Abstract
The coronavirus spike envelope glycoprotein is an essential viral component that mediates virus entry events. Biochemical assessment of the spike protein is critical for understanding structure–function relationships and the roles of the protein in the viral life cycle. Coronavirus spike proteins are typically proteolytically processed and activated by host cell enzymes such as trypsin-like proteases, cathepsins, or proprotein-convertases. Analysis of coronavirus spike proteins by western blot allows the visualization and assessment of proteolytic processing by endogenous or exogenous proteases. Here, we present a method based on western blot analysis to investigate spike protein proteolytic cleavage by transient transfection of HEK-293 T cells allowing expression of the spike protein of the highly pathogenic Middle East respiratory syndrome coronavirus in the presence or absence of a cellular trypsin-like transmembrane serine protease, matriptase. Such analysis enables the characterization of cleavage patterns produced by a host protease on a coronavirus spike glycoprotein.
Key words: Coronavirus, Spike protein, Virus entry, Middle East respiratory syndrome (MERS), Proteolytic processing, Host cell protease, Matriptase, Western blot, Transient transfection
Introduction
The coronavirus spike protein is a type I transmembrane protein that assembles as a trimer and projects outward from viral particles forming the distinctive corona or crown-like appearance of coronaviruses [1]. The spike protein controls to a large extent virus entry events as it accomplishes the critical functions of host cell receptor attachment and membrane fusion [2]. Coronavirus spike envelope glycoproteins are classified as class I viral fusion proteins [3]. Spike monomers are organized into a receptor-binding domain, named S1, and the fusion machinery domain, called S2 [4]. Many coronavirus spike proteins are proteolytically processed by host cell proteases at the junction between the S1 and S2 domains, at a site named S1/S2 (Fig. 1) [5]. An additional cleavage event occurs at a site found within the S2 domain, immediately upstream of the fusion peptide, and termed S2’ (Fig. 1) [6]. For coronaviruses, entry and viral fusion can occur at the plasma membrane or in endosomes, depending on protease availability and other microenvironmental cues [7, 8]. The cleavage event at S2’ is analogous to the proteolytic processing of the prototypical class I viral fusion protein influenza virus hemagglutinin (HA) [9, 10]. Influenza virus entry occurs in the endosomal compartment. The critical HA proteolytic activation step releases the fusion peptide, located at the N-terminal tip of the fusion domain (HA2). Within maturing endosomes, the acidic pH triggers major conformational changes that allow membrane insertion of the fusion peptide and initiation of the merging of viral and host cell membranes [7].
Fig. 1.
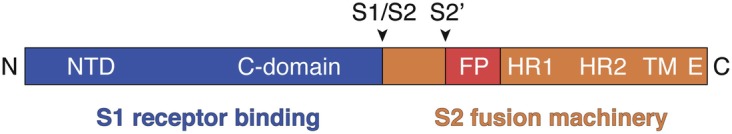
Schematic of the coronavirus spike protein. Shown in the linear diagram are the S1 receptor binding domain and the S2 fusion machinery domain. S1/S2 denotes the proteolytic cleavage site between S1 and S2 domains. S2’ is the cleavage site located within the S2 fusion domain, immediately upstream of the fusion peptide (FP). HR1: heptad repeat 1 region; HR2: heptad repeat 2 region; TM: transmembrane domain; E: endodomain
Biochemical characterization of the spike activation step in the coronavirus life cycle is crucial to understand how this essential event impacts entry into host cells and modulates host cell and tissue tropism, host range and pathogenicity. Slight modifications of viral envelope glycoprotein cleavage sites via mutations can have a profound impact on host protease substrate recognition and activation, leading to changes in pathogenicity. This is typified by highly pathogenic strains of avian influenza (H5N1 strains) where modifications of the HA cleavage motif from a monobasic to a polybasic site switches the proteolytic activation mechanism [11]. Such cleavage site modification is accompanied by a change in host protease requirement from a trypsin-like protease to a more ubiquitously expressed furin-like protease, allowing for systemic infection and increased disease severity. Interestingly, coronavirus spike proteins and in particular the envelope glycoprotein of Middle East respiratory syndrome coronavirus (MERS-CoV) have been shown in cell culture to be cleaved by a number of host cell proteases such as cathepsins [12–14], trypsin-like proteases, notably membrane-bound transmembrane protease, serine 2 ( TMPRSS2) [12–15], and members of the pro-protein convertase family of enzymes such as furin [16, 17]. The latter group of proteases may not be major players for typical routes of virus entry in respiratory epithelial cells [18], but could nonetheless be important activators for extra-pulmonary spread of MERS-CoV [17]. While various proteases have been shown to activate MERS-CoV spike in cell culture systems, it has recently been proposed that TMPRSS2 plays a predominant activating role in virus entry in respiratory epithelial cells [19].
