Abstract
The currently available anti-HIV-1 therapeutics is highly beneficial to infected patients. However, clinical failures occur as a result of the ability of HIV-1 to rapidly mutate. One approach to overcome drug resistance is to target HIV-1 proteins that are highly conserved among phylogenetically distant viral strains and currently not targeted by available therapies. In this respect, the nucleocapsid (NC) protein, a zinc finger protein, is particularly attractive, as it is highly conserved and plays a central role in virus replication, mainly by interacting with nucleic acids. The compelling rationale for considering NC as a viable drug target is illustrated by the fact that point mutants of this protein lead to noninfectious viruses and by the inability to select viruses resistant to a first generation of anti-NC drugs. In our review, we discuss the most relevant properties and functions of NC, as well as recent developments of small molecules targeting NC. Zinc ejectors show strong antiviral activity, but are endowed with a low therapeutic index due to their lack of specificity, which has resulted in toxicity. Currently, they are mainly being investigated for use as topical microbicides. Greater specificity may be achieved by using non-covalent NC inhibitors (NCIs) targeting the hydrophobic platform at the top of the zinc fingers or key nucleic acid partners of NC. Within the last few years, innovative methodologies have been developed to identify NCIs. Though the antiviral activity of the identified NCIs needs still to be improved, these compounds strongly support the druggability of NC and pave the way for future structure-based design and optimization of efficient NCIs.
Keywords: Long Terminal Repeat, Virtual Screening, Nuclear Magnetic Resonance Spectroscopy, Isothermal Titration Calorimetry, Nucleocapsid Protein
Introduction
The clinical implementation of antiretroviral drugs (ARDs) for HIV-infected individuals resulted in shifting an acute and lethal disease, AIDS, to a clinically manageable condition. This combined use of ARDs as a therapy is based on small molecular weight inhibitors targeting either (i) the key viral enzymes required for HIV-1 replication, namely reverse transcriptase (RT), integrase (IN), and protease (PR) or, less commonly, (ii) the key proteins that control viral entry into the host cell. These therapies efficiently lower the systemic viral burden below the detection limit in the blood and strongly favor long-term survival of HIV-infected patients despite the persistence of latently infected cellular reservoirs. Nevertheless, emergence and circulation of multidrug-resistant HIV-1 strains are fueled by the high rates of HIV-1 mutation and recombination, thus emphasizing the continuous need for novel therapies and innovative strategies to overcome drug resistance (DR) (Richman 2014).
Resistant and multidrug-resistant HIV-1 strains have been identified in the clinic for each of commercially available antiretroviral drugs, as well as for most of the drug combinations. Therefore, defining a suitable combination of antiretroviral drugs to maintain viral suppression at the individual patient level, as well as developing novel drug-development strategies with the purpose of providing a cure, constitutes the current research and clinical efforts worldwide. As a consequence, there is a strong need for identifying novel antivirals to crucial viral determinants that are currently not targeted by available therapies. Moreover, to have a global impact, the antivirals should have a sustained effect on different HIV-1 subtypes, including viruses resistant to RT, PR, and IN inhibitors. In this regard, the nucleocapsid (NC) protein is an ideal target due to its strikingly high conservation among all viral clades and its necessary involvement in a succession of key steps of the viral life cycle (Fig. 1). The present review will focus on the biological role and structure–function relationships of NC in the viral life cycle, as well as on the pharmacological strategies that have been recently published identifying novel, active small molecules against NC (Breuer et al. 2012; Goudreau et al. 2013; Mori et al. 2012; Shvadchak et al. 2009). It should be noted that there are numerous reviews focusing on retroviral NC proteins (Darlix et al. 2011; Levin et al. 2010; Mirambeau et al. 2010; Rein et al. 2011), with current, state-of-the-art findings being reported after each International Retroviral Nucleocapsid Conference (http://www.ncsymposium2013.org).
Fig. 1.
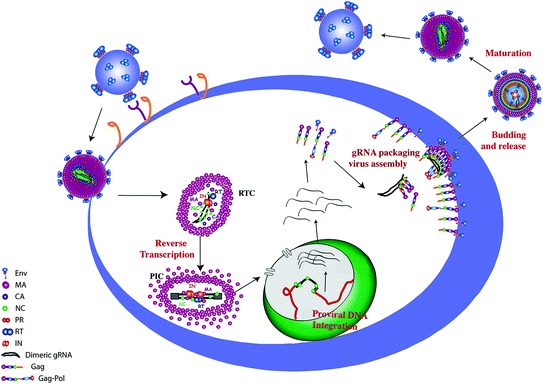
Role of the nucleocapsid protein in the HIV-1 life cycle. The mature NC protein (NCp7) is thought to assist RT in converting the single-strand genomic RNA into the double-stranded proviral DNA flanked by two long terminal repeats and to chaperone the IN-mediated integration of the proviral DNA into the host genome. As a part of the Gag polyprotein, the NC domain selectively recognizes, dimerizes, and packages the full-length genomic RNA during viral assembly. In the inner core of the viral particle, approximately 1500 NCp7 molecules are bound to the dimeric RNA genome. IN integrase; MA matrix; NC nucleocapsid protein; PR protease; RT reverse transcriptase
The past 20 years of research on NC revealed this protein to play a central role in virus replication (Fig. 1) and to be highly conserved in diverse HIV-1 subtypes and drug-resistant viruses (Fig. 2). As a component of the Gag structural polyprotein precursor, the corresponding NC domain (GagNC) selects, dimerizes, and packages the genomic RNA during virus assembly. Then, GagNC–RNA interactions favor transactions with (i) the cellular ESCRT complex to direct viral budding and (ii) the viral protease to direct the viral maturation that includes the processing and maturation of NC, needed for the proper condensation of the ribonucleoprotein architecture. The 55 amino acid mature form of NC (NCp7) exerts architectural and chaperone activities on HIV-1 RNA and DNA in the virion and during reverse transcription. This is done in a close partnership with the cellular tRNALys3 for reverse transcription initiation and with a set of viral RNA/DNA sites and RT itself for the subsequent steps leading to the faithful synthesis of the complete viral DNA, properly embedded within the preintegration complex. Directed mutagenesis in NC zinc fingers has been shown to affect these steps, including viral assembly/budding (Dussupt et al. 2011; Grigorov et al. 2007) and the spatiotemporal coordination of reverse transcription (Didierlaurent et al. 2008), leading to fully noninfectious viruses. These results on NCp7 mutations imply that an NCp7 inhibitor should impede the HIV-1 replicative cycle at its early and late steps, with GagNC being a highly relevant target in addition to the mature protein NCp7 (Breuer et al. 2012).
Fig. 2.
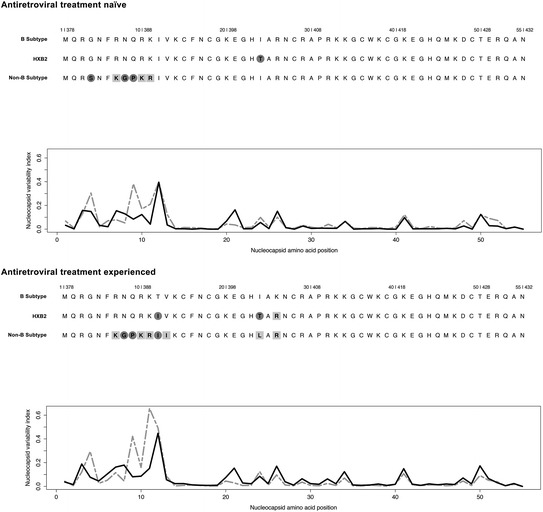
NCp7 sequence is highly conserved across different HIV-1 subtypes as well as in viral isolates obtained from antiretroviral naïve and treated individuals. Top panel, antiretroviral treatment naïve NCp7 consensus sequences from B (594 sequences) and non-B subtypes (4938 sequences) as well as the HXB2 molecular clone (GenBank accession number K03455), which is often considered as a representative subtype B virus, are shown (http://www.hiv.lanl.gov/content/sequence/HIV/REVIEWS/HXB2.htm). Bottom panel, antiretroviral NCp7 consensus sequences from B (7351 sequences), and non-B subtypes (14,286 sequences), as well as the B subtype representative molecular clone HXB2. Gray circles on amino acids indicate non-conservative amino acid substitution, such as charged to hydrophobic, whereas gray blocks indicate conservative amino acid changes. The nucleocapsid variability index reflects the variability of the amino acid changes at each position of NC, the higher the number the more amino acid variability. Black lines are the B subtype sequences, whereas the gray dashed lines are the non-B subtypes. Viral sequence information was obtained from the Los Alamos database (http://www.hiv.lanl.gov/content/index). The nucleocapsid variability index is a modification from the conservation index (Li et al. 2013)
Accordingly, a highly selective inhibition of the interaction of NCp7 and Gag-NC with their nucleic acid (NA) partners should lead to a potent antiretroviral activity, in synergy with common ARDs, and greatly enhance the genetic barrier for resistance. In this context, through the pleiotropic functions of NCp7 in the whole viral life cycle, these NC inhibitors will offer the new possibility to affect the assembly, and budding steps, that have not been targeted so far, in addition to the viral steps already targeted by other ARDs.
Structure and Zinc-binding Properties of the Nucleocapsid Protein
NCp7 is a basic protein of only 55 amino acids that is characterized by two strictly conserved CCHC zinc fingers (ZFs), flanked by small domains rich in basic residues (Fig. 3). The ZFs chelate zinc ions with high affinity (1013–1014 M−1) through three Cys and a His residues (Mely et al. 1996). The zinc-binding mechanism of NCp7 and notably of its distal ZF motif was investigated in depth (Bombarda et al. 2001, 2002, 2005, 2007; Mely et al. 1996). Binding of Zn2+ to the unfolded distal ZF was found to be initiated through the deprotonated Cys36 and His44 residues, resulting in a partly folded intermediate that subsequently converts into the final stable complex through deprotonation of the Cys39 and Cys49 residues and intramolecular substitution of coordinated water molecules. The two zinc-bound ZFs exhibit similar folding patterns (Morellet et al. 1992, 1994; Summers et al. 1992), while the linker between the two ZFs appears responsible for their spatial proximity (Lee et al. 1998; Mely et al. 1994; Morellet et al. 1994; Ramboarina et al. 2002). Importantly, the folding of the ZFs allows the formation on their top of a hydrophobic plateau that includes the hydrophobic residues of the proximal (Val13, Phe16, Thr24, and Ala25) and the distal (Trp37, Gln45, and Met46) ZFs (Fig. 3a, b). This hydrophobic plateau plays a key role in NCp7 functions, since nonconservative single point mutations in this plateau were found to lead to noninfectious viruses (Demene et al. 1994; Dorfman et al. 1993; Wu et al. 2013). Similarly, single point mutations of the zinc-binding residues cause also a complete loss of virus infectivity (Aldovini et al. 1990; Dorfman et al. 1993; Gorelick et al. 1990). These mutations do not prevent the binding of Zn2+ (Bombarda et al. 2002, 2005), but rather lead to an inappropriate folding of the mutated ZFs, so that formation of the hydrophobic plateau is prevented (Stote et al. 2004). This plateau is pivotal for the binding of NCp7 to NAs, through its multiple contacts with the NA bases and backbone (Amarasinghe et al. 2000; Bourbigot et al. 2008; De Guzman et al. 1998; Morellet et al. 1998; Spriggs et al. 2008). Among the residues of the plateau, the Trp37 residue is especially important as it conservatively stacks with exposed guanosines in all the 3D structures of NCp7/oligonucleotide complexes that have been solved (Fig. 3b). Another key feature of NCp7 is its high plasticity, which is required to adapt to the sequence and structure variability of its NA targets (Darlix et al. 2011; Godet et al. 2013).
Fig. 3.
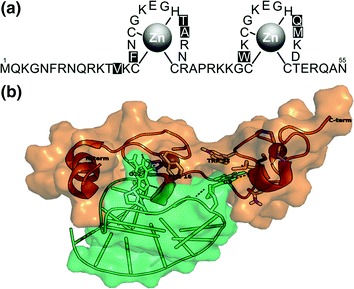
Amino acid sequence and 3D structure of the nucleocapsid protein. a NCp7 sequence showing the amino terminus, cysteine, and histidine amino acids that coordinate Zn and the Val13, Phe16, Thr24 Ala25, Trp37, Gln45, and Met46 amino acids (black boxes) that form the hydrophobic plateau. b 3D structure of NCp7/SL2 complex. On binding to SL2, the N-terminal domain folds into a helix. The Phe16 and Trp37 of the NCp7 hydrophobic plateau, which interact with the guanosine residues of SL2 loop through hydrogen bonding, are highlighted. The structure is from the PDB (1F6U)
In line with the importance of NC for viral function, the requirement to maintain a ZF-binding structure, and ability to interact with NAs, the amino acid sequence is highly conserved across B and non-B subtypes and in viral isolates from treated patients [Fig. 2 and (Darlix et al. 2011; Godet et al. 2012)]. When the variation indexes of NCp7 sequences are scrutinized (Fig. 2), it appears that the key residues Val13, Phe16, Thr24, Ala25, Trp37, Gln45, and Met46 of the hydrophobic plateau are invariant (Amarasinghe et al. 2000; Bazzi et al. 2011, 2012; Bourbigot et al. 2008; De Guzman et al. 1998; Morellet et al. 1998; Spriggs et al. 2008). Moreover, in most mutated sequences, the NCp7 consensus amino acids exchange with an amino acid of a similar profile. This strong requirement for amino acid conservation to maintain the structural integrity for function appears to provide few mutational options to escape inhibitors targeted against NC.
The Nucleocapsid Protein is Necessary for a Large Spectrum of Viral Activities
Once the HIV-1 Gag polyprotein has been translated from the viral unspliced mRNA at the polyribosomes, the Gag is transported via host-cell proteins and interacts with genomic RNA (gRNA) through its NC domain, Fig. 4 [for a review, see (Muriaux and Darlix 2010; Thomas and Gorelick 2008; Waheed and Freed 2012)]. To initiate viral assembly, few GagNC copies efficiently bind gRNA at specific loci within the Psi (Ψ) region, allowing a selective capture of HIV-1 gRNA in a dimeric form from the pool of spliced viral and cellular RNAs (Jouvenet et al. 2011; Kutluay and Bieniasz 2010; Kuzembayeva et al. 2014; Nikolaitchik et al. 2013). An optimized interaction of the NC hydrophobic pocket with the GXG-containing stem-loop sequences (SL1, SL2, and SL3) of the ψ-element has been proposed as a key feature for selectivity (Lu et al. 2011). After this nucleation, nonspecific GagNC–RNA interactions serve to load Gag and Gag-Pol on gRNA in cooperation with interactions directed by the other critical domains of Gag, CA-SP1 for Gag–Gag interactions and MA for Gag–membrane interactions (Datta et al. 2011; Kutluay and Bieniasz 2010; Munro et al. 2014). Deletion or mutations of NC strongly impede proper viral assembly (Ott et al. 2009). Moreover, GagNC also traps tRNALys3 and its cognate tRNA synthetase, and promotes the annealing of tRNALys3 with the the primer-binding site (PBS) of the HIV-1 gRNA (Guo et al. 2009).
Fig. 4.
