Abstract
p53 is known to play a role in iron homeostasis and is required for FDXR-mediated iron metabolism via iron regulatory protein 2 (IRP2). Interestingly, p53 is frequently mutated in tumors wherein iron is often accumulated, suggesting that mutant p53 may exert its gain of function by altering iron metabolism. In this study, we found that FDXR deficiency decreased mutant p53 expression along with altered iron metabolism in p53R270H/− MEFs and cancer cells carrying mutant p53. Consistently, we found that decreased expression of mutant p53 by FDXR deficiency inhibited mutant p53-R270H to induce carcinoma and high grade pleomorphic sarcoma in FDXR+/−; p53R270H/− mice as compared with p53R270H/− mice. Moreover, we found that like its effect on wild-type p53, loss of IRP2 increased mutant p53 expression. However, unlike its effect to suppress cell growth in cells carrying wild-type p53, loss of IRP2 promoted cell growth in cancer cells expressing mutant p53. Finally, we found that ectopic expression of IRP2 suppressed cell growth in a mutant p53-dependent manner. Together, our data indicate that mutant p53 gain-of-function can be suppressed by IRP2 and FDXR deficiency, both of which may be explored to target tumors carrying mutant p53.
Introduction
p53 is often mutated in more than half of human cancers. Many p53 mutants have acquired oncogenic properties, called gain of function, which promote tumor development and progression [1–3]. This concept is supported by the observations that mice expressing a knock-in mutant p53 (R172H, R270H) develop an altered spectrum of tumors that are more metastatic and aggressive than the tumors observed in p53-null mice [4–7]. In addition, recent studies provide compelling evidence that GOF mutant p53 modulates various metabolic pathways and that metabolic pathways normally regulated by wild-type p53 are altered by mutant p53 to facilitate the supply of metabolites required for cell proliferation [8].
Iron is essential for a variety of cellular processes, such as energy production and biogenesis of iron–sulfur (Fe–S) clusters [9–11]. Dysfunction of iron metabolism increases the risk of cancer and promotes tumor growth [12–15]. Indeed, iron overload is associated with cancer development and other pathological conditions, including heart disease, diabetes, and liver cirrhosis [16–20]. Thus, proper control of iron homeostasis is critical for suppressing tumorigenesis.
Systemic iron homeostasis is primarily controlled by the peptide hormone hepcidin [21]. In contrast, cellular iron homeostasis is regulated by a number of factors, especially iron regulatory proteins (IRP1 and IRP2) [22, 23]. As a RNA-binding protein, IRP1/2 bind to an iron-responsive element in their target mRNAs and regulate gene expression through mRNA stability and/or translation, including transferrin receptor-1 (TfR1) and ferritin heavy chain 1 (TFH1) [24–26], both of which are also regulated by p53 [27]. Likewise, IRP1/2 control mRNA stability or translation of iron transporters DMT1 [28] and ferroportin [24–26], both of which are also regulated by iron.
Ferredoxin reductase (FDXR), a mitochondrial flavoprotein, transfers an electron from NADPH to ferredoxin 1 (FDX1) and FDX2 for steroidogenesis and biogenesis of Fe–S clusters and heme A [29–33]. FDXR, a p53 target [34, 35], is found to be a highly consistent internal biodosimetry marker in the peripheral blood following radiation therapy and a potential marker for efficacy of chemotherapy [36–40]. Recently, we found that FDXR deficiency leads to iron overload in mitochondria and that p53 is a mediator of FDXR-dependent iron metabolism, indicating that the FDXR-p53 loop is critical for tumor suppression via iron homeostasis [41]. However, it is largely unknown whether mutant p53 plays a role in iron metabolism. Using both FDXR-deficient cell lines and mouse models, we made novel observations that FDXR deficiency decreases mutant p53 expression and inhibits mutant p53-R270H gain-of-function. We also found that loss of IRP2 increases, whereas ectopic expression of IRP2 suppresses, mutant p53 expression and mutant p53-dependent cell growth. Together, our data indicate that mutant p53 gain-of-function can be suppressed by IRP2 and FDXR deficiency, both of which may be explored to target tumors carrying mutant p53.
Results
Mutant p53 is regulated by FDXR and plays a role in iron metabolism
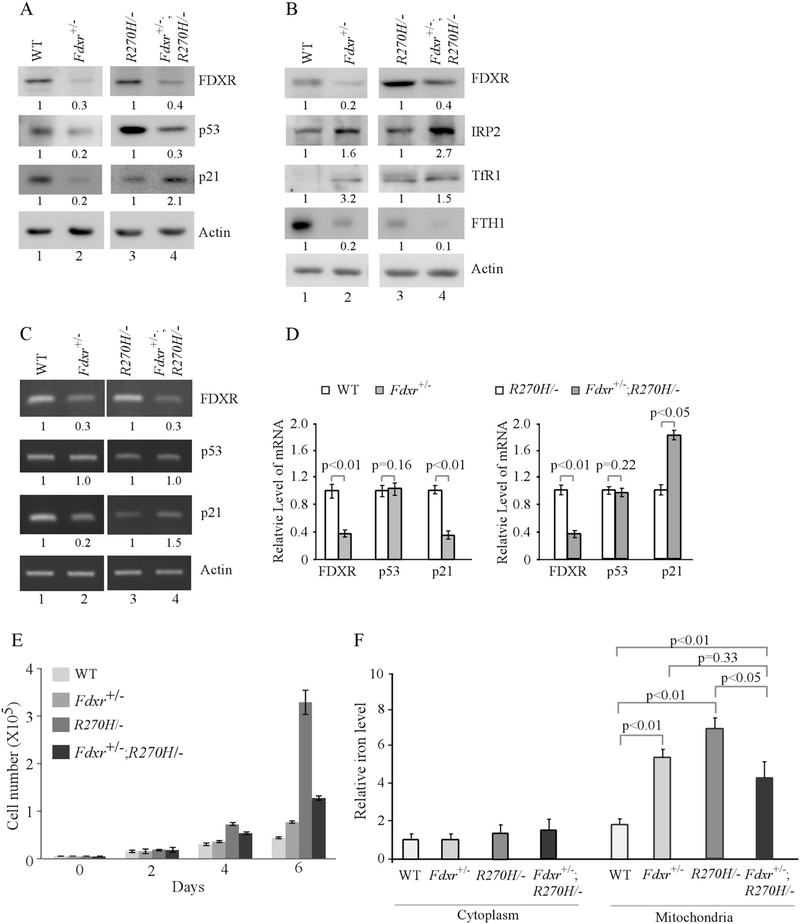
Recently, we found that wild-type p53 regulates iron homeostasis and is required for FDXR-mediated iron metabolism [41]. To determine whether mutant p53 is involved in FDXR-mediated iron metabolism, we generated p53R270H/− and Fdxr+/−;p53R270H/− mice in that endogenous wild-type p53 R270 (equivalent to R273 in humans) was substituted with histidine (R270H) [7]. These mice were used to generate a set of p53R270H/− and Fdxr+/−;p53R270H/− MEFs along with wild-type and Fdxr+/− MEFs as a control. We found that Fdxr deficiency led to decreased expression of both wild-type and mutant p53 (Fig. 1a, compare lanes 1 and 3 with 2 and 4, respectively). We also found that p21 expression was decreased in Fdxr+/− MEFs (Fig. 1a, compare lanes 1 and 2), possibly due to decreased expression of wild-type p53, but increased in Fdxr+/−;p53R270H/− MEFs (Fig. 1a, compare lane 3 with 4), possibly due to decreased expression of mutant p53 [42]. In addition, like in Fdxr deficient MEFs, IRP2 and TfR1 were increased whereas FTH1 was decreased by Fdxr deficiency in Fdxr+/−; p53R270H/− MEFs (Fig. 1b, compare lanes 1 and 3 with 2 and 4, respectively). Next, semiquantitative and quantitative RT-PCR was performed and showed that the levels of Fdxr transcript were decreased as expected in Fdxr+/− and Fdxr+/−;p53R270H/− MEFs as compared with that in WT and p53R270H/− MEFs, respectively (Fig. 1c, compare lanes 1 and 3 with 2 and 4, respectively and Fig. 1d, FDXR columns). However, the levels of p53 transcript remained unchanged (Fig. 1c, compare lanes 1 and 3 with 2 and 4, respectively and Fig. 1d, p53 columns), consistent with our previous report that p53 mRNA translation, but not transcription and mRNA stability, was regulated by FDXR [41]. As a control, we found that the level of p21 transcript was deceased in Fdxr+/− MEFs but increased in Fdxr+/−; p53R270H/− MEFs (Fig. 1c, compare lanes 1 and 3 with 2 and 4, respectively and Fig. 1d, p21 columns), consistent with the levels of p21 protein in these MEFs (Fig. 1a). Furthermore, we found that cell proliferation was increased in Fdxr+/− MEFs possibly due to decreased expression of wild-type p53, but decreased in Fdxr+/−;p53R270H/− MEFs possibly due to decreased expression of mutant p53 (Fig. 1e).
