Abstract
Multiprotein complexes regulate most if not all cellular functions. Elucidating the structure and function of these complex cellular machines is essential for understanding biology. Moreover, multiprotein complexes by themselves constitute powerful reagents as biologics for the prevention and treatment of human diseases. Recombinant production by the baculovirus/insect cell expression system is particularly useful for expressing proteins of eukaryotic origin and their complexes. MultiBac, an advanced baculovirus/insect cell system, has been widely adopted in the last decade to produce multiprotein complexes with many subunits that were hitherto inaccessible, for academic and industrial research and development. The MultiBac system, its development and numerous applications are presented. Future opportunities for utilizing MultiBac to catalyze discovery are outlined.
Keywords: Protein complexes, Baculovirus, Insect cell expression system, MultiBac, Structural biology, X-ray crystallography, Electron microscopy, Virus-like particles (VLPs), Vaccines, Gene transfer, Lepidoptera, Synthetic biology
Introduction: The Baculovirus Expression Vector System (BEVS)
More than 30 years ago, the high level production of a heterologous protein by using an insect specific baculovirus, derived from the Autographa californica multiple nuclear polyhedrosis virus (AcMNPV) was reported. Max Summers and co-workers produced functional human IFN-β in insect cells infected by a recombinant baculovirus [1]. This development was made possible by the previous observations that late in its viral life cycle, baculoviruses express at very high levels a protein, polyhedrin, which is not essential in laboratory culture. Substitution of the polyhedrin gene in the baculoviral polh locus by a foreign gene of interest resulted in comparably high-level expression of the desired gene product, driven by the polh promoter, without compromising virus infectivity and the viral life cycle [2]. Shortly after, a second study by Lois Miller and colleagues demonstrated that another very late promoter p10, showed similar characteristics and could also be used for high-level production of heterologous proteins [3].
These two seminal studies established the baculovirus/insect cell expression system as a powerful means to produce proteins recombinantly. In the three decades since these hallmark contributions, baculovirus expression has become a widely adopted technology for academic and industrial applications, in research and development as well as manufacturing, and a wide range of proteins have been made by baculovirus expression vector systems (BEVS) [2, 4–7]. Multicomponent virus-like particles (VLPs) resembling complex virus shells have been produced successfully with BEVS, including VLPs from bluetongue, rotavirus and others [8–11]. More recently, the first baculovirus produced proteins have been approved in the therapy or prevention of human disease, including vaccines against influenza (Flublok®) and cervical cancer (Cervarix®), and immune-therapeutics against tumors of the prostate (Provenge®) [4]. Moreover, baculovirus itself has emerged as a versatile tool for gene therapy, either as a production system for recombinant adeno-associated viruses [12–14] or as a DNA-based gene delivery vehicle in its own right [15, 16].
The development of BEVS for heterologous protein production and its manifold exploits has been authoritatively reviewed recently in a number of contributions, comprehensively recapitulating the technical aspects of this technology [2, 4, 5]. The subject of this present contribution is MultiBac, a particular baculovirus expression vector system developed and implemented more recently [17–25]. MultiBac was originally conceived to meet the imposing challenge of producing eukaryotic multiprotein complexes, vital cornerstones of biological activity, in high quality and quantity for high-resolution structural and functional analysis. The system has been uniquely successful in catalyzing multiprotein complex research globally. MultiBac, its ongoing development, its numerous applications and future prospects are reviewed in the following.
The MultiBac System for Expressing Eukaryotic Multiprotein Complexes
Protein complexes catalyze key functions in the cell, and as a consequence, are an intense focus of contemporary biological research efforts. Genomics and proteomics studies have underpinned that most if not all proteins in eukaryotic cells are part of larger assemblies, which in humans often comprise ten or more individual subunits. The complex interplay of proteins in these complexes is essential for cell homeostasis, biological activity and development. High-resolution functional and structural characterization of the large number of multiprotein assemblies in the cell is critical to understanding cell biology [20, 26, 27].
Multisubunit complexes may be purified from their native cell environment and their structure and function analyzed successfully at near-atomic resolution, provided they are sufficiently abundant and homogeneous. Well-known examples include RNA polymerases and ribosomes [28–31]. The overwhelming majority of multiprotein complexes in the cell, however, are characterized by low or very low abundance, which considerably complicates or even rules out their purification from native source material. Furthermore, it is becoming increasingly clear that many proteins may exist not only in one, but a number of distinct complexes, carrying out diverse functions depending on the partner molecules they associate with at a given time. Together, this often obstructs obtaining compositionally pure and homogeneous material by classical fractionation of cells and subsequent biochemical purification, notwithstanding significant progress notably in endogenous tagging methods to genomically modify endogenous proteins by powerful extraction aids such as tandem affinity purification (TAP) tags, for instance [32–34]. A solution to these issues is recombinant overproduction, enabled by the development and implementation of powerful overexpression technologies that can achieve high-level production of homogeneous and active eukaryotic complexes for detailed mechanistic analysis at the molecular level.
Recombinant protein overproduction had a profound and game-changing impact on protein science, making previously inaccessible targets readily available. A very large number of proteins, their mutants and variants have been produced recombinantly, and their structure and function determined at high resolution. The availability of entire genomes has made it possible to address the protein repertoire of cells on a system-wide scale, applying high-throughput technologies [35]. Recombinant protein expression in E. coli as a prokaryotic expression host has become prevalent in molecular biology laboratories world-wide. The recombinant production of protein complexes of eukaryotic origin, however, poses a number of challenges which frequently rule out prokaryotic expression hosts. Eukaryotic proteins are often large and can exceed the size range E. coli can overproduce efficiently (typically up to ~100 kDa). Post-translational modifications and processing are commonplace in eukaryotic proteins and can be essential for activity, but are generally not supported by a prokaryotic host. The eukaryotic protein folding machinery differs significantly from the chaperone system in E. coli, further restricting its utility for eukaryotic protein production. Much effort has been and is being devoted to improving prokaryotic host systems for heterologous production [36–38]. However, in many cases eukaryotic proteins and their complexes will likely require a eukaryotic expression host system for their overproduction, and if a eukaryotic system can be applied with comparable ease as E. coli based expression, then this system will likely be a preferred choice. The MultiBac system has been developed precisely with this intention to put in place such a eukaryotic expression system that supports high-level and high-quality production of eukaryotic proteins and their complexes, by using standard operating protocols (SOPs) which make its application comparably facile and routine as E. coli-based expression [18, 39–41].
MultiBac Developments
The baculovirus/insect cell expression system is particularly well-suited for the production of eukaryotic proteins. At the core of this expression technology is a recombinant baculovirus into which the heterologous genes of interest have been inserted. This composite baculovirus is then used to infect insect cell cultures grown in the laboratory. MultiBac is a more recent baculovirus/insect cell system which has been specifically tailored for the overproduction of eukaryotic complexes that contain many subunits [40]. An important prerequisite for the efficient expression of eukaryotic proteins and their complexes is easy-to-use reagents for (multi)gene assembly and delivery. Equally required are robust and standardized protocols for all steps involved in the expression experiment, from gene to purified protein complex. These steps should ideally be implemented as standard operating procedures (SOPs), especially in laboratories where the expression experiment itself and its optimizations are not the primary objective, but rather the protein complex and the determination of its structure and mechanism within a reasonable time-frame. The implementation of such SOPs will then enable non-specialist users to apply the technology with relative ease. The MultiBac system has been designed to meet these requirements [18, 40, 41].