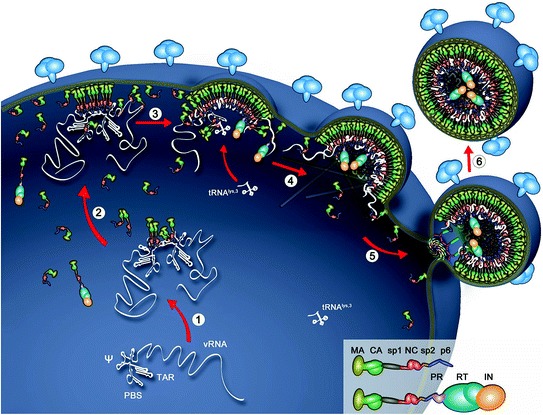
Assembly and budding of HIV-1 particles. Early and specific recognition of viral RNA (1) by Gag binding at the ψ RNA region, which ensures viral RNA dimerization. The Gag-vRNA complexes, free Gag, and Gag-Pol proteins migrate (2) through the cytoplasm toward the plasma membrane. The N-terminal MA domain of Gag binds to the cellular membrane (3) while the C-terminal NC domain projects into the cytoplasm and binds viral RNA. Gag and Gag-Pol units aggregate though MA-lipid, Gag-Gag, and NC/RNA interactions. Other components are loaded onto Gag, one of them being the tRNALys3 required to prime DNA synthesis. RNA scaffolding, growth of the Gag network, presumably GagNC–actin interactions, and finally ESCRT recruitment (4 and 5) mediate membrane curvature and the final budding of the particle (6)
GagNC–actin interactions in relation to actin dynamics likely modify the local curvature of the membrane (Kerviel et al. 2013; Schiralli Lester et al. 2013; Wilk et al. 1999), in order to allow the formation of the budding particle. The cellular ESCRT machinery is recruited to allow the release of the budding particle (Van Engelenburg et al. 2014). GagNC is engaged in this recruitment by interacting with Alix-containing Bro1 domain in cooperation with the neighboring Gagp6 domain that binds to the Alix-V domain (Dussupt et al. 2009; Popov et al. 2008). Moreover, it has been proposed that NC–Bro1 interactions depend on RNA in the cell (Sette et al. 2012). Similar to GagNC-Bro1 interaction, GagNC interacts with Tsg101 in the ESCRT I complex to support budding, which in turn, maintains gRNA integrity for packaging by preventing premature reverse transcription assembly due to budding defects (Chamontin et al. 2015).
During virus maturation (Fig. 5), the NC domain is released from Gag under the first wave of proteolysis leading to the transient species NCp15 (Mirambeau et al. 2010). Subsequent proteolytic cleavage at its C-terminus during two consecutive steps results in the liberation of the p6 protein, leading to NCp9, followed by cleavage and release of the C-terminal 16 amino acid peptide, called SP2, leading to the final product, NCp7. NCp15 processing appears strongly activated by NC–RNA interactions, which drives the condensation within the viral core (de Marco et al. 2012; Mirambeau et al. 2007; Sheng and Erickson-Viitanen 1994).
Fig. 5.
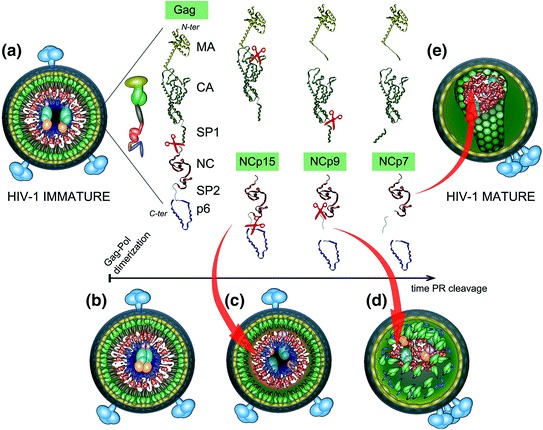
Nucleocapsid maturation Gag processing is sequential and ordered. The first PR to be self-processed from Gag-Pol is thought to direct the sequential Gag and Gag-Pol proteolytic events that will ultimately convert the immature virion into the mature particle (a–e). PR self-activation and cleavage from Gag-Pol is driven by the proper alignment of HIV-1 Gag-Pol precursors within the immature particle. The different protein species generated during the steps of Gag processing are indicated. PR cleavage of Gag initially occurs between SP1 and NC leading to the first NC intermediate form, NCp15 (partial cleavage product containing NC/SP2/p6), cleavage then results in NCp9 (partial cleavage product containing NC/SP2), and finally to the fully processed form, NCp7. The self-assembly properties of CA and NC, after removal of SP1, SP2 and p6, allow assembly of the viral core. Furthermore, SP1-NC cleavage by PR separates the MA-CA from the nucleocapsid complex formed between RNA, NCp15, RT, and IN. Processing of NCp15 by PR into NCp9 leads to a NC/RNA condensed aggregate, in which NCp9 is finally processed into NCp7, allowing the reverse transcription complex to form and be primed for function within the confines of the capsid cone
In the early steps of HIV-1 replication, the mature NCp7 protein is thought to assist RT in converting the single-stranded gRNA into a double-stranded proviral DNA (Fig. 6) (Darlix et al. 2007; Hu and Hughes 2012; Levin et al. 2010; Lyonnais et al. 2013; Thomas et al. 2008). As a first step, NCp7 directs the annealing of the tRNALys3 primer with the PBS (Sleiman et al. 2012; Tisne et al. 2004). In addition, NCp7 chaperones the first strand transfer by annealing cTAR DNA with TAR RNA allowing RT to resume the minus-strand DNA elongation step (Darlix et al. 2011). Moreover, NCp7 ensures the fidelity of plus-strand DNA priming at the two polypurine tracts (PPT) by blocking mispriming by non-PPT RNAs and by removing the 5′-terminal fragments annealed to minus-strand DNA (Hergott et al. 2013). In order for RT to perform the plus-strand synthesis after its pausing, NCp7 must chaperone the second strand transfer (i) by facilitating the RT–RNaseH removal of primer tRNALys3 from the 5´-end of minus-strand DNA, and (ii) by promoting the annealing of the PBS DNA copy at the 3´-end of plus-strand DNA with the complementary PBS at the 3´-end of minus-strand DNA (Darlix et al. 2011). NCp7 also increases RT processivity during reverse transcription, including at the termination steps where DNA synthesis coupled with strand displacement is necessary for long terminal repeat (LTR) duplication and the generation of the DNA central flap (Grohmann et al. 2008; Hameau et al. 2001). Finally, NCp7 is thought to play a possible role during integration by stimulating LTR DNA integration by IN (Buckman et al. 2003; Carteau et al. 1997; Poljak et al. 2003; Thomas and Gorelick 2008).
Fig. 6.
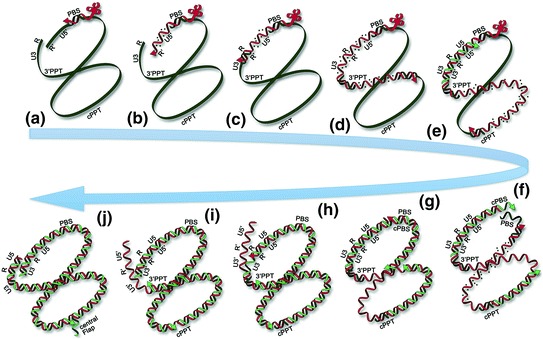
Reverse transcription a Reverse transcription initiation from the tRNALys3 primer at the PBS site. b Synthesis of minus-strand DNA and RNA digestion. c minus-strand transfer by cTAR-TAR hybridation and RT elongation. d minus-strand DNA synthesis, with RNAse-H activity releasing the 3′PPT. e Release of the cPPT upon minus-strand DNA synthesis and plus-strand synthesis from the 3′PPT. f Removal of tRNALys3 upon plus-strand synthesis and plus-strand synthesis starting from the cPPT. g plus-strand transfer by base pairing of the minus-strand PBS and plus-strand PBS sequences, elongation of the plus-strand strand DNA. h and i Synthesis of plus-strand DNA, with strand displacement of the U5 extremity. j Termination of plus-strand synthesis with LTR duplication and strand displacement to generate the central DNA flap. The two NA ends are in close proximity throughout reverse transcription. RNA fragments released by the RNaseH activity of RT are shown as dashed points behind RT along the elongating plus-strand DNA template. NC assists RT all along the process
The Nucleocapsid Protein Interacts with Self and Host-cell Proteins
The viral proteins RT (Druillennec et al. 1999; Lener et al. 1998), Vif (Bouyac et al. 1997), Vpr (de Rocquigny et al. 1997; Li et al. 1996), and Tat (Boudier et al. 2010) have been proposed to interact with NC. In the case of Vif, GagNC is likely the main target, while the main partner of RT is NCp7. Within Gag, the NC domain is also suspected to interact with its neighboring domain, p6. GagNC has also been shown to interact with cellular factors such as the actin cytoskeleton (Liu et al. 1999), the dsRNA-binding protein Staufen (Chatel-Chaix et al. 2007, 2008), the IGF-II mRNA-binding protein 1 (Zhou et al. 2008), the cellular ATP-binding protein ABCE1 (also termed HP68) (Lingappa et al. 2006), and Alix (Popov et al. 2008). These protein–protein interactions, notably with Alix, are thought to participate in HIV-1 assembly and budding. Moreover, most of these cellular proteins are packaged into viral particles (Alce and Popik 2004; Mouland et al. 2000; Ott et al. 1996; Zhou et al. 2008). In the case of Alix, a ternary complex has been recently proposed to form between GagNC, RNA, and the Bro domain of Alix, suggesting that GagNC–RNA interactions could be useful to recruit cellular proteins (Sette et al. 2012).
The Nucleocapsid Protein is Key for HIV-1 Nucleic Acids Regulation
NCp7 binds both specifically and nonspecifically to a large panel of NA sequences of sufficient length (5–8 nt.), with a reverse binding polarity between RNA and ssDNA [for a review, see (Darlix et al. 2011)]. The binding constants can vary by several orders of magnitude depending on the nature, the sequence, and the folding of the interacting sequences (Fisher et al. 1998; Vuilleumier et al. 1999), so that NCp7 can exert different functions, depending on the respective concentrations of the protein and the NA sequences. As a consequence of its basic character and its millimolar range concentration in the virus, NCp7 molecules can likely coat the complete gRNA (Chen et al. 2009a, b; Chertova et al. 2006), ensuring its protection against cellular nucleases (Krishnamoorthy et al. 2003). NCp7 also exhibits sequence-specific binding properties to defined single-stranded sequences. These specific and strong binding properties notably play a critical role in the recognition by the NC domain of Gag of the Ψ-encapsidation signal of the gRNA, enabling its specific recognition and selection among a large excess of cellular RNAs during virus assembly (Aldovini and Young 1990; Cimarelli and Darlix 2002; Lever et al. 1989; Muriaux and Darlix 2010; Muriaux et al. 2004).
Through its binding to NA, NCp7 exerts a role as a NA chaperone, which allows the protein to direct the rearrangement of NAs into their most stable conformation, and to promote the annealing of complementary sequences (Godet and Mely 2010; Levin et al. 2005; Rein et al. 1998). These NA chaperone properties rely on the ability of NCp7 to transiently destabilize the NA secondary structure (Azoulay et al. 2003; Beltz et al. 2003, 2004; Bernacchi et al. 2002; Cosa et al. 2006; Egele et al. 2004; Godet et al. 2011, 2013; Liu et al. 2005; Williams et al. 2001). This destabilization is mainly mediated by the hydrophobic region located on the top of the folded ZFs and strongly depends on the NA stability and structure, suggesting a co-evolutionary relationship between NCp7 and its NA targets (Beltz et al. 2003, 2005; Godet et al. 2011, 2013; Hergott et al. 2013). Guanosine is the pivotal nucleoside to be trapped (Grohman et al. 2013). This destabilization is further accompanied by the exposure and freezing of the local mobility of the bases where NCp7 is bound (Avilov et al. 2008; Bourbigot et al. 2008; Godet et al. 2011, 2013), a feature which is thought to be critical for the recognition of the complementary oligonucleotide sequence in the annealing reaction. A second major component of the NCp7 chaperone properties relies on its ability to promote the rapid annealing of complementary NA sequences (Darlix et al. 1993; Godet et al. 2006; Hargittai et al. 2004; Liu et al. 2007; Ramalanjaona et al. 2007; Vo et al. 2006, 2009; You and McHenry 1994). This component mainly depends on the N-terminal basic domain and its NA aggregation properties, which provide the highly dynamic macromolecular context to favor efficient strand exchange (Mirambeau et al. 2006; Stoylov et al. 1997). The ZFs and the hydrophobic plateau are also instrumental in the annealing reaction, by promoting specific pathways which are notably required to faithfully and specifically chaperone the two obligatory strand transfers, during reverse transcription (Godet et al. 2011, 2013). Effective strand annealing activity is further correlated with NCp7’s ability to rapidly bind and dissociate from NAs. Indeed, NC variants with slow on/off rates are poorly efficient in rearranging NAs, even though they are still capable of promoting aggregation of NAs (Cruceanu et al. 2006a, b; Stewart-Maynard et al. 2008). Comparison of the various forms of NC further revealed that Gag is a less efficient NA chaperone than NCp7 (Cruceanu et al. 2006a, b) and that NCp15 appears much weaker for NA aggregation compared to NCp9 and NCp7 (Mirambeau et al. 2006, 2007; Wang et al. 2014).
Zinc Ejectors as Nucleocapsid Protein Inhibitors
Due to their key involvement at many critical points in the HIV-1 replication cycle and their strong conservation among HIV-1 strains, the ZFs of NCp7 were naturally selected as the primary target for the development of inhibitors. To properly exert their functions, the ZFs of NC crucially rely on the binding of zinc atoms that are required to fold them into their highly constrained structures. As a consequence, molecules able to eject the zinc atoms from the fingers were naturally developed as the first NC inhibitors. As anticipated, these molecules were found to induce NC unfolding as well as a full loss of HIV-1 infectivity.
Since the development of the first zinc ejectors in 1993 (Rice et al. 1993), a number of different classes of compounds were designed [for a review, see (de Rocquigny et al. 2008; Goldschmidt et al. 2010; Musah 2004; Turpin et al. 2008)]. Most of these compounds exhibited strong antiviral activity and elicited little viral resistance, clearly underlining the relevance of NC as an appropriate target for an antiviral therapy. Unfortunately, these compounds appeared also quite toxic, so that their use for systemic administration was prevented. Currently, efforts are underway to use them as topical microbicides, in order to prevent HIV-1 transmission.
Zinc Ejectors: Structure and Mechanism of Action
Various classes of compounds able to alter the coordination of the strongly bound zinc ions to NC and subsequently cause Zn ejection were developed. Figure 7 shows several illustrative examples of these compounds, which include 3-nitrosobenzamide (NOBA) as a representative of C-nitroso-compounds (Rice et al. 1993), 2,2′-dithiobisbenzamide disulfides (DIBA) (Rice et al. 1996), cyclic 2,2′-dithiobisbenzamide (SRR-SB3) (Witvrouw et al. 1997), benzisothiazolones (Loo et al. 1996), dithiaheterocyclic molecules such as 1,2-dithiane-4,5-diol-1,1-dioxide (Rice et al. 1997a, b), pyridinioalkanoyl thioesters (PATE) (Turpin et al. 1999), S-acyl-2-mercaptobenzamide thioesters (SAMT) (Jenkins et al. 2005), azodicarbonamide (ADA) as a α-carbonyl azoic compound (Vandevelde et al. 1996), trans-chlorobispyridine (9-ethylguanine) platinum(II) (Anzellotti et al. 2006; Quintal et al. 2011), and the most recently identified N,N′-bis(4-ethoxycarbonyl-1,2,3-thiadiazol-5-yl)benzene-1,2-diamine (NV038) (Pannecouque et al. 2010) and 2-methyl-3-phenyl-2H-[1,2,4]thiazol-5-yideneamine (WDO-217) (Vercruysse et al. 2012).
Fig. 7.
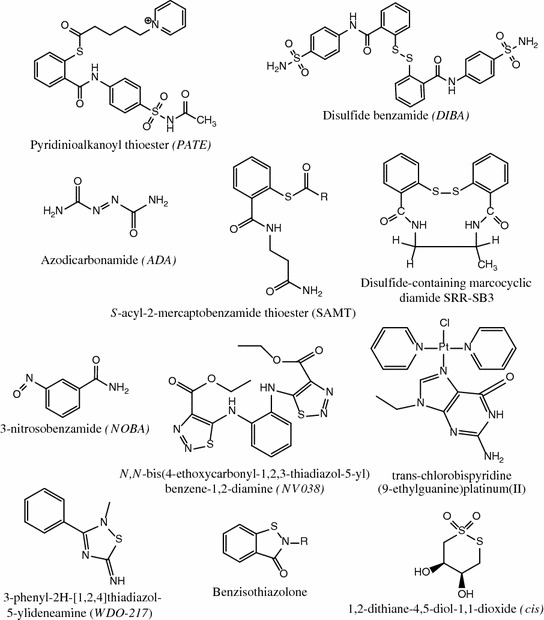
Structures of zinc ejectors of various chemical classes
The mechanism of action of several of these compounds was carefully investigated to identify the NC chemical groups targeted by these compounds and the sequence of chemical reactions that results in zinc ejection. The mechanisms of inactivation of NC ZFs by these compounds can be classified into three main groups: (i) electrophilic attack of the zinc fingers, (ii) zinc ejection through chelation, and (iii) covalent binding of the Cys residues by Pt.
