Abstract
Intrinsically disordered proteins have been reported to undergo ‘disorder to order’ transitions upon binding to their partners in the cell. The extent of the ordering on binding and the lack of order prior to binding is difficult to visualize with classical structure determination methods. Binding of p27 to the Cdk2/cyclin A complex is accompanied by partial folding of p27 in the KID domain, with the retention of dynamic behaviour for function, particularly in the C-terminal half of the protein, positioning it as an exemplary system to probe conformational diversity. Here we employ native ion mobility with mass spectrometry (IM-MS) to measure the intrinsic dynamic properties of p27, both in isolation and within the trimeric complex with Cdk2/cyclin A. This stepwise approach reveals the conformational distributions of the constituent proteins and how they are restructured on complex formation; the trimeric Cdk2/cyclin A/p27-KID complex possesses significant structural heterogeneity cf. Cdk2/cyclin A. These findings support the formation of a fuzzy complex in which both the N and C termini of p27 interact with Cdk2/cyclin A in multiple closely associated states.
Keywords: ion mobility mass spectrometry, protein structure, intrinsically disordered proteins
The protein p27Kip1 is a cell cycle regulator that binds to the Cdk2/cyclin A complex, thereby inhibiting the kinase activity of Cdk2 (cyclin-dependent kinase 2), and blocking the transition of a cell from G1 to S-phase.[1,2] p27Kip1 (termed p27 hereon) in isolation is an intrinsically disordered protein (IDP), lacking resolvable secondary structure on the timescale of an NMR experiment, instead, existing in a plethora of transient 3-dimensional conformations. IDPs can bind to multiple proteins with high specificity, which often involves folding, or a loss of conformational diversity upon interaction with a given binding partner. These specific interactions ensure prompt control and reversibility in signalling cascades.
The p27 protein is composed of two distinct domains; the N-terminal kinase-inhibitory domain (p27-KID) and the disordered C-terminal domain (p27-C). In the presence of Cdk2/cyclin A, p27-KID sequentially folds into a conformation that blocks substrate binding to cyclin A and independently inhibits ATP binding to Cdk2 and substrate phosphorylation.[3] p27-C, which contains several sites for post-translational modification, is highly disordered both in isolation and when p27-KID is bound to Cdk2/cyclin A.[3] The flexibility of p27-C within the complex persists with phosphorylation and subsequent ubiquitin-dependent proteolysis of p27 and reactivation of the Cdk2/cyclin A complex leading to cell cycle progression.[3] Molecular dynamics simulations demonstrate that the segment of p27-C immediately following the KID protrudes at an abrupt angle perpendicular to the surface of Cdk2/cyclin A.[3] This is thought to arise due to electrostatic repulsion between residues 110–140 of p27 and the negatively charged surface of Cdk2/cyclin A/p27-KID. Analytical ultracentrifugation and small angle X-ray scattering experiments confirm a highly extended conformation of p27-C in the ternary complex.[4]
The objectives of this study were to examine the dynamic behaviour of each domain of p27, of the intact wild-type protein and as part of a trimeric complex with Cdk2/cyclin A. In parallel, an investigation of the C-terminal charge permutants of p27 has been conducted.[5] In this work, we examine the conformational behavior of the N-terminal KID which folds upon binding and the C-terminal domain which remains disordered. The conformational distributions of full-length p27 (p27-FL) p27-KID and p27-C were measured. We show that the binding of p27-KID to Cdk2/cyclin A significantly increases the conformational flexibility of the protein complex and that the binding of p27-FL has an even greater effect of the dynamic behaviour of the complex. Figure 1 depicts a cartoon representation of the complexes analyzed.
Figure 1.
Schematic representation of Cdk2/cyclin A dimer, Cdk2/cyclin A/p27-KID trimer and Cdk2/cyclin A/p27-FL trimer.
