).
Benchtop centrifuge (Allegra X-15R, Beckman Coulter) with microplate carriers (SX4750μ, Beckman Coulter).
PCR for Groups 1 and 2 Coronaviruses
AmpliTaq Gold DNA polymerase, 5 U/μL (Applied Biosystems).
10× Gold PCR buffer (Applied Biosystems).
dNTP.
25 mM MgCl2 solution (Applied Biosystems).
10 μM PCR forward primer, 5′-GGTTGGGACTATCCTAAGTGTGA-3′.
10 μM PCR reverse primer, 5′-CCATCATCAGATAGAATCATCAT-3′.
Themocycler (GeneAmp 9700, Applied Biosystems).
Methods
RNA Extraction
- For a new kit, perform the following procedures before specimen processing (see Note 6):
- Add 1 mL of AVL buffer to a tube of lyophilized carrier RNA (310 μg). Dissolve the carrier RNA thoroughly, then transfer to the buffer AVL bottle and mix thoroughly. Store the AVL buffer at 4°C for up to 6 months (see Note 7).
- For every 19 mL of AW1 buffer, add 25 mL of ethanol (96–100%). Mix it well. Store the AW1 buffer at room temperature for up to 12 months.
- For every 13 mL of AW2 buffer, add 30 mL of ethanol (96–100%). Mix it well. Store the buffer AW1 at room temperature for up to 12 months.
Equilibrate all reagents to room temperature before use.
Transfer 140 μL of the sample into a 1.5 mL microcentrifuge tube (see Note 8).
Add 560 μL of prepared buffered AVL with carrier RNA to the microcentrifuge tube.
Briefly vortex the tubes for 15 s and incubate at room temperature for 10 min.
Briefly centrifuge the microcentrifuge tube. Add 560 μL ethanol (96–100%) and mix by pulse-vortexing for 15 s.
Briefly centrifuge the microcentrifuge tube.
Transfer 630 μL of the solution from the tube to a QIAamp spin column placed in a provided 2 mL collection tube. Centrifuge at 6,000 × g (8,000 RPM) for 1 min at room temperature/4°C. Place the spin column in a clean 2 mL collection tube. Discard the tube containing the filtrate.
Open the spin column and repeat step 8.
Add 500 μL buffer AW1. Centrifuge at 6,000 × g (8,000 RPM) for 1 min. Place the spin column in a clean 2 mL collection tube. Discard the tube containing the filtrate.
Add 500 μL buffer AW2. Centrifuge at 20,000 × g (14,000 RPM) for 3 min. Place the spin column in a clean 2 mL collection tube and centrifuge at 20,000 × g for another 1 min. Place the spin column in a clean 1.5 mL microcentrifuge tube. Discard the tube containing the filtrate.
Apply 50 μL buffer AVE equilibrated to room temperature directly on the membrane of the column. Close the cap and incubate at room temperature for 1 min.
Centrifuge at 6,000 × g (8,000 RPM) for 1 min. Collect the filtrate for cDNA synthesis. Store the RNA at −20 or −70°C.
Reverse Transcription
-
Prepare a reverse transcription master mix sufficient for the designated number of samples in a sterile 1.5 mL microcentrifuge tube as shown below:
Reagent Volume per reaction Volume for N reactions Final concentration 5× First strand buffer 4 μL 4 × N μL 1× 0.1 mM DTT 2 μL 2 × N μL 0.01 mM 10 mM dNTP 1 μL N μL 0.5 mM Random primers (150 ng/μL) 1 μL N μL 7.5 ng/μL Reverse transcriptase (200 U/μL) 1 μL N μL 200 U/reaction Ribonuclease inhibitor (optional) 1 μL N μL 40 U/reaction Total volume of master mix 10 μL 10 × N μL − Vortex and centrifuge the tube briefly. Keep the tube on ice.
Add 10 μL of master mix solution into separate 0.5 microcentrifuge tubes. Label the tube accordingly and keep these tubes on ice.
Add 10 μL of purified RNA samples into these tubes accordingly.
Vortex and centrifuge the tubes briefly.
Stand the tubes at room temperature for 10 min and then incubate at 42°C for 50 min.
Inactivate the transcription reaction by incubating the tubes at 95°C for 5 min and then chill the samples on ice. Store the cDNA samples at −20°C (see Note 9).
