Abstract
Despite heroic efforts to prevent the emergence of an influenza pandemic, avian influenza A virus has prevailed by crossing the species barriers to infect humans worldwide, occasionally with morbidity and mortality at unprecedented levels, and the virus later usually continues circulation in humans as a seasonal influenza virus, resulting in health-social-economic problems each year. Here, we review current knowledge of influenza viruses, their life cycle, interspecies transmission, and past pandemics and discuss the molecular basis of pandemic acquisition, notably of hemagglutinin (lectin) acting as a key contributor to change in host specificity in viral infection.
Key words: Influenza, Replication, Transmission, Host range, Pandemic, Hemagglutinin, Sialylglycoconjugate
Introduction
An influenza pandemic is grim as it is unpredictable, rapidly spreads throughout the world, and is mostly associated with severe clinical disease and death in humans, leading to the serious socioeconomic problems. Influenza is an infectious respiratory illness epidemically caused by human influenza A, B, and C viruses (classified on the basis of serologic responses to matrix proteins and nucleoproteins), which are single-stranded, negative-sense RNA viruses of the family Orthomyxoviridae [1]. Type A viruses have the greatest genetic diversity, harboring numerous antigenically distinct subtypes of the two main viral surface glycoproteins; so far, 18 hemagglutinin (HA) and 11 neuraminidase (NA) subtypes have been identified. All possible combined 16 × 9 subtypes, except for H17–18 and N10–11 subtypes, which are detected only in bats (mammals) and exhibit functions completely different from those of the other subtypes [2–4], are maintained in waterfowls, mainly in wild ducks. These avian viruses are occasionally transmitted to other species that are immunologically naïve, in which they may only cause outbreaks or may acquire mutations so as to be efficiently transmitted between new hosts, and they can lead to a pandemic in human populations. An influenza pandemic has continued to circulate as an epidemic with an antigenic variant each year, and two human influenza A subtypes, H3N2 Hong Kong/68 and H1N1 2009 variants, have been seasonally found among humans.
The unexpectedly rapid emergence of A/H1N1 2009 swine pandemic recently, ongoing outbreaks of avian influenza viruses in humans, circulating avian/human/swine influenza viruses in pigs, and the emergence of influenza A viruses that are resistant to currently available anti-influenza virus drugs have raised a great concern that a pandemic could spread rapidly without time to prepare a public health response to stop the illness spread and could threaten human health and life throughout the world, becoming a major impediment to socioeconomic development. We have collected available information in the influenza A virus field, especially factors playing important roles in determining viral transmission, in order to know how best to perform surveillance, prevent, slow, or limit a future pandemic.
Influenza A Virus Infection and Replication
Influenza A virus contains eight (−) ssRNA genomic segments that encode at least ten proteins; nine are structural proteins and 1–4 depending on the virus strain and host species are nonstructural proteins (see Fig. 1; also see Table 1). Once inside the host, the virus is able to escape the host’s innate immune responses in two ways: mainly by viral nonstructural protein 1 (NS1) attacking multiple steps of the type I IFN system, resulting in evasion of type I IFN responses [20], and by viral NA removing decoy receptors on mucins (see Figs. 2 and 3), cilia, and cellular glycocalyx and preventing self-aggregation of virus particles [22]. Also, the virus is capable of evasion of adaptive immune responses: evasion of the preexisting humoral or neutralizing antibodies and seasonal vaccines by antigenic variation in HA and NA antigens [23], and evasion of cellular immune response by amino acid substitutions in cytotoxic T-lymphocyte (CTL) epitopes of viral proteins, resulting in a decrease in CTL response [24]. Furthermore, although the precise functions of PB1-F2 remain unclear and are virus strain-specific and host-specific, it has been thought that PB1-F2 plays roles in both innate and adaptive immune responses in order to support influenza virus infection [25].
Fig. 1.
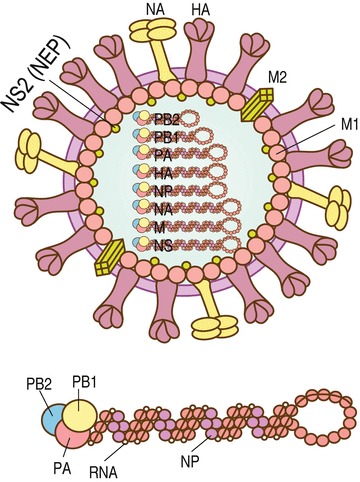
Influenza A virus structure. Influenza A virus particles are roughly spherical in shape with sizes ranging from 80 to 120 nm in diameter, and each particle is enveloped by a lipid bilayer derived from the host cell membrane. Inside the envelope, each of eight (−) ssRNA genomic segments are wrapped with multiple nucleoproteins (NPs) and bound to RNA polymerases (PB1, PB2, PA) forming the vRNPs. The inner layer of the lipid envelope is attached to M1 molecules bound to vRNPs and to NS2. The outer layer of the lipid envelope is spiked with HA, NA, and M2 molecules with a ratio of about 5/2/1. See color figure in the online version
Table 1.
Influenza A virus proteins encoded by each viral RNA segment and their functions
| RNA segment (no. of nucleotides)a | Gene product (no. of amino acids)a | Molecules per virion | Function |
|---|---|---|---|
| Structural proteins | |||
| Polymerase complex | Viral mRNA transcription and viral RNA replication | ||
| 1 (2,341) | PB2 (759) | 30–60 | 1. Recognition of caps of host mRNAs, endonucleolytically cleaved by PA for use as primers for viral mRNA transcription (cap-snatching mechanism) |
| – A host range determinant [5] | |||
| 2. Nuclear import | |||
| 3. Inhibition of expression of interferon-β [6] | |||
| 2 (2,341) | PB1 (757) | 30–60 | 1. Nucleotide addition |
| 3 (2,233) | PA (716) | 30–60 | 1. Endonuclease activity that cleaves host mRNA 10-13 nucleotides |
| 4 (1,778) | Hemagglutinin (566) | 500 | 1. Major antigen |
| 2. Proteolytically cleaved to be a fusion-active form | |||
| – A pathogenic determinant (dependent on virus and host) | |||
| 3. Sialic acid and linkage binding | |||
| – A major determinant of host range and tissue tropism | |||
| 4. Fusion | |||
| 5 (1,565) | Nucleoprotein (498) | 1,000 | 1. Nucleocapsid protein (viral RNA coating) |
| 2. Intracellular trafficking of viral genome, viral transcription/replication and packaging [7] | |||
| 3. Induction of apoptosis [8] | |||
| 6 (1,413) | Neuraminidase (454) | 100 | 1. Sialidase activity that prevents virus aggregation and facilitates viral entry into and budding from the host cell |
| – Variations in the stalk lengths of NAs between residues 36-90 (N2 numbering) may play roles in pathogenesis, transmission, and host range | |||
| – H274Y mutation in N1 subtype, E119V in N1/N2, N294S in N1/N2, and R292K in N2/N9 are associated with decrease in NAI susceptibility | |||
| 2. Antigen | |||
| 7 (1,027) | Matrix protein M1 (252) | 3,000 | 1. Matrix protein that controls direction of viral RNPs transport [9] (dissociated from incoming vRNPs allowing their import into host nucleus and associated with newly assembled viral RNPs promoting their export from the nucleus) |
| 2. Binding to vRNPs, M1 inhibits viral RNA polymerase activity [10] | |||
| 3. Required for virus assembly and budding [11] | |||
| Spliced | Matrix protein M2 (97) | 20–60 | 1. Ion channel to modulate pH of the virion during viral entry allowing M1/NP dissociation and that of the Golgi during transport of viral integral membrane proteins |
| – S31N mutation decreases adamantane susceptibility | |||
|
8 (890) Spliced |
Nonstructural protein2 (NS2) (121) | 130–200 | 1. Nuclear export protein that is involved in the nuclear export of viral RNPs |
| Nonstructural proteins | |||
|
2 (2,341) Ribosomal leaky scanning |
PB1-F2 (up to 90 amino acids depending on virus strains) | – | 1. Pro-apoptotic protein [12] |
| 2. Inhibition of induction of type I interferon synthesis | |||
| – N66S mutation increases virulence [13] | |||
| N40 (lack of 40 residues in N-terminal) | 1. Interacts with the viral polymerase complex with an unclear function | ||
| Note: Both proteins are nonessential for virus replication, but their expression level changes are detrimental to virus replication [14] | |||
|
8 (890) Spliced |
NS1 (strain specific length of 230–237 amino acids) [15] | – | 1. Inhibition of the nuclear export of cellular mRNAs to promote cap-snatching |
| 2. Inhibition of splicing of cellular mRNAs is also for use of host splicing apparatus for viral splicing [16] | |||
| 3. Inhibition of the nuclear export of viral mRNAs until the appropriate time for their expression | |||
| 4. Inhibition of host innate and adaptive immune responses | |||
| – P42S mutation increases in virulence [17] | |||
| 5. Anti-apoptotic and pro-apoptotic protein (control of host lifespan in order to complete its replication cycle) (After 13 h.p.i., pro-apoptotic signaling ≫ anti-apoptotic signaling) [15, 18] | |||
| NS3b (a deletion of NS1 amino acids 126–168) | – | Unknown [19] | |
aThe number of nucleotides and that of amino acids may vary depending on the host and strain of influenza A virus
bNS3 was experimentally found during the adaptation of a human virus within a mouse host [19]
Fig. 2.
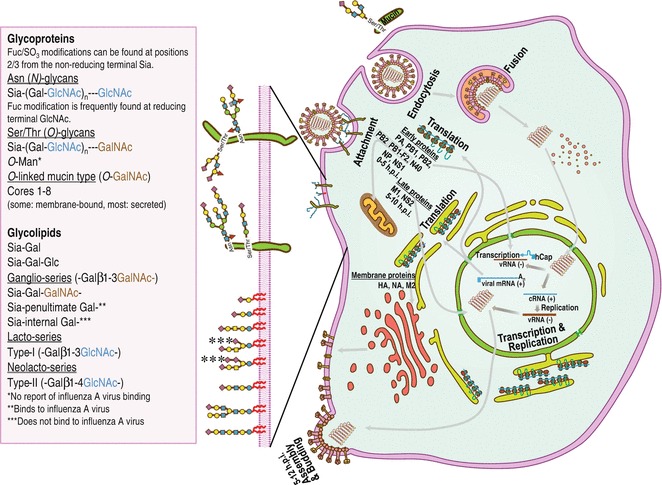
Replication cycle of influenza A virus. The viral HAs can bind to sialylglycoconjugated proteins or lipids (with cartoon representations in the left panel; symbol and text nomenclature used according to the Nomenclature Committee of the Consortium for Functional Glycomics) including mucin (an O-GalNAc glycoprotein with Siaα2-3Gal linkage consisting of mucus shielding the epithelial surface for cellular protection from both physical and chemical damage and pathogen infection). The virus particles adsorbed to mucins can be released by viral NAs that preferentially cleave sialic acid moieties with preference to Siaα2-3Gal linkage over Siaα2-6Gal linkage [21]. Right panel: Schematic of replication cycle of influenza A virus, which can be divided into six distinct parts: (1) attachment, (2) receptor-mediated endocytosis, (3) fusion, (4) transcription and replication, (5) translation (protein synthesis), and (6) assembly, budding, and release. See the text for details. See color figure in the online version
Fig. 3.
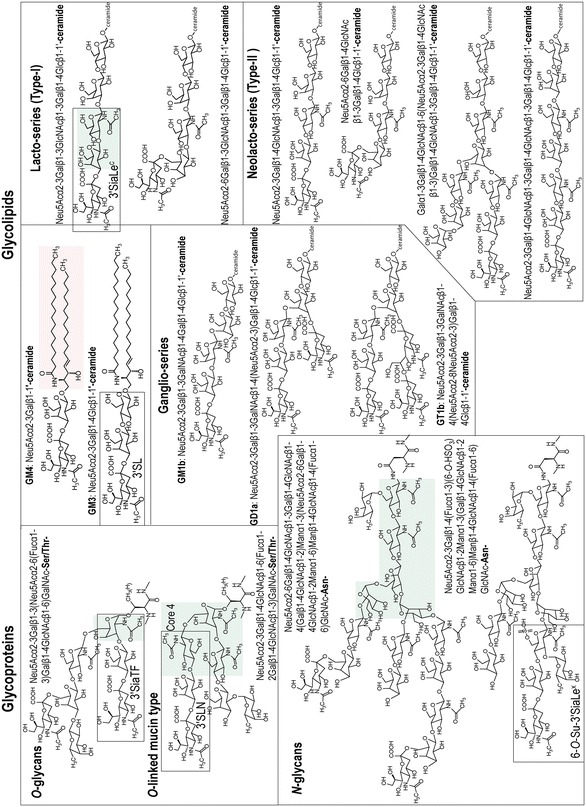
Examples of chemical structures of glycans recognized by influenza A viruses. Glycoproteins are classified into N-linked glycans attached to Asn-X-Ser/Thr, X being any amino acid except proline, and O-linked glycans attached to Ser/Thr. Glycolipids are divided into simple gangliosides GM3, gala-series (GM4), globo-series (not shown here), ganglio-series, lacto-series, and neolacto-series. Core structures are highlighted by green shading. Ceramide structure is highlighted by pink shading. Rectangles show terminal trisaccharides with abbreviation names: 3′SiaTF for Neu5Acα2-3Galβ1-3GalNAc, 3′SLN for Neu5Acα2-3Galβ1-4GlcNAc, 6-O-Su-3′SiaLeX for Neu5Acα2-3Galβ1-4(Fucα1-3)(6-O-HSO3)GlcNAc, 3′SL for Neu5Acα2-3Galβ1-4Glc, and 3′SiaLec for Neu5Acα2-3Galβ1-3GlcNAc. See color figure in the online version
The replication cycle of an influenza A virus (see Fig. 2) starts from attachment of viral HAs to sialic acid (Sia, 5-amino-3,5-dideoxy-d-glycero-d-galacto-2-nonulosonic acid or neuraminic acid (Neu)) receptors on the host cell surface (see Subheading 5.3). This attachment mediates internalization of the virus into the cell by receptor-mediated endocytosis. While an early endosome gradually matures, the acidity in the endosome gradually increases. The low pH activates the integral membrane protein M2 of influenza virus, which is a pH-gated proton channel in the viral lipid envelope, conducting protons into the virion interior. Acidification of the virus interior causes weakening of electrostatic interaction, leading to dissociation of M1 proteins from the viral RNP complexes (unpacking of the viral genome). The low pH in late endosomes also triggers a conformational change in HAs, resulting in exposure of their fusion peptides that immediately bind hydrophobically to the endosomal membrane (fusion), followed by release of vRNPs into the cytoplasm. During the course of the endocytic pathway, sialidase of NAs has been shown to be active [26], possibly in order to promote HA-mediated fusion [27]; however, further studies are needed to determine the exact mechanisms of this NA function. It should be noted that HA fusion will not occur if the HA protein (HA0) is not cleaved to form HA1 and HA2 by a membrane-bound host protease either before or during the release of progeny virions or with incoming viruses prior to endocytosis at the cell surface (see Subheading 5.2).