MultiBac consists of an engineered baculovirus that has been optimized for multiprotein complex expression [17] (Fig. 13.1). The MultiBac baculovirus exists as a bacterial artificial chromosome (BAC) in E. coli cells (DH10MultiBac or DH10MB). The replicon (F-factor) present on the BAC restricts its copy number to (typically) one [42]. The MultiBac genome has been modified by deleting proteolytic and apoptotic functionalities from the baculoviral genome that were found to be detrimental for the quality of the heterologous target complexes produced [17–19, 23, 41]. The MultiBac system furthermore comprises an array of small custom-designed DNA plasmid modules that facilitate the assembly of multigene expression cassettes and their integration into the baculoviral genome (Fig. 13.1). Integration of the multigene expression cassette constructions into the baculoviral genome occurs via two sites (Fig. 13.1). One is a mini-Tn7 attachment site embedded in a LacZcbα gene that is used for blue/white selection and is accessed by the Tn7 transposase which is expressed in the DH10MB cells from a helper plasmid as described previously [43]. Upon integration into this Tn7 site, the LacZα gene is disrupted; white colonies indicate successful transposition. A second entry site is formed by a short imperfect inverted repeat, LoxP, at a location distal from the Tn7 attachment site (Fig. 13.1). It can be accessed by means of the Cre enzyme, a site-specific recombinase that targets the LoxP imperfect inverted repeat (Fig. 13.1). Cre integration occurs by fusing LoxP sites present on the MultiBac genome on the one hand, and on a DNA plasmid module on the other. Successful Tn7 and also Cre integration is imposed by antibiotic selection against the resistance markers encoded by the DNA plasmid modules integrated into the MultiBac genome. The integration sites can be used to integrate genes encoding for one or several multiprotein complexes of choice, but also for additional functionalities that may be required to activate or inactivate the complex (kinases, phosphatases, acetylases, deacetylases, others), support its folding (chaperones) or post-translational processing such as glycosylation [19, 23, 44–46].
Fig. 13.1.
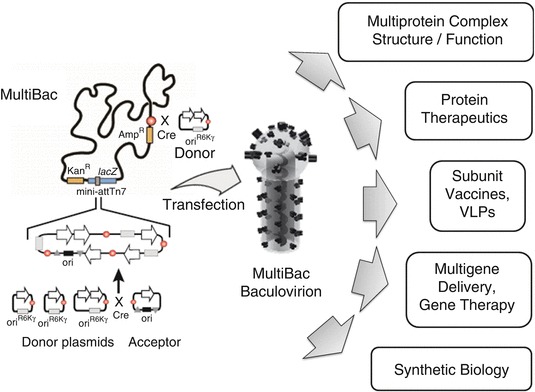
The MultiBac baculovirus/insect cell expression system. The MultiBac system is shown in a schematic view (left). MultiBac consist of an engineered baculovirus optimized for protein complex production. This MultiBac baculoviral genome exists as a bacterial artificial chromosome (BAC) in E. coli cells. It contains two integration sites for foreign genes, by Tn7 transposition or, alternatively, by site-specific recombination mediated by the Cre enzyme. MultiBac further consists of an array of plasmids called Acceptors and Donors that facilitate multigene assembly. MultiBac baculovirions (center) are generated by transfecting composite MultiBac BAC in insect cells. MultiBac is successfully used for a wide variety of applications in basic and applied research and development (right). Genes of interest are shown as arrows filled in white. Circles filled in red indicate LoxP sites. Origins of replication appear as rectangular shapes. R6Kγ phage-derived replicon, VLP virus-like particle, Kan kanamycin marker, Amp Ampicillin marker, Cre Cre-LoxP fusion reaction, LacZα blue/white selection cassette, attn7 Tn7 attachment site
The composite MultiBac baculoviral genome which contains all desired heterologous genes is then purified from small bacterial cultures using standard alkaline lysis protocols and applied to small insect cell cultures, typically in six-well plates, with a lipidic or non-lipidic transfection reagent [24, 41]. The resulting live baculovirions are harvested and applied to larger insect cell cultures for heterologous protein production and purification. Production baculovirus is then stored for example at 4 °C in the dark to avoid degeneration of viral titers. A more secure long-term storage method is provided by freezing small aliquots of baculovirus infected insect cells (BIICs) that are then stored in liquid nitrogen [47].
A centerpiece of the MultiBac system is the facilitated assembly of genes into multiexpression cassettes (Figs. 13.1 and 13.2). Originally, this was addressed by creating two different DNA plasmids that contained a so-called Multiplication Module each. This module allowed iterative assembly of single or dual expression cassettes, each fitted with a promoter (p10 or polh) by restriction/ligation utilizing compatible sites that would be destroyed upon ligation [17]. This functional unit-based plasmid configuration later was popularized as ‘BioBrick’ in the context of synthetic biology.
Fig. 13.2.
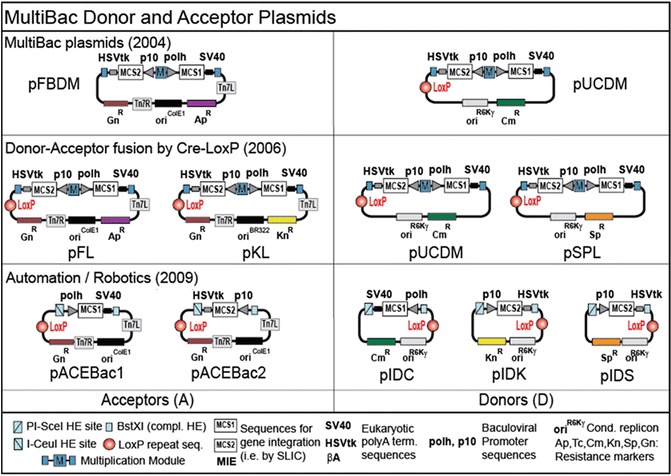
MultiBac tool-kits. A variety of entry plasmids to integrate heterologous genes into the MultiBac baculoviral genome have been created since the introduction of the system in 2004, each with its own merits. Functional modules contained in the plasmids are listed (bottom). All plasmids are compatible with each other and can be used in various combinations to generate recombinant MultiBac baculoviral genomes for multiprotein expression and/or multigene delivery. Expression cassettes are in ‘BioBrick’ format and enable iterative multiplication
One plasmid (pFBDM) accessed the Tn7 site, the second plasmid (pUCDM) accessed the LoxP site on the MultiBac genome. Both plasmids could be fitted with one to many foreign genes by means of multiplication. The pUCDM plasmid contains a conditional origin of replication derived from the phage R6Kγ. Its survival therefore hinges on the presence of a particular gene (pir) in the genome of specific E. coli strains. If a pUCDM plasmid is transformed into DH10MB cells which are pir negative, it will only survive if it is productively fused to the MultiBac genome by Cre-LoxP reaction [17].
The plasmid module repertoire of the MultiBac system was subsequently expanded by fitting out the pFBDM plasmid with a LoxP site, giving rise to pFL. A further plasmid was created, pKL, which in contrast to the high-copy number pFL plasmid is propagated at low copy numbers, and thus could accommodate difficult or very large genes that had turned out to be unstable in pFL. A new version of pUCDM was designed with a different resistance marker (spectinomycin), pSPL. pSPL or pUCDM could now be fused either with the MultiBac genome in vivo in DH10MB if Cre was expressed, or in vitro with pFL or pKL. All plasmids retained the Multiplication Module and therefore could be outfitted with several to many genes in an iterative way. pUCDM and pSPL were now denoted Donor plasmids (D), while pFL and pKL were denoted Acceptors (A) (Fig. 13.2) [18, 41]. One Acceptor could be fused with one or two Donors by Cre-LoxP reaction in vitro, and productive fusions were identified by the combination of resistance markers present on the fusion. The fused AD or ADD constructs carrying several to many heterologous genes, could then be inserted into the Tn7 attachment site of the MultiBac genome by transposition in DH10MB cells as before. In vitro fusion of AD or ADD plasmids prior to integration into the MultiBac genome via the Tn7 site did not rule out use of the LoxP site present on the viral backbone to additionally incorporate a Donor. Integration into the viral LoxP site simply had to precede the transposition reaction [18].