Fig. 1.
Mutant p53 is regulated by FDXR deficiency and plays a role in iron metabolism. a, b Western blots were prepared using extracts from WT, Fdxr+/−, p53R270H/−, and Fdxr+/−; p53R270H/− littermate MEFs. The blots were probed with antibodies against FDXR, p53, p21, IRP2, TfR1, FTH1, and actin, respectively. The level of proteins was normalized to that of actin, and the relative fold change is shown below each pair. c The levels of FDXR, p53, p21 and actin transcripts were measured in WT, Fdxr+/−, p53R270H/−, and Fdxr+/−; p53R270H/− littermate MEFs. The level of transcripts was normalized to that of actin, and the relative fold change is shown below each pair. d Quantitative RT-PCR was performed to measure the levels of FDXR, p53 and p21 transcripts in WT, Fdxr+/−, p53R270H/−, and Fdxr+/−; p53R270H/− littermate MEFs. e The number of WT, Fdxr+/−, p53R270H/−, and Fdxr+/−; p53R270H/− littermate MEFs over a 6-day period was counted and presented as mean ± SD from three separate experiments. f The levels of cytosolic and mitochondrial iron (Fe2+) were measured by QuantiChrom iron assay in WT, Fdxr+/−, p53R270H/−, and Fdxr+/−; p53R270H/− littermate MEFs. The level of cytosolic iron in WT MEFs (the first left column) was set at 1.0. The data represent mean ± standard deviation from three independent experiments
To determine whether iron metabolism is altered by mutant p53, QuantiChrom iron assay was performed. We found that Fdxr deficiency markedly increased the level of iron in mitochondria but not in cytoplasm in MEFs (Fig. 1f), consistent with our previous report [41]. We also found that the level of iron in mitochondria was highly increased in p53R270H/− MEFs as compared with that in wild-type MEFs. Surprisingly, the abnormal mitochondrial iron overload by Fdxr deficiency or mutant p53 was not further increased, but instead slightly decreased, by Fdxr deficiency together with mutant p53-R270H in Fdxr+/−;p53R270H/− MEFs (Fig. 1f).
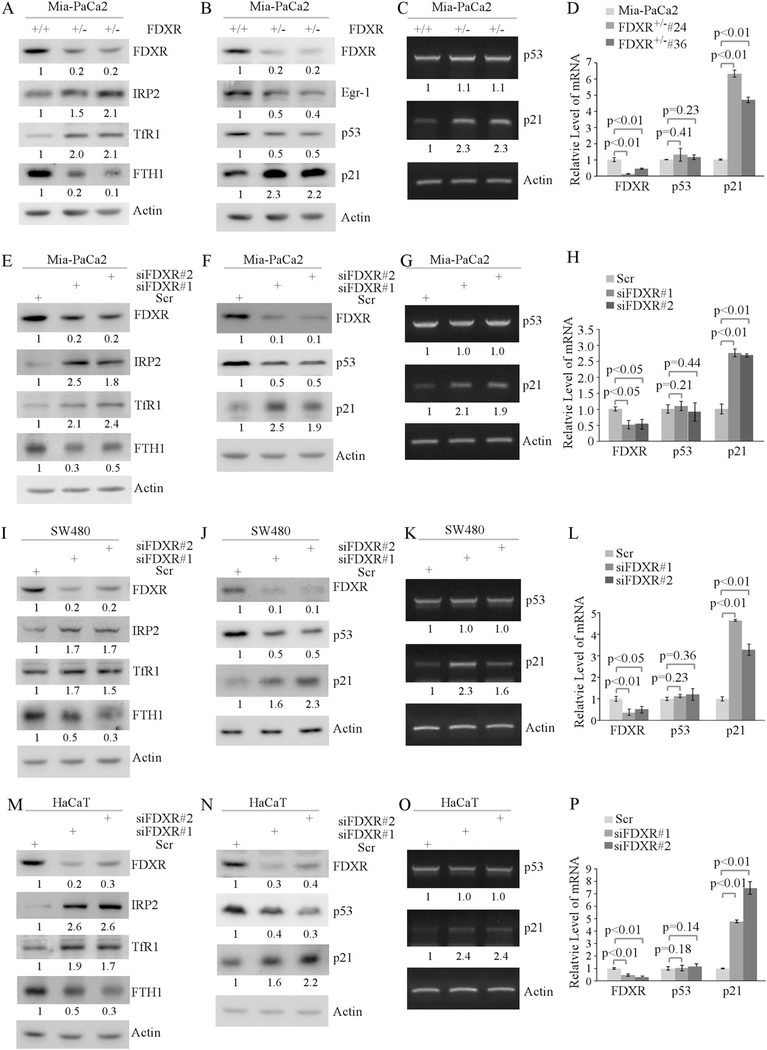
To confirm the regulation of mutant p53 expression by FDXR in MEFs, we determined whether FDXR deficiency modulates mutant p53 expression and iron metabolism in multiple human cancer cell lines carrying a mutant p53. These include MIA-PaCa2 cell line, which expresses a mutant p53-R248W; SW480 cell line, which expresses a mutant p53 with two amino acid substitutions (R273H/P309S); and HaCaT cell line, which also expresses a mutant p53 with two amino acid substitutions (H79Y/R282W). To test this, multiple FDXR-deficient MIA-PaCa2 cell lines were generated by CRISPR-cas9. We found that FDXR deficiency led to increased expression of IRP2 and TfR1, but decreased expression of FTH1 (Fig. 2a), consistent with the observations in HCT116 cells that contain wild-type p53 [41]. In addition, we found that FDXR deficiency led to decreased expression of mutant p53 and early growth responsive protein 1 (Egr-1), which has been confirmed as a target of mutant p53 [43, 44] (Fig. 2b). In contrast, p21 expression was markedly increased (Fig. 2b), which is likely due to decreased expression of mutant p53. Next, semiquantitative and quantitative RT-PCR was performed and showed that the level of mutant p53 transcript was not changed in FDXR+/− MIA-PaCa2 cells (Fig. 2c, d), consistent with the above observation in Fdxr+/− MEFs (Fig. 1c, d). In addition, the level of p21 transcript was found to be increased in FDXR+/− cells (Fig. 2c, d), likely due to decreased levels of mutant p53 and consistent with the level of p21 protein (Fig. 2b). To further confirm this, mutant p53 and iron regulatory proteins were examined in MIA-PaCa2, SW480, and HaCaT cells in which FDXR was knocked down by siRNA (Fig. 2e–p). We found that like knockout of FDXR (Fig. 2a–d) knockdown of FDXR had similar effects on IRP2, TfR1, FTH1, mutant p53, and p21 in MIA-PaCa2, SW480, and HaCaT cells (Fig. 2e–p). In addition, we found that FDXR deficiency led to decreased cell proliferation (Supplemental Fig. S1a), but had no effect on apoptosis as the levels of cleaved PARP and caspase 3 were not increased by FDXR deficiency in three mutant p53-expressing cell lines (Supplemental Fig. S1b–e). These data indicate that WT and mutant p53 are similarly regulated by FDXR and that mutant p53 disrupts normal iron metabolism.