Western blot analyses of spike protein cleavage products produced by host proteases is an important investigative tool allowing a better understanding of the roles of cleavage activation and proteases in the life cycle of coronaviruses. The aim of this chapter is to present a detailed protocol allowing the characterization by western blot of the proteolytic processing of the MERS-CoV spike protein, by transient co-transfection of a plasmid encoding MERS-CoV spike and increasing amounts of a plasmid encoding a host protease (see Note 1). An advantage of this transient transfection-based method compared to MERS-CoV infection-based experiments is that it does not require a biosafety level 3 facility to be performed. We will explain some possible difficulties of the method as well as provide tips and troubleshooting guides. The protease chosen to illustrate the method is matriptase or suppression of tumorigenicity 14 (ST-14) [20], an enzyme belonging to the type II transmembrane serine protease (TTSP) family, which also groups members such as human airway trypsin-like protease (HAT), TMPRSS2, and corin. The activity of matriptase is tightly regulated by the binding of hepatocyte growth factor activator inhibitor type 1 (HAI-1) [21], a Kunitz-type transmembrane serine protease inhibitor . Matriptase is broadly expressed by epithelial cells and throughout the respiratory tract, in particular in airway epithelial cells [22]. While it is synthesized as a membrane-bound protease, the catalytic domain of matriptase can be shed and act extracellularly. Importantly, while matriptase was found to activate the envelope glycoprotein of other viruses such as influenza virus HA [22, 23], its role in proteolytic processing of coronavirus spike protein has not been characterized. The analysis illustrated here allows to characterize the cleavage pattern induced by co-expression of MERS-CoV spike and matriptase in transiently transfected cells. In the absence of matriptase expression, the assay confirms processing of MERS-CoV spike at the S1/S2 site by an endogenous protease, likely furin or a furin-like enzyme as demonstrated previously [17]. Intriguingly, further cleavage, mediated by matriptase expression releases an as-yet uncharacterized spike fragment migrating at 25 kDa.
Materials
All cell culture materials should be kept sterile and manipulated within a biosafety cabinet. When not in use they should be stored at 4 °C. All liquid and solid waste materials should be discarded and/or properly inactivated in appropriate disposable waste containers. Solutions diluted in water should be prepared with ultra-purified water with a resistivity of 18.2 MΩ·cm at 25 °C.
Plasmids and Antibodies
pcDNA3.1-OPT-MERS-wt-S-C9. This plasmid encodes a full-length, wild-type (wt), mammalian codon-optimized sequence of the MERS-CoV spike gene from the EMC/2012 strain fused with a C9 bovine rhodopsin epitope tag at the C-terminus.
pcDNA3.1-hMatriptase. This plasmid contains the coding sequence of the human matriptase gene.
pcDNA3.1. This plasmid is used as an empty vector control plasmid.
Rabbit polyclonal antibody against MERS-CoV strain EMC/ 2012 spike protein .
Mouse monoclonal antibody (IgG1) against the extracellular domain of human matriptase (clone D-7).
Horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG antibodies.
HRP-conjugated goat anti-mouse IgG antibodies.
Cell Culture Reagents and Materials
Dulbecco’s phosphate buffered saline (DPBS) with calcium and magnesium.
Dulbecco’s Modified Eagle Medium (DMEM).
Heat-inactivated fetal calf serum (FCS).
1 M N-2-hydroxyethylpiperazine-N′-2-ethanesulphonic acid (HEPES).
100× penicillin-streptomycin (PS) solution.
Human embryonic kidney (HEK) HEK-293 T/17 cells were obtained from the American Type Culture Collection. The/17 numbering refers to a clone that has been specifically selected to obtain higher transfection efficiencies. Cells were cultured in a 37 °C, 5% CO2 incubator in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% (vol/vol) FCS, 10 mM HEPES, 100 IU/mL penicillin, and 100 μg/mL streptomycin. For long-term storage, the cells can be frozen and stored in liquid nitrogen.
1× Trypsin solution. 0.25% trypsin, 2.21 mM ethylenediaminetetraacetic acid (EDTA).
Cell counting slide with 10 counting grids.
Gibco™ Opti-minimal essential medium (Opti-MEM™) reduced serum medium (for transfections).
Cell culture flasks (75 cm2) with vented caps.
6-Well plate culture vessel.
Transfection and Cell Lysis Reagents
Lipofectamine® 2000 transfection reagent.
10× Radioimmunoprecipitation assay (RIPA) buffer. 0.5 M Tris–HCl, 1.5 M NaCl, 2.5% deoxycholic acid, 10% NP-40, 10 mM EDTA, pH 7.4.
Protease inhibitor cocktail.
Sterile cell scrapers.
Ice bucket with fresh ice.
Rocking device.
Benchtop centrifuge for microcentrifuge tubes.
Protein Gel Migration Reagents and Materials
Electrophoresis System with appropriate power adaptor, or equivalent.