A sine qua non of contemporary structural biology is automation, to increase the throughput of expression experiments by robotic approaches. As a consequence, the multigene assembly technology of the MultiBac system was adapted to automation by using a liquid handling robot [24, 25]. For this, the tandem recombineering technology (TR) originally developed for prokaryotic complex expression [38] was adapted to the MultiBac system [48]. TR combines sequence-and-ligation independent gene insertion (SLIC) with Cre-LoxP fusion to generate multigene expression constructs in high-throughput in a robotic setup. Adaptation of the MultiBac plasmids to TR required subtle adjustments of the plasmids resulting in Acceptor plasmids pACEBac1 and pACEBac2 as well as Donor plasmids pIDK, pIDS and pIDC that are robotics-compatible [22, 24, 40]. Concomitantly, with the objective to simplify the monitoring of protein production by measuring fluorescence levels, a new baculovirus genome, EMBacY, was created expressing a yellow fluorescent protein (YFP) from its backbone, with fluorescence intensity increasing in parallel with the quantity of the heterologous protein complex expressed at the same time.
All Acceptor and Donor plasmids developed for the MultiBac system over time are compatible with each other, and also with the MultiBac and EMBacY genomes (and other MultiBac genome derivatives) that were and are being developed (see also Sects. 13.2.3 and 13.2.4).
A MultiBac User Access Platform
The MultiBac system rapidly developed into a sought after tool following its introduction and original publication, compellingly underscoring the current need for an accessible expression technology for eukaryotic multiprotein complexes. The number of laboratories using MultiBac approaches a thousand at the time this review is written. Research groups in academia as well as the biotech and pharma industries are implementing MultiBac to produce their specimens of interest. Moreover, biotech spin-offs were founded based on MultiBac developments, including a preclinical vaccine development company, Redvax GmbH, which in 2015 was acquired by the global pharma enterprise, Pfizer.
The high demand for accessing this technology resulted in the establishment of a dedicated research and training platform at the European Molecular Biology Laboratory (EMBL) in the Eukaryotic Expression Facility (EEF) established at the EMBL Outstation in Grenoble (Fig. 13.3). The EEF has been supported since 2008 by the European Commission (EC) through infrastructure grants (P-CUBE, BioStruct-X, INSTRUCT) and by the national (French) research agency (ANR) through the Investissement d’Avenir program. More than a hundred projects per year, by local national and transnational users, including academic research projects as well as industrial contracts have been processed in the facility, covering a wide range of applications in basic and applied research and development. Academic project access is based on the sole criteria of excellence, determined by an independent panel reviewing research proposals. The facility has implemented a SOP-based procedure for routinely and rapidly moving projects through the MultiBac platform pipeline [23, 24].
Fig. 13.3.
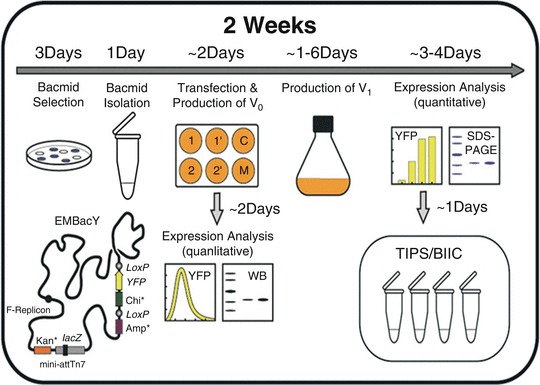
MultiBac expression platform at the EMBL Grenoble. The standard operating protocol (SOP) implemented is illustrated, to express proteins and their complexes by MultiBac. The entire process takes 2 weeks from generation of the composite MultiBac BAC (bacmid) to quantitative expression analysis. A MultiBac baculovirus variant called EMBacY is used in the platform, producing yellow fluorescent protein (YFP) to track virus performance and heterologous protein production. In addition, protein production is monitored by Western blot (WB) analysis or by gel electrophoresis (SDS-PAGE). Production virus is stored long-term in frozen aliquots of baculovirus in insect cells (BIICs)
The entire process from preparing the multigene expression construct to (small-scale) purification of the specimens of interest requires around 2 weeks according to this protocol. A large number of constructs can be processed in parallel. Virus amplification as well as heterologous protein production is monitored by measuring the signal of yellow fluorescent protein (YFP) in small (one million cells) aliquots withdrawn from the cell cultures at defined intervals. The YFP signal reaching a plateau indicates maximal production of the desired complex specimen and the expression culture is harvested at this point for further processing and purification. Particular care is taken to assure highest quality virus production during virus amplification—early (budded) virus is harvested throughout the amplification process to avoid accumulation of defective viral particles that would compromise virus performance. Initial virus is stored at 4 °C, while production virus is stored as BIICs in liquid nitrogen [21–24] (Fig. 13.3).
OmniBac: Universal Multigene Transfer Vectors
Two approaches for foreign gene integration into the baculoviral genome dominate the field. One approach depends on the baculoviral genome present as a BAC in E. coli cells, and relies on gene integration by transposition catalyzed by the Tn7 transposase which is constitutively expressed from a dedicated (helper) plasmid in the cells harboring the BAC. The foreign genes are provided by transforming a transfer plasmid into these E. coli cells. Selection for recombinant BACs occurs based on the resistance marker that is present on the transfer plasmid and also integrated, as well as blue/white selection; productive transposase mediated integration destroys a LacZα gene. The MultiBac system retains this strategy, extended by the option to provide in addition to the Tn7 transposase also the site-specific Cre recombinase transiently from a second dedicated helper plasmid, to fuse an additional Donor construct into a LoxP site present on the MultiBac baculoviral genome [17, 40, 41].
The alternative, original approach is based on homologous recombination, mediated by DNA regions in the transfer plasmid that are also present in the baculoviral genome, flanking the polh locus that had been inactivated by destruction of the polh gene. These regions of homology correspond to the open reading frames Orf1620 and lef2/603. By using this method, insertion occurs by co-transfecting the transfer plasmid and the purified baculoviral genome into insect cells. The baculoviral genome is typically linearized in the region between Orf1629 and lef2/603, to suppress formation of virus devoid of the heterologous DNA of interest. Fusion of the plasmid DNA with the baculoviral DNA to create a replicating genome is then achieved via the homologous recombination system of the insect cells. The genome is thus re-circularized, concomitantly inserting the gene of interest. Live virions are then produced that express the desired protein(s).