Fig. 2.
Iron metabolism and mutant p53 pathway are regulated by FDXR deficiency. a FDXR regulates the iron homeostasis pathway. Cell lysates were collected from isogenic control and FDXR+/− Mia-PaCa2 cells, and subjected to western blot analysis with antibodies against FDXR, IRP2, TfR1, FTH1, and actin, respectively. The level of proteins was normalized to that of actin, and the relative fold change is shown below each lane. b Loss of FDXR downregulates mutant p53 and Egr-1. Cell lysates were collected from isogenic control and FDXR+/− Mia-PaCa2 cells, and subjected to western blot analysis with antibodies against FDXR, p53, p21, Egr-1 and actin, respectively. The level of proteins was normalized to that of actin, and the relative fold change is shown below each lane. c, d FDXR deficiency has no effect on the level of mutant p53 transcript. The levels of p53, p21, and actin transcripts were measured in isogenic control and FDXR+/− Mia-PaCa2 cells by semiquantitative (c) and quantitative (d) PCR. The level of transcripts was normalized to that of actin, and the relative fold change is shown below each lane. e–h The experiments were performed as in a–d except that Mia-PaCa2 cells were transfected with scrambled siRNA (Scr) as a control and with siFDXR#1 or siFDXR#2 to knock down FDXR. i–l The experiments were performed as in a–d except that SW480 cells were transfected with Scr siRNA as a control and with siFDXR#1 or siFDXR#2 for 72 h to knock down FDXR. m–p The experiments were performed as in a–d except that HaCaT cells were transfected with Scr siRNA as a control and with siFDXR#1 or siFDXR#2 for 72 h to knock down FDXR
Fdxr deficiency suppresses mutant p53 gain-of-function via decreased mutant p53 expression in Fdxr+/−;p53R270H/− mice
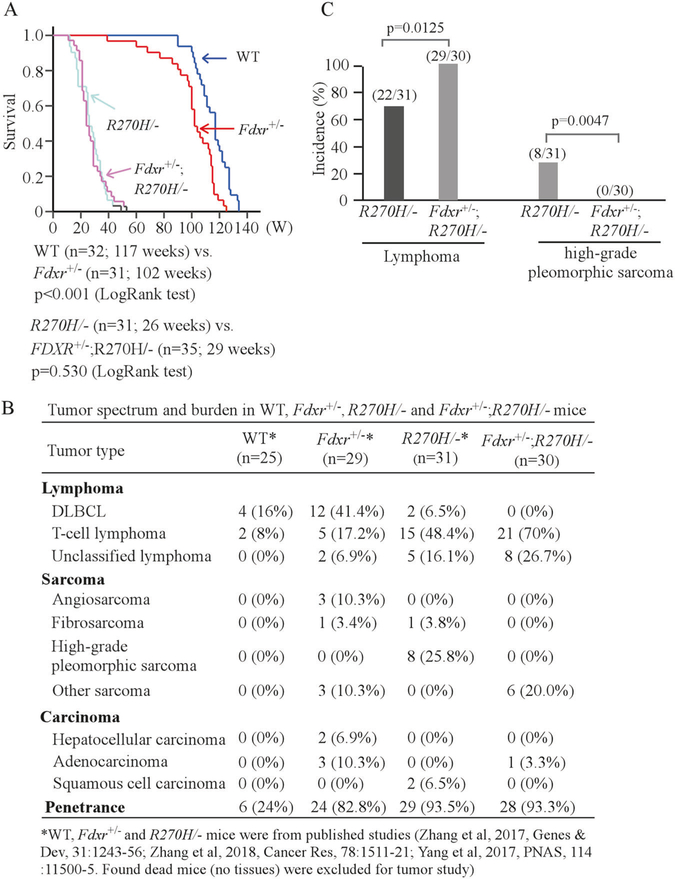
To determine whether mutant p53 cooperates with FDXR deficiency to modulate tumorigenesis, a cohort of Fdxr+/−; p53R270H/− (n = 35) mice was generated and compared with a cohort of previously reported WT (n = 32), Fdxr+/− (n = 31), and p53R270H/− (n = 34) mice throughout their life span (Supplemental Tables S1–S4). We would like to mention that all the mice were derived from the same strain, maintained in the same environment and generated within last few years [41, 44]. We previously showed that the median survival for Fdxr+/− mice (102 weeks) was significantly shorter than that for WT mice (117 weeks) (P < 0.001 by LogRank test) and Fdxr+/− mice were prone to a broad spectrum of spontaneous tumors [41] (Fig. 3a, b; Supplemental Table S2). We also showed that p53R270H/− mice succumbed to tumors, except two p53R270H/− mice died due to an unknown reason [44] (Supplemental Table S3). Here, we found that the median survival for Fdxr+/−; p53R270H/− mice was 29 weeks, which was similar to 26 weeks for p53R270H/− mice (P = 0.530 by LogRank test), but significantly shorter than that for WT and Fdxr+/− mice (Fig. 3a).
Fig. 3.
Fdxr deficiency suppresses mutant p53 gain-of-function via decreased mutant p53 expression in Fdxr+/−; p53R270H/− mice. a Kaplan–Meier survival curve for WT, Fdxr+/−, p53R270H/−, and Fdxr+/−; p53R270H/− mice.b Tumor spectrum, tumor burden and penetrance in a cohort of WT, Fdxr+/−, p53R270H/−, and Fdxr+/−; p53R270H/− mice. c The incidence of lymphoma and high-grade pleomorphic sarcoma in p53R270H/− and Fdxr+/−; p53R270H/− mice
One hallmark for mutant p53 GOF is characterized by the observations that p53R270H/− mice are prone to malignant carcinoma and sarcoma [7, 44]. Thus, histopathological analysis was performed. We found that Fdxr deficiency altered the tumor spectrum in p53R270H/− mice (Fig. 3b; Supplemental Tables S3, S4) and Fdxr+/−;p53R270H/− mice were highly prone to lymphoma as compared with p53R270H/− mice(Fig. 3c; Supplemental Tables S3, S4). Interestingly, several types of malignant tumors, such as fibrosarcoma and squamous cell carcinoma that developed in p53R270H/− mice were not found in Fdxr+/−;p53R270H/− mice (Fig. 3b; Supplemental Tables S3, S4). In addition, high-grade pleomorphic sarcoma was not detected in Fdxr+/−;p53R270H/− mice (0 out of 30) as compare with that in p53R270H/− mice (8 out of 31, P= 0.0047 by LogRank test) (Fig. 3b, c, Supplemental Tables S3, S4). These observations suggest that Fdxr deficiency suppresses p53-R270H GOF in promoting malignant tumor development via decreased mutant p53 expression.