Conformational distribution analysis of the isolated p27 constructs and their Cdk2/cyclin A trimeric complex was performed using an ion mobility mass spectrometer which allows direct measurement of rotationally averaged collision cross sections (CCS).[6] IM-MS is a hybrid method permitting separation of gas phase ions according to their mobility and subsequently their mass-to-charge ratio. Ions were generated using nanoelectrospray ionization (nESI) which allows retention of aspects of the native fold of the protein as they are transferred into the gas phase; following desolvation, ions are separated in the ion mobility cell according to their velocity under a weak electric field while they experience collisions with an inert buffer gas. The mobility of an ion is dependent upon its size, shape and charge. In contrast to X-ray crystallography and NMR spectroscopy, IM-MS cannot provide atomistic detail about the samples; however, it can be employed to isolate conformationally dynamic systems and heterogeneic stoichiometries in a single experiment.
To assess the level of disorder exhibited by the p27 proteins, we compare the charge states in the native mass spectra to the theoretical maximum charge state of a globular protein of the same molecular weight (MW). The maximum number of positive charges a globular spherical protein can accommodate can be calculated using the following equation[7]
where ZR is the maximum (Rayleigh) charge and m is the molecular weight of the protein; here it identifies the transition between compact and extended states.[8] The value of ZR is known to deviate for proteins that are not inherently globular and maybe over-estimated for proteins with molecular weights below 100 kDa.[9] Focusing on the p27-C domain first, MS analysis reveals a broad charge state distribution Δz =11 where 6 ≤ z ≤17, with most intensity in the lowest two charge states, [M+6H]6+ and [M+7H]7+ (Figure 2b). Such a charge state distribution is indicative of a protein that contains both compact (perhaps structured) as well as extended regions.[6] ZR for p27-C is 8.24, suggesting that the maximum net charge a globular form of the protein could carry is 8 and therefore that charge states above this threshold must be extended to some degree. In addition to the high Δz, CCS distributions from ion mobility experiments confirm that charge states below the ZR limit have a compact geometry, with ΔCCS = 7 nm2, and a range from 8–15 nm2. Ions with charge states above [M+7H]7+ increase in size, up to 28 nm2 for [M+17H]17+. The overall peak width (baseline to baseline) is 8–28 nm2 (20 nm2). In comparison to p27-C, the charge state range and CCS distribution of p27-KID (Figure 2c) suggest a conformational ensemble as flexible as that of p27-C. This is surprising considering that the KID domain, which folds upon binding to Cdk2/cyclin A, has been shown to adopt a partially populated α-helical structure prior to binding.[10,11] The low charge states [M+6H]6+ and [M+7H]7+ of p27-KID have CCS distributions resolved into two distinct populations, with the larger conformational family of [M+6H]6+ overlapping in size with the smaller family of [M+7H]7+. The ZR of p27-KID is 7.95, suggesting that all charge states above [M+7H]7+ arise from partially extended and disordered conformations. A relatively high proportion of ions are present above the ZR limit, which results in a broad CCS distribution, ranging from 7.5–25 nm2 (17.5 nm2).
Figure 2.
(a) A representative sequence of the p27-FL components p27-KID, p27-C and p27-FL. Native mass spectra and CCS distributions of individual proteins are shown in (b) p27-C (MWcalc 11234 Da, MWexp 11234 Da); (c) p27-KID (MWcalc 10447 Da, MWexp 10444 Da); and (d) p27-FL (MWcalc 22340 Da, MWexp 22338 Da), respectively. The mass spectra show the charge state distributions of each protein whilst the ion mobility profiles highlight the structural heterogeneity of individual charge states, mapping the conformational landscape of each protein.