In both ZFs, the nucleophilic cysteine thiolates appear as the primary targets for electrophilic attack. Though both fingers contain the same CysX2CysX4HisX4Cys motif, zinc ejectors were found to preferentially react with the distal finger motif. Computational studies (Loo et al. 1996; Maynard and Covell 2001) indicated that this increased reactivity was at least partly related to the better accessibility of the Cys residues in this finger. Electrophilic attack may be accompanied by either formation of intra- or inter-molecular disulfide bonds or acylation of cysteine and then lysine residues. The oxidative mechanism leading to disulfide bonds was observed for compounds of the NOBA and DIBA families (Loo et al. 1996; Yu et al. 1995). For instance, when NCp7 was incubated with NOBA, three intermolecular disulfide bonds, Cys15-Cys18, Cys28-Cys36, and Cys39-Cys49, formed (Yu et al. 1995). Similarly, DIBA was found to initiate the formation of intra- and inter-molecular disulfide bonds by preferentially attacking Cys36 and Cys49 residues (Loo et al. 1996). Formation of three disulfide bridges was also observed with the recently discovered WDO-217 compound, though in this case, the preferential sites of attack were not identified (Vercruysse et al. 2012). An acylation mechanism is observed with PATEs and SAMTs. It involves the nucleophilic attack by a zinc-coordinated cysteine of the carbonyl carbon of the inhibitor. This results in the covalent modification of the cysteine sulfur via an acyl transfer mechanism. Subsequently, additional acyl transfer reactions occur with other cysteine and lysine residues of NCp7 that will further decrease the affinity for zinc and finally lead to zinc ejection. Cys36 and Cys49 are the primary targets of PATEs, while Cys36 is the primary target of SAMT analogs (Basrur et al. 2000; Miller Jenkins et al. 2007). The preferential susceptibility of the Cys49 residue to electrophilic attack is likely related to its rather high pKa value in the zinc-bound protein, which confers it a role of a switch in the dissociation of zinc (Bombarda et al. 2002).
A different mechanism was inferred for NV038. Indeed, based on its structure, this compound is likely unable to allow acyl transfer or thiol-disulfide interchange. In fact, molecular modeling suggests that NV038 may act as a zinc chelator that binds one zinc ion through the two carbonyl oxygens of its ester groups (Pannecouque et al. 2010).
The third mode of action is represented by platinum nucleobase compounds that act through a two-step mechanism (Anzellotti et al. 2006; Quintal et al. 2011). They first recognize the Trp37 residue of NCp7 through π–π stacking and then form a Zn-S–Pt covalent bond, which results in zinc ejection. As for electrophilic zinc ejectors, the primary target of platinum nucleobase compounds is Cys49 in the C-terminal zinc finger.
Antiviral Activity In Vitro
The antiviral activity of zinc ejectors was tested on HIV-1 infected cells (Table 1). To comparatively evaluate their activity as well as their cellular toxicity, their EC50 (concentration of inhibitor required for 50 % inhibition of viral replication), CC50 (concentration that kills 50 % of cells), therapeutic index, and in vivo stability were determined. NOBA exhibited potent anti-HIV-1 activity (Rice et al. 1993), but also high cellular toxicity, which prevented its further use (Huang et al. 1998). DIBA-1, dithiane, PATE-45, and SAMT-19 were found to be highly active as well, but showed far less cytotoxicity, so that their therapeutic indexes were ≥30. ADA, NV038, and platinum nucleobases were found to be somewhat less active. Finally, WDO-217 showed strong activity, but had a rather low therapeutic index, which makes it useful for topical applications. Most of these compounds were shown to be active in both acutely and chronically HIV-1-infected cells, as well as on cell-free HIV-1 virions (Rice et al. 1995; Srivastava et al. 2004; Turpin et al. 1999; Vercruysse et al. 2012). Moreover, these compounds were also active against HIV-2 and SIV strains (Huang et al. 1998; Pannecouque et al. 2010; Srivastava et al. 2004; Vercruysse et al. 2012), as well as against drug-resistant HIV-1 strains, including clinical HIV-1 isolates (Pannecouque et al. 2010; Turpin et al. 1996; Vercruysse et al. 2012). The potent, long-term activity against a large spectrum of HIV-1 strains is a hallmark of zinc ejectors that is consistent with the high conservation of NCp7 (Darlix et al. 2011) and the inability to generate viruses resistant to zinc ejectors (Huang et al. 1998). This lack of resistance generation clearly underscores the high potential of NC inhibitors to obtain a sustained inhibition of HIV-1 replication.
Table 1.
Antiviral activity and cytotoxicity of zinc ejectors
| Compound | EC50 (µM) | CC50 (µM) | Therapeutic index | References |
|---|---|---|---|---|
| NOBA | ND | 10.6 | − | Huang et al. (1998) |
| DIBA-1 | 2.3 | >200 | >87 | Huang et al. (1998), Rice et al. (1995) |
| Dithiane | 6.6 | 184 | 30 | Huang et al. (1998) |
| ADA | 38 | >200 | >5 | Huang et al. (1998), Rice et al. (1997a, b) |
| PATE-45 | 6.2 | >316 | >51 | Turpin et al. (1999) |
| SAMT-19 | 2.9 | 461 | 160 | Srivastava et al. (2004) |
| NV038 | 17 | >300 | >17 | Pannecouque et al. (2010) |
| WDO-217 | 7.9 | 72 | 9 | Vercruysse et al. (2012) |
| [SP-4-2]-[PtCl(NH3)(quin)(9-EtGH)] | 41.9 | >200 | >4.75 | Sartori et al. (2000) |
EC50, 50 % effective concentration (concentration of inhibitor for 50 % inhibition of viral replication); CC50, 50 % cytotoxic concentration (concentration of inhibitor that kills 50 % of cells), Therapeutic index = CC50/EC50. ND, not determined
The activity of zinc ejectors is related to their ability to decrease the affinity of NCp7 for its target nucleic acids, as for example the ψ RNA sequence (Huang et al. 1998; Jenkins et al. 2005; Tummino et al. 1996). This effect depends on the concentration of the zinc ejector and on the order of addition of the partners. While SAMTs and PATEs were able to strongly inhibit RNA binding when preincubated with NCp7, they exhibit nearly no effect on metal coordination and RNA binding when they were added to preformed NCp7-RNA complexes. Likely, RNA protects the zinc-coordinating residues of NCp7 from the inhibitors (Chertova et al. 1998; de Rocquigny et al. 2008; Jenkins et al. 2005). Noticeably, WDO-217 appears quite unique in this respect, as it was found to efficiently eject zinc ions from NCp7, even in complexes with nucleic acids (Vercruysse et al. 2012). In addition, WDO-217 was observed to change the binding mode of NCp7 to oligonucleotides, but with no dramatic change in the binding constant. As the result of their reaction with NCp7, zinc ejectors were found to affect reverse transcription (Morcock et al. 2005; Pannecouque et al. 2010; Rice et al. 1995; Rice and Turpin 1996; Sharmeen et al. 2001), likely by altering the nucleic acid chaperone properties of NCp7 (Pannecouque et al. 2010; Vercruysse et al. 2012) that critically depend on the binding of zinc (Avilov et al. 2008; Beltz et al. 2005; Bernacchi et al. 2002; Godet et al. 2011). In addition, zinc ejectors affect also the late steps of the viral life cycle, since DIBAs and PATEs (Turpin et al. 1996, 1999), SRR-SB3 (Mahmood et al. 1998) and SAMTs (Miller Jenkins et al. 2010), but not WDO-217 (Vercruysse et al. 2012), were found to induce accumulation of aggregated and unprocessed Gag polyproteins (Turpin et al. 1996, 1999) that lead to the release of noninfectious virus particles. This aggregation is likely due to intermolecular bridging of the NC domains of neighbor Gag polyproteins. Zinc ejectors also fully inactivate cell-free HIV-1 virions, by promoting NCp7 oligomerization (Rice et al. 1995) or acylation (Basrur et al. 2000; Jenkins et al. 2005). Furthermore, WDO-217 was found to relieve the protection of the viral RNA from the NCp7 proteins in cell-free virions, through a still unknown mechanism (Vercruysse et al. 2012). Finally, zinc ejectors were also shown to inhibit HIV-1 transmission from infected cells to uninfected ones (Srivastava et al. 2004; Vercruysse et al. 2012).
Cytotoxicity of zinc ejectors is likely related to their limited selectivity for NCp7 over zinc finger-containing host proteins, such as poly(ADP-ribose) polymerase (PARP) (with two CCHC zinc fingers), SP1 (with three CCHH-type Zn fingers), and GATA-1 (with two CCCC-type Zn fingers). For instance, NOBA shows only poor selectivity for NCp7, as it inhibits the enzymatic activity of PARP and blocks GATA-1 binding to their target DNA sequences (Huang et al. 1998). On the contrary, DIBA, ADA, and dithiane did not show any significant reactivity on either PARP or SP1 and GATA-1, which may likely explain their lower cytotoxicity (Huang et al. 1998). Likewise, the poorly cytotoxic PATE compounds did not show any reactivity on SP1 (Turpin et al. 1999). Finally, SAMTs did not react with CCHH zinc finger proteins and RING-like zinc-binding domains, but showed some reactivity toward Friend of GATA-1 (FOG-1) and GATA-1 (Jenkins et al. 2006).
Evaluation of Zinc Ejectors for Therapeutic Applications
Due to their potent antiviral activity in vitro, several attempts were made to evaluate the potential therapeutic use of zinc ejectors in vivo. To our knowledge, only two zinc ejectors, namely ADA and benzisothiazolone, were tested in clinical studies. Due to its toxicity, assays with the second compound were rapidly stopped (Turpin 2003). Preclinical tolerance assays showed that oral doses of 1.5 g ADA daily for 1 month were well tolerated, with no evidence of adverse effects (Vandevelde et al. 1996). Then, ADA was administrated three times daily during 3 months in addition to other antiviral therapy to fifteen individuals with advanced AIDS within a Phase I/II clinical trial. Unfortunately, serious nephrotoxicity as well as glucose intolerance appeared during the treatment, a serious enough event so that several patients dropped out of the clinical trial (Goebel et al. 2001). Moreover, ADA showed only a modest efficacy, as evidenced by an increase in T cell CD4+ counts and a reduction in the viral load in less than half of the treated patients (Goebel et al. 2001). On a more positive note, no ADA resistant virus could be isolated from ADA-treated patients. Unfortunately, the clinical trial was not conclusive, most likely since ADA is clearly not the most efficient antiviral compound in vitro (Table 1) and shows a number of off-target effects, such as inhibition of lymphocyte cytokine production (Rice et al. 1997a, b; Tassignon et al. 1999) and ribonucleotide reductase activity (Fagny et al. 2002). The systemic activity of zinc ejectors was also tested with SAMT compounds on an HIV-1 transgenic mouse model (Schito et al. 2003). These compounds reduced by 2–3 logs the infectivity of viruses expressed from the spleen cells of the transgenic mice and had no effect on immune cell cytokine production. Furthermore, sub-dermal delivery of a SAMT lead compound in cynomolgus macaques infected with SIV/DeltaB670 virus lowered the levels of infectious virus in peripheral blood mononuclear cells, but did not affect the virus load (Schito et al. 2006). Importantly, the SAMT lead compound was well tolerated and did not alter liver, kidney, or immunologic function of the treated monkeys. Though these data suggest that SAMT compounds may be safe in a primate model, it still remains to be demonstrated whether, due to their limited selectivity, zinc ejectors could be reasonably used as a long-term systemic therapeutics in patients.
Due to their potent activity and potential safety concerns, the application of zinc ejectors as topical microbicides appears more promising. The proof of concept for this application was demonstrated with SAMTs, which were shown to prevent HIV transmission from infected cells to uninfected cells, with EC50 values below 0.1 µM (Srivastava et al. 2004). Later, SAMTs were shown in the cervical explant model to inhibit the infection of target cells in the explant tissue and the dissemination of the infection by immune cells migrating out of the explant (Wallace et al. 2009). Interestingly, no virus infectivity was observed up to one week after SAMTs removal. Moreover, SAMTs antiviral activity was retained in both synthetic cervical mucous and human seminal plasma. Finally, the SAMT compounds were shown to induce no significant histology changes and irritation in the rabbit vaginal irritation model (Tien et al. 2005; Wallace et al. 2009). The SAMTs were further evaluated in rhesus macaques to determine their ability to prevent vaginal transmission of the simian-human immunodeficiency virus (SHIV) (Wallace et al. 2009). The monkeys were treated vaginally with 1 % SAMT in hydroxyethylcellulose universal placebo gel 20 min prior to challenge with a mixed CXCR4-tropic and CCR5-tropic SHIV virus inoculum (Wallace et al. 2009). Five out of six macaques were protected from infection, while only one infected animal expressed the CCR5-tropic SHIV. These findings strongly support the use of SAMTs as potential topical microbicides to prevent HIV transmission. Since WDO-217 at low micromolar concentrations was recently shown to inactivate HIV-1 captured by DC-SIGN-expressing cells and prevent their transmission to CD4+ T lymphocytes (Vercruysse et al. 2012), it is anticipated that WDO-217 may also be a valuable candidate for the development of topical microbicide formulations.
In conclusion, zinc ejectors show potent antiviral activity against a large spectrum of HIV-1 strains, without eliciting resistance. However, their limited selectivity raises toxicity concerns, limiting this class of NC inhibitors to microbicide formulations. Alternatively, due to their ability to inactivate HIV-1 efficiently without compromising viral surface antigens, they may have promise for use in vaccine strategies (Arthur et al. 1998; Chertova et al. 1998, 2006).
Inhibitors Targeting Nucleocapsid Protein Interaction with Nucleic Acids
In addition to zinc ejectors, a number of non-covalent NC inhibitors (NCIs) were identified during the past decade and used both as tools to increase our understanding of the biological and pathological functions of NC, as well as hit/lead candidates for the development of potential innovative antiretroviral therapeutics. However, the discovery of NCIs that demonstrate potent antiretroviral activity in vitro and in vivo still remains a considerable challenge. Indeed, only a few of the NCIs disclosed to date were found to inhibit HIV-1 replication in cell-based antiretroviral assays and none reached yet the preclinical phases of pharmaceutical evaluation. Since non-covalent NCIs are thought to show a greater specificity than zinc ejectors, and thus be presumably less toxic, these properties may well be superior for clinical translation, which makes this class of NCIs a desirable pharmaceutical goal. Since pioneering studies on the discovery and preliminary characterization of non-covalent NCIs have been reviewed recently (de Rocquigny et al. 2008; Goldschmidt et al. 2010; Mori et al. 2011a, b), we will mainly focus on novel strategies undertaken since 2009 that have identified small molecules endowed with two different mechanisms of action: (i) non-covalent NCIs binding to NC and (ii) non-covalent NCIs binding to nucleic acid partners of NC.
Non-covalent NCIs Binding to the Nucleocapsid Protein
A report by Shvadchak and colleagues had a major impact on the establishment of small molecule search strategies for NCIs (Shvadchak et al. 2009). The authors developed a high-throughput screening (HTS) assay to identify small molecules that inhibit the NCp7 chaperone activity and notably the NCp7-promoted destabilization of nucleic acid secondary structure (Shvadchak et al. 2009). The assay was based on the use of cTAR DNA labeled at its 3′ and 5′ ends with a fluorophore (Rh6G) and a fluorescence quencher (DABCYL), respectively. The addition of the 12–55 amino acid fragment of NC [NC(12–55)] provoked a partial melting of cTAR DNA, which was easily monitored as an increase of fluorescence with respect to cTAR alone. Positive small molecule hits that compete with the binding of NC(12–55) to the cDNA TAR halt the melting of the labeled cTAR and restored Rh6G fluorescence. This assay was developed to be highly specific and was validated by screening a custom library of about 4800 chemical substances (Shvadchak et al. 2009). Five low molecular weight fragments were identified as inhibitors of the NC chaperone activity, A10, CO7, EO3, HO2, and HO4 (Fig. 8), showing K i values in the micromolar range. Further analyses suggested that these NCI fragments compete with cTAR for binding to NC(12–55), representing therefore the first example of NCIs targeting NC chaperone activity, as well as valuable compound starting points for further chemical optimization.
Fig. 8.