4× Lithium dodecyl sulfate (LDS) sample loading buffer. 106 mM tris(hydroxymethyl)aminomethane (Tris)–HCl, 141 mM Tris Base, 2% LDS, 10% glycerol, 0.51 mM EDTA, 0.22 mM SERVA blue G250, 0.175 mM phenol red.
10× Reducing reagent. 500 mM dithiothreitol (DTT).
PageRuler™ Plus Prestained Protein Ladder.
NuPAGE® 4–12% gradient Bis-Tris polyacrylamide pre-cast protein gels.
20× NuPAGE® 3-(N-morpholino)propanesulfonic acid (MOPS) sodium dodecyl sulfate (SDS) running buffer. 50 mM MOPS, 50 mM Tris Base, 0.1% SDS, 1 mM EDTA, pH 7.7.
Transfer Reagents and Materials
Electrophoretic Transfer Cell with gel holder cassettes and with appropriate power adaptor, or equivalent.
Polyvinylidene fluoride (PVDF) blotting membrane.
Whatman® cellulose filter paper cutouts to the size of the area of gel to transfer. For one transfer, prepare 2 sets of 3 Whatman cutouts.
Transfer fiber pads. Their size should fit the gel holder cassette and should be similar to that of the Whatman paper cutouts. For 1 transfer, 1 set of 2 pads are needed.
20× NuPAGE™ Transfer Buffer. 25 mM Bicine, 25 mM Bis-Tris (free base), 1 mM EDTA, pH 7.2.
Methanol.
Frozen ice pack to maintain cool temperatures during transfer.
Ice bucket with fresh ice.
Immunoblotting Reagents and Materials
Tris-buffered saline (TBS). 50 mM Tri–HCl, 150 mM NaCl, pH 7.5.
TBS-Tween-20 (TBS-T). TBS with addition of 0.05% Tween-20.
TBS-T-bovine serum albumin 2% (TBS-T-BSA2). TBS-T with addition of 2% (2 g for 100 mL) bovine serum albumin (for blocking step).
TBS-T-bovine serum albumin 1% (TBS-T-BSA1). TBS-T with addition of 1% (1 g for 100 mL) bovine serum albumin (for antibody incubations steps).
Small container for membrane incubations.
Chemiluminescence Western Blot Imaging
Enhanced chemiluminescence (ECL) reagents kit.
Chemiluminescence imager.
Methods
Perform steps involving opened sterile solutions or cell culture vessels under sterile conditions using a biosafety cabinet.
Cell Seeding
Observe under a light microscope the HEK-293 T/17 cells cultured in a 75 cm2 flask and check that they are near-confluent, at around 80–90% (see Note 2).
Aspirate spent cell medium and wash cells once with 5 mL of pre-warmed (37 °C) DPBS.
Aspirate supernatant.
Trypsinize cells by adding 1 mL of trypsin solution directly to cells.
Place flask at a 37 °C 5% CO2 incubator for 2–3 min (see Note 2).
Place flask back under biosafety cabinet and add 4 mL of pre-warmed (37 °C) FCS-containing DMEM.
Perform repetitive up-down pipetting to dissociate cells thoroughly.
Transfer dissociated cells to a sterile 50 mL conical tube.
Add 5 mL of pre-warmed (37 °C) FCS-containing DMEM to cells and vortex tube thoroughly.
Transfer 10 μL of the cell solution to a chamber of a cell counting slide.
Count cells under a light microscope.
Dilute cells using pre-warmed (37 °C) FCS-containing DMEM to a concentration of 5 × 105 cells/mL.
Seed cells by adding 2 mL of diluted cell solution to each well of a 6-well plate.
Perform repetitive side-to-side, front-and-back plate movements on a horizontal surface, and gently tap on sides to evenly distribute cells (see Note 3).
Incubate cells overnight in a 37 °C 5% CO2 incubator.
Transfection and Expression of MERS-CoV Spike and Matriptase
Observe HEK-293 T/17 cells in 6-well plate under a light microscope and check that they are approximately 70–80% confluent.
Dilute MERS-CoV S-encoding and human matriptase-encoding plasmid preparations with Opti-MEM reduced serum medium according to volumes shown on Table 1. For this assay, in each transfected well, the MERS-CoV spike plasmid amount remains constant (500 ng of plasmid DNA) while the quantities of plasmid encoding human matriptase is increased (0–200 ng of plasmid DNA). Include a condition using mock-transfected control (pcDNA3.1 empty vector control).
Dilute Lipofectamine 2000 solution with Opti-MEM reduced serum medium according to volumes shown on Table 2 for one well of a 6-well plate. Increase amounts proportionally to the number of wells to transfect and include extra “safety” wells to mitigate pipetting errors. Always add Lipofectamine 2000 to Opti-MEM solution and not the other way around (see Note 4).
Incubate plasmid DNA-Opti-MEM and Lipofectamine 2000-Opti-MEM solutions at room temperature for 5 min.