The vRNPs in the cytoplasm are immediately imported into the nucleus most probably by nuclear localization signals in proteins composed of vRNPs, and the viral RNA polymerase transcribes the (−) vRNAs primed with 5′-capped RNA fragments, which are derived from cellular mRNAs by a cap-snatching mechanism, to viral mRNAs and replicates the unprimed (−) vRNAs to complementary RNAs, (+) cRNAs, used as templates to generate (−) vRNAs (transcription and replication). The viral mRNAs are subsequently exported to the cytoplasm for translation into viral proteins by the cellular protein-synthesizing machinery. Viral proteins needed for viral replication and transcription are transported back to the nucleus. The newly synthesized vRNPs are exported from the nucleus to the plasma membrane, mediated by M1 and NS2 (NEP). Viral HA, M2, and NA proteins are synthesized and glycosylated in the rough endoplasmic reticulum (RER) and the Golgi apparatus and are transported to the cell surface via the trans-Golgi network (TGN). Within the acidic TGN, M2 transports H+ ions out of the TGN lumen to equilibrate pH between the TGN and the host cytoplasm in order to maintain an HA metastable configuration [28]. At the site of budding, HA and NA cluster in lipid rafts enriched in sphingomyelin and cholesterol to provide a sufficient concentration of HA and NA in the budding virus [29, 30]. Cytoplasmic tails of HA and NA proteins bind to M1 proteins, which interact with nucleoproteins of vRNPs. M1 proteins also attach to M2 proteins to form viral particles (assembly). M2 proteins mediate alteration of membrane curvature at the neck of the budding virus, leading to membrane scission. Viral HAs retain the new virions on the cell surface due to binding to cellular sialylglycoconjugates, and thus sialidase activity of viral NAs is required to destroy these bonds, resulting in release and spread of virions (release). The viral HAs attach to other host cells and begin the process anew. The entire influenza A virus replication process (from viral attachment to progeny viruses burst from the infected cell) generally takes about 5–12 h, resulting in the production of as many as 100,000–1,000,000 progeny viruses; only about 1 % of progeny viruses can infect other cells [31].
Influenza A Virus Transmission
Wild waterfowls are the main reservoir of H1–H16 and N1–N9 influenza A viruses [32] (see Fig. 4). Influenza A viruses replicate in the gut of wild waterfowls, which are usually asymptomatic. Infected wild waterfowls excrete viruses in feces and spread viruses mainly via virus-contaminated water and fomites (fecal–contaminated–water–oral route). Influenza A viruses have been thought to able to survive in water for several days [33] and thus migratory wild waterfowls can spread the virus around the world, normally in a north–south direction. Some H5 and H7 subtypes crossing to poultry have acquired mutations converting them into highly pathogenic avian influenza (HPAI) viruses. Low pathogenic avian influenza (LPAI) viruses replicate mainly in respiratory and intestinal organs of poultry and cause epidemics of mild disease, whereas HPAI viruses replicate systemically and cause fatal influenza. Avian influenza viruses may be directly transmitted from infected birds or virus-contaminated environments or indirectly transmitted through mixing with another virus(es) in an intermediate host, such as pigs, to mammals. Influenza viruses typically replicate in the respiratory system of mammals, including humans, pigs, and horses, usually entering through the eyes, nose, mouth, throat, bronchi, and lungs, and are transmitted through the air by coughs or sneezes or through secretions or fomites.
Fig. 4.
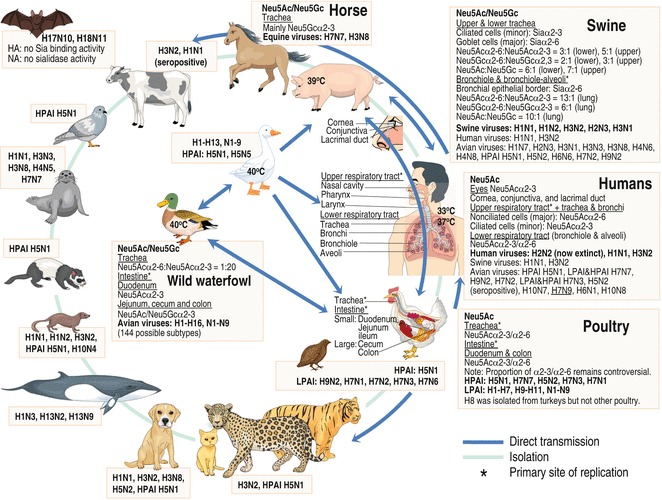
Host range of influenza A viruses. Wild waterfowls are the natural hosts of influenza A viruses of H1–H16 and N1–N9 subtypes. Influenza A viruses from one host are sometimes transmitted to and continue circulating in other host species if they can change and adapt to the new hosts, and once adapted to the new host, they usually lose the capacity to circulate in the previous host. Influenza viruses from wild waterfowl are often transmitted to and from the domestic ducks using the same aquatic areas, and infected domestic ducks spread the viruses to others, including poultry, pigs, and farmers, in a local area. Influenza A viruses have been isolated from various animals as shown in the figure, indicating their capacity to cross the species barrier. Cell surface receptors with Neu5Ac/Neu5Gcα2-6/α2-3 specifically recognized by viral HAs, which initiate viral infection, being a major determinant restricting the host range of influenza A viruses are shown. See color figure in the online version
Emergence of Influenza Pandemics
Having (1) numerous wild waterfowl species as natural reservoirs, (2) various animal hosts, (3) RNA polymerase without proofreading, and (4) segmented genome (3 and 4 causing a high mutation rate), influenza A viruses have been difficult to control and/or eradicate. Efforts for prevention of the next pandemic, either by minimization of cross-infection between species or rapid identification of novel strains, constitute an essential and primary step for preventing influenza infection in human beings. Interspecies transmission of influenza A viruses between animal hosts including pigs, horses, and birds, as well as humans, has occasionally been detected, but successful propagation and transmission in their new host have been restricted. In the past 95 years, only four influenza A virus strains led to sustained outbreaks in human populations and started pandemics (see Table 2; also see Fig. 5).
Table 2.
Pandemic phases by WHO in 2009 [34]
| Phase | Description |
|---|---|
| 1 | No human infection by a circulating animal influenza virus |
| 2 | Human infection by a circulating animal influenza virus |
| 3 | Sporadic cases or small clusters of disease in humans by an animal or human-animal influenza reassortant virus without sufficient human-to-human transmission |
| 4 | Human-to-human transmission of a new virus able to sustain community-level outbreaks |
| Pandemic period | |
| 5 | Sustained community-level outbreaks in two or more countries in one WHO region |
| 6 | Sustained community-level outbreaks in at least one other country in another WHO region |
| Post-peak | Levels of pandemic influenza below peak levels |
| Possible new wave | Levels of pandemic influenza rising again |
| Seasonal period | |
| Post-pandemic | Levels of influenza activity as seen for seasonal influenza |
Fig. 5.
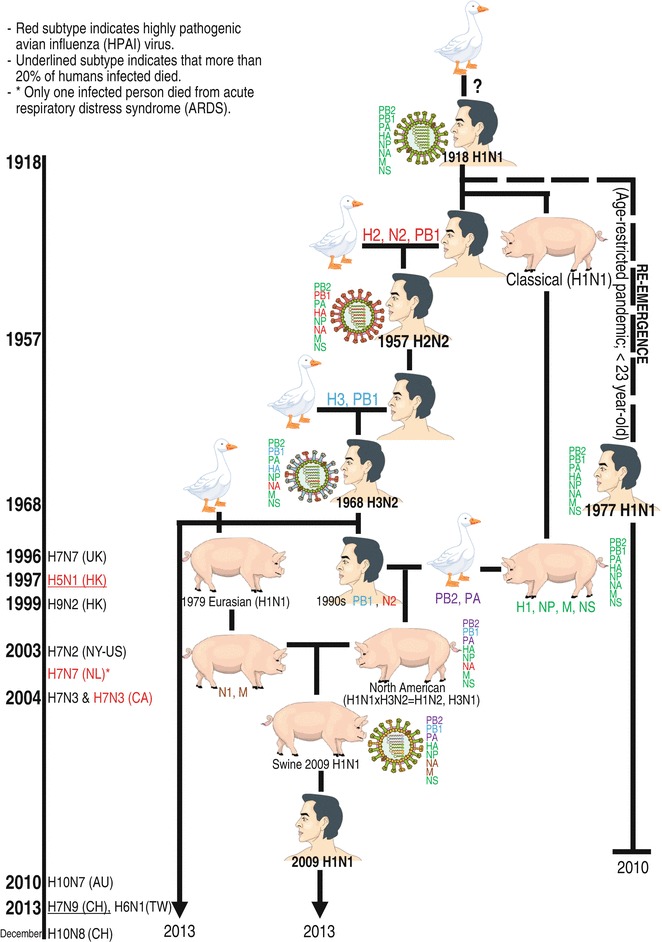
A family tree of human influenza A viruses. Based on influenza history, the 1918 H1N1 virus provides descendants of influenza A viruses that have continued to circulate in human populations by (1) generation of a novel pandemic virus by reassortment between human-avian viruses (1957 and 1968 pandemics) or Fig. 5 (continued) between human-avian-swine viruses (2009 pandemic), (2) recurrence of a previous pandemic virus in new group of populations from a frozen refrigerator (an H1N1 Russian flu in 1977), (3) seasonal viral evolution by intrasubtypic reassortment: for example, A/Fujian/411/2002 (H3N2) having a major antigenic variant due to reassortment between two distinct clades of co-circulating H3N2 viruses [35] (not shown in this chart), and (4) seasonal viral evolution by adaptation associated with point mutations. The left panel shows the timeline of direct transmission of the first reported avian influenza A virus subtypes (H7N7 1996 [36], HPAI H5N1 1997 [37, 38], H9N2 1999 [36], H7N2 2002 (only serologic evidence)—2003 [36], HPAI H7N7 2003 [39], LPAI and HPAI H7N3 2004 [36, 40], H10N7 2010 [41], and H7N9 [42], H6N1 [43], and H10N8 [44] 2013) from avians to humans. Human infections with some of these avian influenza virus subtypes, especially HPAI H5N1, LPAI H9N2, and LPAI H7N9, have occasionally continued to be reported until now (January 2014) [36, 44]. See color figure in the online version
Past Pandemics
H1N1 Spanish Influenza Pandemic (1918–1919): The Greatest Parental Influenza
The Spanish influenza pandemic resulted from an avian-descended H1N1 virus. It killed at least 40 million people globally in 1918–1919, with almost 50 % of the deaths occurring in healthy young adults of 20–40 years of age, although its clinical symptoms and pathological manifestations were mainly in the respiratory tract [45]. This could be due to a too-strong and damaging response to the infection by healthy immune systems [46]; it is by far the most devastating influenza pandemic. The pandemic apparently originated during World War I in the USA at the beginning of 1918 before it appeared in France and then in Spain at the end of 1918 and thus being named Spanish influenza. Despite extensive investigations but a lack of available pre-1918 influenza samples, the emergence pathway of the Spanish pandemic strain either by direct avian viral mutations alone or by reassortment with another virus(es) in an intermediate host is still a mystery.