Both methods have advantages and disadvantages. The Tn7/BAC based integration approach is the method of choice in many laboratories, mainly due to its simplicity. Since the baculoviral genome exists as a BAC in E. coli, it can be, in theory, propagated indefinitely and used for many experiments after obtaining the initial aliquot. Moreover, it can also be manipulated by gene editing technologies in its E. coli host cell. In contrast, the linearized baculoviral DNA for homologous recombination has to be obtained from the supplier for every experiment de novo and cannot be propagated at will. Furthermore, the homologous recombination method is arguably somewhat more involved and may require specialist knowledge. On the other hand, this approach is more amenable to automation as compared to the BAC-based method which is characterized by many steps some of which (such as blue/white screening) cannot be readily scripted into robotics routines. A further disadvantage of the BAC-based system may be found in the relative instability that was described for BAC-derived baculoviruses in insect cells, presumably originating from the presence of extended bacterial DNA elements (selection marker, F-replicon, LacZα gene) [4, 49], limiting its use in human applications mainly to preclinical studies [4]. In fact, baculoviruses used in commercial production to date are still being made almost exclusively by applying the classical homologous recombination technique [4].
Available BEVS all relied on either one method or the other, which were mutually exclusive, and the choice of the transfer plasmid decided which virus would be used for protein production. This situation is unsatisfactory given that both systems (BAC/Tn7-based and classical) provide unique opportunities. Moreover, numerous baculoviruses with customized functionalities have been created for both applications, each with its own merit. We therefore designed and created the “OmniBac” transfer plasmids which combine the DNA elements required for Tn7 transposition in the BAC-based system and also the homology regions for baculovirus generation following the classical method, and thereby are universally applicable (Fig. 13.4). These OmniBac transfer plasmids function as Acceptors in the MultiBac system (Fig. 13.4). They can be combined with a variety of Donors to yield multigene Acceptor-Donor fusions that can then be funneled into any baculovirus of choice [50].
Fig. 13.4.

OmniBac – Universal transfer plasmids for every BEVS. (a) Acceptor plasmids pOmniBac1 and pOmniBac2 are shown schematically, functional modules are same as listed above (Fig. 13.2, bottom). These Acceptors can be combined with the Donor plasmids by Cre-LoxP recombination. (b) OmniBac plasmids comprise elements for homologous recombination as well as Tn7-based transposition. Multigene constructions based on OmniBac plasmids can therefore access all available baculovirus genomes. Thus, with the same plasmids, composite baculovirus for preclinical studies as well as manufacturing can be produced efficiently
The ComplexLink Polyprotein Expression Technology
A challenge frequently encountered when overexpressing protein complexes relates to imbalanced expression levels of the individual protein subunits that are to assemble into the biological target superstructure. If a particular subunit is badly made in a co-expression experiment, it will limit the overall yield of the fully assembled protein complex dramatically. This challenge can be addressed within limits by co-infection approaches using several viruses, or the choice of promoters. In contrast to very late promoters such as polh and p10, earlier promoters are expressed at lower levels. However, it may be often impractical to resort to co-infection or to tuning protein expression levels by promoter choice, also due to the fact that the timing of the production of protein subunits will then be altered as well, with often unpredictable consequences. A solution to such problems can derive from observing the strategies certain viruses utilize to realize their proteome. Coronavirus, the agent that causes severe acute respiratory syndrome (SARS), produces its complete proteome from two long open reading frames (ORFs) that give rise to polyproteins in which the individual protein specimens are covalently linked. A highly specific protease, also encoded by the ORF, then liberates the individual proteins by cleaving them apart.
The ComplexLink technology implements this strategy for recombinant polyprotein production from polygenes [22, 40, 51] (Fig. 13.5). In ComplexLink, genes encoding for a protein complex of choice are conjoined to yield a single ORF. This ORF gives rise to a polyprotein in which the individual proteins are linked by short amino acid sequences representing a cleavage site for the NIa protease from tobacco etch virus (TEV) which is also encoded by the ORF and is the first protein produced. In addition, a cyan fluorescent marker protein (CFP) is present at the C-terminal end of the polyprotein construction. Upon translation, TEV protease liberates itself, and all other proteins including CFP by cleaving the TEV-specific proteolytic sites. The ComplexLink plasmids, pPBac, pKL-PBac and pOmni-PBac function as Acceptors in the MultiBac system and can be fused to Donors which may contain further genes encoding for polyproteins. The ComplexLink technology proved to be highly successful in producing difficult-to-express protein complexes in high quality and quantity, including the physiological core complex of human general transcription factor TFIID [52–54]. A notable exploit is influenza polymerase, an important drug target to combat the flu, which has remained inaccessible for 40 years since its discovery. Influenza polymerase has been produced, for the first time, successfully using ComplexLink in conjunction with MultiBac, enabling elucidation of its structure and mechanism by X-ray crystallography at near-atomic resolution [55, 56] (Figs. 13.5 and 13.6).
Fig. 13.5.
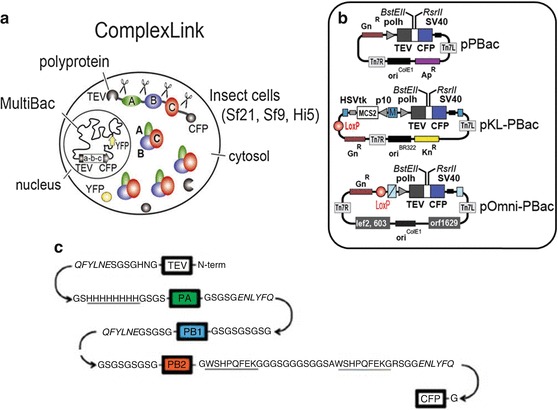
ComplexLink technology. (a) The ComplexLink technology was created to produce multiprotein complexes from self-processing polyprotein constructs as shown here schematically. The polyprotein contains a TEV protease and a fluorescent protein, at the N- and C-termini, respectively. Polyproteins are processed into the individual protein entities by the highly specific TEV protease. (b) Polyprotein expression plasmids pPBac, pKL-PBac and pOmni-PBac are shown. DNA modules are marked as above (Fig. 13.2, bottom). c Schematic representation of a self-processing polyprotein encoding for influenza polymerase, before TEV-mediated cleavage. TEV stands for tobacco etch virus NIa protease, PA, PB1 and PB2 are subunits of the trimeric influenza enzyme complex, CFP stands for cyan fluorescent protein. BstEII and RsrII are asymmetric restriction enzymes that can be used to access polyprotein expression plasmids for restriction/ligation-based heterologous gene integration
Fig. 13.6.
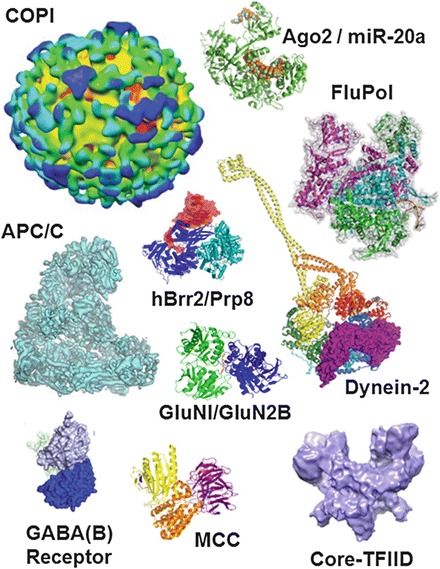
MultiBac complex structure gallery. A selection of recent high impact structures of MultiBac-produced biological specimens are shown. Examples include cryo-EM architectures of COPI-coated Vesicles (EMD-2084 to EMD-2088), the complete human APC/C complex with coactivator and substrate (EMD-2651 to EMD-2654) and the human core-TFIID complex (EMD-2229 to EMD-2231). Notable structures that were determined by X-ray crystallography (PDB identifiers are provided in brackets) include influenza polymerases A and B bound to the viral RNA promoter (PDB identifier 4WSA, 4WRT), human Argonaute Ago2 in complex with miR-20a RNA (4F3T), the spliceosomal complex Brr2HR/Prp8Jab1 (4KIT), human GABA(B) receptor (PDB 4MQE), the dynein-2 motor bound to ADP (4RH7), the mitotic checkpoint complex MCC (4AEZ) and the GluN1/GluN2B N-methyl-D-aspartate receptor (3QEL). Molecular illustrations were prepared with PyMOL (www.pymol.org) and Adobe Photoshop Version CS6
Applications
The MultiBac system in its original form was introduced in 2004, and has become the method of choice for a wide range of applications. Primarily developed for accelerating structural biology of multiprotein complexes, it has since then been modified and improved to benefit also other fields, in basic and applied research. We have followed these developments with interest and have occasionally highlighted them in invited review articles and commentaries [40, 53, 57, 58]. In the following, we intend to summarize these developments without being exhaustive, focusing on recent exploits by researchers who adopted the system we had developed, to catalyze their research.
