Abstract
Protein histochemistry is a tissue-based technique that enables the analysis of viral attachment patterns as well as the identification of specific viral and host determinants involved in the first step in the infection of a host cell by a virus. Applying recombinantly expressed spike proteins of infectious bronchitis virus onto formalin-fixed tissues allows us to profile the binding characteristics of these viral attachment proteins to tissues of various avian species. In particular, sialic acid-mediated tissue binding of spike proteins can be analyzed by pretreating tissues with various neuraminidases or by blocking the binding of the viral proteins with specific lectins. Our assay is particularly convenient to elucidate critical virus–host interactions for viruses for which infection models are limited.
Key words: Protein histochemistry, Spike protein, Neuraminidase, Lectin, Formalin-fixed tissues, Infectious bronchitis virus (IBV), Attachment, Glycan
Introduction
Infectious bronchitis virus (IBV), an avian coronavirus belonging to the genus Gammacoronavirus, is the major cause of contagious respiratory disease or infectious bronchitis in poultry. Many IBV serotypes have been isolated so far and some serotypes induce pathological changes in organs other than the respiratory tissues [1]. This variable tissue tropism is likely due to tissue-specific factors resulting in differences in binding or entry of the virus. Although a specific protein receptor for IBV is yet to be revealed it has been shown by removing sialic acids from the susceptible cell surface, that α2, 3-linked sialic acids are a determinant of cell attachment and entry of IBV [2, 3]. Further elucidation of host–virus interactions is, however, hampered due to limitations in in vitro infection model systems for pathogenic IBV strains.
For IBV the initial cell attachment and entry is mediated by a glycoprotein called spike protein residing in the viral envelope. By swapping the gene encoding for spike protein between different IBV serotypes it has been shown that the spike determines the tissue tropism [4]. The spike protein is cleaved into an S1 and an S2 subunit [5, 6]; while S1 mediates the first step in infection via the initial virus–cell binding, S2 is responsible for cell entry [7]. Analyzing the binding of S1 to tissues with our protein histochemistry protocol enables us not only to profile the attachment of avian coronavirus S1 proteins to various avian tissues but also to elucidate glycan binding specificities of IBV S1 [8] as well as determinants within S1 for tissue attachment [9]. Thereby, this method aids to understand the in vivo tissue tropism of avian coronaviruses.
Materials
The amounts of buffers or chemicals prepared are described such to result in a convenient volume. Any other required volume can be calculated from this.
Components for Expression of Spike Protein (S1) of IBV
Expression plasmid harboring a CMV promoter, signal sequence, GCN4 trimerization domain and Strep-tag for purification and detection: Use codon-optimized IBV S1 sequence of the serotype of interest (see Note 1) and clone S1 into for example pCD5 expression plasmid (see Note 2) in frame with CMV, GCN4, and Strep-tag (Fig. 1).
Polyethylenimine (PEI): Dissolve the powder at a concentration of 1 mg/ml at 50–60 °C (see Note 3). Test the efficiency by transfecting human embryo kidney cells (HEK) 293T with pCMV-EGFP-N1 or any other vector expressing a fluorescent protein (see Note 3).
Supplemented Dulbecco’s modified eagle medium (DMEM): DMEM, 2 % glutamine, 10 % fetal calf serum, 0.1 mg/ml gentamicin.
Supplemented 293 SFM II expression medium: 293 SFM II, 3.7 g/l sodium bicarbonate, 2.0 g/l glucose, 3.0 g/l Primatone RL-UF, 0.1 mg/ml gentamicin, 1× GlutaMAX, 1.5 % dimethyl sulfoxide. Sterilize the medium by filtering.
T175 culture flasks.
Strep-Tactin sepharose 50 % suspension or Strep-Tactin gravity flow columns
Elution buffer: Biotin elution buffer 10×, dilute 10× concentrated to 1× in distilled water (working solution).
Vivaspin 10 or 50 MWCO 3000.
Tube roller.
Fig. 1.