The pandemic virus induced humans to develop immunity that provides selective pressure and drives the virus to evolve their antigenicity. The resulting 1918-derived H1N1 virus, which had antigenic change annually, caused an epidemic with lower death rates and triggered human immunity to the virus over time. Somehow the 1918-derived H1N1 virus underwent dramatic genetic change with acquisition of three novel gene segments, avian-like H2, N2, and PB1 gene segments, resulting in emergence of the H2N2 pandemic in 1957 and disappearance of the 1918-derived H1N1 virus from circulation. However, the 1918-derived H1N1 virus from the pre-1957 period reappeared in 1977, its reemergence believed to be from a laboratory in Russia or Northern China, and caused a (low-grade) pandemic mostly affecting young people less than 20 years of age due to immunological memory to the virus in most elderly people. After that, H1N1 Russian/77 variant became epidemic yearly until the 2009 H1N1 pandemic emerged, and the virus disappeared from humans. The disappearance of the preexisting H1N1 seasonal virus was suggested to be due in part to stalk-specific antibodies boosted from infection with the 2009 H1N1 pandemic virus [47].
H2N2 Asian Influenza Pandemic (1957–1958)
In early 1956, an influenza outbreak of a new H2N2 strain occurred in China and spread worldwide, resulting in a pandemic in 1957. It has been believed that the pandemic H2N2 virus evolved via reassortant between avian H2N2 strain and the preexisting circulating human 1918 H1N1 strain; it consisted of three gene segments coding HA (H2), NA (N2), and PB1 derived from an avian virus, with the other five gene segments derived from a previously circulating human virus. New HA and NA surface antigens to human immunity for protection resulted in the Asian influenza pandemic virus infecting an estimated 1–3 million people worldwide with approximately two million deaths. The virus became seasonally endemic and sporadic and it disappeared from the human population after the next pandemic appeared in 1968 [48].
H3N2 Hong Kong Influenza Pandemic (1968–1969)
In July 1968, a new influenza A virus was detected in Hong Kong. It was identified as H3N2, which was believed to be a result of reassortment between avian and human influenza A viruses: the HA and PB1 gene segments were derived from avian influenza virus and the other six gene segments, including the NA N2 gene segment, were derived from the 1957 H2N2 virus. The virus killed up to one million humans varying widely depending on the source, less deaths than those in previous pandemics. The relatively small number of death is thought to be due to exposure of people to the 1957 virus, who apparently retained anti-N2 antibodies, which did not prevent 1968 infection but limited virus replication and reduced the duration and severity of illness. Although the next pandemic occurred in 2009, the virus is still in circulation globally (as of 2013) as a seasonal influenza strain.
H1N1 Swine Influenza Pandemic (2009–2010)
In April 2009, a widespread outbreak of a new strain of influenza A/H1N1 subtype referred to as swine flu was reported in Mexico. By June 2009, the virus had spread worldwide, starting the first influenza pandemic of the twenty-first century. In August 2010, the influenza activity returned to a normal level as seen for seasonal influenza; at least 18,000 laboratory-confirmed deaths from the pandemic 2009 were reported, but infection with the virus resulted in mild disease with no requirement of hospitalization [49]. To date, the influenza A viruses identified among people are 1968 H3N2 and 2009 H1N1 viruses.
Genetic composition of the pandemic 2009 viruses isolated from initial cases indicated that the viruses are composed of PB2 and PA gene segments of North American avian virus origin, PB1 segment of human H3N2 virus origin, HA (H1), NP and NS segments of classical swine virus origin (their genes having been found to circulate in pigs since 1997/1998 known as avian/human/swine triple reassortant H1N2 swine viruses), and NA (N1) and M segments of Eurasian avian-like swine H1N1 virus origin (which emerged in European pigs in 1979); hence, their original description was “quadruple” reassortants (see Fig. 5). Similarity between the HA sequence patterns of the 1918 pandemic and 2009 pandemic viruses and their cross-antibody neutralization [50–53] suggested that H1 HA of the 2009 pandemic virus may have originated from or derived from the same origin of the 1918 pandemic virus; probably, the 1918-like pandemic H1N1 virus was transmitted and established in domestic pigs between 1918 and 1920 referred to as the classical swine lineage that circulated continuously in pigs in the USA. Domestic pigs having a short lifespan (4- to 6-month-old pigs are killed for their flesh, and female breeding pigs (sows) remain in the farm until the age of 4–5 years and then taken to be slaughtered for sausages and bacon) are a frozen-like source for the influenza virus due to a lack of selection pressure in pigs. Consequently, the HA sequence of the 2009 swine origin derived from the classical swine lineage is not much different from its origin.
Pandemic Mechanisms
The past pandemics and ongoing direct transmission of avian influenza A viruses into humans suggest two plausible mechanisms that would permit influenza A viruses to overcome selective pressure and subsequently become established in human populations (see Fig. 5; also see Subheading 5.3.3). As shown in Fig. 6, one mechanism is an adaptation mechanism, in which a nonhuman virus acquires a mutation(s) (a mutation(s) in the HA gene to recognize the Neu5Acα2-6Gal receptor considered to be an essential prerequisite for the beginning of a pandemic) during adaptation to an intermediate host leading to sufficient human-to-human transmission. The other mechanism is a reassortment mechanism, in which a nonhuman virus reassorts with a nonhuman virus(es) and/or a human virus(es) in an intermediate host producing 28 possible reassortants (if two viruses are reassorted), by which the reassortant with ability leading to sufficient human-to-human transmission becomes dominant in the intermediate host. Then the adapted/reassorted virus can cause influenza outbreaks in immunologically naïve humans, allowing it to adapt (fine tune) for efficient and sustained human-to-human transmission and finally causing a global outbreak (pandemic). The first recorded H1N1 pandemic in 1918 is of an unknown origin, the H2N2 pandemic in 1957 and the H3N2 pandemic in 1968 arose from genetic reassortment between avian virus and human virus with an unknown intermediate host, and the latest H1N1 pandemic in 2009 arose from genetic reassortment among avian virus, swine virus, and human virus with another swine virus in pigs as intermediate hosts. It is not known when and how a future pandemic will emerge, by an adaptation or reassortment mechanism, and it is also impossible to predict which virus subtype will be the next pandemic. In the post-pandemic period, most people develop immunity to the pandemic strain and thus the pandemic virus can continue to cause seasonal outbreaks (epidemics) if it can change its surface antigens (antigenic variation) to evade host immunity.
Fig. 6.
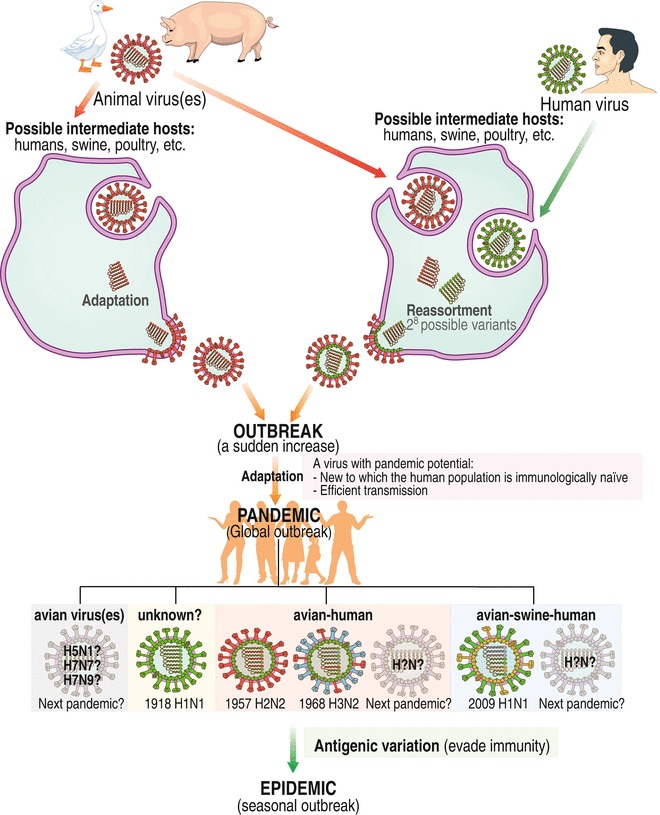
Proposed mechanisms of emergence of an influenza pandemic. The left panel is an adaptation mechanism and the right panel is a reassortment mechanism. See text for details. See color figure in the online version
Hemagglutinin
HA was so named due to its ability to agglutinate red blood cells via binding to Sia on red blood cells. Phylogenetically, 18 HA subtypes are classified into group 1 and group 2 (see Fig. 7a). Notable differences in structure between the two groups of HAs are in the region involved in HA conformational change required for membrane fusion; group 1 HAs contain an additional turn of the helix at residues 56–58, blocking accessibility of tert-butyl hydroquinone, which is accessible to group 2 [54]. HA is encoded by the fourth segment of the influenza A viral genome and is assembled as a homo-trimeric precursor (HA0) (see Fig. 7b). HA is a major target of neutralizing antibodies, plays a pivotal role in avian influenza virus pathogenicity, and is a major determinant of host range restriction. It is a lectin that contains one or more carbohydrate recognition domains that determine host specificity [55] and plays a crucial role in fusion of the viral envelope and cellular endosomal membrane for release of the viral genome into host cells (see Fig. 7c–f).
Fig. 7.
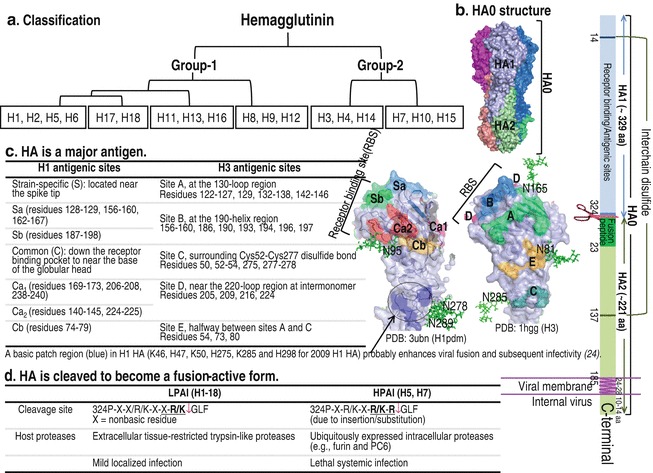
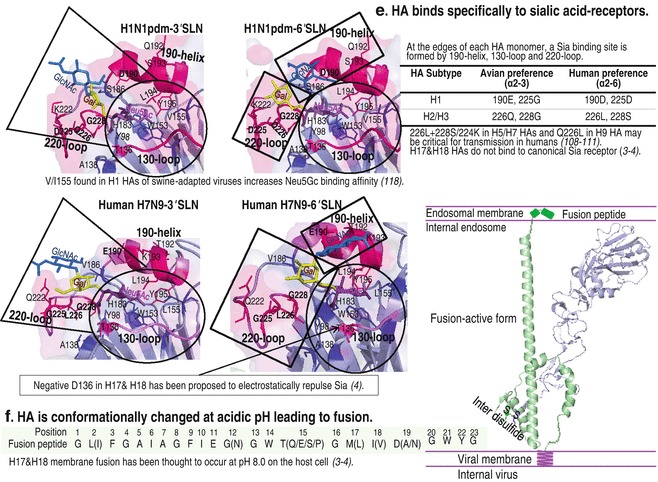
HA structure and its functions. (a) 18 HA subtypes are phylogenetically classified into six clades segregated into two groups as indicated in the chart. (b) HA forms a homo-trimeric precursor (HA0) by which each monomer consists of HA1 and HA2 (PDB: 1rd8) and the C-terminus of HA2 anchors each monomer in the viral membrane. It provides four events important for the entry of influenza virus into the host cell. (c) Antigenicity. HA1 is a major viral antigen with five important antigenic sites as indicated. (d) Cleavage. HA0 with a single basic residue is cleaved by extracellular proteases into HA1 (~329 amino acids) and HA2 (~221 amino acids). HA0 with multiple basic amino acids at the cleavage site (residues 324–329) in HPAI viruses is cleaved intracellularly in the trans-Golgi compartment by ubiquitous proteases. Two amino acids in the cleavage sites that are critical for the recognition by proteases are underlined. (e) Attachment. HA1 carries a receptor binding site at the membrane-distal tip, which is formed by 190-helix, 130-loop, and 220-loop. Amino acid substitutions in this binding site and virus-receptor binding assays revealed two amino acid residues critical for determinants of virus binding preference: at positions 190 and 225 for H1 virus (E190 and G225 HA preferring α2-3 receptors, and D190 and D225 HA favoring α2-6 receptors) and at positions 226 and 228 for H2 and H3 viruses (Q226 and G228 HA preferring α2-3 receptors, and L190 and S225 HA preferring α2-6 receptors). Left panel: Crystal structures of HAs of A/California/04/2009 (H1N1) interactions with 3′SLN (PDB: 3ubq) and with 6′SLN (PDB: 3ubn) and those of A/Anhui/1/2013 (H7N9) interactions with 3′SLN (PDB: 4BSD) and with 6′SLN (PDB: 4BSC). Circular broken lines indicate amino acids that interact with Neu5Ac and are highly conserved among different H1–H16 HA subtypes, except position 155 (V/I for increased binding to Neu5Gc). The other broken lines indicate amino acids involved in interactions with internal sugars of the glycan receptor. (f) Fusion. At acidic pH, the cleaved HA in HA1 (gray)-S-S-HA2 (green) form is conformationally changed. The N-terminal HA2 fusion peptide (residues 1–23) buried inside the interior of the HA molecule at neutral pH pops out and is quickly inserted into the host endosomal membrane, leading to membrane fusion (modeled in right panel). See color figure in the online version
HA1 Is a Major Target of Neutralizing Antibodies
Change in HA antigen is responsible for epidemics and pandemics. Change in HA antigen either by accumulation of mutations in HA1 of five proposed antigenic sites (based on amino acid sequence comparison among viruses isolated from different years or among variants grown in the presence of mouse monoclonal antibodies) as shown in Fig. 7c [56–59] or by intrasubtypic reassortment between distinct clades of co-circulating influenza A viruses [35] is responsible for evasion of recognition by the host antibodies and thus continuous circulation of the virus in host populations. Amino acid sequencing studies of HAs of avian and animal viruses isolated from different periods of time have shown that the HAs of avians and animals that have shorter lifespans have higher conservation of amino acid sequences than that of human virus isolates, suggesting that avian/animal viruses are subjected to little immune pressure, resulting in less antigenic variation than that of human virus strains [60, 61].