Add Lipofectamine 2000-Opti-MEM solution to plasmid DNA-Opti-MEM solutions at a 1:1 vol/vol ratio (e.g., for one well of a 6-well plate add 50 μL of Lipofectamine 2000-Opti-MEM solution to 50 μL of plasmid DNA-Opti-MEM solution).
Incubate transfection solutions at room temperature for 20 min.
Aspirate spent medium of each cell culture well.
Add 1 mL of pre-warmed (37 °C) Opti-MEM to each well.
Add 100 μL per well of transfection solutions in a dropwise manner.
Rock plate gently.
Incubate plate in a 37 °C, 5% CO2 incubator for 4–6 h.
Add 1 mL per well of FCS-containing DMEM without antibiotics (see Note 5).
Incubate plate in a 37 °C, 5% CO2 incubator for 24 h.
Table 1.
Quantities of plasmid DNA and Opti-MEM for transfection of one well of a 6-well plate
| Reagent | Quantity for 1 well of a 6-well plate |
|---|---|
| pcDNA3.1-OPT-MERS-wt-S-C9 | 500 ng |
| pcDNA3.1-hMatriptase/pcDNA3.1-empty vector | 0, 1, 2, 5, 10, 20, 50,100, and 200∗ ng |
| Opti-MEM | To 50 μL |
Increase amounts proportionally to the number of replicate wells to transfect. We recommend adding an extra “safety” well in calculations to mitigate pipetting errors. For the different conditions tested in this protocol, the spike protein encoding plasmid quantity remains constant at 500 ng while the quantities of human matriptase encoding plasmid gradually increases from 0 to 200 ng. The asterisk (∗) denotes that for each matriptase encoding plasmid quantity a reciprocal amount of pcDNA3.1-empty vector should be added so that the total amount plasmid DNA remains constant at 200 ng (e.g., for the 10 ng matriptase encoding plasmid condition, there should be 200–10 = 190 ng of pcDNA3.1 empty vector added)
Table 2.
Quantities of Lipofectamine™ 2000 transfection reagent and Opti-MEM for transfection of one well of a 6-well plate
| Reagent | Quantity for 1 well of a 6-well plate |
|---|---|
| Lipofectamine™ 2000 | 3 μL |
| Opti-MEM | 47 μL |
Make a master mix by increasing proportionally the amounts of reagents to the numbers of wells to transfect and add additional “safety” wells in calculations to mitigate pipetting errors
Cell Lysis and Preparation of Protein Samples
To preserve protein sample integrity, all steps should be performed on ice (tubes and plates). Prechill all buffers/solutions on ice until the sample boiling step. The temperature of the microcentrifuge should also be set at 4 °C.
Prepare 50 mL of 1× RIPA buffer by diluting stock solution with ultrapure water.
Add 1 tablet of protease inhibitor cocktail (PIC) to the RIPA buffer.
Place tube on rocker until tablet dissolves completely.
Chill tube of RIPA buffer on ice.
Place plate of transfected cells on ice.
Aspirate gently the spent media of cells.
Wash cells gently by adding 1 mL per well of prechilled DPBS, avoiding pipetting directly onto cells.
Aspirate supernatants.
Lyse cells by adding 300 μL per well of prechilled RIPA buffer supplemented with PIC.
Incubate cells on ice with rocking for 10 min.
Use sterile scrapers for each well to completely detach lysed cells.
Transfer each cell lysate to a chilled microcentrifuge tube placed on ice.
Centrifuge samples at 15,000 × g on a benchtop microcentrifuge set at 4 °C for 20 min.
Transfer supernatants of each sample to a new set of chilled microcentrifuge tubes placed on ice. The pellets can be discarded.
Prepare LDS loading buffer and DTT reducing agent solution (Table 3) (see Note 6).
Add 161 μL of LDS-DTT solution to each sample.
Heat samples at 95 °C for 5 min (see Note 6).
Place tubes on ice to cool down for 1 min.
Perform a quick microcentrifugation step to spin down evaporated water on microcentrifuge tube caps.
Store samples at −20 °C (see Note 7).
Table 3.
Quantities of reagents to add to 300 μL of lysate sample
| Reagent | Quantity for each sample |
|---|---|
| Protein sample | 300 μL |
| 4× LDS | 115 μL |
| 10× DTT | 46 μL |
We suggest to first prepare a master mix composed of proportionally increased amounts of 4× LDS and 10× DTT corresponding to the number of samples being prepared. As mentioned previously, include an extra “safety” sample in calculations to mitigate pipetting errors, and then add 161 μL of the mix to each sample of 300 μL
Polyacrylamide Gel Electrophoresis
Make 1 L of 1× Bis-Tris gel running buffer by diluting buffer stock solution in ultrapure water.
Prepare pre-cast gel (Bis-Tris 4–12% gradient) by removing the comb and adhesive tape, rinse exterior casing with ultrapure water, and gently wash each lane well with ultrapure water (see Note 8).