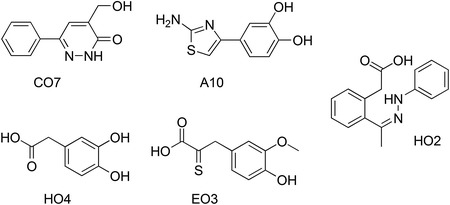
Chemical structures of the five fragment NCIs identified from HTS targeting NC chaperone activity (Shvadchak et al. 2009)
In an attempt to provide structural hints on the binding of these fragments to NC, an in-depth molecular modeling study was performed by Mori and colleagues (Mori et al. 2011a, b). NCI fragments were docked toward two computationally refined structures of NCp7 (Mori et al. 2010), showing that these molecules may preferentially bind to the Trp37 residue on the ZF hydrophobic platform (Fig. 3). The good correlation between experimental and theoretical findings corroborated the reliability of the computational model, thus paving the way for possible structure-based drug design approaches.
The HTS assay methodology, discussed above, was also used to characterize a methylated oligoribonucleotide NCI (Avilov et al. 2012; Grigorov et al. 2011). Although modified oligoribonucleotides may be considered at the boundary between small molecules and biomolecules, the findings of this study have significantly contributed to the understanding of the molecular basis of NC inhibition and theoretical design of NCIs. Based on the evidence that NCp7 chaperones reverse transcription, methylated oligoribonucleotides (mODNs) mimicking the long terminal repeat end sequences of proviral DNA were synthesized and evaluated in vitro and ex vivo. Inhibition of the NCp7 chaperone activity was monitored through the fluorescence of the Rh6G-5′-cTAR-3′-Dabcyl DNA sequence (Shvadchak et al. 2009). Further tests revealed that mODN-11, having the sequence 2′-O-Me-(GGUUUUUGUGU-NH2), was the most potent oligoribonucleotide among the test set, inhibiting HIV-1 replication in MT4 cells at sub-nanomolar concentrations (IC50 = 0.3 nM) and also showing low cytotoxicity (CC50 = 7.7−13.4 µM). Time of addition experiments further revealed that mODN-11 inhibited HIV-1 replication with the same time frame as the reverse transcriptase (RT) inhibitor AZT, thus suggesting that the reverse transcription complex may be the target of the oligoribonucleotide. In fact, AZT and mODN-11 provided a synergistic inhibition of HIV-1 replication, further reinforcing the hypothesis, already verified in vitro, that mODN-11 targets NCp7 and that NCp7 is an indispensable partner of RT. The mechanism of action of mODNs was further investigated by isothermal titration calorimetry and fluorescence-based techniques and compared to unmodified oligoribonucleotides (Avilov et al. 2012). Interestingly, this study showed that mODNs bearing repeats of GU or GT pairs tightly bind to NCp7 through nonelectrostatic interactions and compete with NAs for the binding to the NCp7 hydrophobic pocket, suggesting that the mODNs may impair the RT-directed viral DNA synthesis by sequestering NCp7 molecules.
Based on these results, one may speculate that the methylation of the GU- or GT-rich oligoribonucleotides improves their lipophilicity and, therefore, their affinity for the small hydrophobic pocket of NCp7. Indeed, although NCp7 is a highly basic protein that interacts with NAs by means of electrostatic interactions, hydrophobicity appears as a key feature for potent and effective NCIs. In agreement with the several studies that emphasized the crucial role of NCp7 aromatic residues Trp37 and Phe16, the ideal NCI should be able to compete for the binding of NAs by interacting with the NCp7 hydrophobic platform. Consistent with this hypothesis, in recent medicinal chemistry-oriented studies, a number of NCIs endowed with hydrophobic/aromatic groups have been discovered by means of different techniques, including virtual screening and HTS. Moreover, the three-dimensional structure of NCp7 in complex with a NCI confirmed the key role of the aromatic residues in the interaction. Highlights of these studies are reported below.
Botta’s group studied the structure and potential druggability of NCp7 by means of molecular dynamics simulations (MD) and molecular modeling studies performed on two nuclear magnetic resonance (NMR) structures of NCp7 in complex with oligonucleotides (Mori et al. 2010, 2011a, b). The aim of these theoretical studies was the understanding of NCp7 flexibility and the subsequent identification of pharmacophoric hot spots for small molecules able to compete with NAs for binding sites on the NCp7. Outcomes of these studies were then incorporated in a virtual screening protocol, which was used to identify possible NCp7 binders among the Asinex database (about 390,000 chemical compounds) (Mori et al. 2012). Ten virtual hits endowed with significant chemical diversity were selected and tested in vitro for their ability to bind to NC(11–55) and inhibit HIV-1 replication in infected cells. Preliminary binding affinity measurements identified two small molecules, namely 6 and 8 (Fig. 9), that are able to interact with NC(11–55) without promoting zinc ejection, which is an essential requisite for non-covalent NCIs. Moreover, biophysical studies with NC(11–55) labeled with fluorescent amino acid analogs at different positions suggested that 6 binds tighter than 8 and that these NCI hits may bind in proximity to the hydrophobic pocket of NC(11–55), as predicted by molecular modeling. The binding affinity of 6 was estimated within the micromolar range (5.6 ± 0.9 µM). Though the strong intrinsic fluorescences of 6 and 8 seriously limited the possibility to perform functional tests on NC(11–55) in vitro, antiretroviral assays on P4.R5 MAGI cells showed that 6 inhibited HIV-1 replication with an IC50 of about 2 µM, which is consistent with its binding affinity to NC(11–55). Overall, this work provided the first example of small molecules with non-covalent NCI activity in HIV-1 infected cells discovered by using computational methods.
Fig. 9.
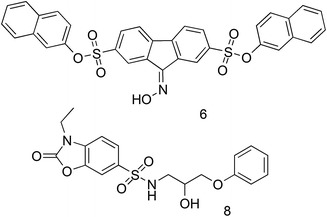
Chemical structure of the NCIs identified by means of virtual screening (Mori et al. 2012)
In 2012, a new HTS assay to search for NCIs interacting with the NC was developed at the Scripps Research Institute (Breuer et al. 2012). The assay consisted of a two-step screen, with the first screen based on fluorescence polarization to identify small molecules able to disrupt the interaction between NC and DNA. Next, positive hits from the first assay were screened by differential scanning fluorimetry for hits that bound to NC. Similar in concept of Shvadchak and co-workers in the use of a DNA tracer (Shvadchak et al. 2009), the first screen relied on a fluorescently labeled stem-loop-2 (SL2) DNA tracer that bound to the p2-NC protein. The displacement of the p2-NC–SL2 DNA interaction by small molecules was monitored by changes in fluorescence polarization. To identify compounds from the first screen that bound directly to p2-NC and disrupt SL2 DNA binding, differential scanning fluorimetry was utilized in the second screen to identify which compounds altered p2-NC melting temperature as the result of compound binding over that of p2-NC only. The two-step assay was used to screen a drug-like subset of the Maybridge Library collection consisting of 14,400 small molecules. Five compounds (CMPD-1, CMPD-5, CMPD-8, CMPD-9, and CMPD-10) as shown in Fig. 10 were selected by fluorescence polarization and differential scanning fluorimetry for their ability to disrupt p2-NC–SL2 DNA interaction via p2-NC binding. Notably, these NCIs were found to have K i in the nanomolar range and their ability to disrupt the p2-NC–SL2 DNA interaction was further verified in vitro by an electrophoretic mobility shift assay (EMSA) with p2-NC. Of these five compounds three, CMPD-5, CMPD-9 and CMPD-10, were found to have significant cell cytotoxic effects at 0.1 and 1 µM, whereas CMPD-1 and CMPD-8 were not cytotoxic and found to have anti-HIV-1 activity with EC50s of 3.5 and 0.32 µM, respectively, ex vivo, in CD4 T cells. The mode of action of the NCIs appears to be inhibition at later stages of HIV replication.
Fig. 10.
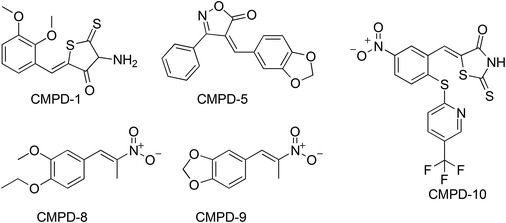
Chemical structure of the five highly active NCIs identified by a two-step HTS (Breuer et al. 2012)
Overall, the study provided a new HTS for identifying NCIs with a specific mechanism of action, which was exemplified by the identification of two low molecular weight NCIs with modest antiretroviral activity in ex vivo cell assays. These compounds provide a starting point from which to rationally optimize their NCI efficacy through directed medicinal chemistry effort. Notably, CMPD-8 shares a significant pharmacophoric similarity with EO3 and HO2 fragments previously identified (Shvadchak et al. 2009), thus suggesting that this molecular scaffold may be highly promising for the development of effective NCIs.
The optimization of the above-discussed NCIs for increased antiviral efficacy is hampered by the lack of structural details on their respective adducts with NCp7. This could be partially attributed to the high flexibility of NCp7, which makes it not suitable for high-throughput techniques such as X-ray crystallography. To this point, all published structures of NCp7 to date have been solved by NMR spectroscopy. Although the conformation of NCp7 in complex with a small hydrophobic NCI was unknown at the time of these works, molecular modeling studies have generally assumed that NCp7 in complex with a small hydrophobic NCI may be similar to the conformation adopted in binding to NAs.
Recently, the three-dimensional structure of the non-covalent adduct between NCp7 and NCI has been solved by means of NMR spectroscopy by Goudreau and co-workers from the Boehringer Ingelheim Ltd Company (Canada) (Goudreau et al. 2013). Although the initial hit (1—Fig. 11) was uncovered through an assembly assay screen for identification of HIV-1 capsid (CA) inhibitors, analysis of the mechanism of action revealed that the molecule binds to the full-length NC and competes with NA binding. Rational optimization provided two additional NCIs, 2 and 3 (Fig. 11), endowed with a sub-micromolar affinity for NCp7, as shown by isothermal titration calorimetry.
Fig. 11.
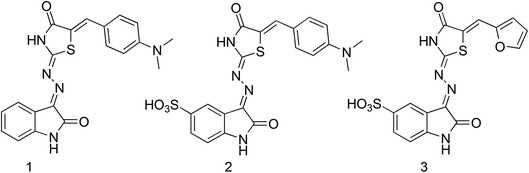
Chemical structure of the NCIs discovered by the capsid assembly assay. The complex between 3 and the NC has been characterized by NMR spectroscopy (Goudreau et al. 2013)
The use of 13C- and 15N-double-labeled NCp7 allowed the NMR-based characterization at high resolution of its adduct with 3 (PDB ID: 2M3Z—Fig. 12). This solution structure showed that 3 binds preferentially within the hydrophobic pocket of NCp7 performing a π–π stacking interaction with the side chain of Trp37, thus behaving as a mimetic of the guanosine nucleobase of NC nucleic acid partners. Moreover, this class of NCIs likely forms a 2:1 complex with the protein, with a second NCI molecule binding in a non-covalent manner to NCp7, and connecting the hydrophobic pocket with the N-terminal region (Fig. 12a). Moreover, although the NCp7 is highly flexible, residues that interact with, or are proximal to the NCI, were observed to be rigid, whereas residues not involved in binding to the NCI keep their intrinsic flexibility (Fig. 12b). Comparison with other NMR structure of NCp7/nucleic acid complexes finally confirmed that the protein is able to adopt a conformation that is highly dependent upon the chemical nature of the binding partner. This conformation of NCp7 in complex with a small molecule has been recently used in the rational design in silico of AN3 (Fig. 13), a 2-amino-4-phenylthiazole NCI that has been optimized starting from the A10 fragment disclosed by Shvadchak et al. (2009), and has been characterized by biophysical methods, such as mass spectrometry, fluorescence spectroscopy, and NMR, as well as by antiretroviral assays in infected cells. Interestingly, AN3 proved to be an efficient non-toxic and non-zinc-ejecting NCI, binding to the NCp7 hydrophobic platform and providing antiretroviral activity in cells (Mori et al. 2014).
Fig. 12.
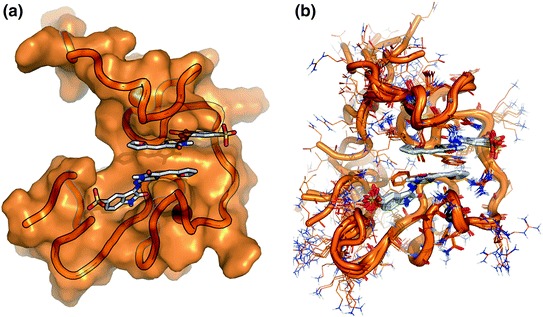
NMR structure of the complex between NCp7 and the NCI 3 (2:1 stoichiometry) (Goudreau et al. 2013). a NCp7 is shown as a transparent surface, the NCI as sticks, and Zn ions are showed as spheres. The best NMR model included in PDB ID:2M3Z is shown. b Superimposition of the best 10 NMR models of PDB ID: 2M3Z. Residues contacted by the NCI conserve their position in all models, whereas most of residues not involved in binding to the NCI are highly flexible
Fig. 13.

Chemical structure of AN3, a 2-amino-4phenylthiazole NCI active in infected cells, and designed by rational optimization in silico of A10 (Mori et al. 2014; Shvadchak et al. 2009)
In summary, these reports provide both an important step forward bettering the understanding of the molecular basis for NC inhibition by small molecules as well as strongly supporting the druggability of NCp7. Moreover, the high-resolution details of NCp7 in complex with a guanosine mimicking NCI may be used for future structure-based design and optimization of more efficient and drug-like NCIs.
Non-covalent NCIs Binding to Nucleic Acid Partners of the Nucleocapsid Protein
In the attempt to identify non-covalent NCIs, another strategy is to design small molecules that bind to the NA partners of NC, in order to prevent the interaction between NC and NAs or to disrupt the already formed complexes. As a proof of concept, in 2009, Turner and colleagues used a series of non-covalent molecular probes to investigate the structural features involved in the NC-mediated dimerization of HIV-1 genomic RNA (Turner et al. 2009). To this end, the authors used general intercalators, minor groove binders, mixed-mode intercalator/groove binders, and multifunctional polycationic aminoglycosides that, notably, have shown to not bind NC. The polycationic aminoglycosides were found to prevent the NC chaperone activity by binding to specific sites of the RNA stem loop 1 (SL1) mostly by mimicking the RNA-binding properties of the NC through electrostatic interactions, whereas all other molecules reduced the efficiency of NC-mediated isomerization by stabilizing double-stranded RNA structures. Although these studies were performed with molecular probes that are rather far from being considered as candidate therapeutics, these findings point out that inhibition of NC chaperone activity in vitro could be accomplished using small molecules binding to nucleic acids partners of NC, an important precedent.
While searching for small molecules that would inhibit NC-mediated SL1 dimer maturation, Chung and collaborators identified an activator of the SL1 dimer maturation (KA-AMC—Fig. 14) (Chung et al. 2008) that, together with three chemical derivatives (RR-AMC, R-AMC and R-MHQ—Fig. 14), was further studied by means of NMR, fluorescence emission, and molecular modeling studies (Chung et al. 2010). These three small molecules share a modified coumarine ring connected to either basic amino acids or dipeptides, which mimics the multiple interaction sites of NC for SL1 binding. Structure–activity relationship (SAR) studies further highlighted the role of the coumarine oxygen in accepting H-bonds from nucleobases, as its replacement with a NH (hydroxyquinoline R-MHQ) provoked a 3-fold decrease of activity. With respect to the amino acidic portion, the positive charge was found to be crucial for mimicking NC, allowing a strong interaction with SL1. Indeed, RR-AMC provided the tightest binding affinity, also suggesting that H-bond interactions may be relevant to stabilize the complex of the small molecule with SL1. Although the anti-NC and anti-HIV activities of these molecules have not yet been evaluated in ex vivo cell culture, the SAR data provided in the report should allow for the rational design of NCIs which can bind SL1. This is an important target as SL1 is required for the HIV-1 replication cycle, namely at the RNA dimer maturation and packaging stage.
Fig. 14.