Introduction of a novel HA antigen, resulting from genetic reassortment during mixed infection, from direct introduction of a nonhuman influenza virus, or from reintroduction of human influenza viruses that had disappeared from circulation, into immunologically naïve human populations is a key factor of an influenza virus with pandemic potential.
HA Cleavage Is a Critical Determinant of Pathogenicity in Gallinaceous Poultry
To enable HA conformational changes that lead to membrane fusion, which is critical for viral infectivity and dissemination, HA0 must be cleaved by a host cell protease into subunits HA1 and HA2; thereby, the host protease is a determinant of tissue tropism of the virus. Influenza virus HA0 usually contains a monobasic cleavage site (see Fig. 7d), which is recognized by extracellular trypsin-like proteases, such as tryptase Clara from rat bronchiolar epithelial Clara cells [62] and mast cell tryptase from the porcine lung, found only in a few organs, and thus virus infection is localized in a limited number of organs, such as the respiratory and intestinal tracts, resulting in mild or asymptomatic symptoms (including ruffled feathers and decreased egg production); hence, the causative viruses are called LPAI viruses [63]. Multiplication of H5 or H7 LPAI viruses in chickens and turkeys generates HPAI viruses having multiple basic amino acids (see Fig. 7d) at the HA cleavage site recognized by intracellular ubiquitous subtilisin-like proteases, such as furin and proprotein convertase 6 (PC6), which are present in a broad range of organs, allowing the virus to infect multiple internal organs with a mortality rate as high as 100 % within 48 h (lethal systemic infection or fowl plaque typically being characterized by cyanosis of combs and wattles, edema of the head and face, and nervous disorders [64–67]). Sequence analysis of the HA cleavage site showed that some LPAI H5 and H7 subtypes contain a purine-rich sequence, and thus a direct duplication (unique insertion) in this region could lead to lysine (K) and/or arginine (R)-rich codons (codon AAA or AAG specifying lysine and codon AGA or AGG specifying arginine); this is a reason why HPAI viruses have been derived only from subtypes H5 and H7 [68]. Not only the basic cleavage site sequence but also a carbohydrate side chain near the cleavage site contributes to determination of pathogenicity (virulence) if it interferes with the host protease accessibility.
Since the first report of a HPAI H5N1 progenitor strain in 1996 from a farmed goose in Guangdong Province, China (A/gs/Guangdong/1/96 designated as Clade 0), the world has intermittently experienced HPAI virus outbreaks, both recurrence and new HPAI virus outbreaks, in domestic birds classified into groups or clades (20 clades having been recognized at present) and subdivided into subclades and lineages based on their phylogenetic divergence as the virus continues to evolve rapidly [69, 70]. Although they are generally restricted to domestic poultry on farms with high mortality rates and substantial economic losses, HPAI H5N1 viruses have occasionally been isolated from some species of wild waterfowls, including wood ducks and laughing gulls, with varying degrees of severity [71–75]; thus, migration of susceptible waterfowls could spread HPAI H5N1 viruses over long distances, leading to difficulties for avian influenza control. The significant species-related variation in susceptibility to and clinical disease caused by H5N1 virus infection has been determined not only in wild birds but also in other animals. For example, pigs can be infected with HPAI H5N1 viruses, but they have almost no or very weak disease symptoms or only slight respiratory illness. Without influenza-like symptoms, the virus may adapt to mammalian hosts in the respiratory tract of this potential intermediate host [76], which contains gradual increases in Neu5Acα2-6Gal, a human receptor, over Neu5Acα2-3Gal, an avian receptor, from upper and lower parts of the porcine trachea towards the porcine lung, a primary target organ for swine-adapted virus replication [77]. Humans can be infected with HPAI H5N1 virus (first report in 1997) with severe disease and high death rate. The ecological success of this virus in crossing the species barrier from poultry to infect diverse species including wild migratory birds and other mammals including pigs, cats, and dogs with sporadic infections in humans often with fatal outcomes [78, 79] highlighted the possibility of HPAI H5N1 development to a pandemic strain either by gradual modification of existing structures or rapid modification by reassortment with a human epidemic strain. Although the world has been at phase 3 in WHO’s six phases of pandemic alert since 2006 [80], HPAI H5N1 viruses should not be neglected in efforts to perform surveillance and health management planning.
In addition to HPAI H5N1 viruses, other avian influenza viruses including LPAI H5N1, HPAI and LPAI H7N7, HPAI H7N3, and LPAI H9N2 viruses have occasionally crossed the species barrier to infect other mammals including humans (see Fig. 4) and have caused generally mild disease (with conjunctivitis or mild respiratory symptoms) in humans. Recently, LPAI H7N9 virus (a novel avian–avian reassortant virus: HA from wild-duck H7N3 virus, NA from wild-bird H7N9 virus, PA, PB1, PB2, NP, and M from chicken H9N2 virus, and NS from another chicken H9N2 virus) identified in humans in February 2013 in China has killed 45 (due to severe pneumonia) of the 139 laboratory-confirmed cases (case-fatality ratio of about 32 %) according to WHO data in November 2013 [42, 81]. This evidence indicated that HPAI viruses primarily infect poultry and cause severe illness and high death rates in poultry and that they occasionally infect other non-poultry species with variation in severity depending on the virus strain and host. LPAI viruses spread silently in poultry and occasionally spread to other non-poultry species and often cause mild illness but are capable of causing severe disease, such as disease caused by LPAI H7N9 virus infection in humans. Thus, more studies are needed to understand differences in pathogeneses of these viral infections.
Receptor Specificity Is Responsible for the Host Range Restriction of Influenza Virus
Influenza viruses enter the body and search for cells among the host cells in which they can replicate and grow. Influenza virus homing is triggered by interactions between viral HA spikes and sialylglycoconjugates on host cell surface [82], which play roles in a wide variety of host biological processes, including cell proliferation, apoptosis, and differentiation [83]. More than 50 types of sialic acids are found in nature, with N-acetylneuraminic acid (Neu5Ac) and N-glycolylneuraminic acid (Neu5Gc) being the most prevalent forms. Not only sialic acid type but also glycosidic linkage type (the most common terminal linkages being α2-3 and α2-6 linkages), substructure (such as GalNAc or GlcNAc), and other modifications (such as fucosylation and sulfation) cause diversity in sialylglycoconjugates (see Figs. 2, left panel and 3) [84, 85]. The sialic acid type and the glycosidic linkage type on the host cell surface are the principal determinants of host range restriction of influenza viruses, although other glycan modifications may be involved in the virus-receptor binding preference. Therefore, the distribution of sialylglycoconjugates among animal species and tissues, a crucial factor for influenza A infection and transmission, has been extensively investigated either by lectin histochemical analysis with Maackia amurensis agglutinin (MAA-I specific for Siaα2-3Galβ1-4GlcNAc-, MAA-II for Siaα2-3Galβ1-3GalNAc) and Sambucus nigra agglutinin (SNA specific for Siaα2-6Galβ1-4GlcNAc-) or by structural characterization using sequential glycosidase digestion in combination with HPLC and mass spectrometry. Figure 4 shows sialic acid-containing receptors in main target organs in important host species of influenza A viruses.
Wild Waterfowls
So far (2013), all H1–H16 and N1–N9 avian influenza viruses have been reported in 12 bird orders, most having been isolated from the order Anseriformes, especially in the family Anatidae (ducks, swans, and geese), and the order Charadriiformes (shore birds) in the family Laridae (gulls, terns, and relatives). It should be noted that the newest H17N10 and H18N11 viruses recognized in 2012 and 2013, respectively, were found only in bats, the little yellow-shouldered bat Sturnira lilium for H17N10 and the flat-faced fruit bat Artibeus planirostris for H18N11, in the family Phyllostomidae, a family of frugivorous bats that are abundant in Central and South America [2, 3]. Ducks in the Anatinae subfamily belonging to the family Anatidae are the most common source of influenza A virus isolation and risk for virus transmission [86]. Almost all ducks are naturally attracted to aquatic areas including wetlands, lakes, and ponds for resting, feeding, and breeding in their course of migration, allowing influenza viruses to be transmitted to and from domestic duck populations. Infected domestic ducks spread the virus to other avian species in a local area [73]. The duck tracheal and intestinal epithelium was shown to predominantly express Siaα2-3Gal oligosaccharides (the ratio of Siaα2-6Gal to Siaα2-3Gal in the duck trachea being approximately 1:20) [87, 88]. Not only Neu5Acα2-3Gal but also Neu5Gcα2-3Gal (not found in chickens) glycans are present in the epithelium of the duck jejunum, cecum, and colon [89]. Correlated with the duck hosts, the duck-isolated influenza viruses preferentially bind to Neu5Ac/Neu5Gcα2-3 receptors (avian receptors) [82, 89–91]. This also agrees with the finding that avian influenza virus isolates replicate efficiently in chorioallantoic cells of 10-day-old chicken embryonated eggs that contain N-glycans, which are essential for entry into host cells of influenza virus infection [92], with molar percents of α2-3 linkage and α2-6 linkage of 27.2 and 8.3, respectively [93].
Studies on sialic acid substructure binding specificity of influenza viruses revealed that although most avian viruses share their preferential binding to terminal Neu5Acα2-3Gal, duck-isolated influenza viruses prefer the β1-3 linkage between Neu5Acα2-3Gal and the next sugar residue such as 3′SiaLec and 3′SiaTF, whereas gull-isolated influenza viruses show high affinity for the β1-4 linkage such as 3′SLN, for fucosylated receptors such as 3′SiaLex, and for sulfated receptors such as Neu5Acα2-3Galβ1-4(6-O-HSO3)GlcNAc (6-O-Su-3′SLN) and 6-O-Su-3′SiaLeX (see Fig. 3) [90]. These receptor-binding specificity data of influenza viruses are correlated well with intestinal epithelial staining with SNA and MAA lectins showing that the duck intestinal epithelium expressed a high level of Siaα2-3Galβ1-3GalNAc-moieties (preferential to MAA-II), whereas the gull intestinal epithelium dominantly expressed Siaα2-3Galβ1-4GlcNAc-moieties (preferential to MAA-I).
Screening using a virus-receptor binding assay together with molecular modeling revealed that gull-viral HAs with 193R/K displayed increased affinity for 6-O-Su-3′SLN and 6-O-Su-3′SiaLeX due to favorable electrostatic interactions of the sulfate group of the receptor and positively charged side chain of 193R/K [94, 95]. The gull-viral HAs with 222Q exhibited binding affinity for the fucosylated receptor 3′SiaLex similar to binding affinity for the nonfucosylated counterpart 3′SLN, while duck influenza viruses showed inefficient binding to the fucosylated receptor due to steric interference between its bulky 222K on the HA and the fucose moiety of the receptor [94, 95]. Only some gull-isolated influenza viruses have potential to infect ducks, indicating that there is a host-range restriction between avian species [96].
Poultry
Several influenza A viruses including H1–H13 and N1–N9 subtypes have been isolated from domesticated poultry in the family Phasianidae of the order Galliformes, including turkeys, chickens, quails, and guinea fowls [97, 98]. Adapted avian influenza viruses in domestic poultry can be divided into two main forms according to their capacity to cause low or high virulence in the infected poultry (see Subheading 5.2). Both forms of avian isolates from poultry before 2002 mainly bind to α2-3 sialyl linkages using either synthetic sialyloligosaccharides or erythrocytes as molecular probes for influenza virus binding specificity [99–102], but since 2002, some of the isolates have shown an increase in binding to α2-6 sialyl linkages (see Subheading 5.3.3). Tissue staining with avian and human influenza viruses and with MAA and SNA lectins has shown the presence of Siaα2-3Gal- and Siaα2-6Gal-terminated sialyloligosaccharides in respiratory and intestinal epithelia of gallinaceous poultry, including chickens and quails [87, 103–108]. However, the proportion of α2-3 and α2-6 sialyl linkages in respiratory and intestinal epithelia of the poultry is still controversial [87, 107, 109]. Thus, more investigations of the structure and distribution of receptors in the replication sites of influenza A viruses are needed to understand the basis of viral infection and transmission.