Diagrammatic representation of S1 expression cassette. S1 was cloned into pCD5 expression plasmid in frame with signal sequence (SS), trimerization motif (GCN4), and Strep-tag (ST2). The promoter sequence was from Cytomegalovirus (CMV)
Components for Protein Histochemistry
Xylene.
Ethanol at 100, 96, and 70 %.
Tissue section slides of 3–4 μm on Superfrost Plus or KP plus glasses (see Note 4).
Citrate buffer (pH 6.0): Add 2.1 g of citrate buffer monohydrate to 800 ml of distilled water and while stirring adjust the pH to 6 at room temperature by adding 10 N NaOH drop wise. Then add up the total volume to 1,000 ml by adding distilled water.
Strep-Tactin HRP conjugated.
PBS 10×: Add 35.6 g of Na2HPO4⋅2H2O and 6.24 g of NaH2PO4⋅2H2O into 2.4 l of distilled water. Check if the pH is 7.4–7.5 and add 216 g of NaCl.
PBS: Dilute stock PBS 10× in distilled water to prepare 1× working solution.
PBS–Tween 0.1 %: Dilute 500 ml 10× stock PBS into 4,500 ml of distilled water. Add 5 ml of Tween 20.
PBS (pH 5.0): Adjust the pH to 5.0 by adding 6 N HCl into PBS.
1 % hydrogen peroxide: Add 2.85 ml of 35 % hydrogen peroxide to 97.15 ml of absolute methanol.
VECTASTAIN ABC Kit (Vector Laboratories Inc.): Add 10 μl of solution A to 240 μl of PBS and add 10 μl of solution B to 240 μl of PBS. Mix and incubate for 30 min at RT.
3-Amino-9-ethylcarbazole (AEC).
Normal goat serum: Dilute goat serum in PBS to reach 10 %.
Neuraminidases: Add 1 mU of Vibrio cholera neuraminidase or Arthrobacter ureafaciens neuraminidase to 100 μl of PBS (pH 5.0).
Lectins: Dilute MALI and MALII at a concentration gradient from 64 to 256 μg/ml in PBS (see Note 5).
Hematoxylin.
Aquatex mounting medium.
Dako or Immunopen.
Coverslips (24 × 32 mm).
Coplin jar.
Humidity chamber.
Methods
Carry out all procedures at room temperature unless otherwise specified. Centrifuge 50 ml tubes in a benchtop centrifuge and Eppendorf tubes in microcentrifuge.
Expression of Recombinant IBV S1 Protein
Amounts are shown for expression of S1 protein in one T175 flask.
Day 1: Seed a T175 culture flask with 1 × 107 HEK 293 T cells in a total volume of 25 ml of DMEM + medium. Incubate the cells at 37 °C for 24 h until the cells reach a confluence of 50–60 % (see Note 6).
Day 2: Prepare reaction mix. For one T175 flask first pipette 15 μg of the expression vector pCD5 containing IBV S1 into DMEM and then pipette PEI into the DMEM. The total volume of DNA, PEI and DMEM should be 1.5 ml per flask to be transfected. Incubate the reaction mix for 15 min. Remove 5 ml of the medium from the cells and add the reaction mix into the medium with the T175 flask in an upright position then gently agitate before repositioning the flask horizontally to incubate at 37 °C for 24 h (see Note 2).
Day 3: Replace DMEM with 20 ml of the 293 SFM + and continue incubation at 37 °C.
Day 8: Collect the supernatant into 50 ml tube (usually 7 days after transfection) and centrifuge at 300 × g for 10 min. Transfer into new 50 ml tube and centrifuge another 10 min at 800 × g. Transfer the supernatant into a new tube. The supernatant can now be stored at −20 °C or directly proceed to Subheading 3.2, step 1.
Purification of Recombinant IBV S1 Protein
Add Strep-Tactin sepharose 50 % suspension (see Note 7) to the supernatant and incubate overnight at 4 °C on a tube roller.