Accelerating Complex Structural Biology
The first users which implemented MultiBac were structural biologists interested in elucidating the architecture and mechanics of important multiprotein machines in cell biology at the molecular, near-atomic level. This is also the field where an impressive flurry of highest impact MultiBac-enabled contributions was achieved to date. We had highlighted some of these exciting structures, including important drug targets such as the LKB1-STRAD-MO25 complex [59], and the first structure of a nucleosome-bound chromatin remodeler, Isw1 [60] in a contribution just over 2 years ago in Trends in Biochemical Sciences [40]. In the short time since then, spanning a mere 2 years, a large number of new structural studies were carried out using material produced with MultiBac. A selection of these exploits is presented in Fig. 13.6.
Landmark achievements are the recent crystal structures of influenza polymerase [55, 56]. This success was catalyzed by applying the ComplexLink and MultiBac technologies in combination, to produce this trimeric protein complex which had remained elusive for decades. The structures describe fluA and fluB variants of the polymerase bound to its RNA ligand, and provide important structural insights into cap-snatching and RNA synthesis by this enzyme complex, opening up new avenues for pharmaceutical development to combat flu. Further crystallographic exploits include the structures of human cytoplasmic dynein-2 primed for its power stroke [61], the human argonaute-2/RNA complex [62], the structure of the spliceosomal protein Prp8 bound to an RNA helicase, Brr2 [63, 64], and structures of PI4KIIIβ kinase complexes [65] among numerous others. Highlights achieved by using MultiBac produced material for electron microscopic studies include the structures of COP1-coated vesicles, revealing alternate coatomer conformations and interactions [66, 67], the architecture of the physiological core of human general transcription factor TFIID [52, 58], or the elucidation of the molecular mechanisms of the anaphase promoting complex APC/C at sub-nanometer resolution [68]. Recently, the entire human Mediator transcription factor holo-complex has been successfully assembled by using MultiBac, and functionally characterized [69]. MultiBac reagents have been incorporated into pipelines for producing membrane proteins and their complexes [70]. We anticipate that many more exciting structures of important protein assemblies will be determined in the future, by using the MultiBac system as a production tool.
MultiBac in Pharma and Biotech
The baculovirus/insect cell system has had a major impact on the production of high-value protein targets, for pharmacological characterization, structure-based drug design, diagnostics, biosensor engineering and high-throughput proteomics [2, 4, 5, 71]. Notably human proteins, virus-like particles (VLPs) and vaccines have been successfully expressed by using BEVS [2, 4, 5]. Glycoproteins are sought-after biologics in the pharma and biotech sector, and insect cells have proven to be well suited for the expression of biologically active and immunogenic specimens. The MultiBac system has been engineered to enable high-quality production of glycoproteins and their complexes. The original MultiBac baculoviral genome was already lacking the v-cath and chiA genes, which are encoding for cathepsin-like protease and chitinase, respectively. Both v-cath and chiA have been shown to be detrimental to glycoprotein production [72–74]. The glycosylation pattern of secreted proteins in insect cells differs from mammalian patterns which involve more complex N-glycans [75, 76]. These differences can have adverse effects on human glycoproteins produced in insect cells. To overcome this impediment, a new MultiBac-derived baculovirus, SweetBac, was constructed, which includes glycosyltransferases in the backbone, resulting in mammalianized glycosylation patterns of SweetBac-produced glycoproteins [23, 44, 45]. More recently, improved MultiBac variants were introduced to minimize fucosylation in insect cell derived glycoproteins to reduce binding to antibodies from the sera of patients with allergies [46].
The BEVS has demonstrated its aptitude to produce complex multicomponent assemblies such as virus-like particles (VLPs) [4, 9–11, 77]. VLPs are promising candidates for vaccination. VLPs resemble natural virus shells, but are lacking genetic material and therefore are safe and not infectious. VLPs can be proteinaceous, such as for example papilloma VLPs used to prevent cervical cancer. More recently, enveloped viruses have been successfully produced using BEVS, including influenza and chikungunya vaccine candidates [4, 77–80]. The MultiBac system was already successfully used to produce a number of VLPs including an array of papilloma serotypes [40]. Complex VLPs representing highly pathological virus strains were produced safely [81]. In particular the availability of OmniBac plasmids, which are part of the MultiBac vector suite, may provide unique opportunities for VLP vaccine development, as they are equally useful for exploratory preclinical studies in high-throughput as well as pharmacological manufacturing, by choosing the most appropriate viral backbone for large scale expression (Fig. 13.4).
Baculoviruses not only infect insect cells, but can also transduce mammalian cells efficiently [15, 82–84]. By choosing mammalian-active promoters instead of polh, p10 or other baculoviral promoters, proteins of interest can be produced from a baculovirus that has entered a mammalian host cell. Baculoviruses do not replicate in mammalian cells, therefore, the current consensus is that this BacMam approach can be performed safely in laboratories. A particular benefit of BacMam is that large DNA insertions including multicomponent signaling cascades or entire metabolic pathways can be transduced into mammalian cells by the baculovirus which can tolerate very large gene insertions, and can be amplified and produced in large amount in a straightforward manner in insect cell cultures. Baculovirus is thus emerging as a highly promising gene delivery tool into mammalian cells, for a multitude of applications [14, 15]. Already, multigene MultiBac constructions were successfully used to produce recombinant adeno-associated viruses (AAVs) for gene therapy [12, 13, 40].
Synthetic Biology: Rewiring the Genome
The AcMNPV genome has a size of around 130 kilobases and contains numerous functionalities, which are essential in the natural life cycle of the virus, but dispensable in laboratory culture. Moreover, in the laboratory, it has been recognized that the genome has a tendency to undergo multiple deletions in its genome during amplifications, notably in regions containing foreign gene cargo that has been inserted for overproduction [4, 50]. This can have severely detrimental consequences for heterologous target protein production, especially for fermenter-scale manufacturing of biologics which require large foreign DNA insertions in the viral genome and several rounds of amplification, until sufficient volumes of production virus are obtained to charge the fermenter. We have shown that some of these limitations can be overcome at least on laboratory-scale by stringently adhering to virus generation protocols that avoid or limit the occurrence of widespread deletions [18]. Moreover, it appears that there is considerable scope for improving virus performance by eliminating mutational or deletion hot-spots in the viral genome, and by removing DNA regions which are not required in cell culture. The baculoviral genome, in particular when present as a BAC, lends itself excellently to genome manipulation and editing techniques. We and others have exploited this avenue to remove unnecessary or undesired functionalities from the virus, such as the polh, p10, v-cath and chiA genes among (many) others.