The relationship between charge and CCS for p27-KID is unusual but reproducible. The smaller conformational family of [M+6H]6+ is centered around 9.5 nm2, which is smaller than the average CCS of [M+5H]5+ (11.3 nm2). This decrease in CCS upon the addition of a proton could indicate stabilization of compact conformations. Also surprising is the gradual decrease in average CCS for increasing charge states above [M+10H]10+. The KID domain can form a helix, and a plausible explanation for the lack of extension in the higher charge states would be a helical geometry. In such a conformation, protonated residues would be held away from each other, minimizing coulombic repulsion, which would result in no or little increase in CCS with respect to increased protonation.[12,13] This would indicate that the lower charge states are more charge solvated forms, which is supported by the substantial adductation of the protein by sodium and potassium for z=5–7, and the fact that a substantial portion of the measured conformational spread comes from charges states that fall below the limit predicted for compact/folded states.[7]
The native mass spectrum of p27-FL (Figure 2d) exhibits a wide charge state range Δz = 24 and the multimodal nature of the distribution reveals the coexistence of multiple distinct conformational families upon desolvation.[14] The majority of these highly charged species fall above the ZR limit of 11.6 and in combination with the broad CCS distribution of 13–55 nm2 (42 nm2), it is evident that p27-FL occupies a myriad of extended states. The wide CCS distribution possessed by individual charge states, in particular 20 ≤ z ≤ 23, indicates that multiple conformers are sampled from solution.
In previous work, we have demonstrated how the linear distribution of charged amino acid residues on the amino acid sequence of p27-C domain substantially affects the populations of stable conformers.[5] Three charge permutants of p27-C with identical amino acid composition but a different distribution of charged residues were studied and were described in terms of the κ parameter, which corresponds to the charge asymmetry of an amino acid sequence. The value of κ ranges from 0 (even distribution of opposite charges) to 1 (opposite charges cluster together).[15] Studying the three charge permutants in isolation revealed that the permutant with evenly dispersed charged residues (termed p27-C-κ14, κ=0.14) populates wider conformational space than the wild-type protein (p27-C-κ31, κ=0.31). In contrast, a second permutant (p27-C-κ56, κ=0.56) in which oppositely charged residues were clustered together, occupied significantly narrower conformational space.[4,5] While considering the effect the distribution of charged residues played on the conformational landscape of the p27-C, we investigated the impact on the p27-FL where the C-terminus of the wild-type protein was replaced with the κ14 and κ56 variants. As shown in Figure S1, the flexibility observed for the p27-C charge permutants was mitigated in the full-length protein, where the wild-type construct (p27-FL-κ31) and two permutants (p27-FL-κ14 and p27-FL-κ56) display a broadly similar charge state distribution and conformational landscapes.[4]
The interaction between the p27 domains and Cdk2/cyclin A was then investigated. Native MS results confirm the stoichiometry of the protein complexes; Cdk2/cyclin A alone exists only as a dimer which is predominantly present in three charge states z = 15–17, all below the ZR limit of 19.75. Upon addition of p27-KID, a trimeric complex is observed in four charge states, (z = 16–19, ZR=21.29). On addition of p27-FL, the Cdk2/cyclin A/p27-FL complex presents in only 4 charges (z =18–21, ZR=22.92), with an overall increase in net charge of only 2 cf. the KID complex.
In the mass spectrum of the p27-FL (WT) complex, a small quantity of the unbound Cdk2/cyclin A dimer is observed, as well as some cyclin A/p27-FL complex, attributed to displacement of a small amount of Cdk2 upon p27 binding to Cdk2/cyclin A. This provides details on the mechanism of complex formation which complements previous results: Lacy et al. reported that binding of p27-KID to cyclin A alone is more thermodynamically favourable than binding Cdk2 alone.[10] Furthermore, binding of p27 to the Cdk2/cyclin A complex occurs at a similar rate to that observed for binding to cyclin A alone. This is justified by slow remodelling of Cdk2 that occurs upon association with p27.
While considering the formation of the Cdk2/cyclin A/p27-FL complex, we also examined the behaviour of the trimeric complex when bound to two charge permutants (p27-FL-κ14 and p27-FL-κ56). In each case, the proteins formed a trimeric complex that is present in [M+18H]18+-[M+21H]21+ charge states, albeit the relative intensity of these has changed (shown in Figure S2). The wild-type p27-FL (p27-FL-κ31) forms the greater proportion of a trimeric complex when compared to the two other variants. The p27-FL-κ31 is the endogenous binding partner of Cdk2/cyclin A, and this suggests that the wild-type C-terminal domain forms interactions that stabilize the trimeric complex that are less favourable or absent in complexes with the two charge permutants.