Human Beings
Human-adapted influenza A viruses that possess efficient human-to-human transmission ability mainly target the human upper respiratory tract, where they can be readily spread with a sneeze or cough. Lectin histochemistry of human respiratory tissues demonstrated that epithelial cells in the upper respiratory tract (nose-larynx) and in the upper part of the lower respiratory tract (trachea and bronchi) are enriched in α2-6 sialylated glycan receptors with a small proportion of α2-3 sialylated glycans [110]; using human airway epithelium (HAE) cells, lectin staining indicated that α2-6-linked sialylated receptors are dominantly present on the surface of nonciliated cells, while α2-3-linked sialylated receptors are present on ciliated cells [111, 112]. In the lower part of the lower respiratory tract (lung), α2-6-sialylated glycans can be found on epithelial cells of the bronchioles and alveolar type-I cells; α2-3-sialylated glycans can be found on nonciliated cuboidal bronchiolar cells and alveolar type-II cells [110, 113]. Recent mass spectromic analysis of glycan structures of human respiratory tract tissues showed that both Sia α2-3 and α2-6 glycans are present in the lung and bronchus [114]. The pattern of lectin localization correlated with the pattern of virus binding and infection: human-adapted viruses bound extensively to bronchial epithelial cells but intensively to alveolar cells, and the opposite results were found for avian viruses [110]; human-adapted viruses and avian viruses preferentially infected nonciliated cells and ciliated cells in the HAE, respectively [111, 112]. Clinically, seasonal influenza viruses mainly infect the upper respiratory tract [115]; however, pulmonary complications of influenza virus infection related to secondary bacterial pneumonia (such as by Staphylococcus aureus infection) rather than primary influenza pneumonia can occur, especially in children less than 2 years of age, adults more than 65 years of age, pregnant women, and people with comorbid illnesses/poor nutrition [115, 116]. The 2009 H1N1pdm viruses mostly attack the upper respiratory tract, resulting in subclinical infections or mild upper airway illness, but some are able to replicate in the lower respiratory tract as seen from diffuse alveolar damage in autopsy tissue samples from patients who died from the 2009 H1N1pdm virus. The viruses probably acquire D222G in HAs, leading to dual receptor specificity for α2-3- and α2-6-linked sialic acids [117], and more than 25 % of samples were co-infected with bacteria [118, 119]. Either HPAI H5 or H7 infection in terrestrial poultry spreads rapidly and causes damage throughout the avian body [120], but HPAI H5N1 infection in humans seems to be restricted to the respiratory tract and intestine, and H5N1 virus mainly replicates in pneumocytes, frequently causing death with acute respiratory distress syndrome (ARDS) (fatality rate of about 60 %) [121]. Either HPAI or LPAI H7 infection or LPAI H9, H10, and H6 infection in humans can result in disease in both ocular tissues that predominantly express α2-3-linked Sia receptors [122] and the respiratory tract with uncomplicated influenza-like illness [40, 41, 123, 124], but one veterinary doctor who was infected with HPAI H7N7 virus, in an outbreak in the Netherlands in 2003, died with ARDS [39] and approximately 32 % of people infected with a novel reassortant LPAI (H7N9) virus died from severe pneumonia and breathing difficulties (dyspnea) [42, 81, 125]. The first human case of avian influenza A (H10N8) virus has recently been detected in China in a 73-year-old immunocompromised female, who visited a live bird market and was hospitalized on November 30, 2013 and died of severe pneumonia on December 6, 2013 [44]. Not only epidemiologic surveillance but also molecular surveillance of influenza virus infection has become strengthened for rapid response to outbreaks of influenza virus having potentially unpredictable changes.
In general, influenza viruses evolve with changes in their environment, such as immune response and receptors, until achieving optimal viral fitness. Since 2002, some H5 and H7 poultry isolates, including A/Ck/Egypt/RIMD12-3/2008 (H5N1) of sublineage A [101], A/Tky/VA/4529/02 (H7N2), A/Ck/Conn/260413-2/03 (H7N2) of the North American lineage, and A/Laughing gull/DE/22/02 (H7N3) of the Eurasian lineage, have displayed significantly increased binding to α2-6 sialyl glycans [95, 126]. It should be noted that after the first outbreak of HPAI H5N1 virus in Egypt in 2006 [127], the virus has continued to undergo mutations, resulting in sublineages A–D; at present (2013), sublineages B and D are dominant in Egypt, while sublineage A has not been detected. Hemagglutination of an H9 human isolate, A/Hong Kong/1073/99 (H9N2), with guinea pig erythrocytes was shown to be inhibited by both α2-3 and α2-6-linked sialic acid containing polymers [128]. This characteristic of avian H5, H7, and H9 viruses highlights the possibility of the potential of these avian influenza viruses for development to infect and spread among humans in the future. Similar to H2 and H3 HAs of pandemic H2N2 in 1957 and H3N2 in 1968 (see Fig. 7e), Q226L and G228S/N224K mutations in H5 HA [129, 130], Q226L and G228S mutations in H7 HA [131], and Q226L mutation in H9 HA [132] have been experimentally shown to be associated with preferential binding to the α2-6 human-type receptor. Notably, the H5 virus harboring either Q226L-G228S [129] or Q226L-N224K [130] mutation in combination with loss of the 158–161 glycosylation site (N158D or T160A) near the receptor binding pocket and T318I or H107Y substitution in the stalk region (believed to increase the stability of the HA variant) has been shown to have preferential binding to Siaα2-6Gal, efficient respiratory droplet transmission in ferrets, and viral attachment to human tracheal epithelia. Nonetheless, another viral factor(s) has been believed to be involved for avian viruses to gain efficient human-to-human transmission (see Subheadings 6 and 7).
Pigs
Pigs serve as intermediate hosts for pandemic generation due to being mixing reservoirs of influenza A viruses, allowing genetic reassortment [133]. Indeed, interspecies transmission of avian and human viruses to pigs and vice versa has been documented in nature [134–136], and the recent pandemic H1N1 2009 has been confirmed to be of swine origin [137]. Lectin staining demonstrated high levels of α2-3 and α2-6 Sia expressed in the porcine respiratory epithelium [88, 133], and HPLC and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) analyses showed gradually increased molar ratios of α2-6/α2-3-linked sialyl glycans of 3.2-, 4.9-, and 13.2-fold for Neu5Ac and 1.8-, 2.7-, and 5.9-fold for Neu5Gc from the upper trachea and the lower trachea towards the lungs (the major replication site of swine-adapted influenza viruses) of a pig, respectively [77]. Neu5Ac/Neu5Gc ratios are 24.8/4.3 in the swine upper trachea, 27.1/4.1 in the swine lower trachea, 40.5/4.2 in the swine lung [77], and 98/2 in the duck intestine [89], whereas normal human tissues carrying nonfunctional hydroxylase to produce Neu5Gc [138] possess only Neu5Ac if Neu5Gc-containing food such as pork has not been eaten. Most duck-derived and swine-derived influenza A viruses displayed marked binding to Neu5Gc, related to the presence of V/I155 in H1 swine-adapted HAs [139], but they preserved preferential binding to Neu5Ac glycoconjugates, whereas human-adapted influenza viruses showed preferential binding to only Neu5Ac glycoconjugates [89, 140]. The swine-origin pandemic H1N1 2009 virus containing V155 rapidly spreads worldwide. Either T155Y or E158G mutation generated by a reverse genetics system in human H3 HA facilitates virus binding to Neu5Gc but retains strong binding affinity to Neu5Ac [141]. HAlo virus (A/Vietnam/1203/04 (H5N1) virus with removal of the multibasic cleavage site, responsible for high pathogenicity) with Y161A mutation generated by a reverse genetics system showed change of preferential binding from Neu5Ac to Neu5Gc with a five- to tenfold growth defect on MDCK cells [142]. It is still uncertain whether different ratios of Neu5Ac/Neu5Gc among animal species affect potential infection of influenza A viruses. Clearly, avian viruses with α2-3 binding preference would not overcome the interspecies barrier for efficient transmission in humans unless its binding preference is switched to α2-6. Thus, findings that classical swine influenza A viruses bind preferentially to Neu5Acα2-6Gal [88, 118, 143, 144] and that avian-like swine viruses acquired higher binding affinity for Neu5Acα2-6Gal over time [88, 118, 143] suggest that pigs provide a great source of natural selection of virus variants with α2-6 receptor-binding HAs, a prerequisite for a human pandemic.
Other Animals
Epithelial cells of the horse trachea showed prevalence of Siaα2-3Gal using lectin staining with Neu5Gc accounting for more than 90 % of Sia by HPLC analysis. Although most equine influenza viruses display high recognition of Neu5Gcα2-3Gal, they still prefer binding to Neu5Acα2-3Gal [82].
Seal and whale lung cells contain predominately Siaα2-3Gal over Siaα2-6Gal by lectin staining, and both seal and whale viruses prefer to recognize Siaα2-3Gal [145].
PB2
Changes in amino acid(s) in the RNA polymerase PB2 subunit resulting in different surface shape and/or charge affecting its protein’s interaction with cellular factors have been thought to contribute to efficient transmission of influenza viruses in humans, a characteristic of an influenza virus in a pandemic outbreak. T271A plays roles in (1) acquisition of HA mutation conferring recognition of a human-type receptor and (2) efficient respiratory droplet transmission [146, 147]. E627K/Q591R/D701N facilitates (3) efficient influenza virus replication in the upper respiratory tract of humans and (4) efficient influenza virus replication at 33 °C in the human upper part airway [102].
Other Influenza Virus Proteins
Changes in amino acids in other viral proteins, such as PA and NS1, interacting with cellular factors could contribute to the emergence of an influenza pandemic, and further studies are therefore needed to clarify viral factors involved in generation of a potential pandemic virus.
Concluding Remarks
Of the three types of influenza viruses, only type A can lead to a pandemic, possibly due to the variety of subtypes originating from wild water fowls that harmoniously interact with the virus in cooperation with the virus’s ability to cross the species barrier to infect a variety of animals (see Fig. 8). A virus crossing the species barrier to infect a new host species must experience a new environment in the host body including cellular receptors, host factors supporting/against virus replication, and local temperature, and thus is limited unless there is transmission evolution to surmount the species barrier. Of the influenza A viruses crossing into and establishing in terrestrial poultry, some of the LPAI H5 and H7 subtypes have evolved into HPAI viruses with a universal pathogenic marker of a multibasic cleavage site causing systemic infection with a mortality rate as high as 100 % in poultry [152]. It was virtually unknown what factors in poultry drive the virus to acquire an HPAI property and why the HPAI viruses have continued to circulate in poultry despite the fact that a rapid and high fatality rate due to the HPAI property could result in a dead end for virus transmission. These questions challenge researchers to unravel the ultimate selection parameter for survival of the fittest during virus-host co-evolution, each of which has evolved to prevail with the host attempting to escape from or restrict the infection by various means including host immunity and apoptosis and the virus evolving strategies to block/evade host clearance mechanisms and to support its propagation including entry to the host cell and use of the host cellular machinery for each step of its life cycle [153]. LPAI and HPAI viruses can sometimes infect other animals but with limited transmission, including wild birds [154], pigs [75], and humans [155], and cause mild to severe and fatal diseases (assessed by the number of severe cases and deaths) depending on the followings: (1) environmental factors, such as cold weather facilitating viral infection, (2) susceptibility and response of each host, which are accounted for by host genetics and other factors including age and health status, and (3) virus strain, with each virus strain having distinct pathogenic profiles in different host species: more research is needed to understand viral pathogenesis. For efficient transmission in humans, a nonhuman virus acquires mutations through either an adaptation or reassortment mechanism for sufficient human-to-human transmission, providing a chance for the virus to further evolve until it can achieve suitable interactions with host factors for efficient replication and transmission in human populations and eventually leading to a pandemic. Viral factors believed to play key roles in generating an influenza virus with pandemic potential are HA and PB2. Homotrimeric HA carries the followings: (1) antigenic sites, which if new, are not recognized by the host immune system, (2) receptor binding sites, which if they have 190D and 225D in H1 HA, 226L together with 228S (or 224K in H5 HA) in H2, H3, H5, or H7 HA, and 226L in H9 HA, are believed to confer virus preferential binding to a human-type receptor, (3) glycosylation sites, which if there is loss of glycosylation at 158–160, are believed to enhance HPAI H5N1 virus binding to a human-type receptor, and (4) stalk domains with T318A or H107Y, possibly being acquired for sustained and efficient human-to-human transmission of the H5 HA variants. Viral PB2 with 271A together with 627K/591R/701N enhances HA binding to human-type receptors, enhances respiratory droplet transmission, supports viral growth in a mammalian host at 33 °C, and facilitates efficient viral replication in the human upper respiratory tract. Investigation should be continued to identify other factors involved in efficient influenza virus replication and transmission in humans for use as viral genetic markers for early surveillance of the emergence of a pandemic.
Fig. 8.