The next day centrifuge at 800 × g for 10 min and carefully remove the supernatant without disturbing the bead pellet. Add 500 μl of PBS onto the beads, stir gently with a pipette tip and transfer the beads into a 2 ml Eppendorf tube (see Note 8).
Wash the beads three times using PBS (bead pellet: PBS is 1:1).
Centrifuge at 1,800 × g for 10 min for each wash.
After the final washing step remove PBS, add elution buffer (see Note 9) and incubate for 5 min, vortexing every 1–2 min.
Centrifuge at 1,800 × g for 10 min and collect the supernatant.
To remove remaining beads in the supernatant centrifuge another 10 min at 1,800 × g and transfer the supernatant into a new Eppendorf tube.
Determine the protein concentration (see Note 10).
Protein Histochemistry (See Figs. 2 and 3)
Fig. 2.
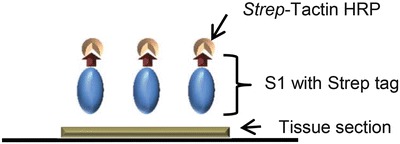
Schematic representation of protein histochemistry. S1 protein was pre-complexed with Strep-Tactin HRP before applying onto tissue section
Fig. 3.

Protein histochemistry for IBV M41-S1. IBV M41-S1 was applied onto (a) untreated chicken trachea and (b) chicken trachea treated with neuraminidase. Positive staining (red ) in cilia and goblet cells is indicated with an arrow and arrowhead, respectively
Deparaffinization and Rehydration of Tissue Sections
Prepare glass dishes with xylene, 100 % ethanol, 96 % ethanol, 70 % ethanol, and distilled water in duplicates.
Arrange the glass slides in a staining rack and immerse slides in xylene to distilled water (xylene, xylene, 100 % ethanol, 100 % ethanol, 96 % ethanol, 96 % ethanol, 70 % ethanol, 70 % ethanol, distilled water, distilled water).
Keep the slides in each dish of xylene for 5 min and in each dish of alcohol and distilled water for 3 min. End with immersing in distilled water.
Antigen Retrieval
Place the staining rack with the slides in a heat-resistant jar or a container and add citrate buffer until the fluid level is at least 2 cm above the slides. Close the container with a lid.
Boil the sections in citrate buffer for 10 min at 900 kW in a microwave (see Note 11).
Leave the slides in the citrate buffer and allow to cool down for 15–20 min.
Transfer the slides into a Coplin jar filled with PBS and keep on a platform rocker for 5 min. Repeat the PBS step twice.
Inactivate Endogenous Peroxidase and Blocking Nonspecific Staining
Remove PBS from the last washing step and add 1 % hydrogen peroxide until the sections are properly covered. Close the jar with a lid and incubate for 30 min.
Discard hydrogen peroxide add PBS–Tween 0.1 % and rinse the slides for 5 min on a platform rocker. Repeat the PBS–Tween 0.1 % wash step twice.
Dry the back of the slides and around the sections using a tissue, and draw lines around the tissues with a Dako or an Immunopen.
Place the slides in a humidity chamber and apply sufficient amounts of 10 % normal goat serum to cover the tissues (usually 50–200 μl depending on the size of the section).
Close the humidity chamber and incubate for 30 min.
Application of Spike Proteins
Premix spike protein to a final concentration of 0.1 mg/ml and Strep-Tactin HRP 1:200 in PBS (see Note 12) in an Eppendorf tube and incubate for 30 min on ice.
Drain goat serum from the sections and apply sufficient amounts (usually 50–200 μl depending on the size of the section) of spike protein–Strep-Tactin HRP complex to tissues.
Incubate the sections with the protein–Strep-Tactin HRP complex overnight at 4 °C.
Visualizing and Counterstaining
Drain the protein–antibody complex and place the slides in a Coplin jar filled with PBS.
Rinse the slides in PBS three times each for 5 min as previously described.
Dry the back of the slides and around the tissues, place the slides in a humidity chamber, and apply AEC dropwise (see Note 13).
Close the chamber and incubate for 15 min.
Dip the sections into a Coplin jar with water and place the glass slides in a staining rack.