The effect of FDXR deficiency on cell growth and iron metabolism in MIA-PaCa2 cells is mutant p53-dependent
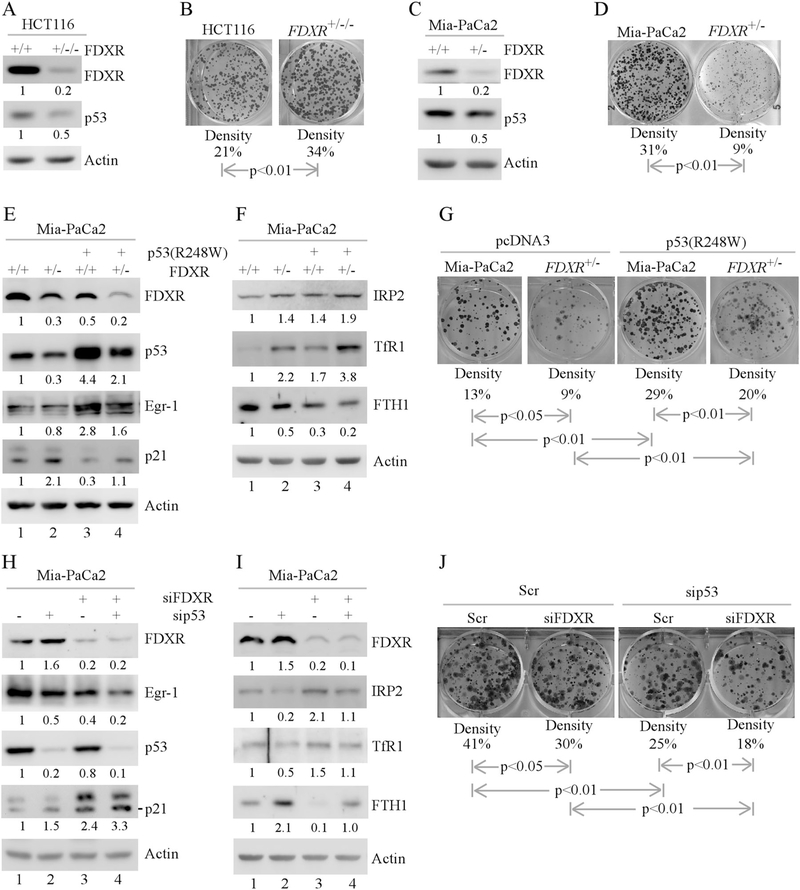
Since WT and mutant p53 have opposing functions in tumorigenesis, we hypothesized that FDXR deficiency would have two distinct biological responses in WT- and mutant p53-dependent manners. To test this, colony formation assay was performed with isogenic controls and FDXR-deficient cells. Indeed, we found that FDXR deficiency decreased WT p53 expression and increased the number of colonies by FDXR+/−/− HCT116 cells Fig. 4a, b). Conversely, we found that FDXR deficiency decreased mutant p53 expression and markedly reduced the number of colonies by FDXR+/− MIA-PaCa2 cells (Fig. 4c, d).
Fig. 4.
The effect of FDXR deficiency on cell growth and iron metabolism in MIA-PaCa2 cells is mutant p53-dependent. a Cell lysates were collected from isogenic control or FDXR+/−/− HCT116 cells and subjected to western blot analysis with various antibodies as indicated. The level of proteins was normalized to that of actin, and the relative fold change is shown below each pair. b Colony formation assay was performed with isogenic control and FDXR+/−/− HCT116 cells. The relative density for colonies was showed below each image. c, d The experiments are performed as in a, b except that isogenic control and FDXR+/− Mia-PaCa2 cells were used. e, f Isogenic control or FDXR+/− Mia-PaCa2 cells were transfected with a control vector or a vector expressing mutant p53 (R248W) for 24 h. Cell lysates were collected and subjected to western blot analysis with various antibodies as indicated. The level of proteins was normalized to that of actin, and the relative fold change is shown below each lane. g Colony formation assay was performed with isogenic control or FDXR+/− Mia-PaCa2 transfected with a control vector or a vector expressing mutant p53 (R248W). The relative density for colonies was showed below each image. h, i Mia-PaCa2 cells were transfected with Scr siRNA or siRNA against p53, followed by cotransfection with Scr siRNA or siRNA against FDXR for 72 h. Cell lysates were collected and subjected to western blot analysis with various antibodies as indicated. The level of proteins was normalized to that of actin, and the relative fold change is shown below each lane. j Colony formation assay was performed with Mia-PaCa2 cells transfected with Scr siRNA or siRNA against p53, followed by cotransfection with Scr siRNA or siRNA against FDXR. The relative density for colonies was showed below each image
To determine whether decreased expression of mutant p53 is responsible for growth suppression, mutant p53 add-back experiments were performed with WT and FDXR+/− MIA-PaCa2 cells (Fig. 4e–g). We found that upon ectopic expression of mutant p53, Egr-1 was induced in both WT and FDXR+/− cells (Fig. 4e, compare lanes 3–4 with 1–2, respectively), suggesting that ectopic mutant p53 is functional in MIA-PaCa2 cells. We also found that the levels of FDXR and p21 were reduced by ectopic mutant p53 in both WT and FDXR+/− cells (Fig. 4e, compare lanes 2–4 with 1–3, respectively). In contrast, the levels of IRP2 and TfR1 were increased, whereas the level of FTH1 was decreased, by ectopic mutant p53 (Fig. 4f, compare lanes 3–4 with 1–2, respectively). Furthermore, ectopic mutant p53 was able to increase colony formation in control MIA-PaCa2 cells (Fig. 4g, compare the 1st column with the 3rd column), which is consistent with our previous report [45]. Most importantly, we found that ectopic mutant p53 was able to restore the ability of FDXR-deficient cells to form colonies (Fig. 4g, compare the 4th column with the 2nd column).
To further determine the requirement of mutant p53 for FDXR to modulate cell growth in MIA-PaCa2 cells, FDXR, mutant p53 or both were knocked down by siRNA (Fig. 4h–j). We found that upon knockdown of mutant p53, the level of Egr-1 was decreased, whereas the levels of FDXR and p21 were increased, in MIA-PaCa2 cells (Fig. 4h, compare lane 1 with 2), consistent with the observations in Fig. 4e, f. We also found that upon knockdown of both FDXR and mutant p53, the levels of mutant p53 and Egr-1 were further decreased, whereas the level of p21 was further increased (Fig. 4h, compare lanes 3–4 with 1–2, respectively). Moreover, we found that upon knockdown of mutant p53, the levels of IRP2 and TfR1 were decreased, whereas the level of FTH1 was increased (Fig. 4i, compare lane 1 with 2). However, upon knockdown of both p53 and FDXR, the levels of IRP2, TfR1, and FTH1 in FDXR-deficient MIA-PaCa2 cells were restored to near normal levels in control cells (Fig. 4i, compare lane 4 with 1 and 2, respectively). Next, colony formation assay was performed and showed that the number of colonies was reduced upon knockdown of FDXR or mutant p53 in MIA-PaCa2 cells (Fig. 4j, compare the 1st column with the 2nd and 3rd columns, respectively), which is consistent with the observations in Fig. 4d. Moreover, the number of colonies was further reduced upon knockdown of both FDXR and mutant p53 as compared to knockdown of FDXR or mutant p53 alone (Fig. 4j, compare the 4th column with the 2nd and 3rd columns, respectively). Together, these data suggest that the level of mutant p53 plays a critical role in cell proliferation and iron homeostasis.