PCR for SARS Coronavirus
-
Prepare a PCR master mix sufficient for the designated number of samples in a sterile 0.5 mL microcentrifuge tube according to the following table. Include at least one positive control and one negative control (water) for each run. Add additional controls (e.g. purified RNA from the studied samples) as necessary:
Reagent Volume per reaction Volume for N reactions Final concentration 10× PCR buffer 5 μL 5 × N μL 1× MgCl2, 25 mM 5 μL 5 × N μL 2.5 mM dNTP, 10 mM 0.5 μL 0.5 × N μL 0.1 mM Forward primers, 10 μM 1.25 μL 1.25 × N μL 0.25 μM Reverse primers, 10 μM 1.25 μL 1.25 × N μL 0.25 μM DNA polymerase (5 U/μL) 0.25 μL 0.25 × N μL 1.25 U/reaction Water 34.75 μL 34.75 × N μL − Total 48 μL 48 × N μL − Vortex and centrifuge the tube briefly. Keep the tube on ice.
Aliquot 48 μL of the master mix into separate 0.5 mL microcentrifuge tubes and label the tube accordingly.
Add 2 μL of cDNA generated from the reverse transcription reactions to these tubes accordingly. For the positive control, add 2 μL of SARS coronavirus cDNA into the reaction. For the negative control, add 2 μL of autoclaved water.
Vortex and centrifuge the tubes briefly.
-
Run the PCR in the following condition:
Step Temperature Time Heat activation 94°C 8 min Thermal cycling (40 cycles) Denaturing step 95°C 30 s Annealing step 50°C 40 s Extension 72°C 15 s Final extension 72°C 2 min Soak 4°C ¥ After the run, analysis the PCR products by gel electrophoresis. Alternatively, the products can be kept at −20°C for short-term storage.
Agarose Gel Electrophoresis
Place a gel-casting tray onto a gel-casting base. Level the base.
Prepare 2% agarose gel by weighing out 1 g of agarose powder. Add it in a 250 mL bottle containing 50 mL 1× TAE buffer. Microwave bottle with a loosened cap until the gel starts to bubble and become transparent (see Note 10).
Cool the melted agarose to about 60°C and pour it into the gel-casting tray. Insert a comb to the tray.
Allow the gel to solidify at room temperature.
Remove the comb from the tray.
Place the tray into the electrophoresis chamber with the wells at the cathode side.
Fill the buffer chamber with 1× TAE buffer at a level that can cover the top of the gel.
Mix 0.5 μL of the DNA markers with 2 μL of 6× gel loading dye and 9.5 μL of water on a parafilm sheet by repeated pepitting.
Mix 10 μL of the PCR products with 2 μL of 6× gel loading dye on a parafilm sheet by pepitting up and down several times.
Apply the mixture to the corresponding well of the gel.
Close the lid of the electrophoresis apparatus and connect the electrical leads, anode to anode (red to red) and cathode to cathode (black to black).
Run the gel at 100 V for 30 min.
Turn off the power, remove the cover and retrieve the gel.
Soak the gel in 1× TAE with 0.5 μg/mL ethidium bromide for 15 min. Wash the gel with water briefly (see Note 11).
Place the gel on top of the transilluminator. Switch on the power of the gel documentary machine (see Note 12).
Adjust the position of the gel and record the results. The size of the expected product for the virus is 182 bp (see Note 13).
Quantitative RT-PCR
- Turn on the quantitative RT-PCR machine. Activate the Detection Manager from the supplied software and confirm the reporter, quencher, passive reference dyes are FAM, NFQ, and ROX, respectively. Set the cycle conditions as follows:
Step Temperature (°C) Time UNG treatment 50 2 min Reverse transcription 60 40 min Heat inactivation 95 5 min Thermal cycling (50 cycles) Denaturing 95 15 s Annealing and extension 55 1 min In the reaction plate template, input the necessary information for the corresponding samples (e.g. positive standard, negative control, or name of the clinical specimen). Include at least one set of tenfold serially diluted positive controls with known copy numbers of the target sequence (e.g. 106 to 10 copies/reaction) and three negative controls (water) in each run. For the positive controls, key in the copy numbers of the target sequence used in the corresponding reactions.
- Prepare a PCR master mix sufficient for the designated number of samples in a sterile 2.5 mL screw cap tube according to following table. Add additional controls (e.g. purified RNA from the studied samples) as necessary.