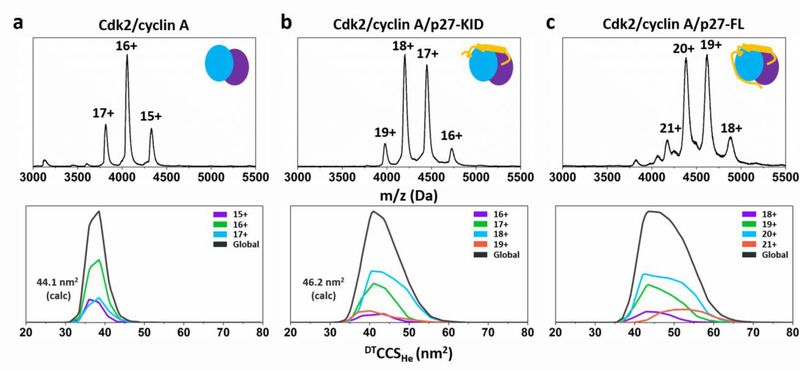
The CCS distributions of individual charge states of the complexes are shown in Figure 3, whilst the sum of the CCS distributions of all protein species investigated is shown in Figure 4 and summarized in Table 1. Viewing the ion mobility data as total CCS distributions (Figure 4) allows visualisation of the global conformational landscape of the Cdk2/cyclin A complex, the isolated p27 domains and how each are modulated in the trimeric complex. Firstly, the Cdk2/cyclin A (64.5 kDa) dimer adopts a highly compact conformation with a narrow CCS range cf. other natively structured proteins of similar molecular weight e.g. transthyretin tetramer (55 kDa) or avidin tetramer (66 kDa) which possess CCS ranges of 22 and 20 nm2, respectively.[16] The apex of the CCS curve marginally increases with each charge state, centered at 38.5 nm2 in the global CCS distribution (Figure 4d), indicating a rigid quaternary structure with minimal conformational dynamics. The upper charge state sits below the limit introduced by de La Mora that can maintain a compact globular geometry.[7]
Figure 3.
MS (top) and IM-MS (bottom) results of Cdk2/cyclin A complex (a, MWcalc 64465 Da, MWexp 64644 Da) and with p27-KID (b, MWcalc 74912 Da, MWexp 74981 Da), or p27-FL (c, MWcalc 86796 Da, MWexp 86822 Da). The global CCS distributions shown in black are the sums of the CCS distributions of individual charges of the complex.
Figure 4.
Global CCS distributions of (a) p27-C, (b) p27-KID, and (c) p27-FL, as well as the protein complexes, (d) Cdk2/cyclin A, (e) Cdk2/cyclin A/p27-KID and (f) Cdk2/cyclin A/p27-FL. The vertical dashed lines in plots a-c represent the theoretical CCS minima and maxima of the p27 constructs, as calculated using a framework model from Beveridge et al.[6] The arrows shown in plots a-c mark the maximum measured CCS for the charge states below the ZR limit, indicative of globular/structured conformations as described in the text; charge states measured in d-f fall below the ZR limit hence are not shown. The global CCS distributions are the sum of the CCS distributions of individual charge states of each protein.
Table 1.
Summary of IM-MS data found for Cdk2/cyclin A, Cdk2/cyclin A/p27-KID and Cdk2/cyclin A/p27-FL complex.
| Species | Peak range (nm2) | Apex of global DTCCSHe distribution (nm2) |
|---|---|---|
| Cdk2/cyclin A | 32 – 47 (15) | 38.5 |
| Cdk2/cyclin A/p27-KID | 32 – 62 (30) | 40.1 |
| Cdk2/cyclin A/p27-FL | 35 – 70 (35) | 43.6 |
In all cases, the peak width was calculated by measuring the width of the CCS distribution at the baseline.