Rinse the slides in tap water for 5 min and immerse in hematoxylin for 40–60 s.
Keep the slides in running water for 10 min.
Finally place a coverslip to cover the tissues using Aquatex (Fig. 2).
Protein Histochemistry on Tissues Pretreated with Neuraminidase
After treating the slides with hydrogen peroxide (Subheading 3.3.3) place the slides in a humidity chamber and circle the tissue regions with Dako or Immunopen.
Dilute 1 mU of neuraminidase (see Note 14) in 100 μl of PBS (pH 5.0) and apply to tissues within the circle.
Close the humidity chamber and keep overnight at 37 °C in an incubator.
The next day rinse the slides in PBS–Tween 0.1 % three times each for 5 min, incubate with 10 % goat serum for 30 min and continue with Subheading 3.3.4.
Protein Histochemistry for Tissues Blocked with Lectins
Notes
The sequences coding for spike were codon-optimized for expression in mammalian cells, resulting in approximately five times higher production of proteins than using non-optimized viral sequences.
Transfection of HEK 293T cells with pCD5 expression vector has been described previously [10, 11].
Dissolving of PEI in distilled water might take up to 1 or 2 days. The solution should be continuously stirred at 50–60 °C and when it is completely dissolved, filter-sterilize, aliquot, and store at −20 °C. The efficiency of PEI for transfecting HEK 293T cells with DNA is tested by using PEI ratios from 1:5 to 1:20. The number of transfected cells is counted using a fluorescence microscope under 10× magnification. The best ratio to use for subsequent transfection is the ratio that gives the highest percentage (usually 40 %) of transfected cells with lowest toxicity or cell death.
Tissues that easily detach during antigen retrieval, including for example trachea, can be mounted onto KP plus slides to reduce the tissue damage.
Concentration gradient ensures reaching the optimum amount of lectins required for complete blocking of the binding of recombinant proteins.
By seeding 1 × 107 cells per T175 flask we were able to reach 50–60 % confluence after 24 h post seeding. When compared to <50 % or >60 % cell confluence, transfection at 50–60 % confluence results in a significantly higher transfection efficiency and thereby higher amounts of recombinant proteins.
Proteins in the supernatant (using 5–10 μl) are analyzed using SDS PAGE followed by western blotting to determine whether the protein is properly produced. In particular we check for any degradation, low or no expression and correct molecular weight (IBV S1 protein is highly glycosylated and migrates around 110 kDa). Upon high amounts of protein in the culture supernatant (usually appearing as thick bands of ≥5 mm in the film) we add 250 μl of 50 % Strep-Tactin sepharose suspension for each 10 ml of supernatant. However, compared to column-based purification minor fraction of proteins were lost with the supernatant after purification with the beads. If necessary, column based purification can be done according to the manufacturer’s instructions.
Since the beads tend to stick on to the surface of the tube, it is important not to disturb the sediment after centrifugation and while transferring to a 2 ml Eppendorf tube. If necessary, to recover more beads from the surface of the tube add PBS for another one or two times, but limit the total volume to no more than 1.8 ml to prevent spilling of the beads while closing the Eppendorf tube.
For every 250 μl of 50 % Strep-Tactin sepharose suspension we use 125 μl of elution buffer. Whenever we obtained low protein yields (<4 mg/ml) the proteins were concentrated using Vivaspin according to the manufacturer’s instructions.
We use ≥2 μl of purified proteins to measure the concentrations in Qubit fluorimeter. We also approximated the protein concentrations compared to a BSA standard after GelCode Blue/Coomassie staining of a SDS PAGE gel.
Performing antigen retrieval in the microwave can destroy some tissues (for example tracheal epithelium and cartilage). In such instances transfer the glass slides into a polypropylene Coplin jar filled with citrate buffer, cover with a lid, and keep in a water bath preheated to 80 °C for 45 min.