Mutant p53 expression is regulated by IRP2
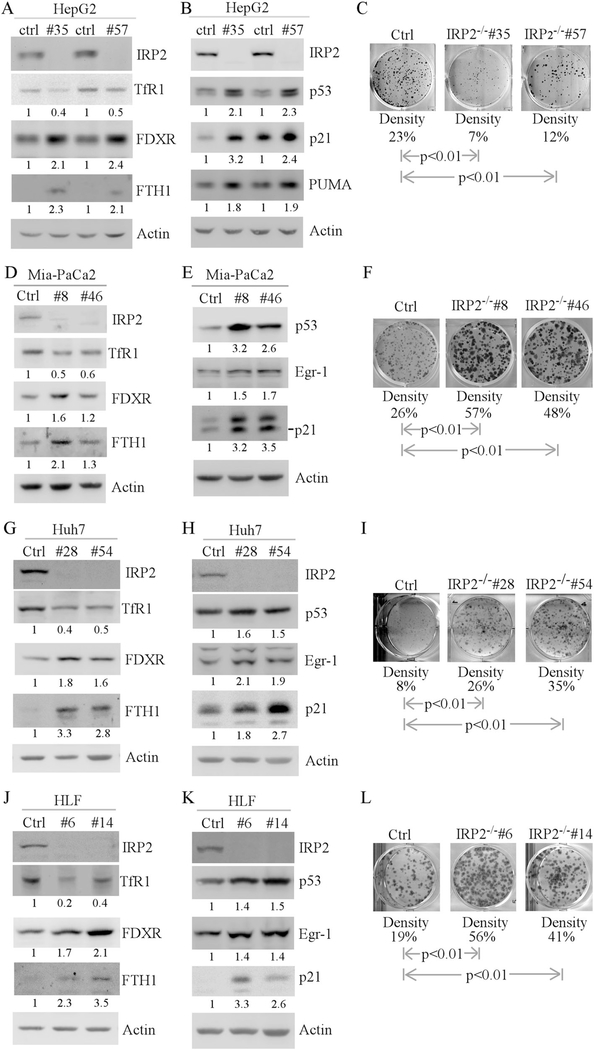
Recently, we found that IRP2 regulates WT p53 translation via an IRE in p53 3′UTR [41]. Indeed, we found that upon knockout of IRP2, WT p53 was increased along with its targets (FDXR, p21 and PUMA) in HepG2 cells (Fig. 5a, b). In addition, knockout of IRP2 led to decreased expression of TfR1 and increased expression of FTH1 (Fig. 5a), consistent with previous report [41]. Moreover, we found that knockout of IRP2 decreased the number of colonies by IRP2−/− HepG2 cells (Fig. 5c), likely due to increased levels of WT p53. Consistently, we found that cell proliferation was decreased by loss of IRP2 in HepG2 cells (Supplemental Fig. S2A). Since mutant p53 also carries an IRE in its 3′UTR, we postulated that mutant p53 should be regulated by IRP2. To test this, IRP2 was deleted by CRISPR/cas9 in MIA-PaCa2, Huh7 and HLF cells, all of which carry mutant p53. As expected, knockout of IRP2 led to decreased expression of TfR1 and increased expression of FTH1 in MIA-PaCa2, Huh7 and HLF cells (Fig. 5d, g, j). Similarly, we found that knockout of IRP2 led to increased expression of mutant p53 along with its target Egr-1 (Fig. 5e, h, k). Interestingly, the levels of FDXR and p21 were induced by loss of IRP2 (Fig. 5d, e, g, h, j, k). Indeed, we found that p21 was regulated by IRP2 independent of p53 (Supplemental Fig. S3). Next, colony formation was performed and showed that knockout of IRP2 increased the number of colonies by MIA-PaCa2, Huh7, and HLF cells (5F, I, L). Consistently, we found that cell proliferation was increased by loss of IRP2 in cells expressing mutant p53 (Supplemental Fig. S2b–d). Thus, although loss of IRP2 increases p21 expression, the level of p21 may not be sufficient to antagonize increased mutant p53 to promote cell growth. Interestingly, loss of IRP2 had no effect on apoptosis as the levels of cleaved PARP were not increased in MIA-PaCa2, Huh7 and HLF cells (Supplemental Fig. S2e–g). These observations suggest that loss of IRP2 exerts two opposing effects on cell growth via WT and mutant p53, respectively.
Fig. 5.
Mutant p53 expression is regulated by IRP2. a, b Iron regulatory proteins and the p53 pathway are regulated by IRP2. Western blots were prepared with lysates from isogenic control or IRP2−/− HepG2 cells (a, b), and then probed with antibodies against IRP2 (a, b), FDXR (a), TfR1 (a), FTH1 (a), p53 (b), p21 (b), PUMA (b) and actin (a, b), respectively. The level of proteins was normalized to that of actin, and the relative fold change is shown below each pair. c Loss of IRP2 inhibits cell growth in HepG2 cell lines. Colony formation assay was performed with isogenic control or IRP2−/− HepG2 cells. The relative density for colonies was showed below each image. d–f The experiments were performed as in a–c except that isogenic control or IRP2−/− Mia-PaCa2 cells were used. g–i The experiments were performed as in a–c except that isogenic control or IRP2−/− Huh7 cells were used. j–l The experiments were performed as in a–c except that isogenic control or IRP2−/− HLF cells were used
Previously, we found that FDXR deficiency modulates ferroptosis in cells carrying wild-type p53 [41]. Thus, we examined whether altered expression of mutant p53 modulates the extent of ferroptosis induced by Erastin and RSL3, both of which are known ferroptosis inducers [46]. We found that FDXR deficiency had little if any effect on Erastin- and RSL-induced ferroptosis in MIA-PaCa2 cells (Supplemental Fig. S4a, b). Interestingly, we found that Erastin-induced ferroptosis was inhibited by loss of IRP2 in MIA-PaCa2 and Huh7 cells (Supplemental Fig. S4c, e). In addition, RSL-induced ferroptosis was inhibited by loss of IRP2 in MIA-PaCa2 but not in Huh7 cells (Supplemental Fig. S4d, f). Thus, further studies are warranted to analyze these intriguing observations.
IRP2 is a suppressor of mutant p53 in tumorigenesis
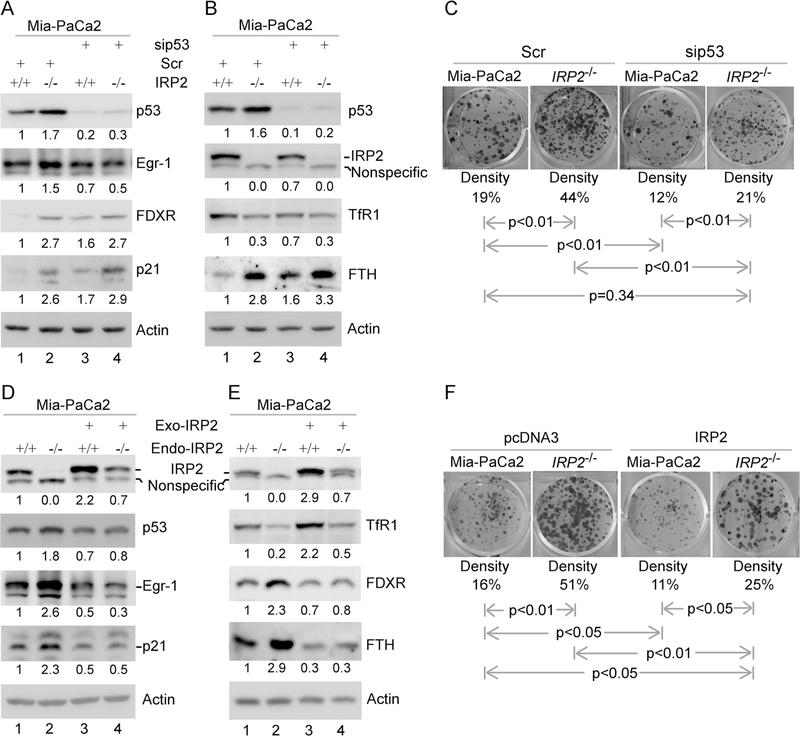
The above studies suggest that loss of IRP2 leads to increased mutant p53 expression and subsequently increased cell growth (Fig. 5 and Supplemental Fig. S2). To determine whether increased expression of mutant p53 is responsible for increased cell growth, mutant p53 was knocked down by siRNA in isogenic control and IRP2−/− MIA-PaCa2 cells. We found that upon knockdown of mutant p53, Egr-1 was decreased whereas the levels of FDXR and p21 were increased in both isogenic control and IRP2−/− cells (Fig. 6a, compare lanes 3–4 with 1–2, respectively), consistent with the above observation (Fig. 5). We also found that upon knockdown of mutant p53, the level of TfR1 was slightly decreased whereas the level of FTH1 was increased in isogenic control cells (Fig. 6b, compare lanes 3 with 1). In addition, the level of IRP2 was slightly decreased by knockdown of mutant p53 (Fig. 6b, compare lanes 3 with 1), consistent with the above observations (Fig. 4i). Furthermore, colony formation assay showed that knockdown of mutant p53 decreased the numbers of colonies by isogenic control cells (Fig. 6c, compare the 1st column with the 3rd column), consistent with the observation in Fig. 4j. Most importantly, the number of colonies increased by loss of IRP2 was reversed to near-normal level by knockdown of mutant p53 (Fig. 6c, compare the 4th column with the 1st and 2nd columns, respectively).
Fig. 6.