The advent of efficient synthetic biology techniques now holds the promise to reverse this somewhat cumbersome top-down approach, and to rationally redesign and rewire the baculoviral genome bottom-up by applying DNA synthesis and assembly methods that have become available more recently. Interestingly, comparison of baculovirus sequences in genome databases indicate that most genes and DNA elements thought to be required for survival of the virus in cell culture are confined to roughly one half of the circular viral genome, while the other half contains DNAs that can be probably largely disposed of in the laboratory [50]. This approach may hold challenges given the complex interplay of baculoviral proteins and their relative expression levels during the different phases of the viral life cycle [4]. Notwithstanding, synthetic approaches, probably best applied in combination with sequential deletions, are exciting and potentially highly rewarding avenues for developing new and minimal baculoviral genomes that can be customized for optimal properties in the research laboratory and also in industrial manufacturing.
Outlook
Since the pioneering first reports more than three decades ago, the baculovirus/insect cell expression system has developed into a mainstream production platform, accelerating a wide range of research projects in academic and industrial laboratories. In the post-genomic era, multiprotein complexes have entered center stage as essential catalysts of cellular activity, and notably the MultiBac BEVS has contributed substantially to make hitherto inaccessible protein complexes available, to unlock their structure and mechanism in molecular detail. Moreover, BEVS has emerged as a remarkably useful tool in the biotech and pharma sector, for the production of complex biologics in disease prevention and therapy. The development of this versatile expression tool is continuing unabated as it is set to benefit markedly from powerful new synthetic biology techniques that are becoming readily available. Fueled by these innovations, BEVS is excellently positioned to play a key and increasing role in the life sciences, in basic and applied research in the future. Exciting times are ahead of us.
Acknowledgments
We thank all members of the Berger laboratory for their support, and in particular Lakshmi S. Vijayachandran (Amrita Center for Nanosciences & Molecular Medicine, India), Yan Nie (Shanghai Institute of Technology, China), Simon Trowitzsch (Goethe University, Frankfurt) and Christoph Bieniossek (Hofmann-La Roche, Basel) for their contribution and helpful discussions. This work was funded by the European Commission (EC) Framework Programme (FP) 7 ComplexINC project (grant number 279039), to DF and IB. The MultiBac user access platform at the EMBL Grenoble received funding from the EC through the FP6 and FP7 projects SPINE2C, P-CUBE, BioStruct-X and INSTRUCT (grant numbers 031220, 227764, 283570 and 211252), and from the French Agence Nationale de Recherhe (ANR) through the Investissement d’Avenir funding scheme. DBTGR was a fellow of the EMBL interdisciplinary post-doctoral opportunities program (EIPOD).
Contributor Information
M. Cristina Vega, Phone: +343434635884266, Email: cristina.vega@strubicib.org.
Imre Berger, Email: iberger@embl.fr.
References
- 1.Smith GE, Summers MD, Fraser MJ. Production of human beta interferon in insect cells infected with a baculovirus expression vector. Mol Cell Biol. 1983;3:2156–2165. doi: 10.1128/MCB.3.12.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Summers MD. Milestones leading to the genetic engineering of baculoviruses as expression vector systems and viral pesticides. Adv Virus Res. 2006;68:3–73. doi: 10.1016/S0065-3527(06)68001-9. [DOI] [PubMed] [Google Scholar]
- 3.Pennock GD, Shoemaker C, Miller LK. Strong and regulated expression of Escherichia coli beta-galactosidase in insect cells with a baculoviral vector. Mol Cell Biol. 1984;4:399–406. doi: 10.1128/MCB.4.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Oers MM, Plijman GP, Vlak JM. Thirty years of baculovirus-insect cell protein expression: from dark horse to mainstream technology. J Gen Virol. 2015;96:6–23. doi: 10.1099/vir.0.067108-0. [DOI] [PubMed] [Google Scholar]
- 5.Contreras-Gomez A, Sanchez-Miron F, Garcia-Camacho F, Molina-Grima E, Chisti Y. Protein production using the baculovirus-inseect cell expression system. Biotechnol Prog. 2014;30:1–18. doi: 10.1002/btpr.1842. [DOI] [PubMed] [Google Scholar]
- 6.Jarvis DL. Baculovirus-insect cell expression systems. Methods Enzymol. 2009;463:191–222. doi: 10.1016/S0076-6879(09)63014-7. [DOI] [PubMed] [Google Scholar]
- 7.Possee RD, King LA. Baculovirus transfer vectors. Methods Mol Biol. 2007;388:55–76. doi: 10.1007/978-1-59745-457-5_3. [DOI] [PubMed] [Google Scholar]
- 8.Perez de Diego AC, et al. Characterisation of protection afforded by a bivaleent virus-like partcile vaccine against bluetongue virus serotypes 1 and 4 in sheep. PLoS One. 2011;6:e26666. doi: 10.1371/journal.pone.0026666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vicente T, Roldao A, Peixeto C, Carrondo MJT, Alves PM. Large-sclae production and purification of VLP-based vaccines. J Invertebr Pathol. 2011;107(Suppl):S42–S48. doi: 10.1016/j.jip.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roy P, Noad R. Bluetongue vaccines. Vaccine. 2009;27(Suppl):D86–D89. doi: 10.1016/j.vaccine.2009.08.037. [DOI] [PubMed] [Google Scholar]
- 11.Roy P, Noad R. Virus-like partciles as a vaccine delivery system: myths and facts. Adv Exp Med Biol. 2009;655:145–158. doi: 10.1007/978-1-4419-1132-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mietzsch M, et al. OneBac: platform for scalable and high-titer production of adeno-associated virus serotype 1–12 vectors for gene therapy. Hum Gene Ther. 2014;25(3):212–222. doi: 10.1089/hum.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marsic D, et al. Vector design Tour de Force: integrating combinatorial and rational approaches to derive novel adeno-associated virus variants. Mol Ther. 2014;22(11):1900–1909. doi: 10.1038/mt.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kotin RM. Large-scale recombinant adeno-associated virus production. Hum Mol Genet. 2011;20(R1):R2–R6. doi: 10.1093/hmg/ddr141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Airenne KJ, et al. Baculovirus: an insect-derived vector for diverse gene transfer applications. Mol Ther. 2013;21(4):739–749. doi: 10.1038/mt.2012.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paul A, Hasan A, Rodes L, Sangaralingam M, Prakash S. Bioengineered baculoviruses as new class of therapeutics using micro and nanotechnologies: principles, prospects and challenges. Adv Drug Deliv Rev. 2014;71:115–130. doi: 10.1016/j.addr.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Berger I, Fitzgerald DJ, Richmond TJ. Baculovirus expression system for heterologous multiprotein complexes. Nat Biotechnol. 2004;22(12):1583–1587. doi: 10.1038/nbt1036. [DOI] [PubMed] [Google Scholar]
- 18.Fitzgerald DJ, et al. Protein complex expression by using multigene baculoviral vectors. Nat Methods. 2006;3(12):1021–1032. doi: 10.1038/nmeth983. [DOI] [PubMed] [Google Scholar]