Since Strep-Tactin HRP is optimized only for western blotting different lots may complex to a different extent with spike proteins. Therefore, every lot number has to be tested using a prior lot number giving positive signals. Moreover, the amount of Strep-Tactin HRP to the total volume (1:200) was optimized for IBV-S1, and has to be optimized accordingly when using a recombinant protein with different molecular weight.
Wear gloves when handling AEC. Apply AEC in a fume hood and discard safely the water drained with AEC. For large tissue sections a coverslip can be used to spread AEC drops gently over the tissues, thus minimizing the required amounts of AEC to sufficiently cover tissues.
We used both Vibrio cholera neuraminidase and Arthrobacter ureafaciens neuraminidase. Compared to Vibrio cholera neuraminidase, Arthrobacter ureafaciens neuraminidase showed more efficient cleaving of sialic acids from tissues embedded in paraffin. It is important to apply sufficient volume of total fluid to prevent drying off the tissues during incubation at 37 °C.
Acknowledgment
We thank Steven van Beurden for critical reading of this chapter.
Contributor Information
Helena Jane Maier, Email: helena.maier@pirbright.ac.uk.
Erica Bickerton, Email: erica.bickerton@pirbright.ac.uk.
Paul Britton, Email: paul.britton@pirbright.ac.uk.
M. Hélène Verheije, Email: m.h.verheije@uu.nl.
References
- 1.Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- 2.Winter C, Schwegmann-Wessels C, Cavanagh D, et al. Sialic acid is a receptor determinant for infection of cells by avian infectious bronchitis virus. J Gen Virol. 2006;87:1209–1216. doi: 10.1099/vir.0.81651-0. [DOI] [PubMed] [Google Scholar]
- 3.Winter C, Herrler G, Neumann U. Infection of the tracheal epithelium by infectious bronchitis virus is sialic acid dependent. Microb Infect. 2008;10:367–373. doi: 10.1016/j.micinf.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casais R, Dove B, Cavanagh D, et al. Recombinant avian infectious bronchitis virus expressing a heterologous spike gene demonstrates that the spike protein is a determinant of cell tropism. J Virol. 2003;77:9084–9089. doi: 10.1128/JVI.77.16.9084-9089.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamada Y, Liu DX. Proteolytic activation of the spike protein at a novel RRRR/S motif is implicated in furin-dependent entry, syncytium formation, and infectivity of coronavirus infectious bronchitis virus in cultured cells. J Virol. 2009;83:8744–8758. doi: 10.1128/JVI.00613-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavanagh D, Davis PJ, Darbyshire JH, et al. Coronavirus IBV: virus retaining spike glycopolypeptide S2 but not S1 is unable to induce virus-neutralizing or haemagglutination-inhibiting antibody, or induce chicken tracheal protection. J Gen Virol. 1986;67:1435–1442. doi: 10.1099/0022-1317-67-7-1435. [DOI] [PubMed] [Google Scholar]
- 7.Bosch BJ, van der Zee R, de Haan CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: Structural and functional characterization of the fusion core complex. J Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wickramasinghe INA, de Vries RP, Gröne A, et al. Binding of avian coronavirus spike proteins to host factors reflects virus tropism and pathogenicity. J Virol. 2011;85:8903–8912. doi: 10.1128/JVI.05112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Promkuntod N, van Eijndhoven REW, de Vrieze G, et al. Mapping of the receptor-binding domain and amino acids critical for attachment in the spike protein of avian coronavirus infectious bronchitis virus. Virology. 2014;448:26–32. doi: 10.1016/j.virol.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bosch B, Bodewes R, de Vries RP, et al. Recombinant soluble, multimeric HA and NA exhibit distinctive types of protection against pandemic swine-origin 2009 A(H1N1) influenza virus infection in ferrets. J Virol. 2010;84:10366–10374. doi: 10.1128/JVI.01035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Vries RP, de Vries E, Bosch BJ, et al. The influenza A virus hemagglutinin glycosylation state affects receptor-binding specificity. Virology. 2010;403:17–25. doi: 10.1016/j.virol.2010.03.047. [DOI] [PubMed] [Google Scholar]