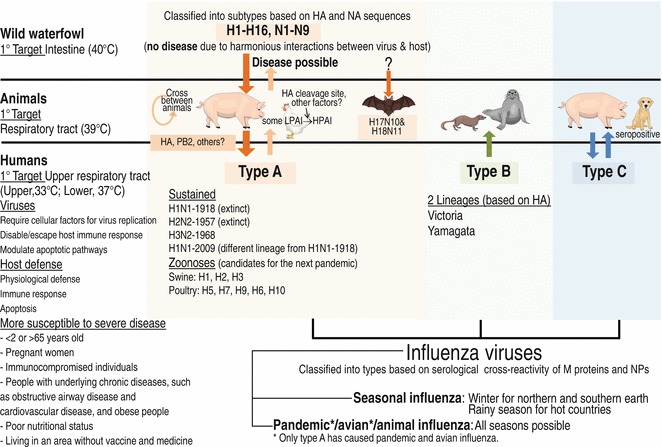
A summary of human infection with influenza virus and emergence of an influenza pandemic. Airborne influenza in humans is caused by type A, B, or C, prevalent in the rainy season in several tropical regions, such as Thailand, Vietnam, and Brazil, and in winter (November–April for the Northern Hemisphere and May–October for the Southern Hemisphere), due to virus stability and host vulnerability to infection. Types A and B cause annual epidemic influenza typically due to virus antigenic variation (minor change); generally, they do not cause severe disease except in people under conditions as indicated [115, 116]. Type C usually causes only mild illness in humans [148]. Type B is restricted to humans, though occasionally found in seals [149] and ferrets [150], type C can be isolated from humans and pigs and is found in seropositive dogs [151], and type A viruses have natural reservoirs in the intestinal tract (40 °C) of wild birds (harmonious interactions with the virus allowing the production of variety of subtypes without selective pressure) and are transmitted via feces to domestic birds (intestinal tract, 40 °C) and from domestic birds to other animals (respiratory tract of pigs, 39 °C) if they are able to fine-tune for transmission to and replication in the new host. So far, only H1N1 Spanish/18, H2N2 Japan/57, H3N2 Hong Kong/68, and H1N1 Swine/09 have been successfully established in humans. Currently only H3N2 Hong Kong/68 and H1N1/09 variants are circulating in humans. Epizootic influenza A viruses still cross to humans occasionally, and thus qualitative surveillance of the next pandemic zoonoses is needed. See color figure in the online version
Although there has been an accumulation of information on influenza; (1) new variants have emerged, (2) avian viruses have occasionally infected other animals including intermediate hosts carrying both avian-type and human-type receptors, such as pheasants, turkeys, quails, and guinea fowls, with pigs in particularly tending to drive the virus binding to α2-6 human-type receptors, and (3) an influenza pandemic is still unpredictable. HPAI H5N1 viruses have caused sporadic human infections since 1997, and the pandemic phase is currently at level 3 (see Table 2, small clusters of disease in people). A new LPAI H7N9 virus reassorted between avian viruses in poultry just crossed species to infect humans in February 2013 and is now (January 2014) at pandemic level 3. Except for the pandemic 1918 virus, past pandemics emerged from an existing human strain picking up new genes (new to human immunity but efficient replication in the human upper respiratory tract) from an avian and/or swine virus(es). This evidence suggested that new variants that have emerged via reassortment acquire major change in their genetic materials fitting with a new host condition more easily and faster than do variants that have emerged via point mutations alone, which have gradual changes in their genetic materials. Thus, avoiding intermediate host infection with more than one influenza virus strain should be important for preventing/delaying the next pandemic. More knowledge of the molecular requirements of reassortment at levels of viral and host factors could lead to a better understanding of how appropriate viruses emerge, leading to strategies for efficient prevention and antiviral interventions. Identifying viral and host factors, especially knowledge gained from their interaction structures, required for efficient replication in each host species may be a key for understanding virus–host determinants and surveillance of viral host jumps and pathogenesis of influenza virus infection leading to the disease.
Available data have suggested that an influenza pandemic has never emerged through direct viral mutations alone. However, highly mutable avian influenza A viruses that have sporadically continued direct transmission to and infection in humans have raised concerns for pandemic potential with unpredictable pathogenesis (depending on virus–host interactions). HPAI H5N1 viruses have continued to infect humans with high morbidity and mortality rates, some isolates showing increased binding to α2-6 human-type receptors, and they are able to infect a variety of animals including wild birds and pigeons, which are responsible for introduction of the viruses they carry into different areas, pigs, which are mixing vessels driving the virus to bind to human-type receptors, and cats, which are in close contact with human beings [156]. Also, novel reassortant LPAI (invisible disease in domestic poultry) H7N9 viruses contain some mammalian flu adaptations, PB2-627K and 226L, and they target upper and lower respiratory tracts of infected primates [157] and cause severe illness with a high death rate in humans. In addition to surveillance of human infection, extensive surveillance of infection of these viruses to other animals and back to migratory birds should be carried out since control of the viral spread into other regions relies on early recognition. Continuing surveillance is important for understanding how a pandemic emerges and establishing strategies for efficient control and treatment if a pandemic arises as well as for prevention and control of the next pandemic. The best way for preventing influenza spread and a pandemic is to avoid direct contact with materials having suspected contamination as well as hygiene in healthcare for both animals and farmers, especially in mixed duck–poultry–pig farms.
Contributor Information
Jun Hirabayashi, Phone: +81029-861-3124, Email: jun-hirabayashi@aist.go.jp.
Yasuo Suzuki, Email: suzukiy@isc.chubu.ac.jp.
References
- 1.Smith FI, Palese P. Variation in influenza virus genes: epidemiological, pathogenic, and evolutionary consequences. In: Krug RM, editor. The influenza viruses. New York: Plenum; 1989. pp. 319–359. [Google Scholar]
- 2.Tong S, Li Y, Rivailler P, et al. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci U S A. 2012;109:4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tong S, Zhu X, Li Y, et al. New world bats harbor diverse influenza a viruses. PLoS Pathog. 2013;9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun X, Shi Y, Lu X, et al. Bat-derived influenza hemagglutinin H17 does not bind canonical avian or human receptors and most likely uses a unique entry mechanism. Cell Rep. 2013;3:769–778. doi: 10.1016/j.celrep.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 5.Foeglein A, Loucaides EM, Mura M, et al. Influence of PB2 host-range determinants on the intranuclear mobility of the influenza A virus polymerase. J Gen Virol. 2011;92:1650–1661. doi: 10.1099/vir.0.031492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graef KM, Vreede FT, Lau YF, et al. The PB2 subunit of the influenza virus RNA polymerase affects virulence by interacting with the mitochondrial antiviral signaling protein and inhibiting expression of beta interferon. J Virol. 2010;84:8433–8445. doi: 10.1128/JVI.00879-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Portela A, Digard P. The influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication. J Gen Virol. 2002;83:723–734. doi: 10.1099/0022-1317-83-4-723. [DOI] [PubMed] [Google Scholar]
- 8.Tripathi S, Batra J, Cao W, et al. Influenza A virus nucleoprotein induces apoptosis in human airway epithelial cells: implications of a novel interaction between nucleoprotein and host protein Clusterin. Cell Death Dis. 2013;4:e562. doi: 10.1038/cddis.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin K, Helenius A. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell. 1991;67:117–130. doi: 10.1016/0092-8674(91)90576-k. [DOI] [PubMed] [Google Scholar]
- 10.Watanabe K, Handa H, Mizumoto K, et al. Mechanism for inhibition of influenza virus RNA polymerase activity by matrix protein. J Virol. 1996;70:241–247. doi: 10.1128/jvi.70.1.241-247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ali A, Avalos RT, Ponimaskin E, et al. Influenza virus assembly: effect of influenza virus glycoproteins on the membrane association of M1 protein. J Virol. 2000;74:8709–8719. doi: 10.1128/jvi.74.18.8709-8719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zamarin D, Garcia-Sastre A, Xiao X, et al. Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PLoS Pathog. 2005;1:e4. doi: 10.1371/journal.ppat.0010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varga ZT, Grant A, Manicassamy B, et al. Influenza virus protein PB1-F2 inhibits the induction of type I interferon by binding to MAVS and decreasing mitochondrial membrane potential. J Virol. 2012;86:8359–8366. doi: 10.1128/JVI.01122-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wise HM, Foeglein A, Sun J, et al. A complicated message: Identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J Virol. 2009;83:8021–8031. doi: 10.1128/JVI.00826-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hale BG, Randall RE, Ortin J, et al. The multifunctional NS1 protein of influenza A viruses. J Gen Virol. 2008;89:2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- 16.Tsai PL, Chiou NT, Kuss S, et al. Cellular RNA binding proteins NS1-BP and hnRNP K regulate influenza A virus RNA splicing. PLoS Pathog. 2013;9:e1003460. doi: 10.1371/journal.ppat.1003460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiao P, Tian G, Li Y, et al. A single-amino-acid substitution in the NS1 protein changes the pathogenicity of H5N1 avian influenza viruses in mice. J Virol. 2008;82:1146–1154. doi: 10.1128/JVI.01698-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhirnov OP, Klenk HD. Control of apoptosis in influenza virus-infected cells by up-regulation of Akt and p53 signaling. Apoptosis. 2007;12:1419–1432. doi: 10.1007/s10495-007-0071-y. [DOI] [PubMed] [Google Scholar]
- 19.Selman M, Dankar SK, Forbes NE, et al. Adaptive mutation in influenza A virus non-structural gene is linked to host switching and induces a novel protein by alternative splicing. Emerg Microbes Infect. 2012;1:e42. doi: 10.1038/emi.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Sastre A. Induction and evasion of type I interferon responses by influenza viruses. Virus Res. 2011;162:12–18. doi: 10.1016/j.virusres.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobasa D, Kodihalli S, Luo M, et al. Amino acid residues contributing to the substrate specificity of the influenza A virus neuraminidase. J Virol. 1999;73:6743–6751. doi: 10.1128/jvi.73.8.6743-6751.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matrosovich MN, Matrosovich TY, Gray T, et al. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J Virol. 2004;78:12665–12667. doi: 10.1128/JVI.78.22.12665-12667.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmolke M, Garcia-Sastre A. Evasion of innate and adaptive immune responses by influenza A virus. Cell Microbiol. 2010;12:873–880. doi: 10.1111/j.1462-5822.2010.01475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rimmelzwaan GF, Berkhoff EG, Nieuwkoop NJ, et al. Functional compensation of a detrimental amino acid substitution in a cytotoxic-T-lymphocyte epitope of influenza a viruses by comutations. J Virol. 2004;78:8946–8949. doi: 10.1128/JVI.78.16.8946-8949.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chakrabarti AK, Pasricha G. An insight into the PB1F2 protein and its multifunctional role in enhancing the pathogenicity of the influenza A viruses. Virology. 2013;440:97–104. doi: 10.1016/j.virol.2013.02.025. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki T, Takahashi T, Guo CT, et al. Sialidase activity of influenza A virus in an endocytic pathway enhances viral replication. J Virol. 2005;79:11705–11715. doi: 10.1128/JVI.79.18.11705-11715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su B, Wurtzer S, Rameix-Welti MA, et al. Enhancement of the influenza A hemagglutinin (HA)-mediated cell-cell fusion and virus entry by the viral neuraminidase (NA) PLoS One. 2009;4:e8495. doi: 10.1371/journal.pone.0008495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeda M, Leser GP, Russell CJ, et al. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc Natl Acad Sci U S A. 2003;100:14610–14617. doi: 10.1073/pnas.2235620100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, Pekosz A, Lamb RA. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J Virol. 2000;74:4634–4644. doi: 10.1128/jvi.74.10.4634-4644.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barry JM. The great influenza: the epic story of the deadliest plague in history. New York: Viking; 2004. [Google Scholar]
- 32.Ito T, Kawaoka Y. Host-range barrier of influenza A viruses. Vet Microbiol. 2000;74:71–75. doi: 10.1016/s0378-1135(00)00167-x. [DOI] [PubMed] [Google Scholar]
- 33.Webster RG, Yakhno M, Hinshaw VS, et al. Intestinal influenza: replication and characterization of influenza viruses in ducks. Virology. 1978;84:268–278. doi: 10.1016/0042-6822(78)90247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.WHO website. Current WHO phase of pandemic alert for Pandemic (H1N1) 2009. http://www.who.int/csr/disease/swineflu/phase/en/. Accessed 26 Nov 2013
- 35.Holmes EC, Ghedin E, Miller N, et al. Whole-genome analysis of human influenza A virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses. PLoS Biol. 2005;3:e300. doi: 10.1371/journal.pbio.0030300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.CDC website. Avian influenza A virus infections of humans. http://www.cdc.gov/flu/avian/gen-info/avian-flu-humans.htm. Accessed 26 Nov 26 2013