IRP2 is a suppressor of mutant p53 in tumorigenesis. a, b Isogenic control and IRP2−/− Mia-PaCa2 cells were transfected with Scr siRNA or siRNA against p53, followed by western blot analysis with various antibodies as indicated. The level of proteins was normalized to that of actin, and the relative fold change is shown below each lane. c Colony formation assay was performed with isogenic control and IRP2−/− Mia-PaCa2 cells transfected with Scr siRNA or siRNA against p53. The relative density for colonies was showed below each image. d, e Isogenic control and IRP2−/− Mia-PaCa2 cells were transfected with control pcDNA3 or a vector expressing HA-tagged IRP2 for 24 h, followed by western blot analysis with various antibodies as indicated. The level of proteins was normalized to that of actin, and the relative fold change is shown below each lane. f Colony formation assay was performed with isogenic control and IRP2−/− Mia-PaCa2 cells transfected with control pcDNA3 or a vector expressing HA-tagged IRP2. The relative density for colonies was showed below each image
Next, to determine whether IRP2 is a suppressor of mutant p53, IRP2 add-back experiments were performed in isogenic control and IRP2−/− MIA-PaCa2 cells. We found that in IRP2-KO cells, the level of TfR1 reduced by IRP2 knockout was increased, whereas the level of FTH1 increased by IRP2 knockout was reduced, by ectopic expression of IRP2 (Fig. 6e, compare lane 4 with 2). These data suggest that ectopic IRP2 is functional in MIA-PaCa2 cells. In addition, mutant p53, p21 and Egr-1 were decreased by ectopic expression of IRP2 in isogenic control cells (carrying endogenous IRP2) (Fig. 5d, compare lane 3 with 1). Moreover, the levels of mutant p53, Egr-1 and p21 increased by IRP2-KO were also reduced by ectopic expression of IRP2 (Fig. 6d, compare lane 4 with 1 and 2, respectively). Furthermore, colony formation assay showed that ectopic expression of IRP2 inhibited the number of colonies by MIA-PaCa2 cells (Fig. 6f, compare the 3rd column with the 1st column), consistent with the effect of IRP2 on mutant p53 expression (Fig. 6d, compare lane 3 with 1). Most importantly, the number of colonies increased by IRP2-KO was markedly reduced by ectopic expression of IRP2 (Fig. 6f, compare the 4th column with the 2nd column, respectively). Taken together, these data suggest that IRP2 represses mutant p53 expression and mutant p53-mediated cell growth.
Discussion
In the current study, we found that FDXR deficiency decreases mutant p53 expression along with altered iron metabolism in p53R270H/− MEFs and cancer cells carrying mutant p53. We also found that loss of IRP2 increases mutant p53 expression as well as increased cell growth in cancer cells expressing mutant p53. Moreover, we found that ectopic expression of IRP2 suppressed cell growth in a mutant p53-dependent manner. These results reveal that both FDXR and IRP2 have two opposing functions in tumorigenesis via WT and mutant p53, respectively.
It is well established that mice carrying knock-in mutant p53-R270H are prone to aggressive tumors, such as sarcoma and carcinoma, as compared with p53−/− mice [7, 47] (also see Fig. 3 and Supplemental Table S3). Since there is no difference in lifespan and tumor incidence between p53R270H/− (~26 weeks) and p53−/− mice (~24 weeks) [7, 44], it is not surprising that the reduced level of mutant p53-R270H by Fdxr deficiency would not make a difference in the lifespan and tumor incidence in Fdxr+/−;p53R270H/− mice as compared with p53R270H/− mice (Fig. 3a, b). However, several types of tumors, especially malignant fibrosarcoma and squamous cell carcinoma that often develop in p53R270H/− mice, were not found in Fdxr+/−; p53R270H/− mice (Fig. 3b and Supplemental Tables S3, S4). In addition, high-grade pleomorphic sarcoma was not detected in Fdxr+/−;p53R270H/− mice (Supplemental Tables S3, S4). These observations suggest that Fdxr deficiency suppresses mutant p53-R270H GOF via decreased expression of mutant p53. Similarly, a previous report showed that FDXR knockdown suppresses T47D mammary tumor cell growth and xenograft [48], potentially due to decreased expression of mutant p53 (L194F) in T47D cells. Moreover, we found that cell growth is suppressed by decreased expression of mutant p53 via FDXR deficiency in multiple cell lines (Figs. 2 and 4; Supplemental Fig. S1). Thus, targeted suppression of FDXR may be explored as an option to kill tumors addicted to mutant p53.
Our data indicate that increased expression of IRP2 in FDXR-deficient cells is responsible for decreased expression of mutant p53 (Fig. 4). In addition, ectopic expression of IRP2 suppresses mutant p53 expression and mutant p53-dependent cell growth (Fig. 6). Moreover, our data suggest that IRP2 expression is indirectly regulated by mutant p53 as knockdown of mutant p53 leads to decreased IRP2 expression (Figs. 4i and 6b) whereas ectopic expression of mutant p53 leads to increased IRP2 expression (Fig. 4f). These observations prompt us to speculate that the mutual regulation between IRP2 and mutant p53 constitutes a feedback loop and that the IRP2-mutant p53 loop plays a critical role in the progression of tumors carrying mutant p53. Thus, activating IRP2 without causing iron overload may be explored as an alternative strategy to kill tumors addicted to mutant p53. However, since p53 is frequently mutated in cancer and IRP2 is ubiquitously expressed, the use of ion chelators to decrease iron overload [49, 50], which is often accompanied with decreased IRP2 expression, may have an unintended off-target effect to increase mutant p53, leading to progression of tumors that carry mutant p53. Therefore, monitoring the status of the p53 gene in tumor patients is recommended prior to the use of iron chelators.
We would like to note that loss of IRP2, which leads to increased expression of mutant p53, de-sensitizes mutant p53-expressing cells to RSL3- and Erastin-induced ferroptosis (Supplemental Fig. S4). Interestingly, a recent report showed that mutant p53 sensitizes tumor cell to ferroptosis [51]. Previously, we found that FDXR modulates RSL3-and Erastin-induced ferroptosis in cells carrying wild-type p53 [41]. Together, these results suggest that the ferroptosis pathway is regulated by p53 modulators, such as FDXR and IRP2, in wild-type and mutant p53-dependent manners.
Materials and methods
Fdxr- and p53-mutant mouse models
Fdxr +/− mice (on C57BL/6N background) were generated by Mouse Biology Program at UC Davis as described previously [41]. The p53+/− mice (on C57BL/6J background) and p53R270H/+ mice (on C57BL/6J background) were purchased from the Jackson Laboratory. The primers used for genotyping Fdxr+/−, p53+/− and p53R270H/+ mice are listed in Supplemental Table S5. All animals were housed and bred and maintained in a specific pathogen-free environment in the experimental mouse facility at the University of California. All animal procedures were approved by UC Davis IACUC and were in adherence to the NIH “Guide for the Care and Use of Laboratory Animals”.
Cell culture
HCT116, SW480, HaCaT, Mia-PaCa2, HepG2, Huh7, HLF, Hep3B, and their derivatives were cultured in DMEM (Dulbecco’s Modified Eagle’s medium, Invitrogen) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT). HCT116, SW480, HaCaT, Mia-PaCa2 cell lines were obtained from ATCC between 2007 and 2018 and used at below passage 20 or within 2 months for this study after reception or thawing. HepG2, Huh7, HLF and Hep3B cells were obtained from Dr. Yuyou Duan at UC Davis Medical Center and were originally purchased from ATCC. Cells were tested negative for mycoplasma and after thawing and used within two months. Since all cell lines from ATCC have been thoroughly tested and authenticated, we did not authenticate the cell lines used in this study. Wild-type, Fdxr +/−, p53R270H/−, and Fdxr +/−;p53R270H/− MEFs were generated as described previously [41, 52] and cultured in DMEM supplemented with 10% fetal bovine serum, 55 μM β-mercaptoethanol, and MEM non-essential amino acid solution (Cellgro, Manassas, VA).