Reagent Volume per reaction Volume for N reactions Final concentration Water 6.2 μL 6.2 × N μL – 5× TaqMan EZ buffer 5 μL 5 × N μL 1× Manganese acetate, 25 mM 3 μL 3 × N μL 3.0 mM dATP, 10 mM 0.75 μL 0.75 × N μL 0.3 mM dUTP, 10 mM 1.5 μL 1.5 × N μL 0.6 mM dCTP, 10 mM 0.75 μL 0.75 × N μL 0.3 mM dGTP, 10 mM 0.75 μL 0.75 × N μL 0.3 mM Forward primers, 50 μM 0.4 μL 0.4 × N μL 0.8 μM Reverse primers, 50 μM 0.4 μL 0.4 × N μL 0.8 μM Probe, 10 μM 1 μL 1 × N μL 0.4 μM rTth DNA polymerase (2.5 U/μL) 1 μL 1 × N μL 2.5 U/reaction AmpErase UNG (1 U/μL) 0.25 μL 0.25 × N μL 0.25 U/reaction Total 21 μL 21 × N μL – Close the cup. Vortex and centrifuge the tube briefly.
Aliquot 21 μL of the master mix into the corresponding wells of the reaction plate.
Add 4 μL of the samples into the corresponding wells carefully (see Note 14).
Seal the reaction plate with an adhesive cover. Make sure each reaction well is sealed properly.
Briefly centrifuge the reaction plate.
Insert the plate to the quantitative PCR machine and perform the RT-PCR cycle.
After the reaction, examine the threshold cycles (Ct) and the amplification curves of the reactions. For a good experiment, the Ct values deduced from the standards should correlate with the log10 copy numbers of the target sequence used in these reactions (Fig. 1a). Positive clinical samples will generate amplification signals above the threshold (Fig. 1b). By contrast, signals from the water controls and negative samples will below the threshold line. Based on the Ct values from the reference standards, the amounts of input target in the positive reactions will be calculated by the software automatically (see Notes 15 and 16).
Fig. 1.
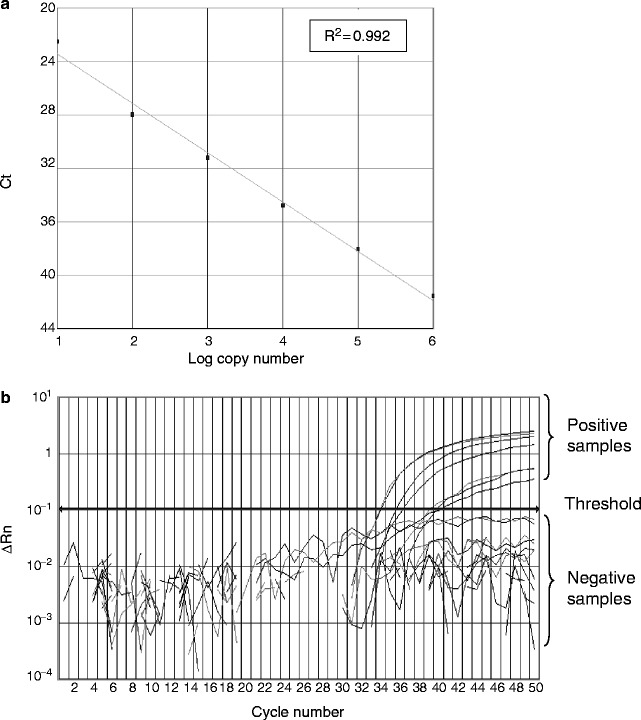
Quantitative RT-PCR assay for SARS-CoV. (a) Standard curve for quantitative analysis of ORF 1b of SARS-CoV. The threshold cycle (Ct) is the number of PCR cycles required for the fluorescent intensity of the reaction to reach a predefined threshold. The Ct is inversely proportional to the logarithm of the starting concentration of the input target. The correlation coefficient (R 2) between these two parameters is shown. (b) An amplification plot of fluorescence intensity against the PCR cycle. The fluorescence signals for positive and negative samples are indicated. SARS-CoV. The X-axis denotes the cycle number of a quantitative PCR assay. The Y-axis denotes the fluorescence intensity. Figures are directly adopted from our previous work (8).