- 19.Fitzgerald DJ, et al. Multiprotein expression strategy for structural biology of eukaryotic complexes. Structure. 2007;15(3):275–279. doi: 10.1016/j.str.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 20.Bieniossek C, Berger I. Towards eukaryotic structural complexomics. J Struct Funct Genomics. 2009;10(1):37–46. doi: 10.1007/s10969-008-9047-6. [DOI] [PubMed] [Google Scholar]
- 21.Trowitzsch S, Bieniossek C, Nie Y, Garzoni F, Berger I. New baculovirus expression tools for recombinant protein complex production. J Struct Biol. 2010;172(1):45–54. doi: 10.1016/j.jsb.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 22.Vijayachandran LS, et al. Robots, pipelines, polyproteins: enabling multiprotein expression in prokaryotic and eukaryotic cells. J Struct Biol. 2011;175(2):198–208. doi: 10.1016/j.jsb.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trowitzsch S, Palmberger D, Fitzgerald D, Takagi Y, Berger I. MultiBac complexomics. Expert Rev Proteomics. 2012;9(4):363–373. doi: 10.1586/epr.12.32. [DOI] [PubMed] [Google Scholar]
- 24.Berger I, et al. The MultiBac protein complex production platform at the EMBL. J Vis Exp. 2013;11(77):e50159. doi: 10.3791/50159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berger I, Chaillet M, Garzoni F, Yau-Rose S, Zoro B. High-throughput screening of multiple protein complexes. Am Lab. 2013;25(8):32–35. [Google Scholar]
- 26.Nie Y, et al. Getting a grip on complexes. Curr Genomics. 2009;10(8):558–572. doi: 10.2174/138920209789503923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson CV, Sali A, Baumeister W. The molecular sociology of the cell. Nature. 2007;450(7172):973–982. doi: 10.1038/nature06523. [DOI] [PubMed] [Google Scholar]
- 28.Ramakrishnan V. The ribosome emerges from a black box. Cell. 2014;159(5):979–984. doi: 10.1016/j.cell.2014.10.052. [DOI] [PubMed] [Google Scholar]
- 29.Fernández-Tornero C, et al. Crystal structure of the 14-subunit RNA polymerase I. Nature. 2013;502(7473):644–649. doi: 10.1038/nature12636. [DOI] [PubMed] [Google Scholar]
- 30.Engel C, Sainsbury S, Cheung AC, Kostrewa D, Cramer P. RNA polymerase I structure and transcription regulation. Nature. 2013;502(7473):650–655. doi: 10.1038/nature12712. [DOI] [PubMed] [Google Scholar]
- 31.Cramer P, et al. Structure of eukaryotic RNA polymerases. Annu Rev Biophys. 2008;37:337–352. doi: 10.1146/annurev.biophys.37.032807.130008. [DOI] [PubMed] [Google Scholar]
- 32.Völkel P, Le Faou P, Angrand PO. Interaction proteomics: characterization of protein complexes using tandem affinity purification-mass spectrometry. Biochem Soc Trans. 2010;38(4):883–887. doi: 10.1042/BST0380883. [DOI] [PubMed] [Google Scholar]
- 33.Li Y. Commonly used tag combinations for tandem affinity purification. Biotechnol Appl Biochem. 2010;55(2):73–83. doi: 10.1042/BA20090273. [DOI] [PubMed] [Google Scholar]
- 34.Janin J, Séraphin B. Genome-wide studies of protein-protein interaction. Curr Opin Struct Biol. 2003;13(3):383–388. doi: 10.1016/S0959-440X(03)00064-2. [DOI] [PubMed] [Google Scholar]
- 35.Almo SC, et al. Protein production from the structural genomics perspective: achievements and future needs. Curr Opin Struct Biol. 2013;23(3):335–344. doi: 10.1016/j.sbi.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haffke M, et al. Characterization and production of protein complexes by co-expression in Escherichia coli. Methods Mol Biol. 2015;1261:63–89. doi: 10.1007/978-1-4939-2230-7_4. [DOI] [PubMed] [Google Scholar]
- 37.Vincentelli R, Romier C. Expression in Escherichia coli: becoming faster and more complex. Curr Opin Struct Biol. 2013;23(3):326–334. doi: 10.1016/j.sbi.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 38.Bieniossek C, et al. Automated unrestricted multigene recombineering for multiprotein complex production. Nat Methods. 2009;6(6):447–450. doi: 10.1038/nmeth.1326. [DOI] [PubMed] [Google Scholar]
- 39.Abdulrahman W, et al. The production of multiprotein complexes in insect cells using the baculovirus expression system. Methods Mol Biol. 2015;1261:91–114. doi: 10.1007/978-1-4939-2230-7_5. [DOI] [PubMed] [Google Scholar]
- 40.Bieniossek C, Imasaki T, Takagi Y, Berger I. MultiBac: expanding the research toolbox for multiprotein complexes. Trends Biochem Sci. 2012;37(2):49–57. doi: 10.1016/j.tibs.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bieniossek C, Richmond TJ, Berger I (2008) MultiBac: multigene baculovirus-based eukaryotic protein complex production. Curr Protoc Protein Sci 51:5.20.1 - 5.20.26 [DOI] [PubMed]
- 42.Tsutsui H, Matsubara K. Replication control and switch-off function as observed with a mini-F factor plasmid. J Bacteriol. 1981;147(2):509–516. doi: 10.1128/jb.147.2.509-516.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luckow VA, Lee SC, Barry GF, Olins PO. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J Virol. 1993;67:4566–4579. doi: 10.1128/jvi.67.8.4566-4579.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palmberger D, Wilson IB, Berger I, Grabherr R, Rendic D. SweetBac: a new approach for the production of mammalianised glycoproteins in insect cells. PLoS One. 2012;7(4):e34226. doi: 10.1371/journal.pone.0034226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palmberger D, Klausberger M, Berger I, Grabherr R. MultiBac turns sweet. Bioengineered. 2013;4(2):78–83. doi: 10.4161/bioe.22327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palmberger D, et al. Minimizing fucosylation in insect cell-derived glycoproteins reduces binding to IgE antibodies from the sera of patients with allergy. Biotechnol J. 2014;9(SI):1206–1214. doi: 10.1002/biot.201400061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wasilko DJ, et al. The titerless infected-cells preservation and scale-up (TIPS) method for large-scale production of NO-sensitive human soluble guanylate cyclase (sGC) from insect cells infected with recombinant baculovirus. Protein Expr Purif. 2009;65(2):122–132. doi: 10.1016/j.pep.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 48.Haffke M, Viola C, Nie Y, Berger I. Tandem recombineering by SLIC cloning and Cre-LoxP fusion to generate multigene expression constructs for protein complex research. Methods Mol Biol. 2013;1073:131–140. doi: 10.1007/978-1-62703-625-2_11. [DOI] [PubMed] [Google Scholar]
- 49.Plijman GP, van Schijndel JE, Vlak JM. Spontaneous excision of BAC vector sequences from bacmid-derived baculovirus expression vectors upon passage in insect cells. J Gen Virol. 2003;84:2669–2678. doi: 10.1099/vir.0.19438-0. [DOI] [PubMed] [Google Scholar]
- 50.Vijachandran LS, et al. Gene gymnastics: synthetic biology for baculovirus expression vector system engineering. Bioengineered. 2013;4(5):279–287. doi: 10.4161/bioe.22966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nie Y, Bellon-Echeverria I, Trowitzsch S, Bieniossek C, Berger I. Multiprotein complex production in insect cells by using polyproteins. Methods Mol Biol. 2014;1091:131–141. doi: 10.1007/978-1-62703-691-7_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bieniossek C, et al. The architecture of human general transcription factor TFIID core complex. Nature. 2013;493(7434):699–702. doi: 10.1038/nature11791. [DOI] [PubMed] [Google Scholar]