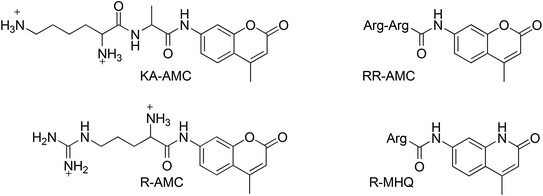
Chemical structure of coumarine derivatives binding to the SL1 RNA (Chung et al. 2010)
Baranger and co-workers performed a docking-based virtual screening of the NCI diversity set library in searching for small molecules that may bind to the stem-loop-3 RNA (SL3) of the HIV-1 packaging element Ψ (Warui and Baranger 2009). The binding affinity of virtual hits toward SL3 was monitored using fluorescence, isothermal titration calorimetry, UV-melting, circular dichroism, and footprinting techniques. Nine molecules, endowed with scaffolds that have not been previously shown to bind RNA, demonstrated micromolar affinity for SL3, with compounds 5 and 9 (Fig. 15) showing the highest affinity. Compound 9 also showed selectivity for SL3 over double- and single-stranded RNA sequences as well as SL2 and SL4. Analysis of the mechanism of action further suggested that 5 and 9 bind the stem region of SL3 without intercalating into the RNA bases. One positive outcome of this study was to pave the way for further medicinal chemistry effort to identify more potent SL3 binders. More recently, the same research group performed another virtual screening using the Chembridge database (about 700,000 small molecules), flanked by a HTS of a representative collection of the same database (about 150,000 molecules) (Warui and Baranger 2012). Although different hits were selected, both methods led to the identification of small molecules able to bind to SL3. From the sixteen positive hits identified with micromolar affinity, two molecules, namely 7 and 17, showed high selectivity for SL3 with respect to single- and double-stranded RNA sequences (Fig. 15). Noticeably, only molecules 1, 3, 4, and 8 (Fig. 15), identified by the computational protocol, were able to disrupt the NC-SL3 complex with K is between 20 and 200 µM, thus behaving as NCIs in vitro. The antiviral activity of these compounds needs still to be determined, to validate their mechanism of action, and to demonstrate their suitability as potential candidate therapeutics.
Fig. 15.
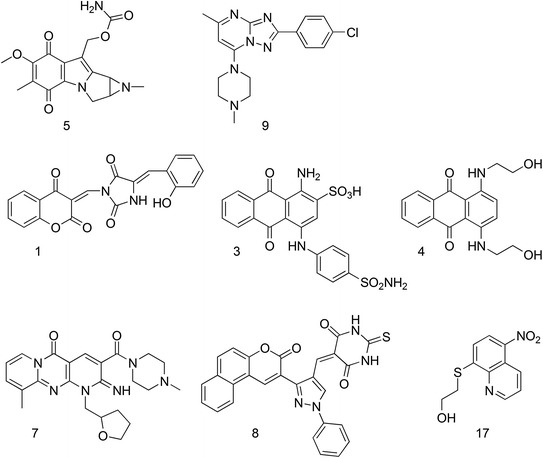
Chemical structure of small molecules binding to SL3 RNA discovered by virtual screening and HTS (Warui and Baranger 2009, 2012)
Unlike the approaches which have focused on the discovery of small molecule binding to the RNA stem-loop sequences, the research group of Gatto recently reported on the discovery and characterization of a NCI binding to the TAR sequence (Sosic et al. 2013). Starting from the anthraquinone derivative 1 (Fig. 16) that was already shown to intercalate between bases and locate its charged side chains in the grooves of NAs, the authors rationally designed and synthesized a number of chemical derivatives by increasing the distance between the positively charged side chain and the anthraquinone core. Among synthesized compounds, two molecules, 5f and 5g (Fig. 16), were found to bind TAR and, to a lesser extent, its complementary sequence cTAR, with higher affinity than other molecules and the reference compound 1. SAR analysis highlighted a linear correlation between TAR-binding affinity and the distance between the anthraquinone core and the positive charge of the side chain, with an optimum distance represented by the ornithine side chain (5g). Moreover, 5f and 5g appeared to be potent inhibitors of the NC-mediated helix destabilization of both TAR and cTAR (IC50 < 10 µM), as well as the NC-mediated TAR/cTAR annealing (IC50 = 44.1 and 21.9 µM, respectively). However, antiretroviral assays performed at 100 µM showed no HIV-1 inhibition detectable ex vivo or cell uptake of these NCIs, suggesting that charged anthraquinones are endowed with limited cell permeation. Nevertheless, these molecules represent a valid example of NCIs showing anti-NC activity in vitro, which may be further optimized as effective antiretroviral agents.
Fig. 16.
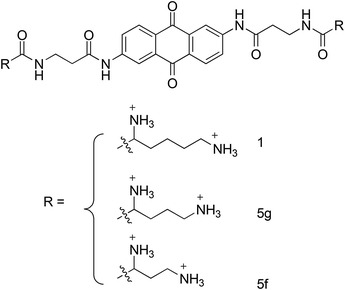
Chemical structure of NCIs binders of TAR and cTAR 5f and 5g rationally designed starting from molecule 1 (Sosic et al. 2013)
Concluding Remarks
We have provided evidence that novel screening methodologies and chemical libraries have resulted in the identification of novel compounds that show inhibitory activity against GagNC/NCp7 (NCIs). Protease inhibitors (PIs) are very effective and demonstrate highly cooperative dose-response curves, which can be explained by the capacity of these inhibitors to independently affect multiple discrete steps in the viral life cycle, such as entry, RT, and post-reverse transcription steps (Rabi et al. 2013). In a parallel capacity to PIs, and as we have discussed in our review, NCIs have the potential to affect multiple discrete viral pathways, similar to PIs. We propose that NCIs will have similar properties to PIs in regard to demonstrating highly cooperative dose-response curves. Most importantly, and in contrast to protease, NCIs should not tolerate mutational changes without considerable loss of function. Therefore, the apparent strong genetic barrier necessary for NCI resistance and the fact that NCIs inhibit a viral protein with multiple key functions throughout the HIV-1 life cycle strongly supports the continued research on identifying and optimizing NCIs as well as investigations into their antiviral mechanisms.
Acknowledgments
This work was supported by the European Project THINPAD “Targeting the HIV-1 Nucleocapsid Protein to fight Antiretroviral Drug Resistance” (FP7—Grant Agreement 601969), ANRS, NIGMS (P50GM103368), and the HIVE Center. G.M. and S.L. are greatly indebted to Jose Maria Gatell for his kind hosting and support of the AIDS Research Group (IDIBAPS).
Abbreviations
- ARDs
Antiretroviral drugs
- CA
Capsid
- CC50
Cytotoxic concentration50
- DR
Drug resistance
- EC50
Effective concentration50
- HIV
Human immunodeficiency virus
- IN
Integrase
- LTR
Long terminal repeat
- MA
Matrix
- NCI
Nucleocapsid inhibitor
- PBS
Primer-binding site
- PI
Protease inhibitor
- PPT
Polypurine tract
- PR
Protease
- Ψ
Packaging element
- RT
Reverse transcriptase
- RTC
Reverse transcriptase complex
- SP1
Spacer peptide 1
- SP2
Spacer peptide 2
- TAR
Trans-activation response element
- Tat
Trans-activator of transcription
- U5
Unique 5′ sequence
- UTR
Untranslated region
- ZF
Zinc finger
Contributor Information
Bruce E. Torbett, Email: betorbet@scripps.edu
David S. Goodsell, Phone: +1858784-2839, Email: goodsell@scripps.edu
Douglas D. Richman, Phone: 858552-8585-7439, Email: drichman@ucsd.edu
Bruce E. Torbett, Phone: 018587849124, Email: betorbet@scripps.edu
Maurizio Botta, Phone: +39 0577234306, Email: botta.maurizio@gmail.com.
Gilles Mirambeau, Phone: +34932275400, Email: gilles.mirambeau@gmail.com.
Yves Mély, Phone: 33368854263, Email: yves.mely@unistra.fr.
References
- Alce TM, Popik W. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J Biol Chem. 2004;279:34083–34086. doi: 10.1074/jbc.C400235200. [DOI] [PubMed] [Google Scholar]
- Aldovini A, Young RA. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarasinghe GK, De Guzman RN, Turner RB, Chancellor KJ, Wu ZR, Summers MF. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J Mol Biol. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- Anzellotti AI, Liu Q, Bloemink MJ, Scarsdale JN, Farrell N. Targeting retroviral Zn finger-DNA interactions: a small-molecule approach using the electrophilic nature of trans-platinum-nucleobase compounds. Chem Biol. 2006;13:539–548. doi: 10.1016/j.chembiol.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Arthur LO, Bess JW, Jr, Chertova EN, Rossio JL, Esser MT, Benveniste RE, Henderson LE, Lifson JD. Chemical inactivation of retroviral infectivity by targeting nucleocapsid protein zinc fingers: a candidate SIV vaccine. AIDS Res Hum Retroviruses. 1998;14(3):S311–S319. [PubMed] [Google Scholar]
- Avilov SV, Piemont E, Shvadchak V, de Rocquigny H, Mely Y. Probing dynamics of HIV-1 nucleocapsid protein/target hexanucleotide complexes by 2-aminopurine. Nucleic Acids Res. 2008;36:885–896. doi: 10.1093/nar/gkm1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilov SV, Boudier C, Gottikh M, Darlix JL, Mely Y. Characterization of the inhibition mechanism of HIV-1 nucleocapsid protein chaperone activities by methylated oligoribonucleotides. Antimicrob Agents Chemother. 2012;56:1010–1018. doi: 10.1128/AAC.05614-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azoulay J, Clamme JP, Darlix JL, Roques BP, Mely Y. Destabilization of the HIV-1 complementary sequence of TAR by the nucleocapsid protein through activation of conformational fluctuations. J Mol Biol. 2003;326:691–700. doi: 10.1016/s0022-2836(02)01430-4. [DOI] [PubMed] [Google Scholar]
- Basrur V, Song Y, Mazur SJ, Higashimoto Y, Turpin JA, Rice WG, Inman JK, Appella E. Inactivation of HIV-1 nucleocapsid protein P7 by pyridinioalkanoyl thioesters. Characterization of reaction products and proposed mechanism of action. J Biol Chem. 2000;275:14890–14897. doi: 10.1074/jbc.275.20.14890. [DOI] [PubMed] [Google Scholar]
- Bazzi A, Zargarian L, Chaminade F, Boudier C, De Rocquigny H, Rene B, Mely Y, Fosse P, Mauffret O. Structural insights into the cTAR DNA recognition by the HIV-1 nucleocapsid protein: role of sugar deoxyriboses in the binding polarity of NC. Nucleic Acids Res. 2011;39:3903–3916. doi: 10.1093/nar/gkq1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzi A, Zargarian L, Chaminade F, De Rocquigny H, Rene B, Mely Y, Fosse P, Mauffret O. Intrinsic nucleic acid dynamics modulates HIV-1 nucleocapsid protein binding to its targets. PLoS ONE. 2012;7:e38905. doi: 10.1371/journal.pone.0038905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltz H, Azoulay J, Bernacchi S, Clamme JP, Ficheux D, Roques B, Darlix JL, Mely Y. Impact of the terminal bulges of HIV-1 cTAR DNA on its stability and the destabilizing activity of the nucleocapsid protein NCp7. J Mol Biol. 2003;328:95–108. doi: 10.1016/s0022-2836(03)00244-4. [DOI] [PubMed] [Google Scholar]
- Beltz H, Piemont E, Schaub E, Ficheux D, Roques B, Darlix JL, Mely Y. Role of the structure of the top half of HIV-1 cTAR DNA on the nucleic acid destabilizing activity of the nucleocapsid protein NCp7. J Mol Biol. 2004;338:711–723. doi: 10.1016/j.jmb.2004.03.019. [DOI] [PubMed] [Google Scholar]
- Beltz H, Clauss C, Piemont E, Ficheux D, Gorelick RJ, Roques B, Gabus C, Darlix JL, de Rocquigny H, Mely Y. Structural determinants of HIV-1 nucleocapsid protein for cTAR DNA binding and destabilization, and correlation with inhibition of self-primed DNA synthesis. J Mol Biol. 2005;348:1113–1126. doi: 10.1016/j.jmb.2005.02.042. [DOI] [PubMed] [Google Scholar]
- Bernacchi S, Stoylov S, Piemont E, Ficheux D, Roques BP, Darlix JL, Mely Y. HIV-1 nucleocapsid protein activates transient melting of least stable parts of the secondary structure of TAR and its complementary sequence. J Mol Biol. 2002;317:385–399. doi: 10.1006/jmbi.2002.5429. [DOI] [PubMed] [Google Scholar]
- Bombarda E, Morellet N, Cherradi H, Spiess B, Bouaziz S, Grell E, Roques BP, Mely Y. Determination of the pK(a) of the four Zn2+-coordinating residues of the distal finger motif of the HIV-1 nucleocapsid protein: consequences on the binding of Zn2+ J Mol Biol. 2001;310:659–672. doi: 10.1006/jmbi.2001.4770. [DOI] [PubMed] [Google Scholar]
- Bombarda E, Cherradi H, Morellet N, Roques BP, Mely Y. Zn(2+) binding properties of single-point mutants of the C-terminal zinc finger of the HIV-1 nucleocapsid protein: evidence of a critical role of cysteine 49 in Zn(2+) dissociation. Biochemistry. 2002;41:4312–4320. doi: 10.1021/bi015956g. [DOI] [PubMed] [Google Scholar]
- Bombarda E, Roques BP, Mely Y, Grell E. Mechanism of zinc coordination by point-mutated structures of the distal CCHC binding motif of the HIV-1 NCp7 protein. Biochemistry. 2005;44:7315–7325. doi: 10.1021/bi047349+. [DOI] [PubMed] [Google Scholar]
- Bombarda E, Grell E, Roques BP, Mely Y. Molecular mechanism of the Zn2+-induced folding of the distal CCHC finger motif of the HIV-1 nucleocapsid protein. Biophys J. 2007;93:208–217. doi: 10.1529/biophysj.106.101378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudier C, Storchak R, Sharma KK, Didier P, Follenius-Wund A, Muller S, Darlix JL, Mely Y. The mechanism of HIV-1 Tat-directed nucleic acid annealing supports its role in reverse transcription. J Mol Biol. 2010;400:487–501. doi: 10.1016/j.jmb.2010.05.033. [DOI] [PubMed] [Google Scholar]
- Bourbigot S, Ramalanjaona N, Boudier C, Salgado GF, Roques BP, Mely Y, Bouaziz S, Morellet N. How the HIV-1 nucleocapsid protein binds and destabilises the (−) primer binding site during reverse transcription. J Mol Biol. 2008;383:1112–1128. doi: 10.1016/j.jmb.2008.08.046. [DOI] [PubMed] [Google Scholar]
- Bouyac M, Courcoul M, Bertoia G, Baudat Y, Gabuzda D, Blanc D, Chazal N, Boulanger P, Sire J, Vigne R, Spire B. Human immunodeficiency virus type 1 Vif protein binds to the Pr55Gag precursor. J Virol. 1997;71:9358–9365. doi: 10.1128/jvi.71.12.9358-9365.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer S, Chang MW, Yuan J, Torbett BE. Identification of HIV-1 inhibitors targeting the nucleocapsid protein. J Med Chem. 2012;55:4968–4977. doi: 10.1021/jm201442t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckman JS, Bosche WJ, Gorelick RJ. Human immunodeficiency virus type 1 nucleocapsid zn(2+) fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA. J Virol. 2003;77:1469–1480. doi: 10.1128/JVI.77.2.1469-1480.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carteau S, Batson SC, Poljak L, Mouscadet JF, de Rocquigny H, Darlix JL, Roques BP, Kas E, Auclair C. Human immunodeficiency virus type 1 nucleocapsid protein specifically stimulates Mg2+-dependent DNA integration in vitro. J Virol. 1997;71:6225–6229. doi: 10.1128/jvi.71.8.6225-6229.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamontin Célia, Rassam Patrice, Ferrer Mireia, Racine Pierre-Jean, Neyret Aymeric, Lainé Sébastien, Milhiet Pierre-Emmanuel, Mougel Marylène. HIV-1 nucleocapsid and ESCRT-component Tsg101 interplay prevents HIV from turning into a DNA-containing virus. Nucleic Acids Research. 2014;43(1):336–347. doi: 10.1093/nar/gku1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatel-Chaix L, Abrahamyan L, Frechina C, Mouland AJ, DesGroseillers L. The host protein Staufen1 participates in human immunodeficiency virus type 1 assembly in live cells by influencing pr55Gag multimerization. J Virol. 2007;81:6216–6230. doi: 10.1128/JVI.00284-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatel-Chaix L, Boulay K, Mouland AJ, Desgroseillers L. The host protein Staufen1 interacts with the Pr55Gag zinc fingers and regulates HIV-1 assembly via its N-terminus. Retrovirology. 2008;5:41. doi: 10.1186/1742-4690-5-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Nikolaitchik O, Singh J, Wright A, Bencsics CE, Coffin JM, Ni N, Lockett S, Pathak VK, Hu WS. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc Natl Acad Sci USA. 2009;106:13535–13540. doi: 10.1073/pnas.0906822106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wu B, Musier-Forsyth K, Mansky LM, Mueller JD. Fluorescence fluctuation spectroscopy on viral-like particles reveals variable gag stoichiometry. Biophys J. 2009;96:1961–1969. doi: 10.1016/j.bpj.2008.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertova EN, Kane BP, McGrath C, Johnson DG, Sowder RC, 2nd, Arthur LO, Henderson LE. Probing the topography of HIV-1 nucleocapsid protein with the alkylating agent N-ethylmaleimide. Biochemistry. 1998;37:17890–17897. doi: 10.1021/bi980907y. [DOI] [PubMed] [Google Scholar]
- Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JW, Jr, Sowder RC, 2nd, Barsov E, Hood BL, Fisher RJ, Nagashima K, Conrads TP, Veenstra TD, Lifson JD, Ott DE. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol. 2006;80:9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J, Mujeeb A, Jiang Y, Guilbert C, Pendke M, Wu Y, James TL. A small molecule, Lys-Ala-7-amido-4-methylcoumarin, facilitates RNA dimer maturation of a stem-loop 1 transcript in vitro: structure-activity relationship of the activator. Biochemistry. 2008;47:8148–8156. doi: 10.1021/bi800230m. [DOI] [PubMed] [Google Scholar]
- Chung J, Ulyanov NB, Guilbert C, Mujeeb A, James TL. Binding characteristics of small molecules that mimic nucleocapsid protein-induced maturation of stem-loop 1 of HIV-1 RNA. Biochemistry. 2010;49:6341–6351. doi: 10.1021/bi100660r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimarelli A, Darlix JL. Assembling the human immunodeficiency virus type 1. Cell Mol Life Sci. 2002;59:1166–1184. doi: 10.1007/s00018-002-8495-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosa G, Zeng Y, Liu HW, Landes CF, Makarov DE, Musier-Forsyth K, Barbara PF. Evidence for non-two-state kinetics in the nucleocapsid protein chaperoned opening of DNA hairpins. J Phys Chem B. 2006;110:2419–2426. doi: 10.1021/jp054189i. [DOI] [PubMed] [Google Scholar]
- Cruceanu M, Gorelick RJ, Musier-Forsyth K, Rouzina I, Williams MC. Rapid kinetics of protein-nucleic acid interaction is a major component of HIV-1 nucleocapsid protein’s nucleic acid chaperone function. J Mol Biol. 2006;363:867–877. doi: 10.1016/j.jmb.2006.08.070. [DOI] [PubMed] [Google Scholar]
- Cruceanu M, Urbaneja MA, Hixson CV, Johnson DG, Datta SA, Fivash MJ, Stephen AG, Fisher RJ, Gorelick RJ, Casas-Finet JR, Rein A, Rouzina I, Williams MC. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res. 2006;34:593–605. doi: 10.1093/nar/gkj458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlix JL, Vincent A, Gabus C, de Rocquigny H, Roques B. Trans-activation of the 5′ to 3′ viral DNA strand transfer by nucleocapsid protein during reverse transcription of HIV1 RNA. C R Acad Sci III. 1993;316:763–771. [PubMed] [Google Scholar]
- Darlix JL, Garrido JL, Morellet N, Mely Y, de Rocquigny H. Properties, functions, and drug targeting of the multifunctional nucleocapsid protein of the human immunodeficiency virus. Adv Pharmacol. 2007;55:299–346. doi: 10.1016/S1054-3589(07)55009-X. [DOI] [PubMed] [Google Scholar]
- Darlix JL, Godet J, Ivanyi-Nagy R, Fosse P, Mauffret O, Mely Y. Flexible nature and specific functions of the HIV-1 nucleocapsid protein. J Mol Biol. 2011;410:565–581. doi: 10.1016/j.jmb.2011.03.037. [DOI] [PubMed] [Google Scholar]
- Datta SA, Temeselew LG, Crist RM, Soheilian F, Kamata A, Mirro J, Harvin D, Nagashima K, Cachau RE, Rein A. On the role of the SP1 domain in HIV-1 particle assembly: a molecular switch? J Virol. 2011;85:4111–4121. doi: 10.1128/JVI.00006-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- de Marco A, Heuser AM, Glass B, Krausslich HG, Muller B, Briggs JA. Role of the SP2 domain and its proteolytic cleavage in HIV-1 structural maturation and infectivity. J Virol. 2012;86:13708–13716. doi: 10.1128/JVI.01704-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rocquigny H, Petitjean P, Tanchou V, Decimo D, Drouot L, Delaunay T, Darlix JL, Roques BP. The zinc fingers of HIV nucleocapsid protein NCp7 direct interactions with the viral regulatory protein Vpr. J Biol Chem. 1997;272:30753–30759. doi: 10.1074/jbc.272.49.30753. [DOI] [PubMed] [Google Scholar]
- de Rocquigny H, Shvadchak V, Avilov S, Dong CZ, Dietrich U, Darlix JL, Mely Y. Targeting the viral nucleocapsid protein in anti-HIV-1 therapy. Mini Rev Med Chem. 2008;8:24–35. doi: 10.2174/138955708783331603. [DOI] [PubMed] [Google Scholar]
- Demene H, Dong CZ, Ottmann M, Rouyez MC, Jullian N, Morellet N, Mely Y, Darlix JL, Fournie-Zaluski MC, Saragosti S, et al. 1H NMR structure and biological studies of the His23–>Cys mutant nucleocapsid protein of HIV-1 indicate that the conformation of the first zinc finger is critical for virus infectivity. Biochemistry. 1994;33:11707–11716. doi: 10.1021/bi00205a006. [DOI] [PubMed] [Google Scholar]
- Didierlaurent L, Houzet L, Morichaud Z, Darlix JL, Mougel M. The conserved N-terminal basic residues and zinc-finger motifs of HIV-1 nucleocapsid restrict the viral cDNA synthesis during virus formation and maturation. Nucleic Acids Res. 2008;36:4745–4753. doi: 10.1093/nar/gkn474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman T, Luban J, Goff SP, Haseltine WA, Gottlinger HG. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1993;67:6159–6169. doi: 10.1128/jvi.67.10.6159-6169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druillennec S, Caneparo A, de Rocquigny H, Roques BP. Evidence of interactions between the nucleocapsid protein NCp7 and the reverse transcriptase of HIV-1. J Biol Chem. 1999;274:11283–11288. doi: 10.1074/jbc.274.16.11283. [DOI] [PubMed] [Google Scholar]
- Dussupt V, Javid MP, Abou-Jaoude G, Jadwin JA, de La Cruz J, Nagashima K, Bouamr F. The nucleocapsid region of HIV-1 Gag cooperates with the PTAP and LYPXnL late domains to recruit the cellular machinery necessary for viral budding. PLoS Pathog. 2009;5:e1000339. doi: 10.1371/journal.ppat.1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussupt V, Sette P, Bello NF, Javid MP, Nagashima K, Bouamr F. Basic residues in the nucleocapsid domain of Gag are critical for late events of HIV-1 budding. J Virol. 2011;85:2304–2315. doi: 10.1128/JVI.01562-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egele C, Schaub E, Ramalanjaona N, Piemont E, Ficheux D, Roques B, Darlix JL, Mely Y. HIV-1 nucleocapsid protein binds to the viral DNA initiation sequences and chaperones their kissing interactions. J Mol Biol. 2004;342:453–466. doi: 10.1016/j.jmb.2004.07.059. [DOI] [PubMed] [Google Scholar]
- Fagny C, Vandevelde M, Svoboda M, Robberecht P. Ribonucleotide reductase and thymidine phosphorylation: two potential targets of azodicarbonamide. Biochem Pharmacol. 2002;64:451–456. doi: 10.1016/s0006-2952(02)01185-1. [DOI] [PubMed] [Google Scholar]
- Fisher RJ, Rein A, Fivash M, Urbaneja MA, Casas-Finet JR, Medaglia M, Henderson LE. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J Virol. 1998;72:1902–1909. doi: 10.1128/jvi.72.3.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godet J, Mely Y. Biophysical studies of the nucleic acid chaperone properties of the HIV-1 nucleocapsid protein. RNA Biol. 2010;7:687–699. doi: 10.4161/rna.7.6.13616. [DOI] [PubMed] [Google Scholar]
- Godet J, de Rocquigny H, Raja C, Glasser N, Ficheux D, Darlix JL, Mely Y. During the early phase of HIV-1 DNA synthesis, nucleocapsid protein directs hybridization of the TAR complementary sequences via the ends of their double-stranded stem. J Mol Biol. 2006;356:1180–1192. doi: 10.1016/j.jmb.2005.12.038. [DOI] [PubMed] [Google Scholar]
- Godet J, Ramalanjaona N, Sharma KK, Richert L, de Rocquigny H, Darlix JL, Duportail G, Mely Y. Specific implications of the HIV-1 nucleocapsid zinc fingers in the annealing of the primer binding site complementary sequences during the obligatory plus strand transfer. Nucleic Acids Res. 2011;39:6633–6645. doi: 10.1093/nar/gkr274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godet J, Boudier C, Humbert N, Ivanyi-Nagy R, Darlix JL, Mely Y. Comparative nucleic acid chaperone properties of the nucleocapsid protein NCp7 and Tat protein of HIV-1. Virus Res. 2012;169:349–360. doi: 10.1016/j.virusres.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godet J, Kenfack C, Przybilla F, Richert L, Duportail G, Mely Y. Site-selective probing of cTAR destabilization highlights the necessary plasticity of the HIV-1 nucleocapsid protein to chaperone the first strand transfer. Nucleic Acids Res. 2013;41:5036–5048. doi: 10.1093/nar/gkt164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel FD, Hemmer R, Schmit JC, Bogner JR, de Clercq E, Witvrouw M, Pannecouque C, Valeyev R, Vandevelde M, Margery H, Tassignon JP. Phase I/II dose escalation and randomized withdrawal study with add-on azodicarbonamide in patients failing on current antiretroviral therapy. Aids. 2001;15:33–45. doi: 10.1097/00002030-200101050-00007. [DOI] [PubMed] [Google Scholar]
- Goldschmidt V, Miller Jenkins LM, De Rocquigny H, Darlix JL, Mely Y. The nucleocapsid protein of HIV-1 as a promising therapeutic target for antiviral drugs. HIV Therapy. 2010;4:179–198. [Google Scholar]
- Gorelick RJ, Nigida SM, Jr, Bess JW, Jr, Arthur LO, Henderson LE, Rein A. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J Virol. 1990;64:3207–3211. doi: 10.1128/jvi.64.7.3207-3211.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudreau N, Hucke O, Faucher AM, Grand-Maitre C, Lepage O, Bonneau PR, Mason SW, Titolo S. Discovery and structural characterization of a new inhibitor series of HIV-1 nucleocapsid function: NMR solution structure determination of a ternary complex involving a 2:1 inhibitor/NC stoichiometry. J Mol Biol. 2013;425:1982–1998. doi: 10.1016/j.jmb.2013.02.022. [DOI] [PubMed] [Google Scholar]
- Grigorov B, Decimo D, Smagulova F, Pechoux C, Mougel M, Muriaux D, Darlix JL. Intracellular HIV-1 Gag localization is impaired by mutations in the nucleocapsid zinc fingers. Retrovirology. 2007;4:54. doi: 10.1186/1742-4690-4-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorov B, Bocquin A, Gabus C, Avilov S, Mely Y, Agopian A, Divita G, Gottikh M, Witvrouw M, Darlix JL. Identification of a methylated oligoribonucleotide as a potent inhibitor of HIV-1 reverse transcription complex. Nucleic Acids Res. 2011;39:5586–5596. doi: 10.1093/nar/gkr117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohman JK, Gorelick RJ, Lickwar CR, Lieb JD, Bower BD, Znosko BM, Weeks KM. A guanosine-centric mechanism for RNA chaperone function. Science. 2013;340:190–195. doi: 10.1126/science.1230715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann D, Godet J, Mely Y, Darlix JL, Restle T. HIV-1 nucleocapsid traps reverse transcriptase on nucleic acid substrates. Biochemistry. 2008;47:12230–12240. doi: 10.1021/bi801386r. [DOI] [PubMed] [Google Scholar]
- Guo F, Saadatmand J, Niu M, Kleiman L. Roles of Gag and NCp7 in facilitating tRNA(Lys)(3) Annealing to viral RNA in human immunodeficiency virus type 1. J Virol. 2009;83:8099–8107. doi: 10.1128/JVI.00488-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hameau L, Jeusset J, Lafosse S, Coulaud D, Delain E, Unge T, Restle T, Le Cam E, Mirambeau G. Human immunodeficiency virus type 1 central DNA flap: dynamic terminal product of plus-strand displacement dna synthesis catalyzed by reverse transcriptase assisted by nucleocapsid protein. J Virol. 2001;75:3301–3313. doi: 10.1128/JVI.75.7.3301-3313.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargittai MR, Gorelick RJ, Rouzina I, Musier-Forsyth K. Mechanistic insights into the kinetics of HIV-1 nucleocapsid protein-facilitated tRNA annealing to the primer binding site. J Mol Biol. 2004;337:951–968. doi: 10.1016/j.jmb.2004.01.054. [DOI] [PubMed] [Google Scholar]
- Hergott CB, Mitra M, Guo J, Wu T, Miller JT, Iwatani Y, Gorelick RJ, Levin JG. Zinc finger function of HIV-1 nucleocapsid protein is required for removal of 5’-terminal genomic RNA fragments: a paradigm for RNA removal reactions in HIV-1 reverse transcription. Virus Res. 2013;171:346–355. doi: 10.1016/j.virusres.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W.-S., Hughes S. H. HIV-1 Reverse Transcription. Cold Spring Harbor Perspectives in Medicine. 2012;2(10):a006882–a006882. doi: 10.1101/cshperspect.a006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Maynard A, Turpin JA, Graham L, Janini GM, Covell DG, Rice WG. Anti-HIV agents that selectively target retroviral nucleocapsid protein zinc fingers without affecting cellular zinc finger proteins. J Med Chem. 1998;41:1371–1381. doi: 10.1021/jm9708543. [DOI] [PubMed] [Google Scholar]
- Jenkins LM, Byrd JC, Hara T, Srivastava P, Mazur SJ, Stahl SJ, Inman JK, Appella E, Omichinski JG, Legault P. Studies on the mechanism of inactivation of the HIV-1 nucleocapsid protein NCp7 with 2-mercaptobenzamide thioesters. J Med Chem. 2005;48:2847–2858. doi: 10.1021/jm0492195. [DOI] [PubMed] [Google Scholar]
- Jenkins LM, Durell SR, Maynard AT, Stahl SJ, Inman JK, Appella E, Legault P, Omichinski JG. Comparison of the specificity of interaction of cellular and viral zinc-binding domains with 2-mercaptobenzamide thioesters. J Am Chem Soc. 2006;128:11964–11976. doi: 10.1021/ja063329e. [DOI] [PubMed] [Google Scholar]
- Jouvenet N, Laine S, Pessel-Vivares L, Mougel M. Cell biology of retroviral RNA packaging. RNA Biol. 2011;8:572–580. doi: 10.4161/rna.8.4.16030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerviel A, Thomas A, Chaloin L, Favard C, Muriaux D. Virus assembly and plasma membrane domains: which came first? Virus Res. 2013;171:332–340. doi: 10.1016/j.virusres.2012.08.014. [DOI] [PubMed] [Google Scholar]