Assemble pre-cast gel in the electrophoresis tank following the manufacturer’s guidelines.
Add 1× Bis-Tris running buffer in the electrophoresis tank making sure that the pre-cast gel assembly is properly sealed and does not leak out into the outer parts of the tank.
Load 25 μL of each sample in individual gel lane wells and include a lane with protein ladder (10 μL).
Connect electrophoresis tank to power supply generator. Turn on power with constant voltage initially set at 100 V (see Note 9).
Check that protein samples are migrating downward by looking at migration front.
Incrementally increase voltage up to 200 V, within a 10–15 min timeframe (see Note 9).
Migrate samples until migration front reaches bottom of gel (approximately where the adhesive tape was located). Migration time typically lasts for a little over an hour.
Turn off power supply and remove gel from electrophoresis tank.
Electrophoretic Transfer of Protein Samples
Prepare 1 L of 1× transfer buffer with methanol (10% final) by diluting the buffer stock solution with ultrapure water (see Note 10).
Prechill transfer buffer on ice.
Incubate PVDF membrane cutouts (the size should cover the area of gel to transfer) in pure methanol for 10 min (see Note 11).
Discard methanol from membrane and immediately replace with transfer buffer.
Soak Whatman paper (6 paper cutouts per transfer) and fiber pads (2 pads per transfer) in transfer buffer.
De-cast carefully the polyacrylamide gel delicately and immediately place it in a container with transfer buffer (see Note 12).
Layer transfer components within a transfer cassette according to diagram shown in Fig. 2.
Roll out bubbles after layering PVDF membrane on gel using a clean serological pipette that has been humidified with transfer buffer (see Note 12).
Lock transfer components within transfer cassette.
Place transfer cassette in transfer electrophoretic tank being mindful of the direction of the electric current in the tank. In the transfer tank used here the black panel of the transfer cassette should directly face the black wall of the electrodes assembly.
Place frozen ice pack in transfer tank.
Add chilled transfer buffer to transfer tank.
Place transfer tank in ice bucket containing fresh ice (see Note 13).
Connect transfer tank with power supply generator.
Turn power on using constant current set at 200 mA for 3 h (see Note 13).
Turn off power supply generator.
Fig. 2.
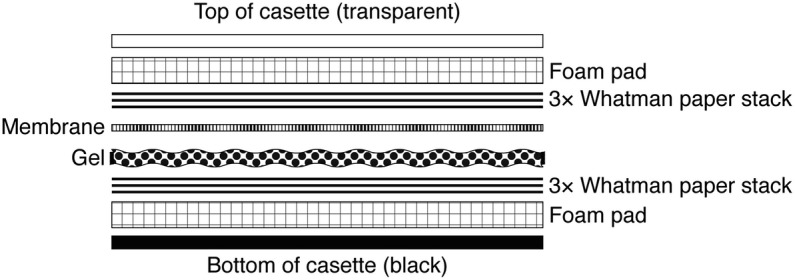
Side-view diagram of transfer component stack within transfer cassette. Displayed in expanded view are the various components of the transfer stack to place in the transfer cassette. The transfer stack should be prepared in a container filled with transfer buffer. The exaggerated gaps between the different components shown here are for clarity only. In the actual transfer stack there should be no gaps or air bubbles between the different layers
Immunoblotting
Prepare 1 L of TBS-T, 50 mL of TBS-T-BSA2 and 50 mL of TBS-T-BSA1 solutions (see Note 14).
Disassemble layered transfer components from transfer cassette avoiding dehydrating PVDF membrane.
Place PVDF membrane in TBS-T buffer in a small container immediately after disassembly of transfer components.
Replace TBS-T with TBS-T-BSA2 blocking buffer (see Note 14).
Incubate membrane for 1 h at room temperature with gentle rocking.
Prepare primary antibody dilutions in TBS-T-BSA1 dilution buffer (1/2000 for anti-MERS-CoV-S antibodies and 1/1000 for anti-matriptase antibodies) (see Note 15). Typically, 8–10 mL of antibody solution is enough to cover surface of a membrane in “mini-gel” format.
Incubate membrane with antibodies overnight at 4 °C with rocking.
Wash the membrane three times by incubating in 10 mL of TBS-T with rocking for 10 min each time.
Prepare secondary antibody dilutions in TBS-T-BSA1 dilution buffer (1/2500 for HRP-anti-mouse IgG antibodies and 1/5000 for HRP anti-rabbit IgG antibodies). Typically, 8–10 mL of antibody solution is enough to cover surface of a membrane in “mini-gel” format.
Incubate membrane in secondary antibodies solution for 1 h at room temperature with rocking.
Wash three times the membrane by incubating in 10 mL of TBS-T for 10 min with rocking each time.