- 37.Claas EC, Osterhaus AD, van Beek R, et al. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 38.Subbarao K, Klimov A, Katz J, et al. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 39.Belser JA, Zeng H, Katz JM, et al. Infection with highly pathogenic H7 influenza viruses results in an attenuated proinflammatory cytokine and chemokine response early after infection. J Infect Dis. 2011;203:40–48. doi: 10.1093/infdis/jiq018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirst M, Astell CR, Griffith M, et al. Novel avian influenza H7N3 strain outbreak, British Columbia. Emerg Infect Dis. 2004;10:2192–2195. doi: 10.3201/eid1012.040743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arzey GG, Kirkland PD, Arzey KE, et al. Influenza virus A (H10N7) in chickens and poultry abattoir workers, Australia. Emerg Infect Dis. 2012;18:814–816. doi: 10.3201/eid1805.111852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao R, Cao B, Hu Y, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 43.Yuan J, Zhang L, Kan X, et al. Origin and molecular characteristics of a novel 2013 avian influenza A(H6N1) virus causing human infection in Taiwan. Clin Infect Dis. 2013;57:1367–1368. doi: 10.1093/cid/cit479. [DOI] [PubMed] [Google Scholar]
- 44.WHO website. Emerging disease surveillance and response (avian influenza). http://www.wpro.who.int/emerging_diseases/AvianInfluenza/en/. Accessed 9 Jan 2014
- 45.Horimoto T, Kawaoka Y. Influenza: lessons from past pandemics, warnings from current incidents. Nat Rev Microbiol. 2005;3:591–600. doi: 10.1038/nrmicro1208. [DOI] [PubMed] [Google Scholar]
- 46.Palese P. Influenza: old and new threats. Nat Med. 2004;10:S82–S87. doi: 10.1038/nm1141. [DOI] [PubMed] [Google Scholar]
- 47.Pica N, Hai R, Krammer F, et al. Hemagglutinin stalk antibodies elicited by the 2009 pandemic influenza virus as a mechanism for the extinction of seasonal H1N1 viruses. Proc Natl Acad Sci U S A. 2012;109:2573–2578. doi: 10.1073/pnas.1200039109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simonsen L, Clarke MJ, Schonberger LB, et al. Pandemic versus epidemic influenza mortality: a pattern of changing age distribution. J Infect Dis. 1998;178:53–60. doi: 10.1086/515616. [DOI] [PubMed] [Google Scholar]
- 49.Dawood FS, Jain S, Finelli L, et al. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360:2605–2615. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- 50.Xu R, Ekiert DC, Krause JC, et al. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328:357–360. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sriwilaijaroen N, Suzuki Y. Molecular basis of the structure and function of H1 hemagglutinin of influenza virus. Proc Jpn Acad Ser B Phys Biol Sci. 2012;88:226–249. doi: 10.2183/pjab.88.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manicassamy B, Medina RA, Hai R, et al. Protection of mice against lethal challenge with 2009 H1N1 influenza A virus by 1918-like and classical swine H1N1 based vaccines. PLoS Pathog. 2010;6:e1000745. doi: 10.1371/journal.ppat.1000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cohen J. Swine flu pandemic. What's old is new: 1918 virus matches 2009 H1N1 strain. Science. 2010;327:1563–1564. doi: 10.1126/science.327.5973.1563. [DOI] [PubMed] [Google Scholar]
- 54.Russell RJ, Kerry PS, Stevens DJ, et al. Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion. Proc Natl Acad Sci U S A. 2008;105:17736–17741. doi: 10.1073/pnas.0807142105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sauter NK, Glick GD, Crowther RL, et al. Crystallographic detection of a second ligand binding site in influenza virus hemagglutinin. Proc Natl Acad Sci U S A. 1992;89:324–328. doi: 10.1073/pnas.89.1.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raymond FL, Caton AJ, Cox NJ, et al. The antigenicity and evolution of influenza H1 haemagglutinin, from 1950–1957 and 1977–1983: two pathways from one gene. Virology. 1986;148:275–287. doi: 10.1016/0042-6822(86)90325-9. [DOI] [PubMed] [Google Scholar]
- 57.Stray SJ, Pittman LB. Subtype- and antigenic site-specific differences in biophysical influences on evolution of influenza virus hemagglutinin. Virol J. 2012;9:91. doi: 10.1186/1743-422X-9-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wiley DC, Wilson IA, Skehel JJ. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 59.Wilson IA, Cox NJ. Structural basis of immune recognition of influenza virus hemagglutinin. Annu Rev Immunol. 1990;8:737–771. doi: 10.1146/annurev.iy.08.040190.003513. [DOI] [PubMed] [Google Scholar]
- 60.Kawaoka Y, Bean WJ, Webster RG. Evolution of the hemagglutinin of equine H3 influenza viruses. Virology. 1989;169:283–292. doi: 10.1016/0042-6822(89)90153-0. [DOI] [PubMed] [Google Scholar]
- 61.Luoh SM, McGregor MW, Hinshaw VS. Hemagglutinin mutations related to antigenic variation in H1 swine influenza viruses. J Virol. 1992;66:1066–1073. doi: 10.1128/jvi.66.2.1066-1073.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sakai K, Kawaguchi Y, Kishino Y, et al. Electron immunohistochemical localization in rat bronchiolar epithelial cells of tryptase Clara, which determines the pneumotropism and pathogenicity of Sendai virus and influenza virus. J Histochem Cytochem. 1993;41:89–93. doi: 10.1177/41.1.8380186. [DOI] [PubMed] [Google Scholar]
- 63.Webster RG, Bean WJ, Gorman OT, et al. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Woo GH, Kim HY, Bae YC, et al. Comparative histopathological characteristics of highly pathogenic avian influenza (HPAI) in chickens and domestic ducks in 2008 Korea. Histol Histopathol. 2011;26:167–175. doi: 10.14670/HH-26.167. [DOI] [PubMed] [Google Scholar]
- 65.Banks J, Speidel ES, Moore E, et al. Changes in the haemagglutinin and the neuraminidase genes prior to the emergence of highly pathogenic H7N1 avian influenza viruses in Italy. Arch Virol. 2001;146:963–973. doi: 10.1007/s007050170128. [DOI] [PubMed] [Google Scholar]
- 66.Webster RG, Rott R. Influenza virus A pathogenicity: the pivotal role of hemagglutinin. Cell. 1987;50:665–666. doi: 10.1016/0092-8674(87)90321-7. [DOI] [PubMed] [Google Scholar]
- 67.Ito T, Goto H, Yamamoto E, et al. Generation of a highly pathogenic avian influenza A virus from an avirulent field isolate by passaging in chickens. J Virol. 2001;75:4439–4443. doi: 10.1128/JVI.75.9.4439-4443.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perdue ML, Garcia M, Senne D, et al. Virulence-associated sequence duplication at the hemagglutinin cleavage site of avian influenza viruses. Virus Res. 1997;49:173–186. doi: 10.1016/s0168-1702(97)01468-8. [DOI] [PubMed] [Google Scholar]
- 69.Abdel-Ghafar AN, Chotpitayasunondh T, Gao Z, et al. Update on avian influenza A (H5N1) virus infection in humans. N Engl J Med. 2008;358:261–273. doi: 10.1056/NEJMra0707279. [DOI] [PubMed] [Google Scholar]
- 70.WHO website. Updated unified nomenclature system for the highly pathogenic H5N1 avian influenza viruses. http://www.who.int/influenza/gisrs_laboratory/h5n1_nomenclature/en/. Accessed 1 Dec 1 2013
- 71.Keawcharoen J, van Riel D, van Amerongen G, et al. Wild ducks as long-distance vectors of highly pathogenic avian influenza virus (H5N1) Emerg Infect Dis. 2008;14:600–607. doi: 10.3201/eid1404.071016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brown JD, Stallknecht DE, Beck JR, et al. Susceptibility of North American ducks and gulls to H5N1 highly pathogenic avian influenza viruses. Emerg Infect Dis. 2006;12:1663–1670. doi: 10.3201/eid1211.060652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim JK, Negovetich NJ, Forrest HL, et al. Ducks: the "Trojan horses" of H5N1 influenza. Influenza Other Respir Viruses. 2009;3:121–128. doi: 10.1111/j.1750-2659.2009.00084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cui Z, Hu J, He L et al (2013) Differential immune response of mallard duck peripheral blood mononuclear cells to two highly pathogenic avian influenza H5N1 viruses with distinct pathogenicity in mallard ducks. Arch Virol 159:339–343 [DOI] [PubMed]
- 75.Chen H, Smith GJ, Zhang SY, et al. Avian flu: H5N1 virus outbreak in migratory waterfowl. Nature. 2005;436:191–192. doi: 10.1038/nature03974. [DOI] [PubMed] [Google Scholar]
- 76.Nidom CA, Takano R, Yamada S, et al. Influenza A (H5N1) viruses from pigs, Indonesia. Emerg Infect Dis. 2010;16:1515–1523. doi: 10.3201/eid1610.100508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sriwilaijaroen N, Kondo S, Yagi H, et al. N-Glycans from porcine trachea and lung: predominant NeuAcα2-6Gal could be a selective pressure for influenza variants in favor of human-type receptor. PLoS One. 2011;6:e16302. doi: 10.1371/journal.pone.0016302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vijaykrishna D, Bahl J, Riley S, et al. Evolutionary dynamics and emergence of panzootic H5N1 influenza viruses. PLoS Pathog. 2008;4:e1000161. doi: 10.1371/journal.ppat.1000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.CDC website. Highly pathogenic avian influenza A (H5N1) in birds and other animals. http://www.cdc.gov/flu/avianflu/h5n1-animals.htm. Accessed 26 Nov 2013
- 80.WHO website. Current WHO phase of pandemic alert for avian influenza H5N1. http://apps.who.int/csr/disease/avian_influenza/phase/en/index.html. Accessed 26 Nov 2013
- 81.WHO website. Human infection with avian influenza A(H7N9) virus – update. http://www.who.int/csr/don/2013_11_06/en/index.html?utm_source=twitterfeed&utm_medium=twitter. Accessed 26 Nov 2013
- 82.Suzuki Y, Ito T, Suzuki T, et al. Sialic acid species as a determinant of the host range of influenza A viruses. J Virol. 2000;74:11825–11831. doi: 10.1128/jvi.74.24.11825-11831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang B, Brand-Miller J. The role and potential of sialic acid in human nutrition. Eur J Clin Nutr. 2003;57:1351–1369. doi: 10.1038/sj.ejcn.1601704. [DOI] [PubMed] [Google Scholar]
- 84.Suzuki Y. Sialobiology of influenza: molecular mechanism of host range variation of influenza viruses. Biol Pharm Bull. 2005;28:399–408. doi: 10.1248/bpb.28.399. [DOI] [PubMed] [Google Scholar]
- 85.Paulson JC, de Vries RP. H5N1 receptor specificity as a factor in pandemic risk. Virus Res. 2013;178:99–113. doi: 10.1016/j.virusres.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gilbert M, Xiao X, Domenech J, et al. Anatidae migration in the western Palearctic and spread of highly pathogenic avian influenza H5N1 virus. Emerg Infect Dis. 2006;12:1650–1656. doi: 10.3201/eid1211.060223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kuchipudi SV, Nelli R, White GA, et al. Differences in influenza virus receptors in chickens and ducks: implications for interspecies transmission. J Mol Genet Med. 2009;3:143–151. doi: 10.4172/1747-0862.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ito T, Couceiro JN, Kelm S, et al. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol. 1998;72:7367–7373. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ito T, Suzuki Y, Suzuki T, et al. Recognition of N-glycolylneuraminic acid linked to galactose by the α2,3 linkage is associated with intestinal replication of influenza A virus in ducks. J Virol. 2000;74:9300–9305. doi: 10.1128/jvi.74.19.9300-9305.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gambaryan A, Yamnikova S, Lvov D, et al. Receptor specificity of influenza viruses from birds and mammals: new data on involvement of the inner fragments of the carbohydrate chain. Virology. 2005;334:276–283. doi: 10.1016/j.virol.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 91.Masuda H, Suzuki T, Sugiyama Y, et al. Substitution of amino acid residue in influenza A virus hemagglutinin affects recognition of sialyl-oligosaccharides containing N-glycolylneuraminic acid. FEBS Lett. 1999;464:71–74. doi: 10.1016/s0014-5793(99)01575-6. [DOI] [PubMed] [Google Scholar]
- 92.Chu VC, Whittaker GR. Influenza virus entry and infection require host cell N-linked glycoprotein. Proc Natl Acad Sci U S A. 2004;101:18153–18158. doi: 10.1073/pnas.0405172102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sriwilaijaroen N, Kondo S, Yagi H, et al. Analysis of N-glycans in embryonated chicken egg chorioallantoic and amniotic cells responsible for binding and adaptation of human and avian influenza viruses. Glycoconj J. 2009;26:433–443. doi: 10.1007/s10719-008-9193-x. [DOI] [PubMed] [Google Scholar]
- 94.Gambaryan A, Tuzikov A, Pazynina G, et al. Evolution of the receptor binding phenotype of influenza A (H5) viruses. Virology. 2006;344:432–438. doi: 10.1016/j.virol.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 95.Gambaryan AS, Matrosovich TY, Philipp J, et al. Receptor-binding profiles of H7 subtype influenza viruses in different host species. J Virol. 2012;86:4370–4379. doi: 10.1128/JVI.06959-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kawaoka Y, Chambers TM, Sladen WL, et al. Is the gene pool of influenza viruses in shorebirds and gulls different from that in wild ducks? Virology. 1988;163:247–250. doi: 10.1016/0042-6822(88)90260-7. [DOI] [PubMed] [Google Scholar]
- 97.Senne DA. Avian influenza in the Western Hemisphere including the Pacific Islands and Australia. Avian Dis. 2003;47:798–805. doi: 10.1637/0005-2086-47.s3.798. [DOI] [PubMed] [Google Scholar]