Upon addition of p27-KID to Cdk2/cyclin A, the lower boundary of the CCS distribution remains the same at 32 nm2, while the upper boundary increases considerably to 62 nm2 (CCS range 30 nm2, Figure 4e). The negligible effect of p27-KID on the size of the structure at the lower end of the CCS range indicates that the IDP is capable of tightly binding to the globular dimer. The apex of the global CCS distribution (40.1 nm2) is only 1.6 nm2 larger than that of the Cdk2/cyclin A alone. This indicates that a high proportion of the Cdk2/cyclin A/p27-KID complex is found in a compact conformation, although the apex of these CCS distributions increases for the complex by 32% (cf. with a 16% increase expected due to the increase in molecular weight). Such an increase in the upper limit of the CCS range is unlikely to be achieved by the bound p27-KID alone, rather, we propose that it is due to structural remodelling exerted by p27 on Cdk2 [10] [17].
When p27-FL forms the ternary complex with Cdk2/cyclin A, the lower boundary of the CCS range is still only marginally larger than that of the Cdk2/cyclin A dimer (+3 nm2), whereas the upper boundary increases to 70 nm2 (CCS range 35 nm2, Figure 4f). Incorporation of p27-FL into the complex increases its size only slightly more than the addition of p27-KID. The apex of the global CCS curve increases by 5.1 nm2 compared to the Cdk2/cyclin A dimer and 3.5 nm2 compared to the Cdk2/cyclin A/p27-KID complex. In addition, the width of the CCS curve of the Cdk2/cyclin A/p27-FL complex is only marginally larger than that of the p27-FL protein alone (CCS range 15–55 nm2, Figure 4c), despite the complex being three-times the molecular weight of the isolated protein. We do not attribute this increase in the width of the CCS curve to an untethered C terminal domain (Figure 1); such a configuration would result in a wider charge state range, with higher charge states due to the increased solvent accessibility of the p27-C similar to that found for lymphotactin.[18]
The two charge permutants, p27-FL-κ14 and p27-FL-κ56 also form a trimeric complex that has wide CCS distributions and occupies a narrow range of charge states. The apex of the CCS distribution shifts to 50.5 nm2 (+12 nm2) and 47.4 nm2 (+8.9 nm2) when compared to the Cdk2/cyclin A dimer, respectively. Despite the increase of the apex of the global conformation of the complex, the width and shape of the CCS distribution is nearly identical (as shown in Figure S3). This suggests the formation of a fuzzy complex upon binding of p27-KID where the C-terminal domain of p27-FL lies on the surface of the globular protein, and interacts whilst retaining conformational flexibility with respect to Cdk2/cyclin A for each permutant, irrespective of the charge patterning of the C terminus – which in isolation is conformationally distinct.[5,19] The narrow range of charge states coupled with a wide spread of conformations is similar to that found for monoclonal antibodies which has been attributed to flexible interactions between the domains.[16][20]
In summary, we have demonstrated that the dynamic propensity of p27-FL is significantly reduced upon binding to the binary complex of Cdk2/cyclin A, in part agreement with a ‘folding-upon-binding’ process.[10] Conversely, our results show the compact conformation of the Cdk2/cyclin A dimeric complex to gain significant conformational flexibility upon formation of the ternary complex containing p27-KID domain, and only slightly more upon binding of the p27-FL protein. Only subtle changes in the conformational dynamics of the complex were observed when bound to the κ14 and κ56 charge variants. Overall, we were able to map the conformational landscape of individual highly disordered proteins (p27-FL and p27-C) as well as the more structured p27-KID and measure how the disorder is managed in complex with CDk2/cyclin A.
Supplementary Material
Acknowledgements
This research was supported by the BBSRC (awards: BB/L015048/1, BB/K017802/1 and BB/H013636/1 and the BBSRC/EPSRC-funded Manchester Synthetic Biology Research Centre, SYNBIOCHEM (BB/M017702/1)). BBSRC, EPSRC, Waters Corp., LGC Ltd. and the Universities of Edinburgh and Manchester are thanked for their support of studentships to RB and LGM. R.B. acknowledges the Austrian Science Fund for the receipt of a Lise Meitner Postdoctoral Fellowship (project number M2334). RWK acknowledges support from the National Cancer Institute (P30CA21765 to St. Jude Children’s Research Hospital) and ALSAC.