Plasmid construction and cell line generation
FDXR, IRP2, and p53 gRNAs were designed using CRISPR design tool (crispr.mit.edu) and listed in Supplemental Table S6. Cells deficient in FDXR, IRP2, or p53 were generated and confirmed as previously described [41]. The primers used for sequencing are listed in Supplemental Table S5.
Western blot analysis
Western blot was performed as described [41]. Antibodies against p53, p21, Egr-1, IRP2 (The anti-IRP2 antibody used in Figs. 3–5 and Fig. 6 have different lot number), FTH1, TfR1, and actin were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-mouse IRP2 antibody was a generous gift of Elizabeth Leibold (University of Utah) [53]. Anti-mouse p53 (1C12) and anti-PARP antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA). Antibodies against FDXR were purchased from Abcam (Cambridge, MA, USA). HRP-conjugated secondary antibodies against rabbit and mouse IgG were purchased from BioRad (Hercules, CA). The immunoreactive bands were visualized by enhanced chemiluminescence (Thermo Fisher Scientific Inc, Carlsbad, CA) and quantified by densitometry with the BioSpectrum® 810 Imaging System (UVP LLC, Upland, CA).
RNA isolation, RT-PCR analysis, and qRT-PCR
Total RNAs were extracted from cells using TRIzol (Invitrogen Life Technologies, Grand Island, NY) according to the manufacturer’s instructions. cDNA was synthesized using RevertAid Reverse Transcriptase Kit (Thermo Fisher Scientific, Grand Island, NY) according to the manufacturer’s protocol. The levels of p53, p21, FDXR, and actin transcripts were measured by PCR with specific primers listed in Supplemental Table S7.
For qRT-PCR analysis, 15-μL reactions were set up using 2 × qPCR SYBR Green Mix (Thermal Fisher) along with 5 μmol/L primers. The reactions were run on a StepOne plus (Invitrogen, Carlsbad, CA) using a two-step cycling program: 95 °C for 15 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 30 s, and 68 °C for 30 s. A melt curve (57–95 °C) was generated at the end of each run to verify the specificity. The primers for murine and human FDXR, p53 and p21 were listed in Supplemental Table S7.
RNA interference
Scrambled siRNA (5′-GCAGUGUCUCCACGUACUAdTdT-3′), siRNAs against FDXR (siFDXR#1:5′-GCTCAGCAGCATTGGG-TAT-3′ and #2: 5′-GCTCAGCAGCATTGGGTAT-3′), siRNA against human p53 (5′-CACCUUGAUCCAGCGGACUUAdTdT-3′) and siRNA against human IRP2 (5′-GCGAUUUC-CAGGCUUGCUUdTdT-3′) were purchased from Dharmacon (Chicago, IL, USA). For siRNA transfection, siRNAMax Lipid Reagent (Thermo Fisher Scientific) was used according to the user’s manual.
Histological analysis
Mouse tissue processing and H&E staining were performed as previously described [41]. All the mouse tissues were blindly diagnosed by pathologists without revealing the genotypes of mice. Hematoxylin, Eosin, and xylene were purchased from Thermo Fisher Scientific (Fisher, Pittsburgh, PA).
Iron measurement
The level of iron in mitochondria and cytoplasm was detected by QuantiChrom™ Iron Assay Kit (Bioassay, Hayward, CA) as previously described [41]. Iron levels in mitochondria and cytosolic fractions were determined by measuring the value of absorption at 590 nm using a microplate reader (Bio-Rad, Hercules, USA).
Colony formation assay
HCT116, MIA PaCa-2, HepG2, HLF, or Huh7 cells and their derivatives (~1000 per well) in six-well plates were cultured for 13–15 days. The clones were fixed with methanol/glacial acetic acid (7:1) and then stained with 0.1% of crystal violet. The color density of clones was scanned and analyzed using Image J [54].
Statistical analysis
For iron concentration, data were presented as Mean ± SD. Statistical significance was determined by two-tailed Student’s t test. Fisher’s exact test was used for comparison between two genotypes. For Kaplan–Meier survival analysis, LogRank test was performed. Values of P < 0.05 were considered significant.
Data availability
The authors declare that all data supporting the findings of this study are available within the article and its supplementary information files.
Supplementary Material
Acknowledgements
This work was supported in part by the National Institutes of Health grants CA224433-01. The authors would like to thank Dr. Elizabeth Leibold and Dr. Kuanyu Li for their generous gifts of IRP2 antibodies.
Footnotes
Conflict of interest The authors declare that they have no conflict of interest.
Compliance with ethical standards
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information The online version of this article (https://doi.org/10.1038/s41388-019-0876-5) contains supplementary material, which is available to authorized users.
References
- 1.Freed-Pastor WA, Prives C. Mutantp53: one name, many proteins. Genes Dev. 2012;26:1268–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol.2013;15:2–8. [DOI] [PubMed] [Google Scholar]
- 3.Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol. 2010;2:a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanel W, Marchenko N, Xu S, Yu SX, Weng W, Moll U. Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis. Cell Death Differ. 2013;20:898–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li–Fraumeni syndrome. Cell. 2004;119:861–72. [DOI] [PubMed] [Google Scholar]
- 6.Lee MK, Teoh WW, Phang BH, Tong WM, Wang ZQ, Sabapathy K. Cell-type, dose, and mutation-type specificity dictate mutant p53 functions in vivo. Cancer Cell. 2012;22:751–64. [DOI] [PubMed] [Google Scholar]
- 7.Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, et al. Mutant p53 gain of function in two mouse models of Li–Fraumeni syndrome. Cell. 2004;119:847–60. [DOI] [PubMed] [Google Scholar]
- 8.Zhou G, Wang J, Zhao M, Xie TX, Tanaka N, Sano D, et al. Gainof-function mutant p53 promotes cell growth and cancer cell metabolism via inhibition of AMPK activation. Mol Cell. 2014;54:960–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hentze MW, Muckenthaler MU, Galy B, Camaschella C. Two to tango: regulation of Mammalian iron metabolism. Cell. 2010;142:24–38. [DOI] [PubMed] [Google Scholar]
- 10.Lawen A, Lane DJ. Mammalian iron homeostasis in health anddisease: uptake, storage, transport, and molecular mechanisms of action. Antioxid Redox Signal. 2013;18:2473–507. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Pantopoulos K. Regulation of cellular iron metabolism.Biochem J. 2011;434:365–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torti SV, Torti FM. Iron and cancer: more ore to be mined. Nat Rev Cancer. 2013;13:342–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fonseca-Nunes A, Jakszyn P, Agudo A. Iron and cancer risk-asystematic review and meta-analysis of the epidemiological evidence. Cancer Epidemiol Biomark Prev. 2014;23:12–31. [DOI] [PubMed] [Google Scholar]
- 15.Kwok JC, Richardson DR. The iron metabolism of neoplasticcells: alterations that facilitate proliferation? Crit Rev Oncol/Hematol. 2002;42:65–78. [DOI] [PubMed] [Google Scholar]
- 16.Beutler E Hemochromatosis: genetics and pathophysiology.Annu Rev Med. 2006;57:331–47. [DOI] [PubMed] [Google Scholar]
- 17.Hann HW, Stahlhut MW, Hann CL. Effect of iron and desferoxamine on cell growth and in vitro ferritin synthesis in human hepatoma cell lines. Hepatology. 1990;11:566–9. [DOI] [PubMed] [Google Scholar]
- 18.Shaheen NJ, Silverman LM, Keku T, Lawrence LB, Rohlfs EM,Martin CF, et al. Association between hemochromatosis (HFE) gene mutation carrier status and the risk of colon cancer. J Natl Cancer Inst. 2003;95:154–9. [DOI] [PubMed] [Google Scholar]