- 98.Chen H, Bu Z, Wang J. Epidemiology and control of H5N1 avian influenza in China. In: Klenk H-D, Matrosovich MN, Stech J, editors. Avian influenza (monographs in virology) Basel: Karger; 2008. pp. 27–40. [Google Scholar]
- 99.Lu X, Qi J, Shi Y, et al. Structure and receptor binding specificity of hemagglutinin H13 from avian influenza A virus H13N6. J Virol. 2013;87:9077–9085. doi: 10.1128/JVI.00235-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Petersen H, Matrosovich M, Pleschka S, et al. Replication and adaptive mutations of low pathogenic avian influenza viruses in tracheal organ cultures of different avian species. PLoS One. 2012;7:e42260. doi: 10.1371/journal.pone.0042260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Watanabe Y, Ibrahim MS, Ellakany HF, et al. Acquisition of human-type receptor binding specificity by new H5N1 influenza virus sublineages during their emergence in birds in Egypt. PLoS Pathog. 2011;7:e1002068. doi: 10.1371/journal.ppat.1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yamada S, Hatta M, Staker BL, et al. Biological and structural characterization of a host-adapting amino acid in influenza virus. PLoS Pathog. 2010;6:e1001034. doi: 10.1371/journal.ppat.1001034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gambaryan A, Webster R, Matrosovich M. Differences between influenza virus receptors on target cells of duck and chicken. Arch Virol. 2002;147:1197–1208. doi: 10.1007/s00705-002-0796-4. [DOI] [PubMed] [Google Scholar]
- 104.Gambaryan AS, Tuzikov AB, Bovin NV, et al. Differences between influenza virus receptors on target cells of duck and chicken and receptor specificity of the 1997 H5N1 chicken and human influenza viruses from Hong Kong. Avian Dis. 2003;47:1154–1160. doi: 10.1637/0005-2086-47.s3.1154. [DOI] [PubMed] [Google Scholar]
- 105.Guo CT, Takahashi N, Yagi H, et al. The quail and chicken intestine have sialyl-galactose sugar chains responsible for the binding of influenza A viruses to human type receptors. Glycobiology. 2007;17:713–724. doi: 10.1093/glycob/cwm038. [DOI] [PubMed] [Google Scholar]
- 106.Pillai SP, Lee CW. Species and age related differences in the type and distribution of influenza virus receptors in different tissues of chickens, ducks and turkeys. Virol J. 2010;7:5. doi: 10.1186/1743-422X-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wan H, Perez DR. Quail carry sialic acid receptors compatible with binding of avian and human influenza viruses. Virology. 2006;346:278–286. doi: 10.1016/j.virol.2005.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu Y, Han C, Wang X, et al. Influenza A virus receptors in the respiratory and intestinal tracts of pigeons. Avian Pathol. 2009;38:263–266. doi: 10.1080/03079450903055363. [DOI] [PubMed] [Google Scholar]
- 109.Kim JA, Ryu SY, Seo SH. Cells in the respiratory and intestinal tracts of chickens have different proportions of both human and avian influenza virus receptors. J Microbiol. 2005;43:366–369. [PubMed] [Google Scholar]
- 110.Shinya K, Ebina M, Yamada S, et al. Avian flu: influenza virus receptors in the human airway. Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 111.Matrosovich MN, Matrosovich TY, Gray T, et al. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc Natl Acad Sci U S A. 2004;101:4620–4624. doi: 10.1073/pnas.0308001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Thompson CI, Barclay WS, Zambon MC, et al. Infection of human airway epithelium by human and avian strains of influenza a virus. J Virol. 2006;80:8060–8068. doi: 10.1128/JVI.00384-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.van Riel D, Munster VJ, de Wit E, et al. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am J Pathol. 2007;171:1215–1223. doi: 10.2353/ajpath.2007.070248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Walther T, Karamanska R, Chan RW, et al. Glycomic analysis of human respiratory tract tissues and correlation with influenza virus infection. PLoS Pathog. 2013;9:e1003223. doi: 10.1371/journal.ppat.1003223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bouvier NM, Lowen AC. Animal models for influenza virus pathogenesis and transmission. Viruses. 2010;2:1530–1563. doi: 10.3390/v20801530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rothberg MB, Haessler SD, Brown RB. Complications of viral influenza. Am J Med. 2008;121:258–264. doi: 10.1016/j.amjmed.2007.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chutinimitkul S, Herfst S, Steel J, et al. Virulence-associated substitution D222G in the hemagglutinin of 2009 pandemic influenza A(H1N1) virus affects receptor binding. J Virol. 2010;84:11802–11813. doi: 10.1128/JVI.01136-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Childs RA, Palma AS, Wharton S, et al. Receptor-binding specificity of pandemic influenza A (H1N1) 2009 virus determined by carbohydrate microarray. Nat Biotechnol. 2009;27:797–799. doi: 10.1038/nbt0909-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shieh WJ, Blau DM, Denison AM, et al. 2009 Pandemic influenza A (H1N1): pathology and pathogenesis of 100 fatal cases in the United States. Am J Pathol. 2010;177:166–175. doi: 10.2353/ajpath.2010.100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Peiris JS, de Jong MD, Guan Y. Avian influenza virus (H5N1): a threat to human health. Clin Microbiol Rev. 2007;20:243–267. doi: 10.1128/CMR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Uiprasertkul M, Puthavathana P, Sangsiriwut K, et al. Influenza A H5N1 replication sites in humans. Emerg Infect Dis. 2005;11:1036–1041. doi: 10.3201/eid1107.041313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Olofsson S, Kumlin U, Dimock K, et al. Avian influenza and sialic acid receptors: more than meets the eye? Lancet Infect Dis. 2005;5:184–188. doi: 10.1016/S1473-3099(05)01311-3. [DOI] [PubMed] [Google Scholar]
- 123.Nguyen-Van-Tam JS, Nair P, Acheson P, et al. Outbreak of low pathogenicity H7N3 avian influenza in UK, including associated case of human conjunctivitis. Euro Surveill. 2006;11:E060504.2. doi: 10.2807/esw.11.18.02952-en. [DOI] [PubMed] [Google Scholar]
- 124.Editorialteam (2007) Avian influenza A/(H7N2) outbreak in the United Kingdom. Euro Surveill 12:E070531.2 [PubMed]
- 125.Morens DM, Taubenberger JK, Fauci AS. Pandemic influenza viruses—hoping for the road not taken. N Engl J Med. 2013;368:2345–2348. doi: 10.1056/NEJMp1307009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Belser JA, Blixt O, Chen LM, et al. Contemporary North American influenza H7 viruses possess human receptor specificity: Implications for virus transmissibility. Proc Natl Acad Sci U S A. 2008;105:7558–7563. doi: 10.1073/pnas.0801259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Balicer RD, Reznikovich S, Berman E, et al. Multifocal avian influenza (H5N1) outbreak. Emerg Infect Dis. 2007;13:1601–1603. doi: 10.3201/eid1310.070558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Saito T, Lim W, Suzuki T, et al. Characterization of a human H9N2 influenza virus isolated in Hong Kong. Vaccine. 2001;20:125–133. doi: 10.1016/s0264-410x(01)00279-1. [DOI] [PubMed] [Google Scholar]
- 129.Herfst S, Schrauwen EJ, Linster M, et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Imai M, Watanabe T, Hatta M, et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Srinivasan K, Raman R, Jayaraman A, et al. Quantitative description of glycan-receptor binding of influenza A virus H7 hemagglutinin. PLoS One. 2013;8:e49597. doi: 10.1371/journal.pone.0049597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wan H, Perez DR. Amino acid 226 in the hemagglutinin of H9N2 influenza viruses determines cell tropism and replication in human airway epithelial cells. J Virol. 2007;81:5181–5191. doi: 10.1128/JVI.02827-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kida H, Ito T, Yasuda J, et al. Potential for transmission of avian influenza viruses to pigs. J Gen Virol. 1994;75(Pt 9):2183–2188. doi: 10.1099/0022-1317-75-9-2183. [DOI] [PubMed] [Google Scholar]
- 134.Brown IH. The epidemiology and evolution of influenza viruses in pigs. Vet Microbiol. 2000;74:29–46. doi: 10.1016/s0378-1135(00)00164-4. [DOI] [PubMed] [Google Scholar]
- 135.Pensaert M, Ottis K, Vandeputte J, et al. Evidence for the natural transmission of influenza A virus from wild ducts to swine and its potential importance for man. Bull World Health Organ. 1981;59:75–78. [PMC free article] [PubMed] [Google Scholar]
- 136.Ottis K, Sidoli L, Bachmann PA, et al. Human influenza A viruses in pigs: isolation of a H3N2 strain antigenically related to A/England/42/72 and evidence for continuous circulation of human viruses in the pig population. Arch Virol. 1982;73:103–108. doi: 10.1007/BF01314719. [DOI] [PubMed] [Google Scholar]
- 137.Smith GJ, Vijaykrishna D, Bahl J, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459:1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 138.Irie A, Koyama S, Kozutsumi Y, et al. The molecular basis for the absence of N-glycolylneuraminic acid in humans. J Biol Chem. 1998;273:15866–15871. doi: 10.1074/jbc.273.25.15866. [DOI] [PubMed] [Google Scholar]
- 139.Matrosovich MN, Klenk HD, Kawaoka Y. Receptor specificity, host-range, and pathogenicity of influenza viruses. In: Kawaoka Y, editor. Influenza virology: current topics. Wymondham: Caister Academic Press; 2006. pp. 95–137. [Google Scholar]
- 140.Suzuki T, Horiike G, Yamazaki Y, et al. Swine influenza virus strains recognize sialylsugar chains containing the molecular species of sialic acid predominantly present in the swine tracheal epithelium. FEBS Lett. 1997;404:192–196. doi: 10.1016/s0014-5793(97)00127-0. [DOI] [PubMed] [Google Scholar]
- 141.Takahashi T, Hashimoto A, Maruyama M, et al. Identification of amino acid residues of influenza A virus H3 HA contributing to the recognition of molecular species of sialic acid. FEBS Lett. 2009;583:3171–3174. doi: 10.1016/j.febslet.2009.08.037. [DOI] [PubMed] [Google Scholar]
- 142.Wang M, Tscherne DM, McCullough C, et al. Residue Y161 of influenza virus hemagglutinin is involved in viral recognition of sialylated complexes from different hosts. J Virol. 2012;86:4455–4462. doi: 10.1128/JVI.07187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Matrosovich M, Tuzikov A, Bovin N, et al. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol. 2000;74:8502–8512. doi: 10.1128/jvi.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gambaryan AS, Karasin AI, Tuzikov AB, et al. Receptor-binding properties of swine influenza viruses isolated and propagated in MDCK cells. Virus Res. 2005;114:15–22. doi: 10.1016/j.virusres.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 145.Ito T, Kawaoka Y, Nomura A, et al. Receptor specificity of influenza A viruses from sea mammals correlates with lung sialyloligosaccharides in these animals. J Vet Med Sci. 1999;61:955–958. doi: 10.1292/jvms.61.955. [DOI] [PubMed] [Google Scholar]
- 146.Bussey KA, Bousse TL, Desmet EA, et al. PB2 residue 271 plays a key role in enhanced polymerase activity of influenza A viruses in mammalian host cells. J Virol. 2010;84:4395–4406. doi: 10.1128/JVI.02642-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhang Y, Zhang Q, Gao Y, et al. Key molecular factors in hemagglutinin and PB2 contribute to efficient transmission of the 2009 H1N1 pandemic influenza virus. J Virol. 2012;86:9666–9674. doi: 10.1128/JVI.00958-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kauppila J, Ronkko E, Juvonen R et al (2013) Influenza C virus infection in military recruits-symptoms and clinical manifestation. J Med Virol 86:879–885 [DOI] [PubMed]
- 149.Bodewes R, Morick D, de Mutsert G, et al. Recurring influenza B virus infections in seals. Emerg Infect Dis. 2013;19:511–512. doi: 10.3201/eid1903.120965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Francis T., Jr A new type of virus from epidemic influenza. Science. 1940;92:405–408. doi: 10.1126/science.92.2392.405. [DOI] [PubMed] [Google Scholar]
- 151.Crescenzo-Chaigne B, van der Werf S. Rescue of influenza C virus from recombinant DNA. J Virol. 2007;81:11282–11289. doi: 10.1128/JVI.00910-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Alexander DJ. A review of avian influenza in different bird species. Vet Microbiol. 2000;74:3–13. doi: 10.1016/s0378-1135(00)00160-7. [DOI] [PubMed] [Google Scholar]
- 153.Konstantinov K, Foisner R, Byrd D, et al. Integral membrane proteins associated with the nuclear lamina are novel autoimmune antigens of the nuclear envelope. Clin Immunol Immunopathol. 1995;74:89–99. doi: 10.1006/clin.1995.1013. [DOI] [PubMed] [Google Scholar]
- 154.Kim HR, Lee YJ, Park CK, et al. Highly pathogenic avian influenza (H5N1) outbreaks in wild birds and poultry, South Korea. Emerg Infect Dis. 2012;18:480–483. doi: 10.3201/eid1803.111490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Peiris JS. Avian influenza viruses in humans. Rev Sci Tech. 2009;28:161–173. doi: 10.20506/rst.28.1.1871. [DOI] [PubMed] [Google Scholar]
- 156.WHO website. Influenza at the human-animal interface (HAI). http://www.who.int/influenza/human_animal_interface/en/. Accessed 28 Nov 2013
- 157.Watanabe T, Kiso M, Fukuyama S, et al. Characterization of H7N9 influenza A viruses isolated from humans. Nature. 2013;501:551–555. doi: 10.1038/nature12392. [DOI] [PMC free article] [PubMed] [Google Scholar]