PCR for Groups 1 and 2 Coronaviruses
- Prepare a PCR master mix sufficient for the designated number of samples in a sterile 0.5 mL microcentrifuge tube according to following table. Include at least one positive control and one negative control (water) for each run. Add additional controls (e.g. purified RNA from the studied samples) as necessary.
Reagent Volume per reaction Volume for N reactions Final concentration 10× PCR buffer 5 μL 5 × N μL 1× 25 mM MgCl2, 25 mM 5 μL 5 × N μL 2.5 mM dNTP, 10 mM 1 μL N μL 0.2 mM Forward primers, 10 μM 1 μL N μL 0.2 μM Reverse primers, 10 μM 1 μL N μL 0.2 μM DNA polymerase (5 U/μl) 0.2 μL 0.2 × N μL 1 U/reaction Water 34.8 μL 34.8 × N μL – Total 48 μL 48 × N μL – Vortex and centrifuge the tube briefly. Keep the tube on ice.
Aliquot 48 μL of the master mix into separate 0.5 mL microcentrifuge tubes and label them accordingly.
Add 2 μL of cDNA generated from the reverse transcription reactions (Subheading 3.2) to these tubes accordingly. For the positive control, add 2 μL of coronavirus cDNA into the reaction. For the negative control, add 2 μL of autoclaved water.
Vortex and centrifuge the tubes briefly.
- Run the PCR using the following conditions:
Step Temperature (°C) Time Heat activation 94 10 min Thermal cycling (45 cycles) Denaturing step 94 30 s Annealing step 48 30 s Extension 72 40 s Final extension 72 2 min Soak 4 ¥ After the run, analysis the PCR products by gel electrophoresis (Subheading 3.4). The size of the expected product is 440 bp (see Note 17). Alternatively, the products can be kept at −20°C for short-term storage.
Notes
In our evaluation, the performance of the quantitative RT-PCR assay is better than the manual RT-PCR assays (3). In addition, the quantitative results generated from the real-time RT-PCR might provide additional data from prognosis (3).
In our patient cohort, respiratory samples (e.g. nasopharyngeal aspirate, throat swab) collected from patients within the first week of disease onset have the highest positive rates for SARS coronavirus. By contrast, fecal samples have the highest positive rate after the first week of onset. However, to increase the chance of identifying SARS patients in a nonepidemic period, we recommend testing multiple specimens available from suspected patients.
For respiratory samples isolated from early disease onset, the detection rates could be enhanced by increasing the initial extraction volume of the NPA sample from 140 to 560 μL (10).
Viral transport medium contains a high concentration of antibiotic to inhibit bacterial growth.
The primers and probe used in these assays are perfectly matched to those sequences deduced from SARS coronaviruses in human and civets, including those isolated in 2004.
Personal protection equipment should be worn by the health-care worker taking specimens from suspect or probable SARS patients (http://www.who.int/csr/sars/infectioncontrol/en/).
AVL Buffer containing carrier RNA might form white precipitates when stored at 4°C. The precipitate can be dissolved in the buffer by heating the bottle in a water bath. Cool the buffer to room temperature before use.
For extracting RNA from suspected infectious samples, the procedure must be handled in a biosafety level (BSL) 2 containment using BSL 3 working practices. (http://www.who.int/csr/sars/biosafety2003_12_18/en/).
General procedures to prevent PCR cross contaminations should be strictly followed. Aerosol-resistant filtered pipette tips could minimize possible carryovers of amplicons. Separate pipettes and areas are used for sampling processing, PCR and post-PCR analysis. It is essential to include multiple positive and negative controls in the PCR reactions when a large number of samples are tested at the same time.
Agarose solutions can be superheated in microwave oven. Do not handle bottle immediately after microwaving. Always wear heat-resistant gloves when handling melted agarose.
Ethidium bromide is a known mutagen and may be carcinogenic. Handle solutions of ethidium bromide with gloves.
UV light can cause severe skin and eye damage. Wear safety glasses and close the photography hood before turning on the UV transilluminator.
The conventional RT-PCR protocol is highly specific to SARS coronavirus isolated from respiratory samples. However, we observed a small number of false-positive results from RNA isolated from stools. To overcome this problem, all of our positive fecal samples were retested by the quantitative RT-PCR as described in Subheading 3.5 or using a SYBR green-based RT-PCR assay (11) for confirmation.
When performing step 6 in Subheading 3.5, the RNA samples, including those positive standards, must be handled with extreme care. Cross contamination might lead to false-positive or unreliable quantitative results.