References
- [1].Hengst L, Dulic V, Slingerland JM, Lees E, Reed SI, Proc. Natl. Acad. Sci. U. S. A 1994, 91, 5291–5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Polyak K, Lee MH, Erdjument-Bromage H, Koff A, Roberts JM, Tempst P, Massagué J, Cell 1994, 78, 59–66. [DOI] [PubMed] [Google Scholar]
- [3].Galea CA, Nourse A, Wang Y, Sivakolundu SG, Heller WT, Kriwacki RW, Heller T, Kriwacki RW, J. Mol. Biol 2008, 376, 827–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Das RK, Huang Y, Phillips AH, Kriwacki RW, Pappu RV, Proc. Natl. Acad. Sci. U. S. A 2016, 113, 5616–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Beveridge R, Migas LG, Das RK, Pappu RV, Kriwacki RW, Barran PE, DOI 10.26434/chemrxiv.7312277.v1. [DOI] [Google Scholar]
- [6].Beveridge R, Covill S, Pacholarz KJ, Kalapothakis JMD, MacPhee CE, Barran PE, Anal. Chem 2014, 86, 10979–10991. [DOI] [PubMed] [Google Scholar]
- [7].de la Mora J. Fernandez, Anal. Chim. Acta 2000, 406, 93–104. [Google Scholar]
- [8].Testa L, Brocca S, Santambrogio C, D’Urzo A, Habchi J, Longhi S, Uversky VN, Grandori R, Intrinsically Disord. proteins 2013, 1, e25068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].De Freitas KCB, J. Am. Soc. Mass Spectrom 2018, 29, 2059–2066. [DOI] [PubMed] [Google Scholar]
- [10].Lacy ER, Filippov I, Lewis WS, Otieno S, Xiao L, Weiss S, Hengst L, Kriwacki RW, Nat. Struct. Mol. Biol 2004, 11, 358–364. [DOI] [PubMed] [Google Scholar]
- [11].Sivakolundu SG, Bashford D, Kriwacki RW, J. Mol. Biol 2005, 353, 1118–1128. [DOI] [PubMed] [Google Scholar]
- [12].Berezovskaya Y, Porrini M, Barran PE, Int. J. Mass Spectrom 2013, 345–347, 8–18. [Google Scholar]
- [13].Kalapothakis JMD, V Berezovskaya Y, Zampronio CG, Faull PA, Barran PE, Cooper HJ, Chem. Commun 2014, 50, 198–200. [DOI] [PubMed] [Google Scholar]
- [14].Borysik AJ, Kovacs D, Guharoy M, Tompa P, J. Am. Chem. Soc 2015, 137, 13807–13817. [DOI] [PubMed] [Google Scholar]
- [15].Das RK, Pappu RV, Proc. Natl. Acad. Sci 2013, 110, 13392–13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Pacholarz KJ, Porrini M, Garlish RA, Burnley RJ, Taylor RJ, Henry AJ, Barran PE, Angew. Chemie - Int. Ed 2014, 53, 7765–7769. [DOI] [PubMed] [Google Scholar]
- [17].Russo AA, Jeffrey PD, Pavletich NP, Nat. Struct. Biol 1996, 3, 696–700. [DOI] [PubMed] [Google Scholar]
- [18].Harvey SR, Porrini M, Konijnenberg A, Clarke DJ, Tyler RC, Langridge-Smith PRR, MacPhee CE, Volkman BF, Barran PE, J. Phys. Chem. B 2014, 118, 12348–12359. [DOI] [PubMed] [Google Scholar]
- [19].Sharma R, Raduly Z, Miskei M, Fuxreiter M, FEBS Lett. 2015, 589, 2533–2542. [DOI] [PubMed] [Google Scholar]
- [20].Stuchfield D, Barran PE, Curr. Opin. Chem. Biol 2018, 42, 177–185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.