- 19.Simcox JA, McClain DA. Iron and diabetes risk. Cell Metab.2013;17:329–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith AG, Carthew P, Clothier B, Constantin D, Francis JE,Madra S. Synergy of iron in the toxicity and carcinogenicity of polychlorinated biphenyls (PCBs) and related chemicals. Toxicol Lett. 1995;82–83:945–50. [DOI] [PubMed] [Google Scholar]
- 21.Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–3. [DOI] [PubMed] [Google Scholar]
- 22.Hentze MW, Seuanez HN, O’Brien SJ, Harford JB, Klausner RD. Chromosomal localization of nucleic acid-binding proteins by affinity mapping: assignment of the IRE-binding protein gene to human chromosome 9. Nucleic Acids Res. 1989;17:6103–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rouault TA, Tang CK, Kaptain S, Burgess WH, Haile DJ,Samaniego F, et al. Cloning of the cDNA encoding an RNA regulatory protein-the human iron-responsive element-binding protein. Proc Natl Acad Sci USA. 1990;87:7958–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Butt J, Kim HY, Basilion JP, Cohen S, Iwai K, Philpott CC, et al. Differences in the RNA binding sites of iron regulatory proteins and potential target diversity. Proc Natl Acad Sci USA. 1996;93:4345–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henderson BR, Menotti E, Kuhn LC. Iron regulatory proteins 1 and 2 bind distinct sets of RNA target sequences. J Biol Chem. 1996;271:4900–8. [DOI] [PubMed] [Google Scholar]
- 26.Rouault TA. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat Chem Biol. 2006;2:406–14. [DOI] [PubMed] [Google Scholar]
- 27.Zhang F, Wang W, Tsuji Y, Torti SV, Torti FM. Posttranscriptional modulation of iron homeostasis during p53-dependent growth arrest. J Biol Chem. 2008;283:33911–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gunshin H, Allerson CR, Polycarpou-Schwarz M, Rofts A,Rogers JT, Kishi F, et al. Iron-dependent regulation of the divalent metal ion transporter. FEBS Letters. 2001;509:309–16. [DOI] [PubMed] [Google Scholar]
- 29.Brandt ME, Vickery LE. Expression and characterization of human mitochondrial ferredoxin reductase in Escherichia coli. Arch Biochem Biophys. 1992;294:735–40. [DOI] [PubMed] [Google Scholar]
- 30.Lange H, Kaut A, Kispal G, Lill R. A mitochondrial ferredoxin is essential for biogenesis of cellular iron-sulfur proteins. Proc Natl Acad Sci USA. 2000;97:1050–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller JJ, Lapko A, Bourenkov G, Ruckpaul K, Heinemann U.Adrenodoxin reductase-adrenodoxin complex structure suggests electron transfer path in steroid biosynthesis. J Biol Chem. 2001;276:2786–9. [DOI] [PubMed] [Google Scholar]
- 32.Sheftel AD, Stehling O, Pierik AJ, Elsasser HP, Muhlenhoff U,Webert H, et al. Humans possess two mitochondrial ferredoxins, Fdx1 and Fdx2, with distinct roles in steroidogenesis, heme, and Fe/S cluster biosynthesis. Proc Natl Acad Sci USA. 2010;107:11775–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi Y, Ghosh M, Kovtunovych G, Crooks DR, Rouault TA. Both human ferredoxins 1 and 2 and ferredoxin reductase are important for iron-sulfur cluster biogenesis. Biochim Biophys Acta. 2012;1823:484–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hwang PM, Bunz F, Yu J, Rago C, Chan TA, Murphy MP, et al. Ferredoxin reductase affects p53-dependent, 5-fluorouracilinduced apoptosis in colorectal cancer cells. Nat Med. 2001;7:1111–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu G, Chen X. The ferredoxin reductase gene is regulated by thep53 family and sensitizes cells to oxidative stress-induced apoptosis. Oncogene. 2002;21:7195–204. [DOI] [PubMed] [Google Scholar]
- 36.Edmondson DA, Karski EE, Kohlgruber A, Koneru H, Matthay KK, Allen S, et al. Transcript analysis for internal biodosimetry using peripheral blood from neuroblastoma patients treated with (131)I-mIBG, a targeted radionuclide. Radiat Res. 2016;186:235–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lacombe J, Sima C, Amundson SA, Zenhausern F. Candidategene biodosimetry markers of exposure to external ionizing radiation in human blood: a systematic review. PLoS ONE. 2018;13:e0198851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Brien G, Cruz-Garcia L, Majewski M, Grepl J, Abend M, Port M, et al. FDXR is a biomarker of radiation exposure in vivo. Sci Rep. 2018;8:684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okumura H, Uchikado Y, Omoto I, Motomura M, Kita Y, Sasaki K, et al. Ferredoxin reductase is useful for predicting the effect of chemoradiation therapy on esophageal squamous cell carcinoma. Anticancer Res. 2015;35:6471–4. [PubMed] [Google Scholar]
- 40.Yu J, Marsh S, Ahluwalia R, McLeod HL. Ferredoxin reductase: pharmacogenomic assessment in colorectal cancer. Cancer Res. 2003;63:6170–3. [PubMed] [Google Scholar]
- 41.Zhang Y, Qian Y, Zhang J, Yan W, Jung YS, Chen M, et al. Ferredoxin reductase is critical for p53-dependent tumor suppression via iron regulatory protein 2. Genes Dev. 2017;31:1243–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elbendary AA, Cirisano FD, Evans AC Jr, Davis PL, Iglehart JD,Marks JR, et al. Relationship between p21 expression and mutation of the p53 tumor suppressor gene in normal and malignant ovarian epithelial cells. Clin Cancer Res. 1996;2:1571–5. [PubMed] [Google Scholar]
- 43.Sauer L, Gitenay D, Vo C, Baron VT. Mutant p53 initiates a feedback loop that involves Egr-1/EGF receptor/ERK in prostate cancer cells. Oncogene. 2010;29:2628–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang HJ, Zhang J, Yan W, Cho SJ, Lucchesi C, Chen M, et al. Ninjurin 1 has two opposing functions in tumorigenesis in a p53-dependent manner. Proc Natl Acad Sci USA. 2017;114:11500–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan W, Chen X. Characterization of functional domains necessary for mutant p53 gain of function. J Biol Chem. 2010;285: 14229–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang L, Kon N, Li T, Wang SJ, Su T, Hibshoosh H, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bertout JA, Patel SA, Fryer BH, Durham AC, Covello KL, Olive KP, et al. Heterozygosity for hypoxia inducible factor 1alpha decreases the incidence of thymic lymphomas in a p53 mutant mouse model. Cancer Res. 2009;69:3213–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J, Wang C, Chen X, Takada M, Fan C, Zheng X, et al. EglN2 associates with the NRF1-PGC1alpha complex and controls mitochondrial function in breast cancer. EMBO J. 2015;34:2953–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hershko C, Link G, Pinson A, Peter HH, Dobbin P, Hider RC.Iron mobilization from myocardial cells by 3-hydroxypyridin-4-one chelators: studies in rat heart cells in culture. Blood. 1991;77:2049–53. [PubMed] [Google Scholar]
- 50.Porter JB, Abeysinghe RD, Marshall L, Hider RC, Singh S.Kinetics of removal and reappearance of non-transferrin-bound plasma iron with deferoxamine therapy. Blood. 1996;88:705–13. [PubMed] [Google Scholar]
- 51.Xie Y, Zhu S, Song X, Sun X, Fan Y, Liu J, et al. The tumor suppressor p53 limits ferroptosis by blocking DPP4 activity. Cell Rep. 2017;20:1692–704. [DOI] [PubMed] [Google Scholar]
- 52.Zhang J, Xu E, Ren C, Yang HJ, Zhang Y, Sun W, et al. Genetic ablation of Rbm38 promotes lymphomagenesis in the context of mutant p53 by downregulating PTEN. Cancer Res.2018;78:1511–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo B, Yu Y, Leibold EA. Iron regulates cytoplasmic levels of anovel iron-responsive element-binding protein without aconitase activity. J Biol Chem. 1994;269:24252–60. [PubMed] [Google Scholar]
- 54.Guzman C, Bagga M, Kaur A, Westermarck J, Abankwa D. ColonyArea: an ImageJ plugin to automatically quantify colony formation in clonogenic assays. PLoS ONE. 2014;9:e92444. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the article and its supplementary information files.