The amplification curves of all positive samples in the quantitative RT-PCR assays must be examined individually. We occasionally find some clinical specimens might yield high backgrounds and the analytical program might misclassify these samples as positive samples.
To exclude negative results due to the poor recovery of RNA, poor performance of the RT-PCR reaction, the presence of PCR inhibitors or human errors, we subsequently modified our quantitative RT-PCR assays to use a duplex assay. The revised test allows simultaneous detection of SARS coronavirus and endogenous 18 S rRNA derived from host cells (12). The primers and probe for 18 S rRNA are commercially available (TaqMan Ribosomal RNA Control Reagents, Applied Biosystems).
The identities of the positive products should be formally confirmed by DNA sequencing.
Acknowledgements
We acknowledge research funding from Public Health Research Grant from the National Institute of Allergy and Infectious Diseases, USA, VCO SARS Research Fund, European Research Project SARS-DTV (contract no: SP22-CT-2004).
Contributor Information
John R. Stephenson, Phone: 0207 759 2764, FAX: 0207 759 2790, Email: John.Stephenson@hpa.org.uk
Alan Warnes, Phone: 02088692011, Email: alan.warnes@nwlh.nhs.uk.
Leo L. M. Poon, Email: llmpoon@hkucc.hku.hk
References
- 1.Gonzalez JM, Gomez-Puertas P, Cavanagh D, Gorbalenya AE, Enjuanes L. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch Virol. 2003;148:2207–2235. doi: 10.1007/s00705-003-0162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weiss SR, Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol Mol Biol Rev. 2005;69:635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poon LL, Guan Y, Nicholls JM, Yuen KY, Peiris JS. The aetiology, origins, and diagnosis of severe acute respiratory syndrome. Lancet Infect Dis. 2004;4:663–671. doi: 10.1016/S1473-3099(04)01172-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poon LL. Emerging nidovirus infections. In: Perlman S, Gallagher T, Snijder EJ, editors. Nidoviruses. Washington, DC: ASM Press; 2008. pp. 409–418. [Google Scholar]
- 5.Poon LL, Leung CS, Tashiro M, Chan KH, Wong BW, et al. Rapid detection of the severe acute respiratory syndrome (SARS) coronavirus by a loop-mediated isothermal amplification assay. Clin Chem. 2004;50:1050–1052. doi: 10.1373/clinchem.2004.032011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang B, Potter SJ, Lin Y, Cunningham AL, Dwyer DE, et al. Rapid and sensitive detection of severe acute respiratory syndrome coronavirus by rolling circle amplification. J Clin Microbiol. 2005;43:2339–2344. doi: 10.1128/JCM.43.5.2339-2344.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keightley MC, Sillekens P, Schippers W, Rinaldo C, George KS. Real-time NASBA detection of SARS-associated coronavirus and comparison with real-time reverse transcription-PCR. J Med Virol. 2005;77:602–608. doi: 10.1002/jmv.20498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peiris JSM, Poon LL. Detection of SARS coronavirus in humans and animals by coventional and quantitative (real time) reverse transcription polymerase chain reactions. In: Cavanagh D, editor. SARS- and Other Coronaviruses: Laboratory Protocols. 1. New York: Springer; 2008. pp. 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poon LL, Peiris JSM. Detection of Group 1 coronaviruses in bats using universial coronavirus reverse transcription polymerase chain reactions. In: Cavanagh D, editor. SARS- and Other Coronaviruses: Laboratory Protocols. 1. New York: Springer; 2008. pp. 13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poon LL, Chan KH, Wong OK, Yam WC, Yuen KY, et al. Early diagnosis of SARS coronavirus infection by real time RT-PCR. J Clin Virol. 2003;28:233–238. doi: 10.1016/j.jcv.2003.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poon LL, Wong OK, Chan KH, Luk W, Yuen KY, et al. Rapid diagnosis of a coronavirus associated with severe acute respiratory syndrome (SARS) Clin Chem. 2003;49:953–955. doi: 10.1373/49.6.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poon LL, Wong BW, Chan KH, Leung CS, Yuen KY, et al. A one step quantitative RT-PCR for detection of SARS coronavirus with an internal control for PCR inhibitors. J Clin Virol. 2004;30:214–217. doi: 10.1016/j.jcv.2003.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]