Enhanced Chemiluminescence Imaging
Prepare ECL solution by mixing the two solutions at a 1:1 ratio (for a typically sized “mini-gel” format membrane ~3–5 mL of ECL solution should be enough to cover area of a mini-gel).
Blot-out excess moisture from the membrane with Kimwipe tissue by placing one edge of the membrane on the tissue. Do not let the surface of a membrane contact directly tissues or other absorbing surfaces.
Place the membrane on a flat surface with the side that has contacted the gel facing up.
Add a few drops of ECL solution mixture to cover the entire surface of the membrane.
Incubate for 1 min at room temperature.
Place membrane in ECL imager (see Note 16).
Turn imager device on and proceed to imaging in normal light mode (for ladder) and chemiluminescence mode (ECL).
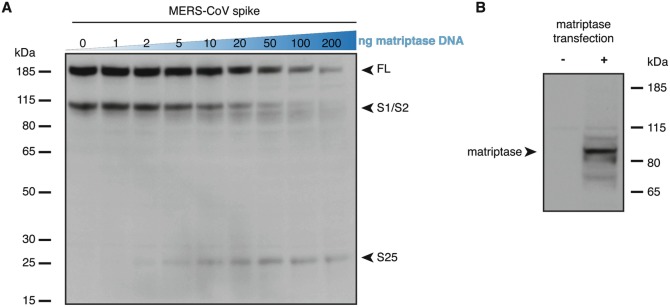
Transfer image files on a computer and proceed with western blot image analyses. Representative results of this method are shown in Fig. 3 and a short summary of results are found in Note 17.
Fig. 3.
Proteolytic processing of MERS-CoV spike protein by matriptase expression. (a) Spike protein western blot analysis. HEK-293 T/17 cells are co-transfected with MERS-CoV spike-encoding and human matriptase-encoding plasmids. The amount of MERS-CoV spike-encoding plasmid is kept constant while the quantity of human matriptase encoding plasmid is gradually increased from 0 to 200 ng as shown on the blue scale triangle. 24 h post-transfection cells are lysed, lysates processed and analyzed by western blot using rabbit polyclonal antibodies raised against MERS-CoV spike protein ectodomain (see note 17 for a summary of the results obtained). FL: full-length spike protein; S1/S2: S1/S2-cleaved fragment. (b) Matriptase expression in transfected cells. To check for protease expression in HEK-293 T/17 cells after transfection of matriptase-encoding plasmid (+, 500 ng) or pcDNA3.1 empty vector control (−, 500 ng), transfected cells were lysed and analyzed by western blot using mouse monoclonal antibodies specific for detecting human matriptase. A strong band was detected for the condition where the human matriptase-encoding plasmid was used for transfection. This band migrated at approximately 90 kDa, which corresponds to the expected apparent molecular weight of human matriptase
Notes
This protocol allows characterization of spike protein proteolytic processing by transient co-transfection of MERS-CoV S-encoding and human matriptase-encoding plasmids. However, there are other methods to analyze the envelope glycoprotein cleavage step. For example, using murine hepatitis virus (MHV) coronavirus infection in susceptible cells, Wicht and colleagues have elegantly engineered a biotinylation-based intracellular labeling assay to selectively label spike proteins that have undergone fusion to analyze their cleavage status [24]. Note that in the case of MERS-CoV infection, the latter experiment would require access to a BSL-3 facility. Another, more biochemical approach is to incubate viral particles such as coronavirus spike-pseudotyped particles with a purified recombinant protease and perform a western blot assay [17, 25].
In our hands, the HEK-293 T/17 cells described in this protocol are found to be only semi-adherent and are prone to easily detach from cell culture surfaces. If this issue is encountered, we recommend to handle the cells with care, being mindful to use pre-warmed (37 °C) DPBS and media as much as possible. We have found that pretreating cell culture plastic surfaces with poly-D-lysine helps the cells adhere better to surfaces. During cell detachment we recommend avoiding incubating cells with trypsin for more than 5 min as this paradoxically often leads to cells aggregating.
To avoid cells clumping in the middle of wells and being sparsely distributed near the edges, we recommend to refrain from circular motion or tilting the plate.
In this protocol we use Lipofectamine 2000 as transfection reagent. We found that it works well for the cell type being used (HEK-293 T/17). Other transfection reagents and/or methods (electroporation) can also be used, if needed. Lipofectamine 2000 tends to bind to plastic materials such as the walls of microcentrifuge tubes. As such, it is recommended to always place Lipofectamine 2000-containing solutions in tubes that already contain a diluent rather than in direct contact with the walls of microcentrifuge tubes.
Transfection reagents such as the one used here (Lipofectamine 2000) increase cell membrane permeability. Because of this effect, antibiotics that are typically added in cell culture media (penicillin and streptomycin) tend to increase in cytotoxicity. It is thus recommended to use cell medium devoid of antibiotics after cells receive transfection reagents such as Lipofectamine 2000.