- Krishnamoorthy G, Roques B, Darlix JL, Mely Y. DNA condensation by the nucleocapsid protein of HIV-1: a mechanism ensuring DNA protection. Nucleic Acids Res. 2003;31:5425–5432. doi: 10.1093/nar/gkg738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutluay SB, Bieniasz PD. Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog. 2010;6:e1001200. doi: 10.1371/journal.ppat.1001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzembayeva M, Dilley K, Sardo L, Hu WS (2014) Life of psi: How full-length HIV-1 RNAs become packaged genomes in the viral particles. Virology [DOI] [PMC free article] [PubMed]
- Lee BM, De Guzman RN, Turner BG, Tjandra N, Summers MF. Dynamical behavior of the HIV-1 nucleocapsid protein. J Mol Biol. 1998;279:633–649. doi: 10.1006/jmbi.1998.1766. [DOI] [PubMed] [Google Scholar]
- Lener D, Tanchou V, Roques BP, Le Grice SF, Darlix JL. Involvement of HIV-I nucleocapsid protein in the recruitment of reverse transcriptase into nucleoprotein complexes formed in vitro. J Biol Chem. 1998;273:33781–33786. doi: 10.1074/jbc.273.50.33781. [DOI] [PubMed] [Google Scholar]
- Lever A, Gottlinger H, Haseltine W, Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol. 1989;63:4085–4087. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- Levin JG, Mitra M, Mascarenhas A, Musier-Forsyth K. Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biol. 2010;7:754–774. doi: 10.4161/rna.7.6.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MS, Garcia-Asua G, Bhattacharyya U, Mascagni P, Austen BM, Roberts MM. The Vpr protein of human immunodeficiency virus type 1 binds to nucleocapsid protein p7 in vitro. Biochem Biophys Res Commun. 1996;218:352–355. doi: 10.1006/bbrc.1996.0061. [DOI] [PubMed] [Google Scholar]
- Li G, Verheyen J, Rhee SY, Voet A, Vandamme AM, Theys K. Functional conservation of HIV-1 Gag: implications for rational drug design. Retrovirology. 2013;10:126. doi: 10.1186/1742-4690-10-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingappa JR, Dooher JE, Newman MA, Kiser PK, Klein KC. Basic residues in the nucleocapsid domain of Gag are required for interaction of HIV-1 gag with ABCE1 (HP68), a cellular protein important for HIV-1 capsid assembly. J Biol Chem. 2006;281:3773–3784. doi: 10.1074/jbc.M507255200. [DOI] [PubMed] [Google Scholar]
- Liu B, Dai R, Tian CJ, Dawson L, Gorelick R, Yu XF. Interaction of the human immunodeficiency virus type 1 nucleocapsid with actin. J Virol. 1999;73:2901–2908. doi: 10.1128/jvi.73.4.2901-2908.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HW, Cosa G, Landes CF, Zeng Y, Kovaleski BJ, Mullen DG, Barany G, Musier-Forsyth K, Barbara PF. Single-molecule FRET studies of important intermediates in the nucleocapsid-protein-chaperoned minus-strand transfer step in HIV-1 reverse transcription. Biophys J. 2005;89:3470–3479. doi: 10.1529/biophysj.105.065326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HW, Zeng Y, Landes CF, Kim YJ, Zhu Y, Ma X, Vo MN, Musier-Forsyth K, Barbara PF. Insights on the role of nucleic acid/protein interactions in chaperoned nucleic acid rearrangements of HIV-1 reverse transcription. Proc Natl Acad Sci USA. 2007;104:5261–5267. doi: 10.1073/pnas.0700166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo JA, Holler TP, Sanchez J, Gogliotti R, Maloney L, Reily MD. Biophysical characterization of zinc ejection from HIV nucleocapsid protein by anti-HIV 2,2’-dithiobis[benzamides] and benzisothiazolones. J Med Chem. 1996;39:4313–4320. doi: 10.1021/jm960253w. [DOI] [PubMed] [Google Scholar]
- Lu K, Heng X, Summers MF. Structural determinants and mechanism of HIV-1 genome packaging. J Mol Biol. 2011;410:609–633. doi: 10.1016/j.jmb.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyonnais S, Gorelick RJ, Heniche-Boukhalfa F, Bouaziz S, Parissi V, Mouscadet JF, Restle T, Gatell JM, Le Cam E, Mirambeau G. A protein ballet around the viral genome orchestrated by HIV-1 reverse transcriptase leads to an architectural switch: from nucleocapsid-condensed RNA to Vpr-bridged DNA. Virus Res. 2013;171:287–303. doi: 10.1016/j.virusres.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood N, Jhaumeer-Lauloo S, Sampson J, Houghton PJ. Anti-HIV activity and mechanism of action of macrocyclic diamide SRR-SB3. J Pharm Pharmacol. 1998;50:1339–1342. doi: 10.1111/j.2042-7158.1998.tb03356.x. [DOI] [PubMed] [Google Scholar]
- Maynard AT, Covell DG. Reactivity of zinc finger cores: analysis of protein packing and electrostatic screening. J Am Chem Soc. 2001;123:1047–1058. doi: 10.1021/ja0011616. [DOI] [PubMed] [Google Scholar]
- Mely Y, Jullian N, Morellet N, De Rocquigny H, Dong CZ, Piemont E, Roques BP, Gerard D. Spatial proximity of the HIV-1 nucleocapsid protein zinc fingers investigated by time-resolved fluorescence and fluorescence resonance energy transfer. Biochemistry. 1994;33:12085–12091. doi: 10.1021/bi00206a011. [DOI] [PubMed] [Google Scholar]
- Mely Y, De Rocquigny H, Morellet N, Roques BP, Gerad D. Zinc binding to the HIV-1 nucleocapsid protein: a thermodynamic investigation by fluorescence spectroscopy. Biochemistry. 1996;35:5175–5182. doi: 10.1021/bi952587d. [DOI] [PubMed] [Google Scholar]
- Miller Jenkins LM, Hara T, Durell SR, Hayashi R, Inman JK, Piquemal JP, Gresh N, Appella E. Specificity of acyl transfer from 2-mercaptobenzamide thioesters to the HIV-1 nucleocapsid protein. J Am Chem Soc. 2007;129:11067–11078. doi: 10.1021/ja071254o. [DOI] [PubMed] [Google Scholar]
- Miller Jenkins LM, Ott DE, Hayashi R, Coren LV, Wang D, Xu Q, Schito ML, Inman JK, Appella DH, Appella E. Small-molecule inactivation of HIV-1 NCp7 by repetitive intracellular acyl transfer. Nat Chem Biol. 2010;6:887–889. doi: 10.1038/nchembio.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirambeau G, Lyonnais S, Coulaud D, Hameau L, Lafosse S, Jeusset J, Justome A, Delain E, Gorelick RJ, Le Cam E. Transmission electron microscopy reveals an optimal HIV-1 nucleocapsid aggregation with single-stranded nucleic acids and the mature HIV-1 nucleocapsid protein. J Mol Biol. 2006;364:496–511. doi: 10.1016/j.jmb.2006.08.065. [DOI] [PubMed] [Google Scholar]
- Mirambeau G, Lyonnais S, Coulaud D, Hameau L, Lafosse S, Jeusset J, Borde I, Reboud-Ravaux M, Restle T, Gorelick RJ, Le Cam E. HIV-1 protease and reverse transcriptase control the architecture of their nucleocapsid partner. PLoS ONE. 2007;2:e669. doi: 10.1371/journal.pone.0000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirambeau G, Lyonnais S, Gorelick RJ. Features, processing states, and heterologous protein interactions in the modulation of the retroviral nucleocapsid protein function. RNA Biol. 2010;7:724–734. doi: 10.4161/rna.7.6.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcock DR, Thomas JA, Gagliardi TD, Gorelick RJ, Roser JD, Chertova EN, Bess JW, Jr, Ott DE, Sattentau QJ, Frank I, Pope M, Lifson JD, Henderson LE, Crise BJ. Elimination of retroviral infectivity by N-ethylmaleimide with preservation of functional envelope glycoproteins. J Virol. 2005;79:1533–1542. doi: 10.1128/JVI.79.3.1533-1542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morellet N, Jullian N, De Rocquigny H, Maigret B, Darlix JL, Roques BP. Determination of the structure of the nucleocapsid protein NCp7 from the human immunodeficiency virus type 1 by 1H NMR. EMBO J. 1992;11:3059–3065. doi: 10.1002/j.1460-2075.1992.tb05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morellet N, de Rocquigny H, Mely Y, Jullian N, Demene H, Ottmann M, Gerard D, Darlix JL, Fournie-Zaluski MC, Roques BP. Conformational behaviour of the active and inactive forms of the nucleocapsid NCp7 of HIV-1 studied by 1H NMR. J Mol Biol. 1994;235:287–301. doi: 10.1016/s0022-2836(05)80033-6. [DOI] [PubMed] [Google Scholar]
- Morellet N, Demene H, Teilleux V, Huynh-Dinh T, de Rocquigny H, Fournie-Zaluski MC, Roques BP. Structure of the complex between the HIV-1 nucleocapsid protein NCp7 and the single-stranded pentanucleotide d(ACGCC) J Mol Biol. 1998;283:419–434. doi: 10.1006/jmbi.1998.2098. [DOI] [PubMed] [Google Scholar]
- Mori M, Dietrich U, Manetti F, Botta M. Molecular dynamics and DFT study on HIV-1 nucleocapsid protein-7 in complex with viral genome. J Chem Inf Model. 2010;50:638–650. doi: 10.1021/ci100070m. [DOI] [PubMed] [Google Scholar]
- Mori M, Manetti F, Botta M. Predicting the binding mode of known NCp7 inhibitors to facilitate the design of novel modulators. J Chem Inf Model. 2011;51:446–454. doi: 10.1021/ci100393m. [DOI] [PubMed] [Google Scholar]
- Mori M, Manetti F, Botta M. Targeting protein-protein and protein-nucleic acid interactions for anti-HIV therapy. Curr Pharm Des. 2011;17:3713–3728. doi: 10.2174/138161211798220972. [DOI] [PubMed] [Google Scholar]
- Mori M, Schult-Dietrich P, Szafarowicz B, Humbert N, Debaene F, Sanglier-Cianferani S, Dietrich U, Mely Y, Botta M. Use of virtual screening for discovering antiretroviral compounds interacting with the HIV-1 nucleocapsid protein. Virus Res. 2012;169:377–387. doi: 10.1016/j.virusres.2012.05.011. [DOI] [PubMed] [Google Scholar]
- Mori M, Nucci A, Lang MC, Humbert N, Boudier C, Debaene F, Sanglier-Cianferani S, Catala M, Schult-Dietrich P, Dietrich U, Tisne C, Mely Y, Botta M. Functional and Structural Characterization of 2-Amino-4-phenylthiazole Inhibitors of the HIV-1 Nucleocapsid Protein with Antiviral Activity. ACS Chem Biol. 2014;9:1950–1955. doi: 10.1021/cb500316h. [DOI] [PubMed] [Google Scholar]
- Mouland AJ, Mercier J, Luo M, Bernier L, DesGroseillers L, Cohen EA. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: evidence for a role in genomic RNA encapsidation. J Virol. 2000;74:5441–5451. doi: 10.1128/jvi.74.12.5441-5451.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Nath A, Farber M, Datta SA, Rein A, Rhoades E, Mothes W. A conformational transition observed in single HIV-1 gag molecules during in vitro assembly of virus-like particles. J Virol. 2014;88:3577–3585. doi: 10.1128/JVI.03353-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriaux D, Darlix JL. Properties and functions of the nucleocapsid protein in virus assembly. RNA Biol. 2010;7:744–753. doi: 10.4161/rna.7.6.14065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriaux D, Darlix JL, Cimarelli A. Targeting the assembly of the human immunodeficiency virus type I. Curr Pharm Des. 2004;10:3725–3739. doi: 10.2174/1381612043382701. [DOI] [PubMed] [Google Scholar]
- Musah RA. The HIV-1 nucleocapsid zinc finger protein as a target of antiretroviral therapy. Current topics in medicinal chemistry. 2004;4:1605–1622. doi: 10.2174/1568026043387331. [DOI] [PubMed] [Google Scholar]
- Nikolaitchik OA, Dilley KA, Fu W, Gorelick RJ, Tai SH, Soheilian F, Ptak RG, Nagashima K, Pathak VK, Hu WS. Dimeric RNA recognition regulates HIV-1 genome packaging. PLoS Pathog. 2013;9:e1003249. doi: 10.1371/journal.ppat.1003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott DE, Coren LV, Kane BP, Busch LK, Johnson DG, Sowder RC, 2nd, Chertova EN, Arthur LO, Henderson LE. Cytoskeletal proteins inside human immunodeficiency virus type 1 virions. J Virol. 1996;70:7734–7743. doi: 10.1128/jvi.70.11.7734-7743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott DE, Coren LV, Shatzer T. The nucleocapsid region of human immunodeficiency virus type 1 Gag assists in the coordination of assembly and Gag processing: role for RNA-Gag binding in the early stages of assembly. J Virol. 2009;83:7718–7727. doi: 10.1128/JVI.00099-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannecouque C, Szafarowicz B, Volkova N, Bakulev V, Dehaen W, Mely Y, Daelemans D. Inhibition of HIV-1 replication by a bis-thiadiazolbenzene-1,2-diamine that chelates zinc ions from retroviral nucleocapsid zinc fingers. Antimicrobial agents and chemotherapy. 2010;54:1461–1468. doi: 10.1128/AAC.01671-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poljak L, Batson SM, Ficheux D, Roques BP, Darlix JL, Kas E. Analysis of NCp7-dependent activation of HIV-1 cDNA integration and its conservation among retroviral nucleocapsid proteins. J Mol Biol. 2003;329:411–421. doi: 10.1016/s0022-2836(03)00472-8. [DOI] [PubMed] [Google Scholar]
- Popov S, Popova E, Inoue M, Gottlinger HG. Human immunodeficiency virus type 1 Gag engages the Bro1 domain of ALIX/AIP1 through the nucleocapsid. J Virol. 2008;82:1389–1398. doi: 10.1128/JVI.01912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintal SM, dePaula QA, Farrell NP. Zinc finger proteins as templates for metal ion exchange and ligand reactivity. Chemical and biological consequences. Metallomics: integrated biometal science. 2011;3:121–139. doi: 10.1039/c0mt00070a. [DOI] [PubMed] [Google Scholar]
- Rabi SA, Laird GM, Durand CM, Laskey S, Shan L, Bailey JR, Chioma S, Moore RD, Siliciano RF. Multi-step inhibition explains HIV-1 protease inhibitor pharmacodynamics and resistance. J Clin Invest. 2013;123:3848–3860. doi: 10.1172/JCI67399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalanjaona N, de Rocquigny H, Millet A, Ficheux D, Darlix JL, Mely Y. Investigating the mechanism of the nucleocapsid protein chaperoning of the second strand transfer during HIV-1 DNA synthesis. J Mol Biol. 2007;374:1041–1053. doi: 10.1016/j.jmb.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Ramboarina S, Srividya N, Atkinson RA, Morellet N, Roques BP, Lefevre JF, Mely Y, Kieffer B. Effects of temperature on the dynamic behaviour of the HIV-1 nucleocapsid NCp7 and its DNA complex. J Mol Biol. 2002;316:611–627. doi: 10.1006/jmbi.2001.5379. [DOI] [PubMed] [Google Scholar]
- Rein A, Henderson LE, Levin JG. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- Rein A, Datta SA, Jones CP, Musier-Forsyth K. Diverse interactions of retroviral gag proteins with RNAs. Trends Biochem Sci. 2011;36:373–380. doi: 10.1016/j.tibs.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WG, Turpin JA. Virus-encoded Zinc Fingers as Targets for Antiviral Chemotherapy. Rev Med Virol. 1996;6(4):187–199. doi: 10.1002/(SICI)1099-1654(199612)6:4<187::AID-RMV176>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Rice WG, Schaeffer CA, Harten B, Villinger F, South TL, Summers MF, Henderson LE, Bess JW, Jr, Arthur LO, McDougal JS, et al. Inhibition of HIV-1 infectivity by zinc-ejecting aromatic C-nitroso compounds. Nature. 1993;361:473–475. doi: 10.1038/361473a0. [DOI] [PubMed] [Google Scholar]