- 53.Barford D, Takagi Y, Schultz P, Berger I. Baculovirus expression: tackling the complexity challenge. Curr Opin Struct Biol. 2013;23(3):357–364. doi: 10.1016/j.sbi.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trowitzsch S, et al. Cytoplasmic TAF2-TAF8-TAF10 complex provides evidence for nuclear holo-TFIID assembly from preformed submodules. Nat Commun. 2015;6:6011. doi: 10.1038/ncomms7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reich S, et al. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature. 2014;516(7531):361–366. doi: 10.1038/nature14009. [DOI] [PubMed] [Google Scholar]
- 56.Pflug A, Gulligay D, Reich S, Cusack S. Structure of influenza A polymerase bound to the viral RNA promoter. Nature. 2014;516(7531):355–360. doi: 10.1038/nature14008. [DOI] [PubMed] [Google Scholar]
- 57.Berger I, Mary LM. Protein production for structural biology: new solutions to new challenges. Curr Opin Struct Biol. 2013;23(3):317–318. doi: 10.1016/j.sbi.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 58.Kandiah E, Trowitzsch S, Gupta K, Haffke M, Berger I. More pieces to the puzzle: recent structural insights into class II transcription initiation. Curr Opin Struct Biol. 2014;24:91–97. doi: 10.1016/j.sbi.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 59.Zeqiraj E, Filippi BM, Deak M, Alessi DR, van Aalten DM. Structure of the LKB1-STRAD-MO25 complex reveals an allosteric mechanism of kinase activation. Science. 2009;326(5960):1707–1711. doi: 10.1126/science.1178377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamada K, et al. Structure and mechanism of the chromatin remodelling factor ISW1a. Nature. 2011;472(7344):448–453. doi: 10.1038/nature09947. [DOI] [PubMed] [Google Scholar]
- 61.Schmidt H, Zalyte R, Urnavicius L, Carter AP. Structure of human cytoplasmic dynein-2 primed for its power stroke. Nature. 2015;518(7539):435–438. doi: 10.1038/nature14023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elkayam E, et al. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012;150(1):100–110. doi: 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Santos KF, et al. Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome. Proc Natl Acad Sci U S A. 2012;109(43):17418–17423. doi: 10.1073/pnas.1208098109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mozaffari-Jovin S, et al. Inhibition of RNA helicase Brr2 by the C-terminal tail of the spliceosomal protein Prp8. Science. 2013;341(6141):80–84. doi: 10.1126/science.1237515. [DOI] [PubMed] [Google Scholar]
- 65.Burke JE, et al. Structures of PI4KIIIβ complexes show simultaneous recruitment of Rab11 and its effectors. Science. 2014;344(6187):1035–1038. doi: 10.1126/science.1253397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sahlmuller MC, et al. Recombinant heptameric coatomer complexes: novel tools to study isoform-specific functions. Traffic. 2011;12(6):682–692. doi: 10.1111/j.1600-0854.2011.01177.x. [DOI] [PubMed] [Google Scholar]
- 67.Faini M, et al. The structures of COPI-coated vesicles reveal alternate coatomer conformations and interactions. Science. 2012;336(6087):1451–1454. doi: 10.1126/science.1221443. [DOI] [PubMed] [Google Scholar]
- 68.Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Molecular architecture and mechanism of the anaphase-promoting complex. Nature. 2014;513(7518):388–393. doi: 10.1038/nature13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cevher MA, et al. Reconstitution of active human core mediator complex reveals a critical role of the MED14 subunit. Nat Struct Mol Biol. 2014;21(12):1028–1034. doi: 10.1038/nsmb.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goehring A, et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat Protoc. 2014;9(11):2574–2585. doi: 10.1038/nprot.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Drugmand JC, Schneider YJ, Agathos SN. Insect cells as factories for biomanufacturing. Biotechnol Adv. 2012;30(5):1140–1157. doi: 10.1016/j.biotechadv.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 72.Hom LG, Ohkawa T, Trudeau D, Volkman LE. Autographa californica M nucleopolyhedrovirus ProV-CATH is activated during infected cell death. Virology. 2002;131:561–565. doi: 10.1006/viro.2002.1378. [DOI] [PubMed] [Google Scholar]
- 73.Hitchman RB, et al. Improved expression of secreted and membrane-targeted proteins in insect cells. Biotechnol Appl Biochem. 2010;56:85–93. doi: 10.1042/BA20090130. [DOI] [PubMed] [Google Scholar]
- 74.Kaba SA, Salceda AM, Wafula PO, Vlak JM, van Oers MM. Development of a chitinase and v-cathepsin negative bacmid of improved integrity of secreted recombinant proteins. J Virol Methods. 2004;122:113–118. doi: 10.1016/j.jviromet.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 75.Harrison RL, Jarvis DL. Protein N-glycosylation in the baculovirus-insect cell system and engineering of insect cells to produce “mammalianized” recombinant glycoproteins. Adv Virus Res. 2006;68:159–191. doi: 10.1016/S0065-3527(06)68005-6. [DOI] [PubMed] [Google Scholar]
- 76.Harrison RL, Jarvis DL. Transforming lepidopteran insect cells for improved protein processing. Methods Mol Biol. 2007;388:341–356. doi: 10.1007/978-1-59745-457-5_17. [DOI] [PubMed] [Google Scholar]
- 77.Fernandes F, Teixeira AP, Carinhas N, Carrondo MJ, Alves PM. Insect cells as a production platform of complex virus-like particles. Expert Rev Vaccines. 2013;12(2):225–236. doi: 10.1586/erv.12.153. [DOI] [PubMed] [Google Scholar]
- 78.Metz SW, et al. Effective chickungunya virus-like particle vaccine produced in insect cells. PLoS Negl Trop Dis. 2013;7:e2124. doi: 10.1371/journal.pntd.0002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Smith GE, et al. Development of influenza H7N9 virus like particle (VLP) vaccine: homologous A/Anhui/1/2013 (H7N9) protection and heterologous A/chicken/Jalisco/CPA/2012 (H7N3) cross-protection in vaccinated mice challenged with H7N9 virus. Vaccine. 2013;31:4305–4313. doi: 10.1016/j.vaccine.2013.07.043. [DOI] [PubMed] [Google Scholar]
- 80.Metz SW, Plijman GP. Arbovirus vaccines: opportunities for the baculovirus-insect cell expression system. J Invertebr Pathol. 2011;107(Suppl):S16–S30. doi: 10.1016/j.jip.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 81.Behzadian F, et al. Baculoviral co-expression of HA, NA and M1 proteins of highly pathogenic H5N1 influenza virus in insect cells. Jundishapur J Microbiol. 2013;6(9):e7665. doi: 10.5812/jjm.7665. [DOI] [Google Scholar]
- 82.Condreay JP, Kost TA. Baculovirus expression vectors for insect and mammalian cells. Curr Drug Targets. 2007;8(10):1126–1131. doi: 10.2174/138945007782151351. [DOI] [PubMed] [Google Scholar]
- 83.Ames RS, Kost TA, Condreay JP. BacMam technology and its application to drug discovery. Expert Opin Drug Discov. 2007;2(12):1669–1681. doi: 10.1517/17460441.2.12.1669. [DOI] [PubMed] [Google Scholar]
- 84.Kost TA, Condreay JP, Ames RS. Baculovirus gene delivery: a flexible assay development tool. Curr Gene Ther. 2010;10(3):168–173. doi: 10.2174/156652310791321224. [DOI] [PubMed] [Google Scholar]