LDS stored at 4 °C partially precipitates. To better pipet correct volumes, we recommend warming LDS at 37 °C to ensure working with a correctly solubilized solution. Also, we have found for other coronavirus spike proteins that reducing conditions (DTT) and/or application of heat (95 °C for 5 min) can negatively impact antibody detection at later steps of the western blot assay. We suggest to test different conditions (with or without DTT and/or with or without heat) to check which ones work best for a given coronavirus spike protein.
At this point, protein samples can be directly analyzed by western blot assay or stored at −20 °C until used. Once frozen and kept at −20 °C, the samples are stable for several months. The protein samples are relatively tolerant to a few freeze-thaw cycles but we would suggest to refrain from multiple freeze-thaw cycles (5 or more). A workaround to avoid this problem is to aliquot samples into smaller volumes prior to the initial freezing.
The polyacrylamide gel used in this protocol is a pre-cast 4–12% gradient gel. We have found that it allows to obtain good separation of full-length coronavirus spike and fragments generated by proteolytic activity. Other kinds of gels can be used as well, including gels made in the lab, which are relatively easy to prepare.
Most of the gel migration is performed at 200 V with constant voltage. However, we have sometimes found that applying the full voltage of 200 V from the start of migration can result in irregularities in the migration pattern of protein bands. To avoid this, we have found that starting at a lower voltage (e.g., 100 V) and incrementally increasing (e.g., 20 V increments) to 200 V over a 10–15 min timeframe allows to obtain more consistent protein migration.
Here, ethanol can be used in place of methanol.
In this chapter a PVDF membrane is used. Alternatively, a nitrocellulose membrane can also be used as we have found that such membrane type can also allow transfer of coronavirus spike protein bands.
Gels and membranes dry easily. As much as possible, avoid leaving them to dry in air and handle them within containers with buffer solution.
Due to their size and heavy glycosylation status, full-length MERS-CoV spike proteins (and most other coronavirus spike proteins) are relatively large with monomers typically migrating with apparent molecular weight of around 180 kDa or more and require relatively long transfer times when performing “wet” transfers as presented here. We found that 3 h allows to properly transfer the protein from the gel to the PVDF membrane. This long transfer time typically leads to heating of the transfer tank and transfer solution. To avoid overheating and potential protein degradation, in addition to placing an ice pack in the transfer tank and using prechilled transfer buffer, we recommend encasing the tank within an ice bucket filled with ice. Another option is to perform the transfer in a refrigerated setting such as within a cold room.
Here, BSA is used for preparing blocking solution (in TBS-T) as well as the diluent for antibody incubations. We have found that BSA at concentrations of 1–2% generally gives clear western blot images with low background signal. An alternative to BSA is to use powdered milk. The concentrations and conditions for incubations with TBS-T supplemented with powdered milk should be tested beforehand to check which one gives the best results in terms of western blot background.
The dilutions and incubation times for the antibodies used in this protocol were chosen after testing different conditions in preliminary experiments. If other antibodies are used, we suggest to also perform such tests to determine which antibody dilutions and incubation times give the best results.
The imaging method presented in this protocol uses a chemiluminescence imager. However, it’s also possible to perform this using X-ray film and developer.
The assay presented here confirms that in the absence of matriptase expression (Fig. 3b, lane -), the MERS-CoV spike protein is cleaved at the S1/S2 site by an intracellular protease (Fig. 3a, lane 0), most likely of the pro-protein convertase family (e.g., furin), as shown previously [17]. The analysis also reveals that upon expression of matriptase, there is dose-dependent increase in the detection of a MERS-CoV spike protein proteolytic fragment (S25) migrating at approximately 25 kDa (Fig. 3a, lanes 1–200). In addition, the band signals corresponding to full-length (FL) and S1/S2-cleaved fragment (S1/S2) appear to decrease upon increased matriptase expression, a result which could indicate that high levels of matriptase expression (e.g., in the 100–200 ng range) are associated with nonspecific degradation of MERS-CoV spike protein. This effect could also be due to the matriptase and MERS-CoV spike co-transfection conditions used here, and it would be interesting to compare these results with the proteolytic processing of matriptase during viral entry. In addition, matriptase is expressed in this system without its natural inhibitor, HAI-1, which potently regulates the enzyme’s activity. It is also noteworthy to point out that even in conditions with very low amounts of transfected matriptase-encoding plasmid (e.g., 2 ng) the S25 band is detected, suggesting that matriptase may be highly processive for the MERS-CoV spike substrate, in the absence of inhibitor. The composition of S25, the associated matriptase cleavage site and the functional consequence of such cleavage await further elucidation.
Acknowledgments
We wish to thank all members of the Whittaker lab for helpful comments. We gratefully acknowledge Marco Straus for providing the plasmid which encodes the sequence for human matriptase. Research funding for this work was provided by the NIH grants R21 AI111085 and R01 AI135270.