- Rice WG, Supko JG, Malspeis L, Buckheit RW, Jr, Clanton D, Bu M, Graham L, Schaeffer CA, Turpin JA, Domagala J, Gogliotti R, Bader JP, Halliday SM, Coren L, Sowder RC, 2nd, Arthur LO, Henderson LE. Inhibitors of HIV nucleocapsid protein zinc fingers as candidates for the treatment of AIDS. Science. 1995;270:1194–1197. doi: 10.1126/science.270.5239.1194. [DOI] [PubMed] [Google Scholar]
- Rice WG, Turpin JA, Schaeffer CA, Graham L, Clanton D, Buckheit RW, Jr, Zaharevitz D, Summers MF, Wallqvist A, Covell DG. Evaluation of selected chemotypes in coupled cellular and molecular target-based screens identifies novel HIV-1 zinc finger inhibitors. J Med Chem. 1996;39(19):3606–3616. doi: 10.1021/jm960375o. [DOI] [PubMed] [Google Scholar]
- Rice WG, Baker DC, Schaeffer CA, Graham L, Bu M, Terpening S, Clanton D, Schultz R, Bader JP, Buckheit RW, Jr, Field L, Singh PK, Turpin JA. Inhibition of multiple phases of human immunodeficiency virus type 1 replication by a dithiane compound that attacks the conserved zinc fingers of retroviral nucleocapsid proteins. Antimicrob Agents Chemother. 1997;41:419–426. doi: 10.1128/aac.41.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WG, Turpin JA, Huang M, Clanton D, Buckheit RW, Jr, Covell DG, Wallqvist A, McDonnell NB, DeGuzman RN, Summers MF, Zalkow L, Bader JP, Haugwitz RD, Sausville EA. Azodicarbonamide inhibits HIV-1 replication by targeting the nucleocapsid protein. Nat Med. 1997;3:341–345. doi: 10.1038/nm0397-341. [DOI] [PubMed] [Google Scholar]
- Richman D. D. Editorial Commentary: Extending HIV Drug Resistance Testing to Low Levels of Plasma Viremia. Clinical Infectious Diseases. 2014;58(8):1174–1175. doi: 10.1093/cid/ciu025. [DOI] [PubMed] [Google Scholar]
- Sartori DA, Miller B, Biebach U, Farrell N. Modulation of the chemical and biological properties of trans platinum complexes: monofunctional platinum complexes containing one nucleobase as potential antiviral chemotypes. J Biol Inorg Chem: JBIC : A Publ Soc of Biol Inorg Chem. 2000;5:575–583. doi: 10.1007/s007750000113. [DOI] [PubMed] [Google Scholar]
- Schiralli Lester GM, Akiyama H, Evans E, Singh J, Gummuluru S, Henderson AJ. Interleukin 2-inducible T cell kinase (ITK) facilitates efficient egress of HIV-1 by coordinating Gag distribution and actin organization. Virology. 2013;436:235–243. doi: 10.1016/j.virol.2012.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schito ML, Goel A, Song Y, Inman JK, Fattah RJ, Rice WG, Turpin JA, Sher A, Appella E. In vivo antiviral activity of novel human immunodeficiency virus type 1 nucleocapsid p7 zinc finger inhibitors in a transgenic murine model. AIDS Res Hum Retroviruses. 2003;19:91–101. doi: 10.1089/088922203762688595. [DOI] [PubMed] [Google Scholar]
- Schito ML, Soloff AC, Slovitz D, Trichel A, Inman JK, Appella E, Turpin JA, Barratt-Boyes SM. Preclinical evaluation of a zinc finger inhibitor targeting lentivirus nucleocapsid protein in SIV-infected monkeys. Curr HIV Res. 2006;4:379–386. doi: 10.2174/157016206777709492. [DOI] [PubMed] [Google Scholar]
- Sette P, Dussupt V, Bouamr F. Identification of the HIV-1 NC binding interface in Alix Bro1 reveals a role for RNA. J Virol. 2012;86:11608–11615. doi: 10.1128/JVI.01260-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharmeen L, McQuade T, Heldsinger A, Gogliotti R, Domagala J, Gracheck S. Inhibition of the early phase of HIV replication by an isothiazolone, PD 161374. Antiviral Res. 2001;49:101–114. doi: 10.1016/s0166-3542(00)00143-1. [DOI] [PubMed] [Google Scholar]
- Sheng N, Erickson-Viitanen S. Cleavage of p15 protein in vitro by human immunodeficiency virus type 1 protease is RNA dependent. J Virol. 1994;68:6207–6214. doi: 10.1128/jvi.68.10.6207-6214.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvadchak V, Sanglier S, Rocle S, Villa P, Haiech J, Hibert M, Van Dorsselaer A, Mely Y, de Rocquigny H. Identification by high throughput screening of small compounds inhibiting the nucleic acid destabilization activity of the HIV-1 nucleocapsid protein. Biochimie. 2009;91:916–923. doi: 10.1016/j.biochi.2009.04.014. [DOI] [PubMed] [Google Scholar]
- Sleiman D, Goldschmidt V, Barraud P, Marquet R, Paillart JC, Tisne C. Initiation of HIV-1 reverse transcription and functional role of nucleocapsid-mediated tRNA/viral genome interactions. Virus Res. 2012;169:324–339. doi: 10.1016/j.virusres.2012.06.006. [DOI] [PubMed] [Google Scholar]
- Sosic A, Frecentese F, Perissutti E, Sinigaglia L, Santagada V, Caliendo G, Magli E, Ciano A, Zagotto G, Parolin C, Gatto B. Design, synthesis and biological evaluation of TAR and cTAR binders as HIV-1 nucleocapsid inhibitors. Med Chem Comm. 2013;4:1388–1393. [Google Scholar]
- Spriggs S, Garyu L, Connor R, Summers MF. Potential intra- and intermolecular interactions involving the unique-5’ region of the HIV-1 5’-UTR. Biochemistry. 2008;47:13064–13073. doi: 10.1021/bi8014373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava P, Schito M, Fattah RJ, Hara T, Hartman T, Buckheit RW, Jr, Turpin JA, Inman JK, Appella E. Optimization of unique, uncharged thioesters as inhibitors of HIV replication. Bioorganic & medicinal chemistry. 2004;12:6437–6450. doi: 10.1016/j.bmc.2004.09.032. [DOI] [PubMed] [Google Scholar]
- Stewart-Maynard KM, Cruceanu M, Wang F, Vo MN, Gorelick RJ, Williams MC, Rouzina I, Musier-Forsyth K. Retroviral nucleocapsid proteins display nonequivalent levels of nucleic acid chaperone activity. J Virol. 2008;82:10129–10142. doi: 10.1128/JVI.01169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stote RH, Kellenberger E, Muller H, Bombarda E, Roques BP, Kieffer B, Mely Y. Structure of the His44 –> Ala single point mutant of the distal finger motif of HIV-1 nucleocapsid protein: a combined NMR, molecular dynamics simulation, and fluorescence study. Biochemistry. 2004;43:7687–7697. doi: 10.1021/bi036137u. [DOI] [PubMed] [Google Scholar]
- Stoylov SP, Vuilleumier C, Stoylova E, De Rocquigny H, Roques BP, Gerard D, Mely Y. Ordered aggregation of ribonucleic acids by the human immunodeficiency virus type 1 nucleocapsid protein. Biopolymers. 1997;41:301–312. doi: 10.1002/(SICI)1097-0282(199703)41:3<301::AID-BIP5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Summers MF, Henderson LE, Chance MR, Bess JW, Jr, South TL, Blake PR, Sagi I, Perez-Alvarado G, Sowder RC, 3rd, Hare DR, et al. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci. 1992;1:563–574. doi: 10.1002/pro.5560010502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassignon J, Ismaili J, Le Moine A, Van Laethem F, Leo O, Vandevelde M, Goldman M. Azodicarbonamide inhibits T-cell responses in vitro and in vivo. Nature medicine. 1999;5:947–950. doi: 10.1038/11392. [DOI] [PubMed] [Google Scholar]
- Thomas JA, Gorelick RJ. Nucleocapsid protein function in early infection processes. Virus Res. 2008;134:39–63. doi: 10.1016/j.virusres.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien D, Schnaare RL, Kang F, Cohl G, McCormick TJ, Moench TR, Doncel G, Watson K, Buckheit RW, Lewis MG, Schwartz J, Douville K, Romano JW. In vitro and in vivo characterization of a potential universal placebo designed for use in vaginal microbicide clinical trials. AIDS Res Hum Retroviruses. 2005;21:845–853. doi: 10.1089/aid.2005.21.845. [DOI] [PubMed] [Google Scholar]
- Tisne C, Roques BP, Dardel F. The annealing mechanism of HIV-1 reverse transcription primer onto the viral genome. J Biol Chem. 2004;279:3588–3595. doi: 10.1074/jbc.M310368200. [DOI] [PubMed] [Google Scholar]
- Tummino PJ, Scholten JD, Harvey PJ, Holler TP, Maloney L, Gogliotti R, Domagala J, Hupe D. The in vitro ejection of zinc from human immunodeficiency virus (HIV) type 1 nucleocapsid protein by disulfide benzamides with cellular anti-HIV activity. Proc Natl Acad Sci USA. 1996;93:969–973. doi: 10.1073/pnas.93.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner KB, Kohlway AS, Hagan NA, Fabris D. Noncovalent probes for the investigation of structure and dynamics of protein-nucleic acid assemblies: the case of NC-mediated dimerization of genomic RNA in HIV-1. Biopolymers. 2009;91:283–296. doi: 10.1002/bip.21107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turpin JA. The next generation of HIV/AIDS drugs: novel and developmental antiHIV drugs and targets. Exp Rev Anti-Infect Ther. 2003;1:97–128. doi: 10.1586/14787210.1.1.97. [DOI] [PubMed] [Google Scholar]
- Turpin JA, Terpening SJ, Schaeffer CA, Yu G, Glover CJ, Felsted RL, Sausville EA, Rice WG. Inhibitors of human immunodeficiency virus type 1 zinc fingers prevent normal processing of gag precursors and result in the release of noninfectious virus particles. J Virol. 1996;70:6180–6189. doi: 10.1128/jvi.70.9.6180-6189.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turpin JA, Song Y, Inman JK, Huang M, Wallqvist A, Maynard A, Covell DG, Rice WG, Appella E. Synthesis and biological properties of novel pyridinioalkanoyl thiolesters (PATE) as anti-HIV-1 agents that target the viral nucleocapsid protein zinc fingers. J Med Chem. 1999;42:67–86. doi: 10.1021/jm9802517. [DOI] [PubMed] [Google Scholar]
- Turpin JA, Schito ML, Jenkins LM, Inman JK, Appella E. Topical microbicides: a promising approach for controlling the AIDS pandemic via retroviral zinc finger inhibitors. Adv Pharmacol. 2008;56:229–256. doi: 10.1016/S1054-3589(07)56008-4. [DOI] [PubMed] [Google Scholar]
- Van Engelenburg SB, Shtengel G, Sengupta P, Waki K, Jarnik M, Ablan SD, Freed EO, Hess HF, Lippincott-Schwartz J. Distribution of ESCRT machinery at HIV assembly sites reveals virus scaffolding of ESCRT subunits. Science. 2014;343:653–656. doi: 10.1126/science.1247786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandevelde M, Witvrouw M, Schmit JC, Sprecher S, De Clercq E, Tassignon JP. ADA, a potential anti-HIV drug. AIDS Res Hum Retroviruses. 1996;12:567–568. doi: 10.1089/aid.1996.12.567. [DOI] [PubMed] [Google Scholar]
- Vercruysse T, Basta B, Dehaen W, Humbert N, Balzarini J, Debaene F, Sanglier-Cianferani S, Pannecouque C, Mely Y, Daelemans D. A phenyl-thiadiazolylidene-amine derivative ejects zinc from retroviral nucleocapsid zinc fingers and inactivates HIV virions. Retrovirology. 2012;9:95. doi: 10.1186/1742-4690-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo MN, Barany G, Rouzina I, Musier-Forsyth K. Mechanistic studies of mini-TAR RNA/DNA annealing in the absence and presence of HIV-1 nucleocapsid protein. J Mol Biol. 2006;363:244–261. doi: 10.1016/j.jmb.2006.08.039. [DOI] [PubMed] [Google Scholar]
- Vo MN, Barany G, Rouzina I, Musier-Forsyth K. HIV-1 nucleocapsid protein switches the pathway of transactivation response element RNA/DNA annealing from loop-loop “kissing” to “zipper”. J Mol Biol. 2009;386:789–801. doi: 10.1016/j.jmb.2008.12.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuilleumier C, Bombarda E, Morellet N, Gerard D, Roques BP, Mely Y. Nucleic acid sequence discrimination by the HIV-1 nucleocapsid protein NCp7: a fluorescence study. Biochemistry. 1999;38:16816–16825. doi: 10.1021/bi991145p. [DOI] [PubMed] [Google Scholar]
- Waheed AA, Freed EO. HIV type 1 Gag as a target for antiviral therapy. AIDS Res Hum Retroviruses. 2012;28:54–75. doi: 10.1089/aid.2011.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace GS, Cheng-Mayer C, Schito ML, Fletcher P, Miller Jenkins LM, Hayashi R, Neurath AR, Appella E, Shattock RJ. Human immunodeficiency virus type 1 nucleocapsid inhibitors impede trans infection in cellular and explant models and protect nonhuman primates from infection. J Virol. 2009;83:9175–9182. doi: 10.1128/JVI.00820-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Naiyer N., Mitra M., Li J., Williams M. C., Rouzina I., Gorelick R. J., Wu Z., Musier-Forsyth K. Distinct nucleic acid interaction properties of HIV-1 nucleocapsid protein precursor NCp15 explain reduced viral infectivity. Nucleic Acids Research. 2014;42(11):7145–7159. doi: 10.1093/nar/gku335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warui DM, Baranger AM. Identification of specific small molecule ligands for stem loop 3 ribonucleic acid of the packaging signal Psi of human immunodeficiency virus-1. J Med Chem. 2009;52:5462–5473. doi: 10.1021/jm900599v. [DOI] [PubMed] [Google Scholar]
- Warui DM, Baranger AM. Identification of small molecule inhibitors of the HIV-1 nucleocapsid-stem-loop 3 RNA complex. J Med Chem. 2012;55:4132–4141. doi: 10.1021/jm2007694. [DOI] [PubMed] [Google Scholar]
- Wilk T, Gowen B, Fuller SD. Actin associates with the nucleocapsid domain of the human immunodeficiency virus Gag polyprotein. J Virol. 1999;73:1931–1940. doi: 10.1128/jvi.73.3.1931-1940.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MC, Rouzina I, Wenner JR, Gorelick RJ, Musier-Forsyth K, Bloomfield VA. Mechanism for nucleic acid chaperone activity of HIV-1 nucleocapsid protein revealed by single molecule stretching. Proc Natl Acad Sci USA. 2001;98:6121–6126. doi: 10.1073/pnas.101033198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witvrouw M, Balzarini J, Pannecouque C, Jhaumeer-Laulloo S, Este JA, Schols D, Cherepanov P, Schmit JC, Debyser Z, Vandamme AM, Desmyter J, Ramadas SR, de Clercq E. SRR-SB3, a disulfide-containing macrolide that inhibits a late stage of the replicative cycle of human immunodeficiency virus. Antimicrob Agents Chemother. 1997;41:262–268. doi: 10.1128/aac.41.2.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Mitra M, McCauley MJ, Thomas JA, Rouzina I, Musier-Forsyth K, Williams MC, Gorelick RJ. Aromatic residue mutations reveal direct correlation between HIV-1 nucleocapsid protein’s nucleic acid chaperone activity and retroviral replication. Virus Res. 2013;171:263–277. doi: 10.1016/j.virusres.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You JC, McHenry CS. Human immunodeficiency virus nucleocapsid protein accelerates strand transfer of the terminally redundant sequences involved in reverse transcription. J Biol Chem. 1994;269:31491–31495. [PubMed] [Google Scholar]
- Yu X, Hathout Y, Fenselau C, Sowder RC, 2nd, Henderson LE, Rice WG, Mendeleyev J, Kun E. Specific disulfide formation in the oxidation of HIV-1 zinc finger protein nucleocapsid p7. Chem Res Toxicol. 1995;8:586–590. doi: 10.1021/tx00046a013. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Rong L, Lu J, Pan Q, Liang C. Insulin-like growth factor II mRNA binding protein 1 associates with Gag protein of human immunodeficiency virus type 1, and its overexpression affects virus assembly. J Virol. 2008;82:5683–5692. doi: 10.1128/JVI.00189-08. [DOI] [PMC free article] [PubMed] [Google Scholar]