Contributor Information
Rahul Vijay, Email: rahul-vijay@uiowa.edu.
Jean K. Millet, Email: jean.millet@inra.fr
References
- 1.Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosch BJ, Rottier P. Nidovirus entry into cells. In: Perlman S, Gallagher T, Snijder EJ, editors. Nidoviruses. Washington, DC: ASM Press; 2008. pp. 157–178. [Google Scholar]
- 3.Bosch BJ, van der Zee R, de Haan CAM, Rottier PJM. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion Core complex. J Virol. 2003;77(16):8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43(3):189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Millet JK, Whittaker GR. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belouzard S, Chu VC, Whittaker GR. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci. 2009;106(14):5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White JM, Whittaker GR. Fusion of enveloped viruses in endosomes. Traffic. 2016;17(6):593–614. doi: 10.1111/tra.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park JE, et al. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc Natl Acad Sci U S A. 2016;113(43):12262–12267. doi: 10.1073/pnas.1608147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lazarowitz SG, Choppin PW. Enhancement of the infectivity of influenza a and B viruses by proteolytic cleavage of the hemagglutinin polypeptide. Virology. 1975;68(2):440–454. doi: 10.1016/0042-6822(75)90285-8. [DOI] [PubMed] [Google Scholar]
- 10.Lazarowitz SG, Compans RW, Choppin PW. Proteolytic cleavage of the hemagglutinin polypeptide of influenza virus. Function of the uncleaved polypeptide HA. Virology. 1973;52(1):199–212. doi: 10.1016/0042-6822(73)90409-1. [DOI] [PubMed] [Google Scholar]
- 11.Klenk HD, Matrosovich M, Stech J. Avian influenza: molecular Mechansims of pathogenesis. In: Mettenleiter T, Sobrino F, editors. Animal Viruses: Molecular Biology. Norfolk, UK: Caister Academic Press; 2008. pp. 253–303. [Google Scholar]
- 12.Gierer S, et al. The spike-protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2 and is targeted by neutralizing antibodies. J Virol. 2013;87(10):5502–5511. doi: 10.1128/JVI.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qian Z, Dominguez SR, Holmes KV. Role of the spike glycoprotein of human Middle East respiratory syndrome coronavirus (MERS-CoV) in virus entry and syncytia formation. PLoS One. 2013;8(10):e76469. doi: 10.1371/journal.pone.0076469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shirato K, Kawase M, Matsuyama S. Middle East respiratory syndrome coronavirus (MERS-CoV) infection mediated by the Transmembrane serine protease TMPRSS2. J Virol. 2013;87(23):12552–12561. doi: 10.1128/JVI.01890-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawase M, Shirato K, van der Hoek L, Taguchi F, Matsuyama S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J Virol. 2012;86(12):6537–6545. doi: 10.1128/JVI.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burkard C, et al. Coronavirus cell entry occurs through the Endo−/Lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 2014;10(11):e1004502. doi: 10.1371/journal.ppat.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Millet JK, Whittaker GR. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc Natl Acad Sci. 2014;111(42):15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuyama S, et al. Middle East respiratory syndrome coronavirus spike protein is not activated directly by cellular Furin during viral entry into target cells. J Virol. 2018;92(19):e00683-18. doi: 10.1128/JVI.00683-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleine-Weber H, Elzayat MT, Hoffmann M, Pöhlmann S. Functional analysis of potential cleavage sites in the MERS-coronavirus spike protein. Sci Rep. 2018;8(1):16597. doi: 10.1038/s41598-018-34859-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oberst MD, et al. Characterization of matriptase expression in normal human tissues. J Histochem Cytochem. 2003;51(8):1017–1025. doi: 10.1177/002215540305100805. [DOI] [PubMed] [Google Scholar]
- 21.Shimomura T, et al. Hepatocyte growth factor activator inhibitor, a novel Kunitz-type serine protease inhibitor. J Biol Chem. 1997;272(10):6370–6376. doi: 10.1074/jbc.272.10.6370. [DOI] [PubMed] [Google Scholar]
- 22.Beaulieu A, et al. Matriptase proteolytically activates influenza virus and promotes multicycle replication in the human airway epithelium. J Virol. 2013;87(8):4237–4251. doi: 10.1128/JVI.03005-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamilton BS, Gludish DW, Whittaker GR. Cleavage activation of the human-adapted influenza virus subtypes by Matriptase reveals both subtype and strain specificities. J Virol. 2012;86(19):10579–10586. doi: 10.1128/JVI.00306-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wicht O, et al. Identification and characterization of a Proteolytically primed form of the murine coronavirus spike proteins after fusion with the target cell. J Virol. 2014;88(9):4943–4952. doi: 10.1128/JVI.03451-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Millet JK, et al. A camel-derived MERS-CoV with a variant spike protein cleavage site and distinct fusion activation properties. Emerg Microbes Infect. 2016;5(12):e126. doi: 10.1038/emi.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]